Abstract
Phosphoinositides are master regulators of multiple cellular processes: from vesicular trafficking to signaling, cytoskeleton dynamics, and cell growth. They are synthesized by the spatiotemporal regulated activity of phosphoinositide-metabolizing enzymes. The recent observation that some protein modules are able to cluster phosphoinositides suggests that alternative or complementary mechanisms might operate to stabilize the different phosphoinositide pools within cellular compartments. Herein, we discuss the different known and potential molecular players that are prone to engage phosphoinositide clustering and elaborate on how such a mechanism might take part in the regulation of intracellular trafficking and signal transduction.
Keywords: Phosphoinositides, membrane organization, trafficking, signal transduction.
Introduction
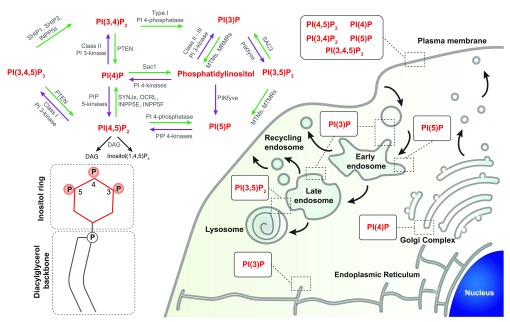
Phosphoinositides (PIs) are essential phospholipids that control, either directly or indirectly, multiple cellular functions including membrane trafficking, signal transduction, cell growth, cytoskeletal dynamics, lipid transport/exchange between organelles, and the regulation of transmembrane proteins 1, 2. PIs are the phosphorylated products of phosphatidylinositol. The reversible phosphorylation of the inositol ring at positions 3, 4, and 5 gives rise to the seven PI isoforms identified in eukaryotic cells ( Figure 1). Inter-conversion of the phosphate group(s) is selectively tuned by numerous kinases and phosphatases, precisely regulated in space and time 3 ( Figure 1). The active metabolism of PIs is intimately linked to their role as precursors of second messengers during signal transduction 4. The accumulation of the different PI species in specific membrane compartments is also directly related to their role in vesicular trafficking including endocytosis and exocytosis, endosome dynamics and trafficking from and towards the Golgi, among many others 5 ( Figure 1). Proteins with multiple trafficking functions are targeted to various membrane compartments based on the selective recognition of their PI-binding motifs. The distribution of protein residues folded in a 3D structure provides the PI-binding motifs with a “PI code”, which is based on the stereospecific recognition of the unique phosphate group’s organization around the inositol ring 6 ( Figure 1). There are at least 11 different structured motifs with a wide range of affinities and specificities for the different PI species. They include the PH ( pleckstrin homology), the FYVE ( Fab1, YOTB, Vac1, and EEA1), the PX ( Phox homology), the ANTH and ENTH ( AP180 and Epsin N- terminal homology), and the FERM (4.1, ezrin, radixin, moesin) modules.
Figure 1. The seven phosphoinositide isoforms identified in eukaryotic cells are phosphorylated derivatives of phosphoinositols, which can be metabolized by different phosphatases and kinases.
Representation of the phosphatidylinositol phospholipid structure: the inositol ring can be phosphorylated in three different positions and is linked to a diacylglycerol backbone by a phosphodiester linker. Schematics of the localization of the different PI isoforms on the cellular compartments.
PIs and the lateral organization of membranes: the needle in a haystack
Cellular membranes are highly heterogeneous composites built of different types of lipids and proteins. For instance, in eukaryotic cells, more than 1000 different lipid species build up the different membrane compartments 7. Lipid molecules freely diffuse in the 2D membrane plane (D ~2.6 × 10 -7 cm 2·s -1) 8 and interact with protein effectors based on their association ( K on) and dissociation ( K off) rates. As a result, lipid-protein interactions are, in general, highly dynamic and thus strongly depend on their respective local concentration.
PIs constitute less than 1% of the steady-state cell lipids 7, yet they work as unique docking sites for the multiple PI effectors on membranes, which in turn either compete or cooperate with each other to interact with downstream partners and elicit specific responses. Thus, what are the driving mechanisms that ensure such a thorough spatiotemporal recognition and membrane association of host PI-binding motifs?
