Abstract
Pseudomonas aeruginosa are noscomially acquired, opportunistic pathogens that pose a major threat to the health of burns patients and the immunocompromised. We sequenced the genomes of P. aeruginosa isolates RNS_PA1, RNS_PA46 and RNS_PAE05, which displayed resistance to almost all frontline antibiotics, including gentamicin, piperacillin, timentin, meropenem, ceftazidime and colistin. We provide evidence that the isolates are representatives of P. aeruginosa sequence type (ST) 235 and carry Tn6162 and Tn6163 in genomic islands 1 (GI1) and 2 (GI2), respectively. GI1 disrupts the endA gene at precisely the same chromosomal location as in P. aeruginosa strain VR-143/97, of unknown ST, creating an identical CA direct repeat. The class 1 integron associated with Tn6163 in GI2 carries a blaGES-5–aacA4–gcuE15–aphA15 cassette array conferring resistance to carbapenems and aminoglycosides. GI2 is flanked by a 12 nt direct repeat motif, abuts a tRNA-gly gene, and encodes proteins with putative roles in integration, conjugative transfer as well as integrative conjugative element-specific proteins. This suggests that GI2 may have evolved from a novel integrative conjugative element. Our data provide further support to the hypothesis that genomic islands play an important role in de novo evolution of multiple antibiotic resistance phenotypes in P. aeruginosa.
Keywords: Pseudomonas aeruginosa, multiple drug resistance, genomic island, class 1 integron, β-lactamases
1. Introduction
Pseudomonas aeruginosa is an opportunistic pathogen and a significant threat to the health of immunocompromised patients hospitalized for periods longer than 7 days [1]. It is a frequent cause of nosocomially derived infections [2,3] and can survive on hospital fomites for extended periods [4–7]. Multiple drug-resistant (MDR), extensively drug-resistant (XDR) and pan-resistant [8,9] P. aeruginosa linages cause life-threatening infections in nosocomial environments and severely restrict drug options to treat infection. As such, MDR P. aeruginosa is expected to contribute significantly to increased rates of morbidity and mortality in hospital environments in the twenty-first century.
The population structure of P. aeruginosa is described as non-clonal and epidemic [10]. Several high-risk clonal lineages including ST111, ST235 and ST175 carry genes that confer resistance to β-lactam antibiotics [10–18] and ST235 clones commonly found in Asia are predominantly associated with the dissemination of IMP type β-lactamase resistance genes [15–17,19–22]. MDR P. aeruginosa that encode resistance to the Carbapenem group of antibiotics via the production of molecular class A β-lactamase and class B β-lactamase (metallo-β-lactamases, MBLs) enzymes pose a serious threat to the health of humans and animals because only a limited number of drugs remain available to counteract infections caused by these pathogens. Genes encoding resistance to the molecular class A β-lactamase, extended-spectrum β-lactamase (ESBLs) and MBL antibiotics are often carried on class 1 integrons.
The acquisition of class 1 integrons plays a significant role in the evolution of strains of P. aeruginosa that are recalcitrant to combination antibiotic therapy, thereby making them extensively drug-resistant (XDR) [23]. However, class 1 integrons are defective transposons, and an association with functional transposons, genomic islands and plasmids is critical for their dissemination. Within the Enterobacteriaceae, plasmids play a key role in mobilizing class 1 integrons. In P. aeruginosa, numerous studies correlate the presence of class 1 integrons with multiple antibiotic resistance, but the genetic context surrounding class 1 integrons is often not investigated. Our earlier studies implicated the important role played by genomic islands in the capture and spread of clustered antibiotic resistance genes within P. aeruginosa [24–26].
In 2009, we described a 25.5 kb transposon Tn6060, which contained two outwardly facing class 1 integrons encoding resistance to eight antibiotics. Tn6060 is found in the res site of a transposon with more than 99% sequence identity to Tn1403 within a genomic island (described as GI1) on the chromosome of P. aeruginosa strain 37308 [25]. Novel class 1 integrons and transposons were subsequently described in at least four distinct chromosomal locations among clinical isolates from Australia and Uruguay [26]. These integrons carried identical cassette arrays comprising aadA6 (encoding resistance to streptomycin and spectinomycin) and gcuD (formerly orfD) within a novel transposon, Tn6162, that is located at precisely the same position as Tn6060 in GI1. Class 1 integrons harbouring the aadA6-gcuD array have been reported from geographically divergent regions including France [27] and in an extensively drug-resistant P. aeruginosa isolate from Thailand [28], but the genetic context of the integron-arrays remains unknown. We also described an Australian P. aeruginosa strain, C79, containing a second-class 1 integron that carries blaGES5 (encoding resistance to extended-spectrum β-lactams), an aacA4-like gene cassette (encoding resistance to aminoglycosides), gcuE15 (unknown function), aphA15 (aminoglycoside resistance) and several IS elements in genomic island 2 (GI2) [26]. The integron in strain C79 was found within a mercury resistance transposon Tn4380 known as Tn6163, also located in a genomic island, described as island 2 or GI2 [26]. While Tn6163 was not detected in isolates from Uruguay, it was identified in seven clonally related clinical isolates of P. aeruginosa collected from Sydney in 2010 [26]. Although the structures of transposons Tn6060, Tn6162 and Tn6163 have been described in detail, the genomic islands harbouring them were not characterized in any of our previous studies.
In P. aeruginosa strain VR-143/97, the blaVIM-1 metallo-β-lactamase gene cassette is associated with a class 1 integron (In70.2) in a defective Tn402-like transposon that is inserted in the urf2 gene of a Tn3-derivate transposon known as Tn6249. Like Tn6060, Tn6249 has a complex structure and carries two class 1 integrons (In90 and In70.2) in a tail-to-tail orientation [29]. Tn6249 is located in a PACS171b-like genomic island, described earlier as GI1 in relation to Tn6060 [25], within the endA gene of the P. aeruginosa chromosome [29]. Tn6249 shares structural identity with Tn6060, although the gene cassettes carried by In90 are different and a part of the Tn3 backbone is missing in Tn6249 [25]. Notably, Tn6162 is also located at an identical position in the genomic backbone of GI1 as Tn6060, but has a simpler complex resistance locus (CRL) consisting of one class 1 integron. Consequently, it has been hypothesized that insertion of a Tn6162-like ancestor into the PACS171b-like genomic island (VR-143/97; 51 424 nt) preceded the genetic events that gave rise to Tn6249 and Tn6060 in clonally dispersed lineages of P. aeruginosa [29].
Here we present an in-depth, whole-genome-based analysis of three MDR ST235 isolates carrying both Tn6162 in GI1 and Tn6163 in GI2. All three isolates are resistant to the entire range of antibiotics used to treat P. aeruginosa infections except colistin (intermediate resistance to colistin) and are part of a larger collection of phylogenetically related MDR isolates recovered in 2006 and 2007 from a burns ward in a hospital in Sydney. The genetic structure of GI1 and GI2 found in P. aeruginosa isolates in Sydney is presented. Our study highlights the roles played by GI1 and GI2 in the capture and dissemination of multiple antibiotic resistance genes in P. aeruginosa ST235 in the Sydney basin.
