Abstract
Post-translational modifications are crucial mechanisms that modulate various cellular signaling pathways, and their dysregulation is associated with many human diseases. Spinocerebellar ataxia type 1 (SCA1) is a dominantly inherited neurodegenerative disease characterized by progressive ataxia, mild cognitive impairments, difficulty with speaking and swallowing, and respiratory failure. It is caused by the expansion of an unstable CAG trinucleotide repeat encoding a glutamine tract in ATAXIN-1 (ATXN1). Although the expansion of the polyglutamine tract is the key determinant of the disease, protein domains outside of the polyglutamine tract and post-translational modifications of ATXN1 significantly alter the neurotoxicity of SCA1. ATXN1 undergoes several post-translational modifications, including phosphorylation, ubiquitination, sumoylation, and transglutamination. Such modifications can alter the stability of ATXN1 or its activity in the regulation of target gene expression, and therefore contribute to SCA1 toxicity. This review outlines different types of post-translational modifications in ATXN1 and discusses their potential regulatory mechanisms and effects on SCA1 pathogenesis. Finally, the manipulation of post-translational modifications as a potential therapeutic approach will be discussed.
Keywords: Ataxin-1, SCA1, polyglutamine disease, post-translational modification, phosphorylation, ubiquitination, sumoylation, transglutamination
Introduction
Post-translational modifications contribute to the structural and functional diversity of proteins through the addition of functional groups such as phosphate, acetate, or various small proteins or lipids [1]. They control protein activity, turnover, localization, and interactions, which in turn regulate a wide variety of signaling pathways. Post-translational modifications are implicated in many human disorders, including neurodegenerative diseases, because of their pivotal roles in cellular processes [2].
Spinocerebellar ataxia type 1 (SCA1, MIM #164400) belongs to a group of diseases caused by the expansion of an unstable CAG trinucleotide repeat encoding a glutamine tract. This family of diseases includes at least nine inherited neurodegenerative disorders, the so-called polyglutamine diseases: Huntington’s disease (HD), spinal and bulbar muscular atrophy (SBMA), dentatorubropallidoluysian atrophy (DRPLA), and six autosomal dominant spinocerebellar ataxias (SCA1, 2, 3, 6, 7, and 17) [3, 4]. SCA1 is caused by the expansion of a polyglutamine tract in ATXN1. The normal ATXN1 alleles in unaffected individuals contain between 6 and 42 CAG repeats, which are usually interrupted by one to four CAT trinucleotides when the number of CAG repeats exceeds 20. By contrast, SCA1-affected individuals have an uninterrupted pure CAG tract ranging from 39 to 83 repeats. SCA1 is characterized by progressive ataxia, cognitive impairments, difficulty with speaking and swallowing, and respiratory failure. The clinical and pathological features of SCA1 result from the degeneration of cerebellar Purkinje cells (PCs), brainstem cranial nerve nuclei, the inferior olive, and spinocerebellar tracts [4].
The polyglutamine tract expansion is the central cause of the disease [5, 6]. In general, longer glutamine tract repeat lengths will result in more severe symptoms and an earlier age of onset of the disease. However, SCA1 pathology does not solely depend on the polyglutamine tract. The polyglutamine-expanded ATXN1 does not induce disease phenotypes in the absence of nuclear localization signals [7], the AXH (ATXN1-HBP1) domain [8], or phosphorylation at serine 776 [9] (Fig. 1). Therefore, it is clear that the protein context and post-translational modifications of ATXN1 can influence the neurotoxicity of SCA1. This review outlines diverse post-translational modifications of ATXN1 and discusses their impact on the pathogenesis of SCA1 (Fig. 2).
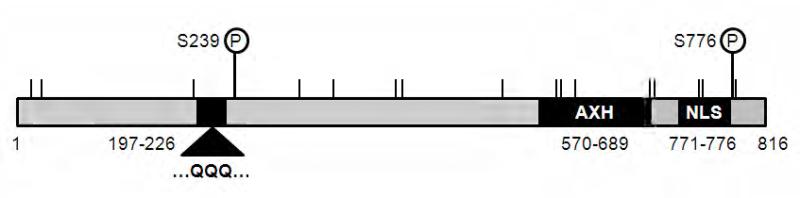
Fig. 1. Functional domains and post-translational modification sites in ATXN1.
Protein domains of a polyglutamine tract (residues 197–226), AXH (residues 570–689), and nuclear localization signal (NLS, residues 771–776) that contribute to SCA1 pathogenesis are indicated as solid boxes. The two in vivo phosphorylation sites are located at serine 239 (S239) and 776 (S776). Seventeen putative sumoylation consensus sites are indicated by vertical lines at lysine (K) residue 16, 29, 194, 309, 347, 414, 421, 530, 590, 595, 610, 692, 697, 746, 750, 782, and 785. The amino acid sequences are numbered based on the full-length wild-type form of ATXN1 with 30 CAG repeat units.
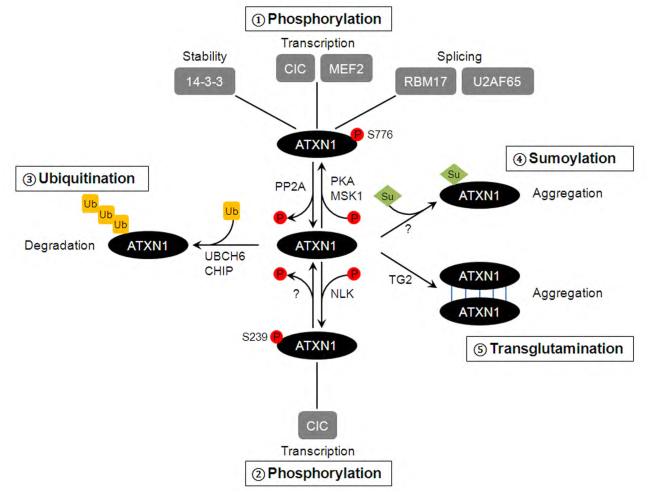
Fig. 2. Scheme depicting the four major post-translational modifications in ATXN1 and their putative impact on ATXN1 function and SCA1 pathology.
Post-translational modifications modulate diverse cellular functions of ATXN1, thereby play key roles in SCA1 pathogenesis: 1) Phosphorylation at S776 influences ATXN1 stability, and transcriptional and/or splicing activity of ATXN1 by regulating physical interactions with various binding partners, such as 14-3-3, CIC, MEF2, RBM17, and U2AF65. This event is controlled by the action of kinases (i.e. PKA and MSK1) and phosphatases (i.e. PP2A). 2) S239 can be phosphorylated by NLK, and it modulates transcriptional activity of ATXN1 and SCA1 toxicity. 3) Ubiquitination of ATXN1 by UBCH6 and CHIP promotes degradation of ATXN1 and controls the steady state levels of the ATXN1 protein. 4) Sumoylation increases aggregation formation of ATXN1. 5) Transglutamination may also promote aggregation formation of ATXN1 proteins by cross-linking them.