An attractive hypothesis is that PIs might be organized as specialized membrane subdomains with distinct organelle localizations 5. PI pools within the same compartment are locally synthesized thanks to the spatiotemporal regulation of different PI-metabolizing enzymes 3, 5. In addition, small GTPases of the ARF and RAB family also contribute to the generation and regulation of PI turnover on membranes 9.
Considering the diffusion coefficient of lipid molecules within the membrane plane, it is likely that complementary mechanisms need to operate in order to spatially preserve the turnover of different PI subdomains. Indeed, several mechanisms have been reported in the literature to play roles as selective and reversible PI sinks by locally sequestering and releasing PIs. This is the case for the myristoylated alanine-rich C-kinase substrate (MARCKS) protein and the growth-associated protein 43 (GAP43) 10. The unstructured basic cluster on the effector domain of the MARCKS protein is able to bind up to at least three PI(4,5)P 2 molecules by means of nonspecific electrostatic interactions at physiologic pH. The Ca 2+/calmodulin complex reversibly controls the association of MARCKS with the plasma membrane 11. Interestingly, a growing number of studies report the local enrichment of PI subdomains independently of the catalytic activity of PI-metabolizing enzymes. Jahn and co-workers have shown that the SNAP receptor protein syntaxin-1A co-clusters with PI(4,5)P 2 via electrostatic interactions with its juxtamembrane polybasic sequence 12. The segregation of PI(4,5)P 2 microdomains by syntaxin-1A has been proposed to work as a molecular beacon at sites of synaptic vesicle docking during exocytosis 13. Similar polybasic clusters to that of the MARCKS protein or syntaxin-1A are found in the cytosolic membrane interface of many plasma membrane proteins 14, 15, including the epidermal growth factor receptor (EGFR) and the NMDA receptor as well as the voltage-gated potassium and calcium ion channels 11. In vitro studies have shown that divalent cations such as Ca 2+ are also capable of clustering together PI(4,5)P 2 molecules, although the exact correlation with the activity of ion channels inside the cell has yet to be established. Following in vitro approaches on giant unilamellar vesicles (GUVs), clustering of PI(4,5)P 2 was initially reported for ezrin 16. Later on, using the yeast endocytic F-BAR/BAR domains, Lappalainen and co-authors have shown that the scaffolding effect of these proteins leads to the formation of stable PI(4,5)P 2 microdomains with reduced lateral diffusion in the membrane plane 17, 18. Since then, the list of proteins involved in the formation of PI(4,5)P 2 clusters has been extended to other endocytic proteins such as Epsin2, AP180, and the N-BAR domain proteins amphiphysin1 and BIN1 19. So far, the formation of PI clusters has been mainly restricted to PI(4,5)P 2, possibly owing to its multiple regulatory functions at the plasma membrane. In addition, PI(4,5)P 2 is more abundant than other more elusive PI isoforms and has therefore been the focus of many studies for several years. However, we recently reported that the monophosphate PIs PI4P and PI5P can also be clustered 19.
PI clustering is a diffusion-driven process
PI clustering has initially been proposed to originate from electrostatic interactions and, to a lesser extent, from hydrogen bonding between PI headgroups. PI molecules appear thus sequestered beneath positively charged surfaces, which results in a significant reduction of lateral diffusion in the membrane plane 17. The number of PI molecules that interact with basic residues is determined by the negative net charge of the PIs at a given pH. For instance, the charge of the PI(4,5)P 2 molecules at pH 3 is −1.5e, whereas at pH 7.4, which is close to the pH of the cytosol (7.2), it is −4e 20. For a N-BAR homodimer of charge +8e, one could estimate that at cytosolic pH, the stoichiometry of PI-interacting molecules per protein module is 2:1, which gives an estimated 1.5-fold increase of local PI(4,5)P 2. However, experimental studies have shown that the binding of the N-BAR module on PI-containing membranes induces a local enrichment of at least 10-fold 19. How could such a difference in the local PIs’ enrichment be explained?