2. Material and methods
2.1. Strains, isolation and culture conditions
The P. aeruginosa isolates examined in this study were obtained during an outbreak of a multiple antibiotic-resistant P. aeruginosa infection at the Royal North Shore Hospital (RNSH) in the burns unit during 2006 and 2007. Isolates RNS_PA1 (rectal swab, November 2006), RNS_PA46 (burn wound, May 2007) and RNS_PAE05 (hand sanitizer, April 2007) were recovered as part of routine microbiological surveillance during the outbreak. The isolates were identified as P. aeruginosa using standard microbiological protocols [30] and stored at −80°C.
For molecular studies, the isolates were routinely grown at 37°C on Luria–Bertani (LB) agar or overnight in LB broth with shaking at 125 r.p.m. For antibiotic sensitivity testing, cells from two to three colonies growing on blood agar were subcultured in tryptone soy broth (TSB) for 3 h at 37°C prior to plating on antibiotic supplemented Columbia agar. For extraction of sequencing quality DNA, cells were grown in 5 ml of M63 minimal salts broth for 20 h at 37°C in a shaking incubator set at 125 r.p.m., harvested by centrifugation and stored at −80°C.
2.2. DNA extraction
Genomic DNA for PCR was extracted from 200 µl of a TSB culture inoculated with several colonies from blood agar plates. DNA was extracted on a BioRobot M48 workstation (Qiagen) at RNSH following the manufacturer's instructions. For next-generation genomic sequencing, DNA samples were extracted using Bioline's ISOLATE-II Genomic DNA extraction kit following recommended protocols.
2.3. RAPD analysis
RAPD analysis was conducted using puRe Taq Ready-To-Go PCR beads (GE Healthcare) with 20 ng of template DNA, using published protocols [31]. The amplicons were resolved and visualized on a QIAxcel (Qiagen) 12-channel capillary electrophoresis system. Data were exported to BioNumerics 6 (Applied Maths) software for comparison and analysis.
2.4. PCR
Routine PCR was conducted in a total volume of 20 µl using Mango Taq (Bioline) master mix. Cycling conditions comprised an initial denaturation step at 94°C for 2 min 30 s, followed by 30 cycles of denaturation (94°C for 30 s) annealing (60°C for 30 s) and extension (72°C for 5 min). An EmeraldAmp Taq polymerase (Master Mix) was used in long-range PCR to bridge Illumina sequence scaffolds. Primer sequences and amplicon sizes are listed in electronic supplementary material, table S1.
2.5. Antibiotic resistance testing
Antibiotic resistance profiles were generated following the CLSI guidelines [32]. Plates were incubated overnight at 37°C. The antibiotic concentration at which colony growth was inhibited was noted as the minimum inhibitory concentration (MIC). Reference strains used in the experiment were P. aeruginosa ATCC27853 and E. coli ATCC25922.
2.6. Sanger sequencing and whole genome sequencing
Promega Wizard SV PCR and Gel Clean Up kits were used to prepare PCR amplicons for sequencing. Sanger sequencing was performed at the Australian Genome Research Facility at Westmead Hospital, Westmead, NSW.
Genome sequencing was performed using a bench top Illumina MiSeq sequencer and MiSeq V3 chemistry at the ithree institute at UTS [33]. Genomes were submitted to GenBank using the following accession numbers: RNS_PA1 (SAMN04038435), RNS_PA46 (SAMN04038437) and RNS_PAE05 (SAMN04038440). 400 nt Illumina reads were assembled using the A5-MiSeq (ngopt_a5pipeline_linux-x64_20130919)) de novo assembly pipeline [34]. Preliminary genome annotations were performed with an online version of RAST [35,36] using FigFAM release 70. Sequences of interest from the preliminary annotation were manually curated using iterative NCBI-ORF finder software coupled with BLASTn and BLASTp analysis. ORFs that returned identity across 100% of the input query sequence in BLASTp analysis were scored.
2.7. Bioinformatic analysis
Phylogenetic comparisons of the three P. aeruginosa genomes (RNS_PA1, RNS_PA46 and RNS_PAE05) with nine completely closed P. aeruginosa genomes available in NCBI (accessed on 3 September 2015) was performed using PhyloSift [37]. Genome sequences of Vibrio cholerae N16961 (NC_002505.1) and Salmonella enterica serovar typhimurium DT104 (NC_022569.1) were used as outgroups in the analysis. FigTree v. 1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/) was used to draw the phylogenetic tree. Two closely related, finished genomes (PA_NCGM2 and PA_PA14) were chosen from the PhyloSift output and used to do a second phylogenetic analysis based on the whole genome alignment protocol in REALPHY [38].
The MLST database (http://pubmlst.org/paeruginosa/) was used to identify the sequence type (ST) of the isolates. Whole genome comparative analysis was performed using MAUVE [39], and regions of interest between the three genomes were further characterized using iterative BLASTn and BLASTp searches [40]. Figures were compiled using BRIG [41].
3. Results
3.1. Characterization of three multiple drug-resistant Pseudomonas aeruginosa isolates from the Royal North Shore Hospital burns unit
The three MDR P. aeruginosa isolates examined in this study were representative of a collection of 36 isolates from RNSH that shared 90% similarity by RAPD analyses (electronic supplementary material, figure S1). PCR analyses indicated that all 36 isolates carried a class 1 integron and a characteristic 1800 bp cassette array amplicon akin to the array present in Tn6162. The amplicon was shown to contain aadA6 and gcuD gene cassettes by nested PCR using a primer pair that internally annealed to the aadA6 (HS513) and gcuD (gcuD-Rv) genes, confirming that these isolates carry Tn6162. Of the 36 isolates, 27 also produced a 2407 bp amplicon using a forward primer (HS1299) in blaGES-5 and reverse primer (HS1236) in aphA15. The 2407 bp amplicon is indicative of the presence of Tn6163 containing blaGES-5–aacA4–gcuE15–aphA15 gene cassettes [26]. Isolates included in this study were tested for sensitivity to antibiotics used at RNSH to treat P. aeruginosa infections, including gentamicin, piperacillin, ticarcillin and clavulanic acid, meropenem, ceftazidime and colistin (table 1). Defining features of the three isolates selected for genome sequence analysis were (i) that they were isolated from different patients/sources at the RNSH over a seventh month period, (ii) that they displayed minor differences in their resistance profile and intermediate resistance to colistin (table 1), a last line treatment option for P. aeruginosa infections, and (iii) a suspicion that they carried Tn6162 and Tn6163 using the PCR strategy described above. As such, these three isolates represent a set that predates (by 4 years) strains from which Tn6162 and Tn6163 were originally described in Australia [26] from a different hospital in the Sydney basin.
Table 1.
Antibiotic resistance profiles of P. aeruginosa isolates. R, resistant to antibiotic; I, intermediate resistance to antibiotic; S, susceptible to antibiotic; number in parentheses is the MIC in µl ml−1. GEN, gentamicin (aminoglycoside); PIP, piperacillin (extended-spectrum β-lactam); TCA, ticracillin [β-lactam]/clavulanic acid [β-lactamase inhibitor]; MPM, meropenem (carbapenem); CAZ, ceftazidime (third-generation cephalosporin); COL, colistin (polymixin E).
| isolate | GENa | PIPb | TCAc | MPMd | CAZe | COLf |
|---|---|---|---|---|---|---|
| RNS_PA1 | R (>32) | R (>256) | R (128/2) | R (64) | R (32) | I (4) |
| RNS_PA46 | R (>32) | R (128) | R (128/2) | R (128) | R (32) | I (4) |
| RNS_PAE05 | R (>32) | R (128) | R (128/2) | R (128) | R (32) | I (4) |
aS ≤ 4, I = 8, R ≥ 16.
bS ≤ 64, R ≥ 128.
cS ≤ 64/2, R ≥ 128/2.
dS ≤ 4, I = 8, R ≥ 16.
eS ≤ 8, I = 16, R ≥ 32.
fS ≤ 2, I = 4, R ≥ 8.