Phosphorylation of ATXN1
Phosphorylation is the addition of a phosphate group to a protein and is a reversible post-translational modification [1]. Protein phosphorylation plays a major role in a broad range of cellular processes, and often alters the function and activity of a protein. In ATXN1, there are at least seven phosphorylation sites identified in vivo [10]. Among them, phosphorylations at the serine 776 (S776) and 239 (S239) residues are suggested to play key roles in SCA1 pathogenesis (Fig. 2).
S776 phosphorylation in SCA1 pathogenesis
S776 was the first endogenous phosphorylation site in ATXN1 identified by mass spectrometry analysis [9]. Extensive in vivo studies in mice show the importance of ATXN1-S776 phosphorylation in the pathogenesis of SCA1. A transgenic mouse (called SCA1-B05) that expresses a polyglutamine-expanded mutant form of human ATXN1 with 82 glutamines (ATXN1[82Q]) under a PC-specific driver (the Pcp2 promoter) develops progressive ataxia and PC degeneration that resembles the features of human SCA1 [11, 12]. By contrast, mice expressing a polyglutamine-expanded mutant ATXN1 with a phosphorylation-defective amino acid substitution at residue 776 (A776, serine to alanine substitution) do not develop neurotoxicity, despite the presence of the polyglutamine-expanded ATXN1 protein [9]. Instead, both behavioral and pathological deficits are dramatically reduced in ATXN1[82Q]-A776 mice. The ATXN1[82Q]-A776 mice are indistinguishable from their wild-type littermates at 19 weeks of age in both their home cage behavior and their performance in a rotating-rod analysis, while ATXN1[82Q]-S776 mice show severely impaired performance at this time point. Cerebellar morphology in ATXN1[82Q]-A776 mice is comparable to that of the wild-type mice, showing no signs of dendritic thinning or PC heterotopia. Only weak pathology is observed in ATXN1[82Q]-A776 mice at very late stages. The thickness of the molecular cell layer is reduced by 30% in comparison to that of the wild-type mice at 37 weeks, without any PC loss or heterotopia. Therefore, this study clearly demonstrates the importance of ATXN1-S776 phosphorylation in the pathogenesis of SCA1. A subsequent study by Duvick et al. (2010) further supports the notion of the critical role of ATXN1-S776 phosphorylation in SCA1 neurotoxicity [13]. To study the importance of S776 phosphorylation in normal ATXN1 function and SCA1 pathology, this group generated a new SCA1 transgenic mouse that overexpresses a wild-type form of ATXN1 (ATXN1[30Q]) with a phosphorylation-mimicking substitution at residue 776 (serine to aspartic acid substitution, D776) under the same PC-specific promoter [13]. ATXN1[30Q]-D776 mice show PC atrophy, a reduction in climbing fiber innervation, and impaired motor performance in the rotating-rod analysis; these observations are similar to those seen in the ATXN1[82Q]-S776 animals. This study is consistent with the notion that S776 phosphorylation in ATXN1 is important for SCA1 pathogenesis. It also raises the question of whether expansion of the polyglutamine tract induces neurotoxicity simply by increasing the phosphorylation level at S776 in ATXN1. Lim et al. (2008) previously investigated this possibility, however, and found that there was no enhancement in the S776 phosphorylation of ATXN1 by polyglutamine expansion [14]. Therefore, it is reasonable to predict that expansion of the polyglutamine tract in the N-terminal region of ATXN1 may alter the conformation of the protein, and that this altered protein conformation may also be induced by S776 phosphorylation in ATXN1.
It is worth mentioning that although the ATXN1[30Q]-D776 and ATXN1[82Q]-S776 mice display similar neuropathology during the earlier stages of development, the ATXN1[30Q]-D776 mice fail to induce PC death at the later stages [13]. This suggests that phosphorylation at S776 is critical for the induction of neuronal dysfunction and disease initiation, while an expanded polyglutamine tract is required for neuronal cell death. Alternatively, aspartic acid substitution may not fully mimic the biological features of serine phosphorylation. In summary, phosphorylation of S776 in ATXN1 is essential for the pathogenesis of SCA1 in animals.
Effects of S776 phosphorylation on ATXN1 stability and the regulation of its interaction with cellular proteins
Although it is clear that phosphorylation of S776 in ATXN1 plays a substantial role in SCA1 pathogenesis in vivo, the molecular mechanisms underlying the effects of ATXN1-S776 phosphorylation on SCA1 pathogenesis are not fully understood. Nonetheless, a growing body of evidence supports the idea that phosphorylation at S776 regulates ATXN1 protein stability and its interactions with other cellular proteins. In this regard, there are at least five proteins known to interact with ATXN1 either directly via the phosphorylated S776 residue itself, or indirectly by interactions that are influenced by the phosphorylation state of S776. These include the cellular signaling protein family 14-3-3 [15], the transcriptional factors Capicua (CIC) [16] and myocyte enhancer factor 2 [17], and the splicing factors RNA-binding motif protein 17 (RBM17) [14] and U2AF65 [18].
14-3-3 proteins were the first proteins to be identified as ATXN1 interactors that depend on the phosphorylation state of S776 [15]. They are a family of highly conserved proteins that bind to phospho-serine-containing motifs in target proteins and regulate many intracellular signal transduction pathways that are essential for differentiation, development, growth, apoptosis, and survival [19]. As phosphopeptide-binding proteins, 14-3-3 proteins only interact with S776-phosphorylated (pS776) ATXN1, and their interaction is enhanced by the expansion of the polyglutamine tract in ATXN1 [15]. The primary outcome of this interaction is the stabilization of ATXN1 proteins. The binding of 14-3-3 to ATXN1 prevents the dephosphorylation of pS776 and protects ATXN1 from subsequent proteolysis [20]. The phosphorylation-mimetic ATXN1 (ATXN1-D776) is unable to bind 14-3-3, but is as stable as pS776-ATXN1 [18, 20]. This suggests that 14-3-3 may stabilize ATXN1 indirectly by protecting the phosphorylation at S776, rather than by the direct physical interaction itself. Moreover, 14-3-3 proteins act as cytoplasmic anchors that control the subcellular distribution of ATXN1, which is critical for SCA1 pathogenesis. The 14-3-3 binding site of ATXN1 (amino acid 774-776) overlaps with the nuclear localization signal (amino acid 771-776) (Fig. 1). Thus, the interaction of 14-3-3 proteins with ATXN1 may prevent ATXN1 from entering the nucleus by masking the nuclear localization signal [20]. The overexpression of 14-3-3 is shown to increase the toxicity of the polyglutamine-expanded ATXN1 in Drosophila [15]. Transgenic flies expressing a polyglutamine-expanded mutant ATXN1 in the Drosophila eye displayed severe retinal degenerative phenotypes, and co-expression of the 14-3-3 epsilon isoform with the mutant ATXN1 strongly enhanced retinal degeneration. Consistent with this, reduced expression of 14-3-3 ameliorates diverse disease symptoms in SCA1 knock-in mice [21]. Specifically, loss of one copy of the 14-3-3 epsilon isoform reduced the steady state levels of the ATXN1 protein in the cerebellum and rescued the cerebellar pathology and motor behavior phenotypes of SCA1 knock-in mice. These results indicate that the phosphorylation state of S776 in ATXN1 is critical for 14-3-3 binding, which leads to ATXN1 stabilization and SCA1 toxicity.