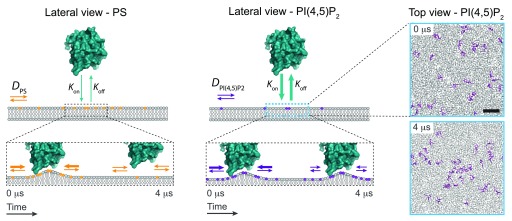
Theoretical studies have shown that the binding of a positively charged protein with a negatively charged membrane induces lipid demixing near to the protein surface 19, 21. This phenomenon is the result of the combination of electrostatic interactions and an entropic effect. Upon protein-membrane binding, charged lipids diffuse in the plane of the membrane towards the protein surface to preserve charge neutrality ( Figure 2). In the case of monovalent lipids such as phosphatidylserine (PS), lipid demixing is almost negligible as a result of the fast K on/ K off rates between the protein and the membrane, which prevents charged lipids to locally segregate 22 ( Figure 2, left panel). However, for multivalent lipids such as some PI species, the transient interaction with a positively charged protein generates an electrostatic potential well, which results in a reduction of the K on/ K off rates and in protein diffusion. Consequently, transient demixing of PI molecules can take place 22 ( Figure 2, right panel). As shown by numerical simulations and consistent with the estimated ~10-fold increase from experimental data, PIs can cluster together up to nine lipid molecules per protein module. The trajectory of PI molecules in the plane of the membrane showed the existence of PI-protein dissociation events, thus pointing out that clustered PI molecules are not sequestered 19. Importantly, this behavior is observed at initial physiological relevant concentrations of 1% PI(4,5)P 2.
Figure 2. Schematic representation on how phosphoinositide (PI)-binding motifs can engage local demixing of PIs on cellular membranes.
As an example, lateral view of the ENTH domain of Epsin (PDB code 1H0A) in cyan upon binding to a membrane that contains monovalent lipids such as phosphatidylserine (PS) (in orange, left panel) or PI(4,5)P 2 (in magenta, right panel). Cyan arrows represent the K on/ K off rates of the ENTH domain binding on membranes, being faster for PS over PI(4,5)P 2. As a result, transient demixing of PI(4,5)P 2 molecules can take place. The diffusion of PS and PI(4,5)P 2 in the plane of the membrane is depicted by orange and magenta arrows, respectively. Right panel shows a top view of PI(4,5)P 2 clustering coarse-grain molecular dynamics simulations (as described in 19) on spontaneous membrane biding of an ENTH domain. The panels are snapshots at t = 0 μs and 4 μs of the individual position of PI(4,5)P 2 molecules (in magenta) along the simulation. Scale bar, 1 nm.
PI demixing has been reported in both flat and curved membranes. In the latter case, the segregation of PI molecules is likely to be amplified by membrane curvature since it is reported to significantly reduce protein diffusion 23 and lipid dynamics 17. This is in agreement with recent molecular simulations that show that clustering of lipids such as PIs and GM3 correlates with membrane curvature 8.
The “PI clustering” toolbox: electrostatic interactions and beyond
Local segregation of PIs into submicron domains has been mostly described for proteins with the intrinsic property to polymerize on membranes, such as the BAR domain family. Proteins of the BAR family can sense and generate membrane curvature, owing to the scaffolding structure that results from the homodimerization of the BAR module. Association of BAR proteins with membranes takes place through electrostatic interactions between positively charged amino acids on the concave/convex face of the dimeric module and acidic phospholipids 24. PI clustering has been reported for proteins with F-BAR, BAR, N-BAR, and I-BAR modules 17– 19. The tendency of multivalent PIs to engage lipid demixing over the monovalent PS provides BAR proteins with some specificity to generate PI subdomains at the plasma membrane, where PI(4,5)P 2 and PI(3,4,5)P 3 are the predominant affected PI isoforms. According to the structural homology within members of the BAR superfamily, it is likely that the formation of PI-enriched microdomains could be a general feature of any protein hosting a BAR module. Combination of the BAR module with PI-binding motifs within the same protein might provide an additional layer of regulation and, possibly, production of monophosphate PI pools in other organelles than the plasma membrane, as observed in the case of BIN1 19. This suggests that the property of PI clustering might be extrapolated to some members of the sorting nexin (SNX) family holding a BAR module and a PX motif 25, although this link has yet to be established.