3.2. Phylogenetic analysis of the three isolates
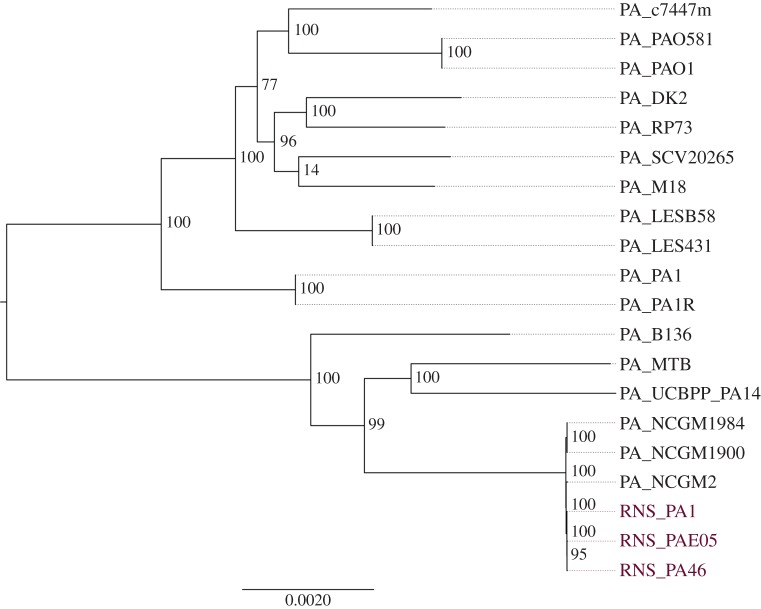
Raw reads were assembled (assembly statistics in electronic supplementary material, table S2) using the A5 pipeline [34] yielding approximately 170 scaffolds with an average of 35–38 × coverage and N50 scores from 99 078 to 111 819 nt. The three P. aeruginosa genomes (RNS_PA1, RNS_PA46 and RNS_PAE05) were compared with 18 finished Pseudomonas genomes deposited in GenBank using PhyloSift. Two of the three isolates (RNS_PA1 and RNS_PA46) clustered in a single clade with P. aeruginosa isolate NCGM2 (electronic supplementary material, figure S2). PhyloSift infers phylogenetic relationships within bacterial populations based on the amino acid sequence identity of 37 conserved core marker genes [37]. The branch that clustered RNS_PA1, RNS_PA46 with NCGM2 displayed a confidence value of 0.92, indicating that P. aeruginosa NCGM2 is the appropriate reference genome for comparative genomic analyses. Isolate RNS_PAE05, from the hand sanitizer bottle, resided on a separate clade. eMLST analysis [42] of NCGM2 and all three of our isolates showed that they belong to ST235. P. aeruginosa isolate PA14 resides in a clade that shares a common ancestral node with RNS_PA1, RNS_PA46 and NCGM2. PA14 belongs to the ST253 clade and is a multiple antibiotic-resistant clinical isolate implicated in a wide variety of diseases [43]. All 20 isolates branching out of PA_PA7 were used to set up a whole genome alignment-based phylogenetic analysis using REALPHY [38]. NCGM2 and PA14 were used as reference genomes in this analysis. The tree generated by aligning whole genomes places RNS_PA1, RNS_PA46 and RNS_PAE05 in a single clade (bootstrap value of 100) and strain NCGM2 as the closest neighbour (figure 1).
Figure 1.
Phylogenetic tree based on whole genome alignments created using REALPHY.
3.3. Characterization of genomic islands GI1, GI2 and exoU in Pseudomonas aeruginosa isolate RNS_PA1
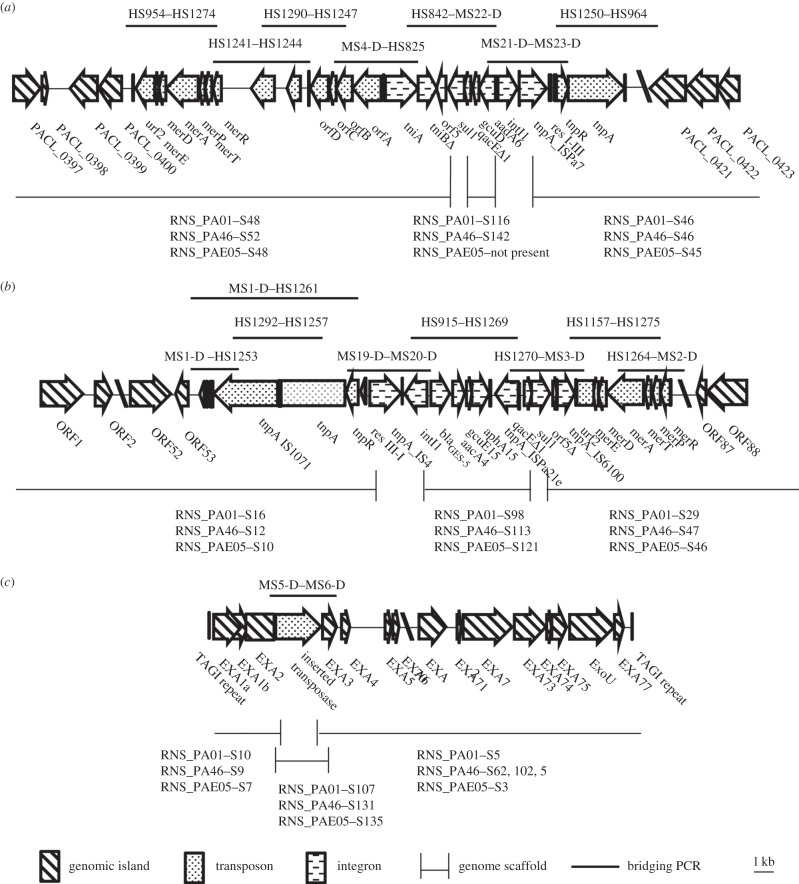
Scaffolds spanning GI1 and GI2 in P. aeruginosa RNS_PA1 were assembled by targeted PCR and Sanger sequencing followed by BLASTn alignments with published sequences. Transposons structures were confirmed using overlapping PCR experiments (figure 2a and 2b, respectively).
Figure 2.
The PCRs used to link scaffold breaks within the three genomic islands found in isolate RNS_PA1. (a) Tn6162 in GI1, (b) Tn6163 in GI2 and (c) exoU island with identified insertion element. PCRs shown with primers above the structure and the scaffolds for MS1 shown below each structure. Amplicons were verified using Sanger sequencing.