There are several reports suggesting that ATXN1 forms at least two independent large protein complexes: one containing the transcriptional factor CIC, and the other containing the splicing factor RBM17 [14, 16, 21, 22]. In the normal mouse cerebellum, the majority of ATXN1 proteins form stable large protein complexes containing CIC [16]. Lam et al. (2006) found that ATXN1 interacts with CIC through the AXH domain, and S776-phosphorylation does not directly affect its interaction with CIC in cell culture systems. However, the phosphorylation state of S776 is required for the formation or maintenance of ATXN1/CIC-containing complexes in the mouse cerebellum in vivo. This concept is supported by the finding that the non-phosphorylated, and thus non-pathogenic form of ATXN1, ATXN1[82Q]-A776, does not incorporate into a large protein complex containing CIC. This also indirectly suggests that ATXN1 must incorporate into a CIC-containing large protein complex to be toxic [16]. Consistent with this, reducing the CIC expression levels also decreases the steady state levels of the mutant ATXN1 protein and suppresses the SCA1 disease features in mice [23]. These studies indicate that the phosphorylation state of S776 indirectly influences ATXN1-CIC complex formation, which is required for the stabilization or maintenance of the ATXN1 protein and the induction of SCA1 neurotoxicity.
Besides the ATXN1/CIC-containing large complexes, as aforementioned, ATXN1 forms another large protein complex containing the splicing factor RBM17. Interestingly, the interaction between ATXN1 and RBM17 is directly dependent on the phosphorylation state of ATXN1-S776 [14]. Furthermore, the interaction between the two proteins is strongly enhanced by the expansion of the polyglutamine tract in ATXN1. Therefore, the formation of ATXN1/RBM17-containing complexes is increased by at least two key pathogenic factors of SCA1, and is relatively more dynamic than the ATXN1/CIC-containing complexes [13, 14]. Overexpression of RBM17 augments the toxicity of the polyglutamine-expanded ATXN1 in Drosophila, while reduced expression of RBM17 suppresses it [14]. This study supports a model in which aberrantly enhanced formation of the ATXN1-RBM17 complexes contributes to SCA1. Unfortunately, the importance of the ATXN1-RBM17-containing complexes on SCA1 neurotoxicity in mice still remains to be elucidated. As there might be a potential fundamental difference between Drosophila and mouse models of SCA1, current data cannot rule out the possibility that the polyglutamine-expanded mutant ATXN1 instead interacts with and inhibits RBM17 activity in a dominant-negative fashion in the mouse cerebellum in vivo.
The S776 residue, and perhaps its phosphorylation, also influences the interaction of ATXN1 with other proteins, such as U2AF65 [18] and myocyte enhancer factor 2 (MEF2) [17]. ATXN1 interacts with and influences the cellular functions of U2AF65, a constitutive component of the spliceosome. Thus, ATXN1 may have an effect on U2AF65-mediated splicing. MEF2 is a transcription factor that regulates neuronal development, synaptic plasticity, and survival [24]. Specifically, normal function of MEF2 is responsible for neuronal survival, whereas its inhibition contributes to neuronal death. MEF2 is known to be highly expressed in the cerebellum, cerebral cortex, and hippocampus, and deregulation of MEF2 activity is associated with neurodegenerative diseases. ATXN1 binds to MEF2 and suppresses its transcriptional activity, whereas the ATXN1-A776 mutant shows a strongly reduced interaction with MEF2 and fails to inhibit MEF2-dependent transcription [17]. Moreover, overexpression of MEF2 partially alleviates the cytotoxicity caused by the polyglutamine-expanded ATXN1 in transfected cell lines [17]. This study suggests that the polyglutamine-expanded mutant ATXN1 inhibits the neuronal survival function of MEF2, and that S776 phosphorylation appears to be a key player in this event. Taken together, the phosphorylation state of S776 in ATXN1 is a key regulator that controls the physical interaction of ATXN1 with its diverse binding partners, and thereby modulates neurotoxicity in SCA1.
Regulation of S776 phosphorylation: kinases and phosphatases
It is of great importance to identify the kinases and phosphatases that are responsible for S776 phosphorylation in ATXN1. Several kinases have been predicted to recognize the sequences surrounding the S776 residue of ATXN1 (KRRWSAP) [25]. An initial study using tissue culture systems and Drosophila models of SCA1 suggested that AKR mouse thymoma (Akt) was the kinase responsible for ATXN1-S776 phosphorylation [15]. However, a later study proposed that cyclic AMP-dependent protein kinase (PKA), not Akt, is the kinase that phosphorylates ATXN1-S776 in the mouse cerebellum in vivo [25, 26]. This finding was based on the following observations: 1) PKA co-fractionates with the peak of ATXN1-S776 phosphorylation by gel-filtration chromatography. 2) Immunodepletion of PKA from cerebellar extracts significantly reduces the phosphorylation state of ATXN1-S776. 3) Treatment with a PKA inhibitor decreases the levels of ATXN1-S776 phosphorylation. And 4) activation of PKA by forskolin treatment enhances ATXN1-S776 phosphorylation in cultured cells and induces the degeneration of PCs in slice cultures. Furthermore, the dopamine receptor D2 (DRD2) signaling pathway is suggested to act upstream of PKA in SCA1 [26]. The expression level of DRD2 is reduced in the cerebellum of SCA1 transgenic mice (B05) [27]. This decrease in DRD2 could theoretically prevent cyclic adenosine monophosphate (cAMP) synthesis, leading to the activation of PKA activity and the subsequent increase in ATXN1-S776 phosphorylation. In contrast, inhibition of Akt either genetically, by expressing a dominant-negative form of the kinase, or pharmacologically, by treatment with an Akt inhibitor, does not decrease ATXN1-S776 phosphorylation in the mouse cerebellum [25]. Thus, these lines of evidence strongly support the idea that PKA is a kinase that phosphorylates ATXN1 at S776 in vivo in the mouse cerebellum.