The clustering of PIs is, however, not necessarily associated with the intrinsic ability of proteins to self-assemble. Indeed, the transient segregation of PIs is likely to generate a positive feedback loop. As a result, proteins that selectively interact with PIs can locally accumulate on PI-enriched areas, independently of their ability to polymerize, as observed for the ENTH and ANTH domains 19. Therefore, PI clustering seems to be a general property of proteins that directly interact with PIs via electrostatic interactions with more or less specificity for a given PI isoform. Accordingly, natively unstructured polybasic protein domains have also been shown to induce local segregation of PIs at the plasma membrane, as observed for MARCKS, GAP43, CAPS23, and syntaxin-1A 10, 13. The number of proteins that associate with acidic lipids at the plasma membrane through polybasic sequences is large 14, 15. For instance, several small GTPases have been shown to interact with plasma membrane PI(3,4,5)P 3 and PI(4,5)P 2 by means of polybasic clusters 26.
PI clustering might be solely limited to ionic protein-lipid interactions, although it is tempting to speculate that alternative or complementary mechanisms might take on the stabilization of PI pools. For instance, recent studies have shown that the pinning of the cytoskeleton on membranes preserves liquid-ordered and liquid-disordered (Lo-Ld) phase coexistence at physiological temperatures (37°C) 27, 28. The polymerization of actin cytoskeleton was also shown to promote segregation of lipid phases in in vitro models 29. These observations are in agreement with the “picket fence” model, which predicts that the cytoskeletal network might act as a diffusion barrier for lipids and proteins 30. The exact partition of PI(4,5)P 2 into Lo-Ld domains is not yet clear, but the depletion of cholesterol with methyl-β-cyclodextrin was shown to reduce PI(4,5)P 2 levels at the plasma membrane 31. The partition of PI(4,5)P 2 to cholesterol-dependent domains was also reported using the targeting of a 5-phosphatase 32. In addition, the sequestration of syntaxin-1A microdomains at sites of synaptic vesicle exocytosis in the plasma membrane was shown to require the formation of cholesterol and PI(4,5)P 2-mediated clusters, which are both distinct from lipid “rafts” 12, 33. An interesting observation is that Ld domains were found to align along actin fibers independently of the lipid phase to which actin was pinned 28. This might be explained by local changes in membrane curvature induced by the actin network. Indeed, Ld domains appear to favor lipid sorting and membrane deformation 34. Recently, numerical simulations have shown that clustering of lipids such as PI(4,5)P 2 correlates with membrane curvature 8. The exact contribution of membrane curvature itself in PI clustering is not yet established, but lipid packing defects associated with membrane curvature might favor a better exposure of PI(4,5)P 2 headgroups 19, 35. Here, one will have to take into account in future experiments the nature of the fatty acids present on PI molecules, which might also impact on the rigidity and shape of the lipid bilayers to which they belong.
PI clustering: a novel regulator of intracellular trafficking and signaling?
Importantly, after PI clustering, protein-PI dissociation can still take place independently of the initial concentration of PIs 19. This suggests that PI clusters are more dynamic than initially anticipated and that a given PI cluster could sequentially interact with different effectors. Thus, PI clustering induced by an upstream protein could favor the recruitment of a downstream PI-binding partner, providing a mechanism to coordinate trafficking or signaling events.
One process that PI clustering could regulate is clathrin-mediated endocytosis (CME). Indeed, the F-BAR, ANTH, ENTH, and N-BAR domains are present in central molecular players involved in CME 36. All of these protein modules have been shown to engage local segregation of PI(4,5)P 2 17, 19, which is the key PI isoform in CME. Therefore, PI clustering could participate in the spatiotemporal regulation of CME based on the affinity constant of the different protein intermediates and their interaction with PI(4,5)P 2. A hypothetical example of how PI clustering might operate in CME is shown in Figure 3, although the number of PI(4,5)P 2 effectors implicated in CME is much larger (see Table 1). The polymerization of the N-BAR module along the bud neck is likely to establish a diffusion barrier 37, highly enriched in PIs, which would thereby be shielded from the activity of kinases and phosphatases. These features might be relevant at different stages of clathrin-coated vesicle biogenesis. Indeed, the metabolic evolution of PIs during CME has been shown to be important for the maturation of clathrin-coated vesicles 38. In addition, the segregation of lipid phases has been reported to generate sufficient line tension to induce membrane scission 39. It is therefore possible that the PI demixing induced by BAR proteins plays an additional role in line tension-mediated fission at the last stage of CME, as suggested by theoretical studies 40.