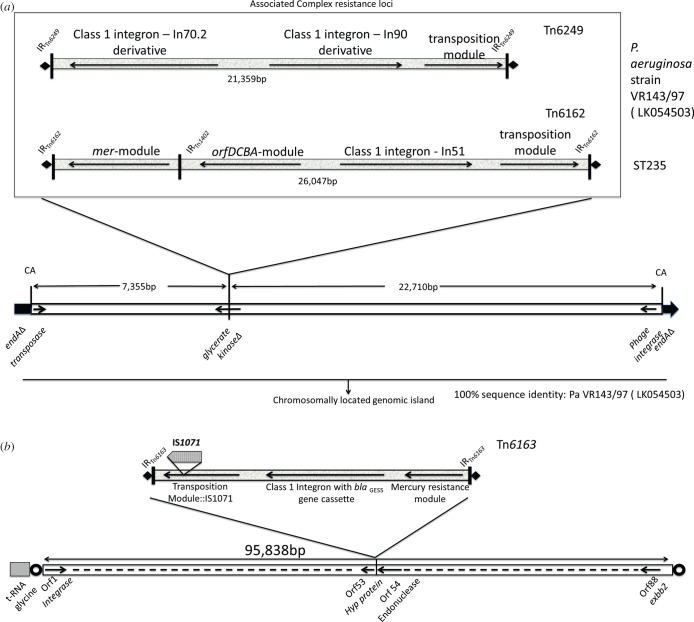
GI1 in RNS_PA1 comprises a 54 840 nt-long DNA segment and harbours Tn6162. GI1 was originally described as part of a genomic fragment in clone fa1389 (GenBank accession: EU595750) from P. aeruginosa isolate PACs171b and was thought to be restricted to isolates from patients with cystic fibrosis [44]. An island that contains significant portions of the fa1389 clone was recently found inserted in the chromosomally located endA gene in strain VR-143/97 isolated from Italy in 1997. We also identified regions within the fa1389 clone in isolates sourced from patients without cystic fibrosis in Australia and South America [25,26]. Insertion of the island creates a signature 2 bp (CA) direct repeat (figure 3a) at the chromosomal insertion site. Tn6162 in our three isolates is 26 056 nt long and has 99% sequence identity over 100% of the query sequence to the first described sequence of Tn6162 (JF826498.1). Our sequence contains five single nucleotide polymorphisms compared with Tn6162 described previously. Beyond the CRL, the sequence of the genomic island is identical at both ends to the GI sequence from strain VR-143/97 (figure 3a).
Figure 3.
Diagrammatic representation of the precise location of the complex resistance loci on the respective genomic islands. (a) Comparison of the complex resistance loci in Genomic Island 1 in P. aeruginosa strain VR-143/97 and in globally disseminated ST235, and their location in the glycerate kinase gene of GI1. (b) Complex resistance region of Genomic Island 2, which includes the precise location of Tn6163. Open circles represent 12 bp direct repeat (AACGCCAGGGAA) generated due to insertion of GI2. Bold diamond depicts 5 bp direct repeat (CTCAA) generated owing to insertion Tn6163.
GI2, which carries Tn6163, was identified on three scaffolds. Gap closing PCRs followed by Sanger sequencing led to the reconstruction of the GI2 sequence (figure 3b; electronic supplementary material, figure S4). In isolate RNS_PA1, GI2 (115 604 nt) comprises 94 368 nt of chromosomal backbone (average G+C content 63.5%) and is located adjacent to a tRNA-gly gene. Tn6163 comprises 21 236 nt and displays 99% sequence identity to Tn6163 (GenBank accession JF826499) with 15 SNPs and eight gaps, and is flanked by the characteristic 5 nt direct CTCAA repeats (figure 3b). A complete integrase gene (WP_003158602.1) resides next to the tRNA insertion site. Manual annotation of the GI2 backbone identified 88 ORFs (electronic supplementary material, figure S4). Details of the gene content in GI2 are presented in electronic supplementary material, table S3. A 12 nt direct repeat with the sequence AACGCCAGGGAA flanks the ends of GI2, suggesting that it is likely to be a recent insertion event at this site. Genes encoding proteins with roles in integration (integrase), conjugative transfer as well as integrative conjugative element-specific proteins are present, indicating that GI2 may be a novel integrative conjugative element.
BLAST analysis of the entire GI2 backbone identified a match (99% sequence identity) to a region in P. aeruginosa strain NCGM 1900 (AP104622) suggesting that this strain contains a variant of GI2 (figure 3). Further analysis of the matching sequence in NCGM 1900 failed to identify any evidence of Tn6163. BLAST alignments also identified a fragment (99% sequence identity) comprising 26 659 nt from ORF63 to the end of GI2 in the NCGM2 genome (NC_017549) indicating the island, like GI1, is not restricted within P. aeruginosa isolates collected in Australia.
The exoU genomic island in RNS_PA1 which harbours the gene encoding the potent cytotoxin ExoU is 83 830 nt in length and shows 99% sequence identity to the exoU island A in GenBank entry DQ437742 (81 751 nt). The sequence in RNS_PA1 differs only by the presence of a 2059 nt insertion element (figure 2c). The type III secretion system (T3SS) in P. aeruginosa plays an important role in pathogenicity by transporting effector molecules such as the cytotoxin ExoU into host cells. The structural components of the T3SS are located on a 23.9 kb region in the PA14 genome and these were aligned to corresponding regions of the three genomes in our study (electronic supplementary material, figure S3). RNS_PA1 had 140 SNPs and seven gaps, RNS_PA46 had 134 SNPs and one gap, and NCGM2 had 140 SNPs and seven gaps, when compared with the T3SS in PA14. The T3SS in isolate RNS_PAE05 had 138 SNPs, one gap and a 52 bp deletion located within a regulatory gene exsC. In Vibrio parahemolyticus, deletion of the exsC gene has been shown to shut down the expression of T3SS [45].
3.4. Comparative genome analysis
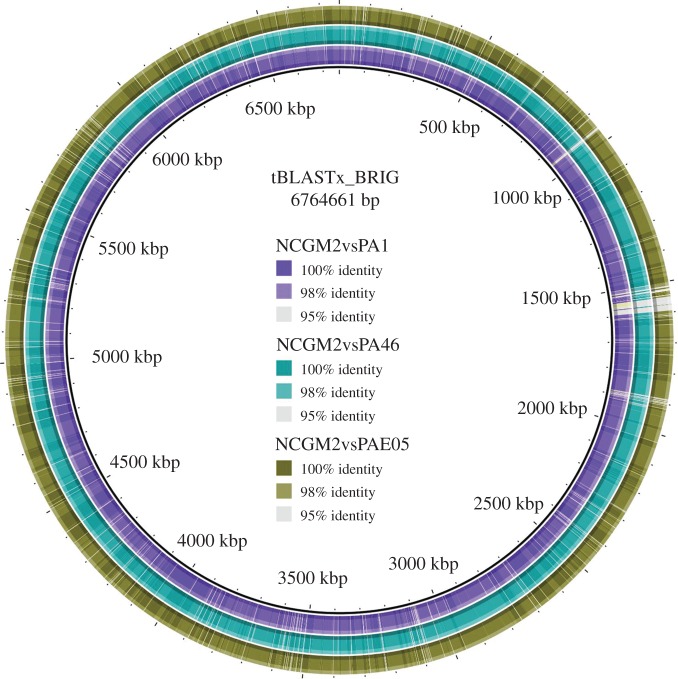
All three genomes were tiled against the closed genome sequence of P. aeruginosa NCGM2. The genome of strain NCGM2 comprises 6 764 661 nt, and is slightly smaller than the genomes from RNS_PA1, RNS_PA46 and RNS_PAE05. tBLASTx analysis comparing whole genomes of RNS_PA1, RNS_PA46 and RNS_PAE05 independently with NCGM2 (figure 4) identified a major region of difference close to the 1500 kbp marker. Bi-directional BLASTp analysis comparing each of our isolates with NCGM2 identified four regions of difference containing phage-derived proteins. These analyses suggest that ST235 isolates derived from different geographical regions have likely been infected with different phage populations and have been subjected to novel phage-mediated genomic rearrangements.