A recent genome-wide RNA interference screen targeting human kinases identified the RAS-MAPK-MSK1 signaling pathway as a modulator of ATXN1 protein levels and SCA1 toxicity [28]. Decreasing the expression levels of components of this signaling pathway reduced ATXN1 protein levels and suppressed the retinal degeneration induced by the polyglutamine-expanded ATXN1 in Drosophila. Furthermore, pharmacological inhibition of the RAS-MAPK-MSK1 pathway decreases ATXN1 levels in both cell culture systems and mouse cerebellar slice cultures. It was therefore suggested that the kinase MSK1 could also phosphorylate the S776 residue of ATXN1. In support of this hypothesis, MSK1 phosphorylates ATXN1 at S776 in in vitro kinase assays and stabilizes the ATXN1 protein in transfected cells. Moreover, the loss of Msk1 decreases mutant ATXN1 (Atxn1[154Q]) levels and is able to rescue the behavioral and pathological phenotypes observed in SCA1 knock-in mice. Collectively, these data support the idea that MSK1 is another kinase that is able to target ATXN1 at S776 in vivo.
In contrast to kinases, phosphatases remove a phosphate group from a protein and are therefore also crucial for the regulation of protein phosphorylation. Protein phosphatase 2A (PP2A) was identified as a potential ATXN1-pS776 phosphatase in the cerebellum by a glutathione S-transferase (GST) pull-down assay [20]. PP2A dephosphorylates ATXN1-pS776 and knock-down of PP2A expression increases the amount of ATXN1-pS776 in transfected cells in vitro. In the mouse cerebellum in vivo, immunodepletion of PP2A reduces the ability of cerebellar extracts to dephosphorylate the endogenous ATXN1-pS776. Collectively, these data support the hypothesis that PP2A is a phosphatase for pS776 in ATXN1 in the mouse cerebellum. Dephosphorylation of ATXN1 at S776 is restricted to the nucleus of cerebellar cells [20]. This compartmentalization can be explained by the fact that ATXN1-pS776 interacts with14-3-3 proteins in the cytoplasm, blocking dephosphorylation of pS776 by PP2A. The addition of synthetic peptides that specifically bind to 14-3-3 proteins to cerebellar cytoplasmic extracts can release this inhibitory effect of 14-3-3 proteins and promote the dephosphorylation of pS776. Interestingly, a recent study suggests that there is a much more complex cross-talk between ATXN1 and PP2A [29]. Sanchez et al. (2013) shows that the polyglutamine-expanded ATXN1 dramatically increases PP2A activity in the cerebellum of SCA1 mice by decreasing the inhibitory phosphorylation of the catalytic subunit of PP2A and by down-regulating the expression levels of ANP32A (acidic leucine-rich nuclear phosphoprotein 32 family, member A), which is a potent and selective PP2A inhibitor. This suggests that ATXN1 is both a substrate and a regulator of PP2A. In addition, PKA is known to phosphorylate and activate PP2A, introducing further avenues of positive and negative regulation in this phosphorylation of ATXN1 [30].
It is worth mentioning that knock-down or depletion of ANP32A expression using siRNA or knockout mouse approaches reverses some of the SCA1 phenotypes in vitro as well as in vivo [31]. Besides its role as a PP2A inhibitor, however, ANP32A also acts a modulator of transcription and cytoskeleton remodeling by inhibiting histone acetyltransferases and interacting with the microtubule-associated protein MAP1B, respectively [32, 33]. Therefore, the role of ANP32A in SCA1 pathogenesis should be carefully evaluated given its multiple cellular functions.
Overall, there exists a complex interplay involving both positive and negative feedback regulation among the diverse signaling components responsible for regulating ATXN1 phosphorylation in the mouse cerebellum. This should be carefully evaluated when targeting kinases or phosphatases for SCA1 therapy.
Phosphorylation of ATXN1 at S239 and its potential role in SCA1 pathogenesis
S239 in ATXN1 is another in vivo phosphorylation site which resides within the consensus sequence motifs of ERK (PXSP), CK1 (TpXXSX), and Cdc2/Cdk5 (SPX) kinases [10, 34]. The functional significance of S239 phosphorylation is largely unknown, but a recent study suggests its potential significance in ATXN1 function and SCA1 pathogenesis [35]. Ju et al. (2013) suggest that Nemo-like kinase (NLK) is able to induce the phosphorylation of ATXN1 at S239, and that this may be important for SCA1 pathogenesis. NLK was initially discovered as an ATXN1 interactor using a yeast two-hybrid screen [36]. It is an evolutionally conserved serine/threonine protein kinase that belongs to the MAPK family [37-40]. Several lines of evidence strongly demonstrate that NLK is a key protein contributing to the pathogenesis of SCA1: 1) NLK physically interacts with ATXN1 and is able to phosphorylate it at S239. 2) NLK enhances SCA1-induced neurodegenerative phenotypes in Drosophila in a kinase activity-dependent manner. 3) Decreased expression of Nlk suppresses cerebellar pathology and motor coordination deficits in SCA1 knock-in mice. Taken together, these findings clearly show that the decreased activity or expression of NLK has beneficial effects against the neurotoxicity induced by the polyglutamine-expanded ATXN1 in diverse model systems of SCA1. However, the molecular mechanism by which NLK affects SCA1 pathology remains to be determined. NLK does not seem to affect the expression levels of ATXN1 or the formation of the large toxic ATXN1 protein complexes in mice, but a cell culture study suggests that NLK may affect the transcriptional activity of the ATXN1/CIC complexes, which may partially explain the mechanistic function of NLK in SCA1 pathogenesis [35]. Interestingly, the effects of NLK on mutant ATXN1-mediated toxicity are dependent on its kinase activity. Since NLK can phosphorylate ATXN1 at S239, this suggests that S239 phosphorylation may be downstream of the NLK-mediated effects on toxicity and may therefore be important for the pathogenesis of SCA1. However, it is still unknown whether reducing or inhibiting ATXN1-S239 phosphorylation can phenocopy the beneficial effects of NLK inhibition/reduction. Furthermore, it remains unclear whether S239 is an in vivo target site of NLK in the cerebellum, or whether NLK is the only kinase responsible for S239 in ATXN1. Protein phosphatases that can dephosphorylate ATXN1-pS239 also remain to be identified. It is also worth noting that NLK may target additional sites in ATXN1, and identifying these residues would also be important for understanding SCA1 pathogenesis. Therefore, further studies are required to fully understand how S239-phosphorylation influences the function and regulation of the polyglutamine-expanded ATXN1 protein and its role in SCA1 pathology.