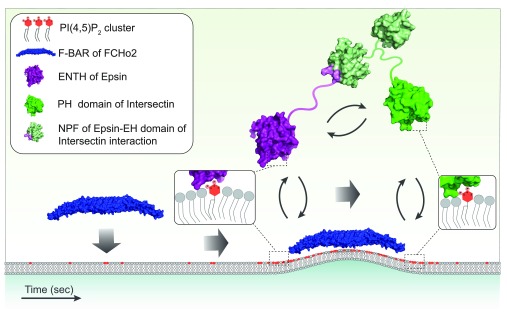
Figure 3. Schematics of the potential role of phosphoinositide (PI) clustering to coordinate cell trafficking events: example of the biogenesis of a clathrin-coated vesicle.
The F-BAR domain (Protein Data Bank [PDB] code 2V0O) of FCHo2 binds to the plasma membrane, driving PI(4,5)P 2 segregation into clusters. The local PI(4,5)P 2 enrichment drives the binding of Epsin through the interaction of its ENTH domain (PDB code 1H0A) with PI(4,5)P 2. The Asn-Pro-Phe (NPF) domain of Epsin can interact with the EH domain (PDB code 3FIA) of Intersectin, which in addition hosts a PH domain (PDB code 1MAI) that binds to PI(4,5)P 2. The dynamics of the system is likely influenced by the affinity constant of the PI(4,5)P 2-binding motifs, which will determine the K on/K off of PI(4,5)P 2-mediated membrane binding, and by the affinity constant between the different protein domains.
Table 1. PI(4,5)P 2 effectors implicated in CME.
The table shows an overview of all the possible options that exist in the PI(4,5)P 2-mediated protein recruitment during the different stages of CME. Notice that although the interaction with PI(4,5)P 2 is mostly electrostatically driven, some effectors hold structured motifs with specific affinities/selectivity for PI(4,5)P 2. In addition, effectors can act as either monomers or larger assemblies, although PI(4,5)P 2 clustering can engage the local accumulation of proteins that typically do not self-assemble as a result of positive feedback 19.
| Mammalia
n protein |
Function | PI(4,5)P
2
interaction |
Self-
assembly |
References |
|---|---|---|---|---|
| FCHo 1/2 | Membrane curvature (F-BAR) | Charge dependent | Yes | 17, 41, 42 |
| AP2 | Adaptor complex | α subunit
C-μ2 subunit |
No | 43, 44 |
| Intersectin | Scaffolding protein | PH domain | Yes | 45– 47 |
| AP180, CALM | Adaptor of AP2 and clathrin | ANTH domain | No | 48, 49 |
| HIP1-HIP1R | Links actin to clathrin | ANTH domain | No | 49, 50 |
| Epsin | Membrane bending | ENTH domain | No | 49, 51 |
| Amphiphysin | Membrane curvature (N-BAR) | Charge dependent | Yes | 19, 24, 52 |
| Endophilin | Membrane curvature (N-BAR) | Charge dependent | Yes | 24, 53 |
| Syndapin | Membrane curvature (F-BAR) | Charge dependent | Yes | 17, 54, 55 |
| SNX 9/18 | Membrane curvature (BAR) | PX domain | Yes | 56, 57 |
| Dynamin | Scission | PH domain | Yes | 58– 60 |
| OCRL | PI 5-phosphatase | PH domain | No | 61 |
| Numb | Cargo adaptor (Notch) | PTB/PI domain | No | 62, 63 |
| Dab2 | Cargo adaptor (LDLR) | DH domain | No | 64– 66 |
| ARH | Cargo adaptor (LDLR) | PTB/PI domain | No | 65, 66 |
It is tempting to propose that the coordinated action of PIs and scaffolding protein complexes, in particular BAR proteins, is a general feature of the biogenesis of transport vesicles 67. For instance, the N-BAR protein Arfaptin 1 has been shown to participate in the biogenesis of secretory storage granules through the interaction with PI4P at the trans-Golgi network 68. The ArfGAP ASAP1 also carries a BAR module along with a PI-binding motif and has been shown to provide a platform to regulate Arf4 and Rab8/Rab11-mediated targeting of rhodopsin transport carriers to cilium 69. Finally, some members of the SNX family also hold a BAR module in addition to the characteristic PX domain, which typically binds to PI3P 6. The SNX-BAR proteins are implicated in tubule-based endosomal sorting 70. This includes the two retromer subunits SNX1 and SNX2, SNX5, and SNX6 or SNX4 among others 71, 72. One may speculate that the formation of PI clustering together with the binding affinity for different PI effectors might be linked to the ability of SNX-BAR proteins to define tubular endosomal subdomains.