Figure 4.
Comparative tBLASTx analysis of the three P. aeruginosa genomes against the reference NCGM2 genome.
Progressive MAUVE alignment of the three genomes was used to identify scaffolds that define the core and accessory genomes in RNS_PA1, RNS_PA46 and RNS_PAE05 (figure 5). Regions coloured in magenta represent shared sequence identity in RNS_PA1, RNS_PA46 and RNS_PAE05. Blue-boxed regions in the core genome of RNS_PA46 are unique to this strain. Scaffold 27 (blue box 1) from strain RNS_PA46 contains genes that encode proteins relating to a ‘mobile genetic element’ like integrase, conjugation proteins and a series of uncharacterized hypothetical proteins. Scaffolds 40 and 77 (boxes 2 and 3, respectively, in figure 5) carry genes encoding phage-associated proteins. Electronic supplementary material, table S4 provides a description of the gene content of the unaligned scaffolds generated by preliminary RAST annotation of regions greater than 1 kb.
Figure 5.
Mauve alignments of the RNS_PA1, RNS_PA46 and RNS_PAE05. Black boxes around the green segment in genome RNS_PA1 and RNS_PAE05 represent parts of scaffold 4 and 8 in the genome sequence, respectively. Blue box 1 in genome RNS_PA46 represents scaffold 27, box 2 represents scaffold 40 and box 3 represents scaffold 77.
In summary, the differences noted in the genomes of RNS_PA1, RNS_PA46 and RNS_PAE05 appear to be phage-related and other laterally acquired regions. RNS_PA46 has the biggest genome, comprising 6672 ORFs, and RNS_PA1 is the smallest genome, with 6564 predicted ORFs. Table 2 shows a summary of SNPs and indels identified at least 100 nt from a scaffold break. RNS_PAE05 contains the largest number of SNPs of the three genomes. These data show that of the functionally defined genes, major SNP differences in the genomes of RNS_PA1, RNS_PA46 and RNS_PAE05 were localized within housekeeping genes. For example, the DNA polymerase gene dnaX, glucose carbohydrate outer membrane porin gene oprB and riboflavin-specific deaminase reductases gene ribD exhibited SNPs. However, differences were also identified in a number of other genes that were annotated as hypothetical proteins by RAST.
Table 2.
Comparison of SNPs identified in P. aeruginosa isolates included in this study.
| SNP |
indel |
||||
|---|---|---|---|---|---|
| individual SNPs | clustered SNP groups | genes/ORFs with clustered SNPs | indels present | large indelsa | |
| RNS_PA1 | 101 | 2 | 61 glucose/carbohydrate outer membrane porin genes 18 riboflavin-specific deaminase/reductases genes |
47 | 0 |
| RNS_PA46 | 119 | 4 | 34 hypothetical protein 13 putative MFS transporter 10 hypothetical protein 19 hypothetical protein (NCGM2_4844) |
49 | 6 |
| RNS_PAE05 | 308 | 11 | 27 hypothetical protein 18 putative glycerol kinase 96 dnaX 10 isochorismatase family hydrolase 15 putative transmembrane sensor 13 hypothetical protein 15 no ORF 18 no ORF 16 hypothetical protein 18 elongation factor Tu 20 putative gamma-glutamyltranspeptidase precursor |
45 | 0 |
aLarge indels defined as larger than 5 kb.
3.5. Genomewide identification of resistance and virulence genes
RNS_PA1, RNS_PA46 and RNS_PAE05 carried identical variants of all the resistance and virulence genes listed in table 3. In addition, we also surveyed the Comprehensive Antibiotic Resistance Database (http://arpcard.mcmaster.ca) with our genome sequences (electronic supplementary material, table S5). The repertoire of antibiotic resistance genes identified in each of the strains is likely to account for their resistance phenotype. Mutations in phoQ and pmrB are known to have a role in colistin resistance [46]. In the genomes of RNS_PA1, RNS_PA46 and RNS_PAE05, an F76Y mutation in phoQ was identified and is unique to our isolates. We also identified a V15I mutation in pmrB. This same mutation was recently reported to alter colistin sensitivity in P. aeruginosa strain P165 [47]. We speculate that these mutations have probably contributed to the intermediate colistin resistance phenotype exhibited in our three Sydney isolates. While the genome of NCGM2 carries a different suite of resistance genes to the ones in RNS_PA1, RNS_PA46 and RNS_PAE05, we cannot comment any further as the resistance profile of NCGM2 has not been reported.
Table 3.
Antibiotic resistance and virulence genes in P. aeruginosa isolates.
| antibiotic resistance genes | RNS_PA1 | RNS_PA46 | RNS_PAE05 | NCGM2 | NCGM 1900 |
|---|---|---|---|---|---|
|
aadA6 (encodes resistance to aminoglycoside) (JF826498.1: 24 514-25 359) |
present 100% nBLAST |
present 100% nBLAST |
present 100% nBLAST |
present 100% nBLAST |
|
|
aphA15 (encodes resistance to aminoglycosides) (JF826499.1: 13 868-14 662) |
present 100% nBLAST |
present 100% nBLAST |
present 100% nBLAST |
||
|
nfxB (encodes resistance to norfloxacin) (CP000438.1: 5 428 030–5 428 593) |
present 98% nBLAST 100% pBLAST |
present 98% nBLAST 100% pBLAST |
present 98% nBLAST 100% pBLAST |
present 98% nBLAST 100% pBLAST |
present 98% nBLAST 100% pBLAST |
|
blaGES-5 (encodes resistance to β-lactam) (JF826499.1: 11 979–12 842) |
present 100% nBLAST |
present 100% nBLAST |
present 100% nBLAST |
||
|
ampC (β-lactam) (CP000438.1: 932 631–933 824) |
present 100% nBLAST |
present 100% nBLAST |
present 100% nBLAST |
present 100% nBLAST |
present 99% nBLAST 100% pBLAST |
|
phoQ
(mutations known to cause colistin resistance) (AE004091.2: 1 278 362–1 279 708) |
present 99% nBLAST 99% pBLAST F76Y |
present 99% nBLAST 99% pBLAST F76Y |
present 99% nBLAST 99% pBLAST F76Y |
present 99% nBLAST 99% pBLAST F76Y |
|
|
pmrB
(mutations known to cause colistin resistance) (AE004091.2|:5 364 760–5 366 193) |
present 99% nBLAST 99% pBLAST V15I |
present 99% nBLAST 99% pBLAST V15I |
present 99% nBLAST 99% pBLAST V15I |
| putative virulence genes | RNS_PA1 | RNS_PA46 | RNS_PAE05 | NCGM2 | NCGM 1900 |
|---|---|---|---|---|---|
|
exoT (effector of T3SS) (AE004091.2: 58786–60 159) |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAT |
|
exoY (effector of T3SS) (AE004091.2: 2 410 344–2 411 480) |
present non-functional 99% nBLAST stop codon present in middle of sequence |
present non-functional 99% nBLAST stop codon present in middle of sequence |
present non-functional 99% nBLAST stop codon present in middle of sequence |
present non-functional 99% nBLAST stop codon present in middle of sequence |
|
|
aprA (extracellular protease) (AE004091.2: 1 355 631–1 357 070) |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
|
lasA (extracellular protease) (AE004091.2: 2 410 344–2 411 480) |
present 98% nBLAST 100% pBLAST |
present 98% nBLAST 100% PBLAST |
present 98% nBLAST 100% pBLAST |
present 98% nBLAST 100% pBLAST |
|
|
lasB (extracellular protease) (AE004091.2: 4 168 987–4 170 483) |
present 98% nBLAST 100% pBLAST |
present 98% nBLAST 100% pBLAST |
present 98% nBLAST 100% pBLAST |
present 98% nBLAST 100% pBLAST |
present 98% nBLAST 100% pBLAST |
|
exoU (acute cytotoxin) (CP000438.1(PA14): 4 580 957–4 583 020) |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
|
ladS (promotes biofilm formation) (AE004091.2: 4 453 289–4 455 676) |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
present 99% nBLAST 100% pBLAST |
4. Discussion
Previously we showed that two genomic islands are implicated in mobilizing transposons that carry multiple antibiotic resistance genes in clinical isolates of P. aeruginosa [26,48]. Here, we describe the genetic architecture of two genomic islands implicated in the dissemination of multiple antibiotic resistance genes in three P. aeruginosa ST235 isolates from Sydney and provide further evidence supporting our hypothesis that genomic islands are major players in the dissemination of multiple antibiotic resistance. We have characterized GI1 in three Sydney isolates which are representatives of ST235, and the structure is very similar to the one described recently from isolate VR-143/97 from Italy. In addition, we define, for the first time, both the boundaries of and the genetic composition of genomic island 2. GI2 was originally proposed to be restricted to isolates collected from the Sydney basin. However, in this study, we provide evidence that suggests that GI2 is not restricted to isolates from a specific geographical location and may be a characteristic of the ST235 clonal lineage.