Ubiquitination
Ubiquitination is a post-translational modification whereby a covalent isopeptidyl bond is formed between a lysine (K) residue on the target protein and the carboxyl terminus of the small protein ubiquitin. Polyubiquitination and the subsequent protein degradation via the ubiquitin-proteasome system (UPS) play a key role in the regulation of protein homeostasis. Since impaired protein degradation and the formation of intracellular protein aggregates are implicated in SCA1 [41], understanding the molecular mechanisms of ATXN1 ubiquitination are critical for the development of SCA1 therapeutics. The role of the UPS in aggregation formation has been assessed by proteasome inhibitor treatments [42]. In transfected cells, polyglutamine-expanded ATXN1 localizes in the nucleus diffusely with little aggregation. Treatment with proteasome inhibitors further promotes the formation of ATXN1 aggregates, which are insoluble in detergent. This result suggests that proteasome-mediated degradation plays a crucial role in aggregation formation. Both the wild-type and polyglutamine-expanded mutant ATXN1 can be polyubiquitinated to similar degrees, but the mutant ATXN1 is somehow more resistant to proteasomal degradation. This implies that the proteasome cannot degrade the polyglutamine-expanded ATXN1 efficiently, despite normal polyubiquitination. This is consistent with in vivo aggregation in human patients and mouse models of SCA1. Nuclear aggregates found in PCs often co-localize with ubiquitin, indicating that the polyubiquitinated mutant ATXN1 is resistant to the proteasome-mediated degradation in vivo.
The ubiquitination system consists of several enzymes, including the ubiquitin-activating enzyme E1, the ubiquitin-conjugating enzyme E2, and the ubiquitin-ligase E3. ATXN1 is known to interact with UBCH6, an E2 ubiquitin-conjugating enzyme, which promotes polyubiquitination and the subsequent degradation of ATXN1 [43]. By contrast, depletion of UBCH6 by siRNA stabilizes ATXN1 expression. Furthermore, UBCH6 modulates the transcriptional repression activity of ATXN1 by affecting ATXN1 degradation [44]. Overexpression of UBCH6 partially inhibits the transcriptional repressor activity of ATXN1, but knock-down of UBCH6 enhances it. Moreover, UBCH6 is more efficient at modulating the transcriptional activity of the wild-type ATXN1 than that of the polyglutamine-expanded ATXN1. As aberrant transcription is implicated in the pathogenesis of SCA1 [23, 45-50], UBCH6-mediated changes in the transcriptional regulation function of ATXN1 may thus contribute to SCA1. Moreover, it suggests that the protein degradation pathway is tightly associated with the toxic gain-of-function of the mutant ATXN1 in transcription.
CHIP is an E3 ubiquitin ligase known to interact with the molecular chaperon Hsp70, and it links the protein folding machinery with the UPS. CHIP and Hsp70 cooperatively re-fold and degrade mis-folded mutant proteins [51, 52]. CHIP interacts and co-localizes with ATXN1 in nuclear inclusions in cultured cells and post-mortem tissues [53]. When overexpressed with E1 and E2 enzymes, CHIP promotes the polyubiquitination and degradation of the mutant ATXN1, and thus suppresses SCA1 toxicity in Drosophila. Hsp70 is associated with CHIP and synergistically controls protein homeostasis. Hsp70 overexpression further promotes CHIP-mediated ubiquitination of ATXN1 in vitro and rescues motor deficits and pathology in SCA1 transgenic mice [54]. In addition, one study found that the E3 ubiquitin ligase UBE3A is also involved in SCA1 pathogenesis [42]. In this study, the loss of Ube3a aggravated cerebellar pathology in SCA1 transgenic mice. Cerebellar pathology, such as molecular cell layer atrophy and loss of dendritic arborization and PC heterotopia, are much more prominent in SCA1 mice with a Ube3a deletion than in age-matched SCA1 mice alone. Furthermore, ubiquitin-like proteins, such as FAT10 and A1Up, are also known to bind and regulate the stability of mutant ATXN1 proteins [55, 56], although the precise mechanisms and its relevancy to the disease are unknown. Taken together, various components of the UPS pathway are involved in the clearance of mutant ATXN1 proteins. Activation of the UPS promotes the degradation of the polyglutamine-expanded mutant ATNX1 and ameliorates SCA1 phenotypes, but its impairment worsens the SCA1 neurotoxicity (Fig. 2).
Sumoylation
ATXN1 is known to be sumoylated [57]. Sumoylation is a post-translational modification that covalently attaches the SUMO (small ubiquitin-like modifier) protein to target proteins. Sumoylation is involved in many different biological processes, such as transcription, apoptosis, protein stability, nuclear-cytoplasmic shuttling, DNA damage response, and cell cycle progression [58]. There are four SUMO genes in humans, SUMO-1, SUMO-2, SUMO-3, and SUMO-4. A similar enzymatic cascade as in ubiquitination is used for sumoylation. The ubiquitin-like modifier activating enzyme 2 (UBA2) and ubiquitin-conjugating enzyme 9 (UBC9) function as E1 and E2 enzymes for sumoylation, respectively. Several known E3 enzymes include PIAS (protein inhibitors of activated STATs), RANBP2 (RAN binding protein 2), and PC2 (polycomb 2). In patients with polyglutamine diseases, such as HD, DRPLA, SCA1, and SCA3, SUMO-1 was detected in affected brain regions, indicating that sumoylation is involved in these diseases [59-62].
There are seventeen consensus sumoylation sites (Ψ -K-X-D/E; Ψ is a hydrophobic residue) in ATXN1 (Fig. 1) [57]. Among them, mutations in K16, K194, K610, K697, and K746 dramatically decrease sumoylation of ATXN1, suggesting that they are primary sumoylation sites. The sumoylation of ATXN1 is closely linked to several known pathogenic factors of SCA1, such as the length of the polyglutamine tract [11], S776 phosphorylation [9], and the nuclear localization [7] of ATXN1. For example, the expanded polyglutamine tract decreases ATXN1 sumoylation, and K194 is located at the start of the polyglutamine tract. In a cell culture system, overexpression of mutant ATXN1 co-localizes with SUMO-1 in the nucleus. In addition, co-overexpression of either SUMO-1 or UBC9 can promote the aggregation of mutant ATXN1 proteins [63]. Induced oxidative stress stimulates, while reduced oxidative stress decreases ATXN1 sumoylation and its aggregation. These data raise the possibility that sumoylation may contribute to SCA1 pathogenesis (Fig. 2). Nevertheless, how sumoylation affects the function and regulation of ATNX1 and its precise role in SCA1 pathogenesis still remains to be elucidated.