In our Sydney isolates, we show that GI1 carries Tn6162 (figure 3) and has inserted into the endA gene at precisely the same position as a PACS171b-like genomic island in multiple antibiotic-resistant P. aeruginosa isolate VR-143/97 isolated in Italy in 1997 [45]. The PACS171b-like genomic island in strain VR-143/97 harbours Tn6249, a complex transposon carrying two different class 1 integrons, one of which houses a VIM-1 gene cassette. The genome sequence for strain VR-143/97 has not been released and as such we cannot determine if VR-143/97 belongs to the ST235 lineage. In Australian isolates of P. aeruginosa, GI2 harbours Tn6163. Tn6163 contains a class 1 integron with blaGES-5–aacA4–gcuE15–aphA15 gene cassettes. It is not known if strain VR-143/97 also harbours Tn6163. Notably, BLAST analysis of the entire GI2 backbone identified (99% sequence identity) a region in ST235 P. aeruginosa strain NCGM 1900 raising the possibility that GI2 may be widely disseminated in the ST235 lineage. While GI2 comprises certain features typical of ICE elements described in other bacteria, it has clearly undergone genetic rearrangements in the three genomes described in this study. Currently, we do not have any evidence to indicate GI2 has the capacity to excise from the genome and transfer elsewhere via conjugation.
The three Australian ST235 isolates carry both GI1 and GI2 and display resistance to almost all frontline antibiotics used to control infections caused by P. aureginosa at RNSH. Tn6163 harbours a class 1 integron that contains the Ambler class A β-lactamase gene cassette blaGES-5 that confers resistance to all β-lactams including carbapenems. It also contains aphA15 that confers resistance to kanamycin and neomycin. The aminoglycoside resistance gene aadA6 that confers resistance to gentamicin forms the first cassette in the array of the class 1 integron in Tn6162.
GI1 appears to have been integrated into the genome by a phage-integrase-mediated site-specific recombination event. The PHAST database was interrogated to identify phage genes that may facilitate movement of the genomic islands (data not shown), but none were identified. However, given the number of phage-related genetic signatures in all three P. aeruginosa genomes, it is possible that parts of GI1 were originally transferred into P. aeruginosa by a phage which has subsequently lost function. The exoU island A in our Australian isolates had 99% sequence identity to the exoU island present in P. aeruginosa strains NCGM1900 and NCGM2 from Japan [49]. We identified a novel insertion sequence in exoU island A that is unique to the Australian isolates of P. aeruginosa described in this study. The closest match to the insertion sequence is found in the genome of Pseudomonas putida strain S12 (CP009974). Further work is needed to determine if the insertion sequence can be used as a marker to identify the exoU island A variant.
RNS_PA1, RNS_PA46 and RNS_PAE05 carry a comprehensive repertoire of putative virulence genes. In P. aeruginosa, the T3SS has four known effectors: ExoU, ExoY, ExoT and ExoS. While clinical isolates have never been shown to carry all four effectors, ExoT has been identified frequently, suggesting it performs a function in pathogenesis [50]. ExoT is known to inhibit host-cell division by interrupting cytokinesis in mammalian cells [50]. In an epithelial cell model, ExoY has adenlylate cyclase activity that leads to increases in intracellular cAMP levels and inhibits cellular invasion [51]. ExoU is an important virulence factor that mediates cell death in macrophages, epithelial cells and fibroblasts [52]. ExoU functions in concert with its chaperone SpcU, and we identified spcU in the exoU island A in the genome sequences of all three Sydney isolates. The T3SS effector proteins play a significant role in causing severe pneumonia [53]. LasA and LasB possess elastolytic activity [54], whereas LadS activates a cascade that decreases cytotoxicity and increases biofilm formation [55]. Finally, AprA encodes an alkaline protease that is secreted by the type 1 secretion system [56].
In conclusion, all three Sydney ST235 isolates have the blaGES-5 gene inserted as a gene cassette within a class 1 integron that forms part of Tn6163 located in GI2. The genomic data presented here and elsewhere (AP014622, AP014646 and [29]) show that genomic islands are playing a seminal role in the dissemination of resistance to a wide range of clinically important antibiotics, including metallo-β-lactams and extended-spectrum β-lactams. It is not known if GI1 and GI2 can be mobilized to other Gram-negative bacteria.
Supplementary Material
Supplementary Material
Acknowledgements
The authors thank the people at the Microbiology laboratory of RNSH for identifying the bacteria and making this project possible.
Data accessibility
Genome sequences described in this manuscript were submitted in GenBank and accession numbers are provided in the Methods section.
Authors' contributions
P.R.C. and M.S. assembled the genome sequences, conceived, analysed and designed the presentation of data in the manuscript, performed comparative genomic analysis and drafted the manuscript. P.H., B.H. and T.K. were involved in the collection of samples and RAPD analysis. P.W. constructed the libraries for genome sequencing. I.G.C. assisted with experimental design and edited the manuscript. S.P.D. managed the project and drafted the final manuscript.
Competing interests
We declare we have no competing interests.
Funding
This work is a product of the AUSGEM partnership.