Transglutamination
Lastly, another protein modification known to occur on the polyglutamine-expanded ATXN1 protein is transglutamination. Transglutamination forms a covalent bond between a free amine group and the acyl group at the end of the side chain of glutamine, thereby cross-linking proteins [64]. Thus, peptides with glutamine repeats can be substrates for the enzyme transglutaminase [65-67]. Consistent with this, transglutamination is suggested to play a role in polyglutamine diseases [68]. Elevated transglutaminase activity was observed in HD patient brains in vivo [69, 70], and transglutaminase-mediated isopeptide bonds were detected in SBMA transgenic mice [71]. Furthermore, transglutamination promotes the aggregation of the polyglutamine-expanded mutant proteins [72], and the cross-linked mutant proteins impair the function of the proteasome by obstructing the proteasome pore [71]. These studies strongly suggest that transglutamination may be an important event in protein aggregation formation and the pathogenesis of polyglutamine diseases. Tissue transglutaminase 2 (TG2) catalyzes the transglutamination reaction in a calcium-dependent manner, and is highly expressed in PCs of the cerebellum [73]. Interestingly, ATXN1 was reported as a substrate of TG2, and elevated nuclear TG2 expression was found in the cerebellar PCs of SCA1 mice [74, 75]. Thus, transglutamination is likely to be involved in the process of mutant ATXN1 aggregate formation (Fig. 2), similar to what was seen in HD and SBMA. Further studies are required to evaluate the relevance of this modification to SCA1 pathogenesis.
Conclusions
The expression and altered activity of the polyglutamine-expanded mutant ATXN1 protein are the major determinants of SCA1 neurotoxicity. Post-translational modifications are able to modulate the stability and activity of the mutant ATXN1 protein (Fig. 2). For example, phosphorylation stabilizes ATXN1 levels and regulates its target gene expression by modulating the formation of native protein complexes. Ubiquitination directs the degradation of the mutant ATXN1 protein, and sumoylation may modulate the aggregation and/or transcriptional activity of ATXN1. Numerous molecules, including kinases, phosphatases, ubiquitin ligases, chaperones, and transcription and splicing factors are involved in diverse aspects of the post-translational modifications that target ATXN1. The comprehensive understanding of the role of post-translational modifications in ATXN1 biology will not only fundamentally advance our understanding of the function of this protein and the pathogenesis of SCA1, but also provide a new target for the development of effective therapeutics.
Acknowledgments
We are grateful to the members of the Lim laboratory for their thoughtful comments on the manuscript and Tiffany Todd for editorial input. This work was generously supported by the National Institutes of Neurological Disorders and Stroke grant NS064146, National Ataxia Foundation, Brain & Behavior Research Foundation (Formerly NARSAD), Alfred P. Sloan Foundation, Charles H. Hood Foundation, National Multiple Sclerosis Society, and Yale Scholar Award Program to J.L.
References
- 1.Karve TM, Cheema AK. Small changes huge impact: the role of protein posttranslational modifications in cellular homeostasis and disease. J Amino Acids. 2011;2011:207691. doi: 10.4061/2011/207691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pennuto M, Palazzolo I, Poletti A. Post-translational modifications of expanded polyglutamine proteins: impact on neurotoxicity. Hum Mol Genet. 2009;18(R1):R40–7. doi: 10.1093/hmg/ddn412. [DOI] [PubMed] [Google Scholar]
- 3.Ross CA. Polyglutamine pathogenesis: emergence of unifying mechanisms for Huntington's disease and related disorders. Neuron. 2002;35(5):819–22. doi: 10.1016/s0896-6273(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 4.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 5.Zoghbi HY, Orr HT. Pathogenic mechanisms of a polyglutamine-mediated neurodegenerative disease, spinocerebellar ataxia type 1. J Biol Chem. 2009;284(12):7425–9. doi: 10.1074/jbc.R800041200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orr HT. SCA1-phosphorylation, a regulator of Ataxin-1 function and pathogenesis. Prog Neurobiol. 2012;99(3):179–85. doi: 10.1016/j.pneurobio.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Klement IA, et al. Ataxin-1 nuclear localization and aggregation: role in polyglutamine-induced disease in SCA1 transgenic mice. Cell. 1998;95(1):41–53. doi: 10.1016/s0092-8674(00)81781-x. [DOI] [PubMed] [Google Scholar]
- 8.Tsuda H, et al. The AXH domain of Ataxin-1 mediates neurodegeneration through its interaction with Gfi-1/Senseless proteins. Cell. 2005;122(4):633–44. doi: 10.1016/j.cell.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 9.Emamian ES, et al. Serine 776 of ataxin-1 is critical for polyglutamine-induced disease in SCA1 transgenic mice. Neuron. 2003;38(3):375–87. doi: 10.1016/s0896-6273(03)00258-7. [DOI] [PubMed] [Google Scholar]
- 10.Huttlin EL, et al. A tissue-specific atlas of mouse protein phosphorylation and expression. Cell. 2010;143(7):1174–89. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Burright EN, et al. SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell. 1995;82(6):937–48. doi: 10.1016/0092-8674(95)90273-2. [DOI] [PubMed] [Google Scholar]
- 12.Clark HB, et al. Purkinje cell expression of a mutant allele of SCA1 in transgenic mice leads to disparate effects on motor behaviors, followed by a progressive cerebellar dysfunction and histological alterations. J Neurosci. 1997;17(19):7385–95. doi: 10.1523/JNEUROSCI.17-19-07385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duvick L, et al. SCA1-like disease in mice expressing wild-type ataxin-1 with a serine to aspartic acid replacement at residue 776. Neuron. 2010;67(6):929–35. doi: 10.1016/j.neuron.2010.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lim J, et al. Opposing effects of polyglutamine expansion on native protein complexes contribute to SCA1. Nature. 2008;452(7188):713–8. doi: 10.1038/nature06731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen HK, et al. Interaction of Akt-phosphorylated ataxin-1 with 14-3-3 mediates neurodegeneration in spinocerebellar ataxia type 1. Cell. 2003;113(4):457–68. doi: 10.1016/s0092-8674(03)00349-0. [DOI] [PubMed] [Google Scholar]