References
- 1.Hirsch EB, Tam VH. 2010. Impact of multidrug-resistant Pseudomonas aeruginosa infection on patient outcomes. Expert Rev. Pharmacoecon. Outcomes Res. 10, 441–451. (doi:10.1586/erp.10.49) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Driscoll JA, Brody SL, Kollef MH. 2007. The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs 67, 351–368. (doi:10.2165/00003495-200767030-00003) [DOI] [PubMed] [Google Scholar]
- 3.Moore NM, Flaws ML. 2011. Epidemiology and pathogenesis of Pseudomonas aeruginosa infections. Clin. Lab. Sci. 24, 43–46. [PubMed] [Google Scholar]
- 4.Kerr KG, Snelling AM. 2009. Pseudomonas aeruginosa: a formidable and ever-present adversary. J. Hosp. Infect. 73, 338–344. (doi:10.1016/j.jhin.2009.04.020) [DOI] [PubMed] [Google Scholar]
- 5.Lister PD, Wolter DJ, Hanson ND. 2009. Antibacterial-resistant Pseudomonas aeruginosa: clinical impact and complex regulation of chromosomally encoded resistance mechanisms. Clin. Microbiol. Rev. 22, 582–610. (doi:10.1128/CMR.00040-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agodi A, Barchitta M, Cipresso R, Giaquinta L, Romeo MA, Denaro C. 2007. Pseudomonas aeruginosa carriage, colonization, and infection in ICU patients. Intens. Care Med. 33, 1155–1161. (doi:10.1007/s00134-007-0671-6) [DOI] [PubMed] [Google Scholar]
- 7.Kramer A, Schwebke I, Kampf G. 2006. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 6, 130 (doi:10.1186/1471-2334-6-130) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ratkai C, Nagy E, Peixe L, Bertalan V, Hajdu E. 2010. Isolation and characterization of an imported extremely-resistant Pseudomonas aeruginosa producing three different extended-spectrum beta-lactamases and hyperproducing two multidrug-efflux pumps. J. Infect. 61, 511–512. (doi:10.1016/j.jinf.2010.10.006) [DOI] [PubMed] [Google Scholar]
- 9.Moore NM, Flaws ML. 2011. Introduction: Pseudomonas aeruginosa. Clin. Lab Sci. 24, 41–42. [PubMed] [Google Scholar]
- 10.Woodford N, Turton JF, Livermore DM. 2011. Multiresistant Gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol. Rev. 35, 736–755. (doi:10.1111/j.1574-6976.2011.00268.x) [DOI] [PubMed] [Google Scholar]
- 11.Pollini S, Antonelli A, Venturelli C, Maradei S, Veggetti A, Bracco S, Rumpianesi F, Luzzaro F, Rossolini GM. 2013. Acquisition of plasmid-borne blaIMP-19 gene by a VIM-1-positive Pseudomonas aeruginosa of the sequence type 235 epidemic lineage. J. Antimicrob. Chemother. 68, 722–724. (doi:10.1093/jac/dks440) [DOI] [PubMed] [Google Scholar]
- 12.Liakopoulos A, Mavroidi A, Katsifas EA, Theodosiou A, Karagouni AD, Miriagou V, Petinaki E. 2013. Carbapenemase-producing Pseudomonas aeruginosa from central Greece: molecular epidemiology and genetic analysis of class I integrons. BMC Infect. Dis. 13, 505 (doi:10.1186/1471-2334-13-505) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glupczynski Y, Bogaerts P, Deplano A, Berhin C, Huang TD, Van Eldere J, Rodriguez-Villalobos H. 2010. Detection and characterization of class A extended-spectrum-beta-lactamase-producing Pseudomonas aeruginosa isolates in Belgian hospitals. J. Antimicrob. Chemother. 65, 866–871. (doi:10.1093/jac/dkq048) [DOI] [PubMed] [Google Scholar]
- 14.Ranellou K, Kadlec K, Poulou A, Voulgari E, Vrioni G, Schwarz S, Tsakris A. 2012. Detection of Pseudomonas aeruginosa isolates of the international clonal complex 11 carrying the blaPER-1 extended-spectrum beta-lactamase gene in Greece. J. Antimicrob. Chemother. 67, 357–361. (doi:10.1093/jac/dkr471) [DOI] [PubMed] [Google Scholar]
- 15.Yoo JS, Yang JW, Kim HM, Byeon J, Kim HS, Yoo JI, Chung GT, Lee YS. 2012. Dissemination of genetically related IMP-6-producing multidrug-resistant Pseudomonas aeruginosa ST235 in South Korea. Int. J. Antimicrob. Agents 39, 300–304. (doi:10.1016/j.ijantimicag.2011.11.018) [DOI] [PubMed] [Google Scholar]
- 16.Kim MJ, et al. 2013. Dissemination of metallo-beta-lactamase-producing Pseudomonas aeruginosa of sequence type 235 in Asian countries. J. Antimicrob. Chemother. 68, 2820–2824. (doi:10.1093/jac/dkt269) [DOI] [PubMed] [Google Scholar]
- 17.Seok Y, Bae IK, Jeong SH, Kim SH, Lee H, Lee K. 2011. Dissemination of IMP-6 metallo-beta-lactamase-producing Pseudomonas aeruginosa sequence type 235 in Korea. J. Antimicrob. Chemother. 66, 2791–2796. (doi:10.1093/jac/dkr381) [DOI] [PubMed] [Google Scholar]
- 18.Pollini S, Maradei S, Pecile P, Olivo G, Luzzaro F, Docquier JD, Rossolini GM. 2013. FIM-1, a new acquired metallo-beta-lactamase from a Pseudomonas aeruginosa clinical isolate from Italy. Antimicrob. Agents Chemother. 57, 410–416. (doi:10.1128/AAC.01953-12) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Sun M, Wang M, Lu Y, Yan Z. 2014. Dissemination of IMP-6-producing Pseudomonas aeruginosa ST244 in multiple cities in China. Eur. J. Clin. Microbiol. Infect. Dis. 33, 1181–1187. (doi:10.1007/s10096-014-2063-5) [DOI] [PubMed] [Google Scholar]
- 20.Kitao T, Tada T, Tanaka M, Narahara K, Shimojima M, Shimada K, Miyoshi-Akiyama T, Kirikae T. 2012. Emergence of a novel multidrug-resistant Pseudomonas aeruginosa strain producing IMP-type metallo-beta-lactamases and AAC(6′)-Iae in Japan. Int. J. Antimicrob. Agents 39, 518–521. (doi:10.1016/j.ijantimicag.2012.01.020) [DOI] [PubMed] [Google Scholar]
- 21.Kouda S, et al. 2009. Increased prevalence and clonal dissemination of multidrug-resistant Pseudomonas aeruginosa with the blaIMP-1 gene cassette in Hiroshima. J. Antimicrob. Chemother. 64, 46–51. (doi:10.1093/jac/dkp142) [DOI] [PubMed] [Google Scholar]
- 22.Koh TH, Khoo CT, Tan TT, Arshad MA, Ang LP, Lau LJ, Hsu L-Y, Ooi EE. 2010. Multilocus sequence types of carbapenem-resistant Pseudomonas aeruginosa in Singapore carrying metallo-beta-lactamase genes, including the novel bla(IMP-26) gene. J. Clin. Microbiol. 48, 2563–2564. (doi:10.1128/JCM.01905-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Walsh TR, Toleman MA, Poirel L, Nordmann P. 2005. Metallo-beta-lactamases: the quiet before the storm? Clin. Microbiol. Rev. 18, 306–325. (doi:10.1128/CMR.18.2.306-325.2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stokes HW, Martinez E, Roy Chowdhury P, Djordjevic S. 2012. Class 1 integron-associated spread of resistance regions in Pseudomonas aeruginosa: plasmid or chromosomal platforms? J. Antimicrob. Chemother. 67, 1799–1800. (doi:10.1093/jac/dks116) [DOI] [PubMed] [Google Scholar]
- 25.Roy Chowdhury P, Merlino J, Labbate M, Cheong EY, Gottlieb T, Stokes HW. 2009. Tn6060, a transposon from a genomic island in a Pseudomonas aeruginosa clinical isolate that includes two class 1 integrons. Antimicrob. Agents Chemother. 53, 5294–5296. (doi:10.1128/AAC.00687-09) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martinez E, Marquez C, Ingold A, Merlino J, Djordjevic SP, Stokes HW, Roy Chowdhury P. 2012. Diverse mobilized class 1 integrons are common in the chromosomes of pathogenic Pseudomonas aeruginosa clinical isolates. Antimicrob. Chemother. 56, 2169–2172. (doi:10.1128/AAC.06048-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naas T, Poirel L, Nordmann P. 1999. Molecular characterisation of In51, a class 1 integron containing a novel aminoglycoside adenylyltransferase gene cassette, aadA6, in Pseudomonas aeruginosa. Biochim. Biophys. Acta 1489, 445–451. (doi:10.1016/S0167-4781(99)00202-X) [DOI] [PubMed] [Google Scholar]