- 16.Lam YC, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127(7):1335–47. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 17.Bolger TA, et al. The neurodegenerative disease protein ataxin-1 antagonizes the neuronal survival function of myocyte enhancer factor-2. J Biol Chem. 2007;282(40):29186–92. doi: 10.1074/jbc.M704182200. [DOI] [PubMed] [Google Scholar]
- 18.de Chiara C, et al. Phosphorylation of S776 and 14-3-3 binding modulate ataxin-1 interaction with splicing factors. PLoS One. 2009;4(12):e8372. doi: 10.1371/journal.pone.0008372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Umahara T, Uchihara T. 14-3-3 proteins and spinocerebellar ataxia type 1: from molecular interaction to human neuropathology. Cerebellum. 2010;9(2):183–9. doi: 10.1007/s12311-010-0158-9. [DOI] [PubMed] [Google Scholar]
- 20.Lai S, et al. 14-3-3 Binding to ataxin-1(ATXN1) regulates its dephosphorylation at Ser-776 and transport to the nucleus. J Biol Chem. 2011;286(40):34606–16. doi: 10.1074/jbc.M111.238527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jafar-Nejad P, et al. Regional rescue of spinocerebellar ataxia type 1 phenotypes by 14-3-3epsilon haploinsufficiency in mice underscores complex pathogenicity in neurodegeneration. Proc Natl Acad Sci U S A. 2011;108(5):2142–7. doi: 10.1073/pnas.1018748108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowman AB, et al. Duplication of Atxn1l suppresses SCA1 neuropathology by decreasing incorporation of polyglutamine-expanded ataxin-1 into native complexes. Nat Genet. 2007;39(3):373–379. doi: 10.1038/ng1977. [DOI] [PubMed] [Google Scholar]
- 23.Fryer JD, et al. Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science. 2011;334(6056):690–3. doi: 10.1126/science.1212673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.She H, Mao Z. Regulation of myocyte enhancer factor-2 transcription factors by neurotoxins. Neurotoxicology. 2011;32(5):563–6. doi: 10.1016/j.neuro.2011.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jorgensen ND, et al. Phosphorylation of ATXN1 at Ser776 in the cerebellum. J Neurochem. 2009;110(2):675–86. doi: 10.1111/j.1471-4159.2009.06164.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hearst SM, et al. Dopamine D2 receptor signaling modulates mutant ataxin-1 S776 phosphorylation and aggregation. J Neurochem. 2010;114(3):706–16. doi: 10.1111/j.1471-4159.2010.06791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goold R, et al. Down-regulation of the dopamine receptor D2 in mice lacking ataxin 1. Hum Mol Genet. 2007;16(17):2122–34. doi: 10.1093/hmg/ddm162. [DOI] [PubMed] [Google Scholar]
- 28.Park J, et al. RAS-MAPK-MSK1 pathway modulates ataxin 1 protein levels and toxicity in SCA1. Nature. 2013;498(7454):325–31. doi: 10.1038/nature12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sanchez I, et al. A novel function of Ataxin-1 in the modulation of PP2A activity is dysregulated in the spinocerebellar ataxia type 1. Hum Mol Genet. 2013;22(17):3425–37. doi: 10.1093/hmg/ddt197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn JH, et al. Protein kinase A activates protein phosphatase 2A by phosphorylation of the B56delta subunit. Proc Natl Acad Sci U S A. 2007;104(8):2979–84. doi: 10.1073/pnas.0611532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cvetanovic M, Kular RK, Opal P. LANP mediates neuritic pathology in Spinocerebellar ataxia type 1. Neurobiol Dis. 2012;48(3):526–32. doi: 10.1016/j.nbd.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kular RK, et al. Neuronal differentiation is regulated by leucine-rich acidic nuclear protein (LANP), a member of the inhibitor of histone acetyltransferase complex. J Biol Chem. 2009;284(12):7783–92. doi: 10.1074/jbc.M806150200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Opal P, et al. Mapmodulin/leucine-rich acidic nuclear protein binds the light chain of microtubule-associated protein 1B and modulates neuritogenesis. J Biol Chem. 2003;278(36):34691–9. doi: 10.1074/jbc.M302785200. [DOI] [PubMed] [Google Scholar]
- 34.Vierra-Green CA, et al. Identification of a novel phosphorylation site in ataxin-1. Biochim Biophys Acta. 2005;1744(1):11–8. doi: 10.1016/j.bbamcr.2004.10.012. [DOI] [PubMed] [Google Scholar]
- 35.Ju H, et al. Polyglutamine Disease Toxicity Is Regulated by Nemo-like Kinase in Spinocerebellar Ataxia Type 1. J Neurosci. 2013;33(22):9328–9336. doi: 10.1523/JNEUROSCI.3465-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim J, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125(4):801–14. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 37.Choi KW, Benzer S. Rotation of photoreceptor clusters in the developing Drosophila eye requires the nemo gene. Cell. 1994;78(1):125–36. doi: 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- 38.Banfi S, et al. Identification and mapping of human cDNAs homologous to Drosophila mutant genes through EST database searching. Nat Genet. 1996;13(2):167–74. doi: 10.1038/ng0696-167. [DOI] [PubMed] [Google Scholar]
- 39.Brott BK, Pinsky BA, Erikson RL. Nlk is a murine protein kinase related to Erk/MAP kinases and localized in the nucleus. Proc Natl Acad Sci U S A. 1998;95(3):963–8. doi: 10.1073/pnas.95.3.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meneghini MD, et al. MAP kinase and Wnt pathways converge to downregulate an HMG-domain repressor in Caenorhabditis elegans. Nature. 1999;399(6738):793–7. doi: 10.1038/21666. [DOI] [PubMed] [Google Scholar]
- 41.Todd TW, Lim J. Aggregation formation in the polyglutamine diseases: protection at a cost? Mol Cells. 2013;36(3):185–94. doi: 10.1007/s10059-013-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cummings CJ, et al. Mutation of the E6-AP ubiquitin ligase reduces nuclear inclusion frequency while accelerating polyglutamine-induced pathology in SCA1 mice. Neuron. 1999;24(4):879–92. doi: 10.1016/s0896-6273(00)81035-1. [DOI] [PubMed] [Google Scholar]
- 43.Hong S, et al. UbcH6 interacts with and ubiquitinates the SCA1 gene product ataxin-1. Biochem Biophys Res Commun. 2008;371(2):256–60. doi: 10.1016/j.bbrc.2008.04.066. [DOI] [PubMed] [Google Scholar]
- 44.Lee S, Hong S, Kang S. The ubiquitin-conjugating enzyme UbcH6 regulates the transcriptional repression activity of the SCA1 gene product ataxin-1. Biochem Biophys Res Commun. 2008;372(4):735–40. doi: 10.1016/j.bbrc.2008.05.125. [DOI] [PubMed] [Google Scholar]
- 45.Lin X, et al. Polyglutamine expansion down-regulates specific neuronal genes before pathologic changes in SCA1. Nat Neurosci. 2000;3(2):157–63. doi: 10.1038/72101. [DOI] [PubMed] [Google Scholar]
- 46.Serra HG, et al. Gene profiling links SCA1 pathophysiology to glutamate signaling in Purkinje cells of transgenic mice. Hum Mol Genet. 2004;13(20):2535–43. doi: 10.1093/hmg/ddh268. [DOI] [PubMed] [Google Scholar]