- 28.Kiddee A, Henghiranyawong K, Yimsabai J, Tiloklurs M, Niumsup PR. 2013. Nosocomial spread of class 1 integron-carrying extensively drug-resistant Pseudomonas aeruginosa isolates in a Thai hospital. Int. J. Antimicrob. Agents 42, 301–306. (doi:10.1016/j.ijantimicag.2013.05.009) [DOI] [PubMed] [Google Scholar]
- 29.Di Pilato V, Pollini S, Rossolini GM. 2015. Tn6249, a new Tn6162 transposon derivative carrying a double-integron platform and involved with acquisition of the blaVIM-1 metallo-beta-lactamase gene in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 59, 1583–1587. (doi:10.1128/AAC.04047-14) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Isenberg HD. 2004. Clinical microbiology procedure handbook, 2nd edn Washington, DC: American Society Microbiology. [Google Scholar]
- 31.Mahenthiralingam E, Campbell ME, Foster J, Lam JS, Speert DP. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34, 1129–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.CLSI. 2012. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically; approved standard, 9th edn. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- 33.Darling AE, Worden P, Chapman TA, Roy Chowdhury P, Charles IG, Djordjevic SP. 2014. The genome of Clostridium difficile 5.3. Gut Pathog. 6, 4 (doi:10.1186/1757-4749-6-4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tritt A, Eisen JA, Facciotti MT, Darling AE. 2012. An integrated pipeline for de novo assembly of microbial genomes. PLoS ONE 7, e42304 (doi:10.1371/journal.pone.0042304) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aziz RK, et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9, 75 (doi:10.1186/1471-2164-9-75) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Overbeek R, et al. 2014. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42(Database issue), D206–DD24. (doi:10.1093/nar/gkt1226) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Darling AE, Jospin G, Lowe E, Matsen F, Bik HM, Eisen JA. 2014. PhyloSift: phylogenetic analysis of genomes and metagenomes. PeerJ 2, e243 (doi:10.7717/peerj.243) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bertels F, Silander OK, Pachkov M, Rainey PB, van Nimwegen E. 2014. Automated reconstruction of whole-genome phylogenies from short-sequence reads. Mol. Biol. Evol. 31, 1077–1088. (doi:10.1093/molbev/msu088) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Darling AE, Mau B, Perna NT. 2010. progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5, e11147 (doi:10.1371/journal.pone.0011147) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J. Mol. Biol. 215, 403–410. (doi:10.1016/S0022-2836(05)80360-2) [DOI] [PubMed] [Google Scholar]
- 41.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. 2011. BLAST ring image generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12, 402 (doi:10.1186/1471-2164-12-402) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jolley KA, Maiden MC. 2010. BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11, 595 (doi:10.1186/1471-2105-11-595) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kidd TJ, Ritchie SR, Ramsay KA, Grimwood K, Bell SC, Rainey PB. 2012. Pseudomonas aeruginosa exhibits frequent recombination, but only a limited association between genotype and ecological setting. PLoS ONE 7, e44199 (doi:10.1371/journal.pone.0044199) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hayden HS, et al. 2008. Large-insert genome analysis technology detects structural variation in Pseudomonas aeruginosa clinical strains from cystic fibrosis patients. Genomics 91, 530–537. (doi:10.1016/j.ygeno.2008.02.005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhou X, Konkel ME, Call DR. 2010. Regulation of type III secretion system 1 gene expression in Vibrio parahaemolyticus is dependent on interactions between ExsA, ExsC, and ExsD. Virulence 1, 260–272. (doi:10.4161/viru.1.4.12318) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front. Microbiol. 5, 643 (doi:10.3389/fmicb.2014.00643) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee JY, Ko KS. 2014. Mutations and expression of PmrAB and PhoPQ related with colistin resistance in Pseudomonas aeruginosa clinical isolates. Diagn. Microbiol. Infect. Dis. 78, 271–276. (doi:10.1016/j.diagmicrobio.2013.11.027) [DOI] [PubMed] [Google Scholar]
- 48.Martinez E. 2013. Genetic context and mobilization of class 1 integrons in Pseudomonas aeruginosa: are plasmids redundant? Sydney, Australia: University of Technology. [Google Scholar]
- 49.Miyoshi-Akiyama T, Kuwahara T, Tada T, Kitao T, Kirikae T. 2011. Complete genome sequence of highly multidrug-resistant Pseudomonas aeruginosa NCGM2.S1, a representative strain of a cluster endemic to Japan. J. Bacteriol. 193, 7010 (doi:10.1128/JB.06312-11) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shafikhani SH, Engel J. 2006. Pseudomonas aeruginosa type III-secreted toxin ExoT inhibits host-cell division by targeting cytokinesis at multiple steps. Proc. Natl Acad. Sci. USA 103, 15 605–15 610. (doi:10.1073/pnas.0605949103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cowell BA, Evans DJ, Fleiszig SM. 2005. Actin cytoskeleton disruption by ExoY and its effects on Pseudomonas aeruginosa invasion. FEMS Microbiol. Lett. 250, 71–76. (doi:10.1016/j.femsle.2005.06.044) [DOI] [PubMed] [Google Scholar]
- 52.Feltman H, Schulert G, Khan S, Jain M, Peterson L, Hauser AR. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147, 2659–2669. (doi:10.1099/00221287-147-10-2659) [DOI] [PubMed] [Google Scholar]
- 53.Schulert GS, Feltman H, Rabin SD, Martin CG, Battle SE, Rello J, Hauser AR. 2003. Secretion of the toxin ExoU is a marker for highly virulent Pseudomonas aeruginosa isolates obtained from patients with hospital-acquired pneumonia. J. Infect. Dis. 188, 1695–1706. (doi:10.1086/379372) [DOI] [PubMed] [Google Scholar]
- 54.Toder DS, Ferrell SJ, Nezezon JL, Rust L, Iglewski BH. 1994. lasA and lasB genes of Pseudomonas aeruginosa: analysis of transcription and gene product activity. Infect. Immun. 62, 1320–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mikkelsen H, McMullan R, Filloux A. 2011. The Pseudomonas aeruginosa reference strain PA14 displays increased virulence due to a mutation in ladS. PLoS ONE 6, e29113 (doi:10.1371/journal.pone.0029113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maunsell B, Adams C, O'Gara F. 2006. Complex regulation of AprA metalloprotease in Pseudomonas fluorescens M114: evidence for the involvement of iron, the ECF sigma factor, PbrA and pseudobactin M114 siderophore. Microbiology 152, 29–42. (doi:10.1099/mic.0.28379-0) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequences described in this manuscript were submitted in GenBank and accession numbers are provided in the Methods section.