- 47.Cvetanovic M, et al. Vascular endothelial growth factor ameliorates the ataxic phenotype in a mouse model of spinocerebellar ataxia type 1. Nat Med. 2011;17(11):1445–7. doi: 10.1038/nm.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cvetanovic M, et al. The role of LANP and ataxin 1 in E4F-mediated transcriptional repression. EMBO Rep. 2007;8(7):671–7. doi: 10.1038/sj.embor.7400983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Crespo-Barreto J, et al. Partial loss of ataxin-1 function contributes to transcriptional dysregulation in spinocerebellar ataxia type 1 pathogenesis. PLoS Genet. 2010;6(7):e1001021. doi: 10.1371/journal.pgen.1001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gatchel JR, et al. The insulin-like growth factor pathway is altered in spinocerebellar ataxia type 1 and type 7. Proc Natl Acad Sci U S A. 2008;105(4):1291–6. doi: 10.1073/pnas.0711257105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qian SB, et al. CHIP-mediated stress recovery by sequential ubiquitination of substrates and Hsp70. Nature. 2006;440(7083):551–5. doi: 10.1038/nature04600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McDonough H, Patterson C. CHIP: a link between the chaperone and proteasome systems. Cell Stress Chaperones. 2003;8(4):303–8. doi: 10.1379/1466-1268(2003)008<0303:calbtc>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Al-Ramahi I, et al. CHIP protects from the neurotoxicity of expanded and wild-type ataxin-1 and promotes their ubiquitination and degradation. J Biol Chem. 2006;281(36):26714–24. doi: 10.1074/jbc.M601603200. [DOI] [PubMed] [Google Scholar]
- 54.Cummings CJ, et al. Chaperone suppression of aggregation and altered subcellular proteasome localization imply protein misfolding in SCA1. Nat Genet. 1998;19(2):148–54. doi: 10.1038/502. [DOI] [PubMed] [Google Scholar]
- 55.Hipp MS, et al. FAT10, a ubiquitin-independent signal for proteasomal degradation. Mol Cell Biol. 2005;25(9):3483–91. doi: 10.1128/MCB.25.9.3483-3491.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davidson JD, et al. Identification and characterization of an ataxin-1-interacting protein: A1Up, a ubiquitin-like nuclear protein. Hum Mol Genet. 2000;9(15):2305–12. doi: 10.1093/oxfordjournals.hmg.a018922. [DOI] [PubMed] [Google Scholar]
- 57.Riley BE, Zoghbi HY, Orr HT. SUMOylation of the polyglutamine repeat protein, ataxin-1, is dependent on a functional nuclear localization signal. J Biol Chem. 2005;280(23):21942–8. doi: 10.1074/jbc.M501677200. [DOI] [PubMed] [Google Scholar]
- 58.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- 59.Ueda H, et al. Enhanced SUMOylation in polyglutamine diseases. Biochem Biophys Res Commun. 2002;293(1):307–13. doi: 10.1016/S0006-291X(02)00211-5. [DOI] [PubMed] [Google Scholar]
- 60.Dorval V, Fraser PE. SUMO on the road to neurodegeneration. Biochim Biophys Acta. 2007;1773(6):694–706. doi: 10.1016/j.bbamcr.2007.03.017. [DOI] [PubMed] [Google Scholar]
- 61.Pountney DL, et al. SUMO-1 marks the nuclear inclusions in familial neuronal intranuclear inclusion disease. Exp Neurol. 2003;184(1):436–46. doi: 10.1016/j.expneurol.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 62.Kang S, Hong S. SUMO-1 interacts with mutant ataxin-1 and colocalizes to its aggregates in Purkinje cells of SCA1 transgenic mice. Arch Ital Biol. 2010;148(4):351–63. doi: 10.4449/aib.v148i4.1201. [DOI] [PubMed] [Google Scholar]
- 63.Ryu J, et al. Oxidative stress-enhanced SUMOylation and aggregation of ataxin-1: Implication of JNK pathway. Biochem Biophys Res Commun. 2010;393(2):280–5. doi: 10.1016/j.bbrc.2010.01.122. [DOI] [PubMed] [Google Scholar]
- 64.Lorand L, Graham RM. Transglutaminases: crosslinking enzymes with pleiotropic functions. Nat Rev Mol Cell Biol. 2003;4(2):140–56. doi: 10.1038/nrm1014. [DOI] [PubMed] [Google Scholar]
- 65.Kahlem P, et al. Peptides containing glutamine repeats as substrates for transglutaminase-catalyzed cross-linking: relevance to diseases of the nervous system. Proc Natl Acad Sci U S A. 1996;93(25):14580–5. doi: 10.1073/pnas.93.25.14580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cooper AJ, et al. Polyglutamine domains are substrates of tissue transglutaminase: does transglutaminase play a role in expanded CAG/poly-Q neurodegenerative diseases? J Neurochem. 1997;69(1):431–4. doi: 10.1046/j.1471-4159.1997.69010431.x. [DOI] [PubMed] [Google Scholar]
- 67.Lai TS, et al. Effect of tissue transglutaminase on the solubility of proteins containing expanded polyglutamine repeats. J Neurochem. 2004;88(5):1253–60. doi: 10.1046/j.1471-4159.2003.02249.x. [DOI] [PubMed] [Google Scholar]
- 68.Bailey CD, Tucholski J, Johnson GV. Transglutaminases in neurodegenerative disorders. Prog Exp Tumor Res. 2005;38:139. doi: 10.1159/000084238. [DOI] [PubMed] [Google Scholar]
- 69.Karpuj MV, et al. Transglutaminase aggregates huntingtin into nonamyloidogenic polymers, and its enzymatic activity increases in Huntington's disease brain nuclei. Proc Natl Acad Sci U S A. 1999;96(13):7388–93. doi: 10.1073/pnas.96.13.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zainelli GM, et al. Calmodulin regulates transglutaminase 2 cross-linking of huntingtin. J Neurosci. 2004;24(8):1954–61. doi: 10.1523/JNEUROSCI.4424-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mandrusiak LM, et al. Transglutaminase potentiates ligand-dependent proteasome dysfunction induced by polyglutamine-expanded androgen receptor. Hum Mol Genet. 2003;12(13):1497–506. doi: 10.1093/hmg/ddg161. [DOI] [PubMed] [Google Scholar]
- 72.Kahlem P, Green H, Djian P. Transglutaminase action imitates Huntington's disease: selective polymerization of Huntingtin containing expanded polyglutamine. Mol Cell. 1998;1(4):595–601. doi: 10.1016/s1097-2765(00)80059-3. [DOI] [PubMed] [Google Scholar]
- 73.Maggio N, et al. Tissue-transglutaminase in rat and human brain: light and electron immunocytochemical analysis and in situ hybridization study. Brain Res Bull. 2001;56(3-4):173–82. doi: 10.1016/s0361-9230(01)00649-9. [DOI] [PubMed] [Google Scholar]
- 74.D'Souza DR, et al. Tissue transglutaminase crosslinks ataxin-1: possible role in SCA1 pathogenesis. Neurosci Lett. 2006;409(1):5–9. doi: 10.1016/j.neulet.2006.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vig PJ, et al. Role of tissue transglutaminase type 2 in calbindin-D28k interaction with ataxin-1. Neurosci Lett. 2007;420(1):53–7. doi: 10.1016/j.neulet.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]