PI clustering could also play an important role in the coordination of signaling events. Interestingly, the juxtamembrane segment of the EGFR, which is implicated in the activation of the receptor, is also composed of a cluster of basic residues that interact with PI(4,5)P 2 73, 74. Indeed, natively unstructured polybasic protein domains have been shown to engage PI(4,5)P 2 clustering 11. The interaction of the EGFR with PI(4,5)P 2 is required for the activation and downstream signaling of the receptor at the plasma membrane and seems also to regulate its fate in the endosomal compartments. The first observation that PI4P 5-kinase activity generating PI(4,5)P 2 pools was associated with the EGFR and required for appropriate activation and downstream signaling originates from the early 90s 75. Later studies demonstrated that PI(4,5)P 2 clustering induced by the binding and antiparallel dimerization of the juxtamembrane segments of two associated EGFRs can lead to the activation of the receptor even in the absence of ligand 76. This property was suggested to be important at a high density of EGFR monomers (>800/µm 2), as is often observed in aberrant activation of the receptor in cancers 73, 77. In this condition, formation of EGFR nanoclusters takes place as a result of the electrostatic interaction between PI(4,5)P 2 molecules at the plasma membrane and the juxtamembrane region of the receptor 78.
Recent evidence demonstrates that PI(4,5)P 2 generated on endosomes is required for the appropriate sorting of active EGFR towards multivesicular bodies and further termination of the signal. This process relies on the recruitment of the endosomal type Iγ PIP kinase, PIPKIγi5, that gets targeted to early endosomes by association with SNX5, an effector of PI(4,5)P 2. The kinase will then increase local pools of PI(4,5)P 2, also required for association of SNX5 with Hrs proteins that will then interact with ubiquitinated EGFR and ensure its proper sorting 79.
It is noteworthy that most of the tyrosine kinase receptors of the EGFR family harbor a polybasic juxtamembrane domain that could play the same role in terms of ligand free activation or sorting and signal transduction (e.g. insulin-like growth factor 1 receptor [IGF1R], vascular endothelial growth factor receptor [VEGFR], platelet-derived growth factor receptor [PDGFR], and fibroblast growth factor receptor 1 [FGFR1], among others) 76. Although PI clustering being a general feature of membrane-associated polybasic domains provides an attractive hypothesis to activate receptors and trigger signaling, work is still needed to define whether it is a broad mechanism or applies only to some specific proteins.
Conclusions
The spatiotemporal remodeling of PI pools within distinct organelles is an intrinsic feature that makes possible the orchestration of PI-mediated cellular functions. Indeed, PIs are constantly subjected to the activity of PI-metabolizing enzymes and must be in addition accessible to effectors. Because the lateral diffusion of lipid molecules within the membrane plane is extremely fast, PI clustering comes up as a realistic mechanism to locally preserve newly metabolized PI pools on cellular membranes. Indeed, Balla and co-workers already anticipated that PI4P replenishment from the Golgi was not essential to preserve the plasma membrane pool, although it does contribute to its formation 80. Irvine and co-authors also showed that the maintenance of the steady-state pool of PI(4,5)P 2 at the plasma membrane does not require localization of its synthetic precursor PI4P on the same cellular compartment 81. It is tempting to speculate that PI clusters might work as potential platforms to coordinate PI-mediated protein interactions or as molecular beacons, as previously proposed 13. Nevertheless, the myriad of protein modules capable of engaging PI clustering is becoming broad. Based on structural homologies, one might predict that the list will progressively increase. An interesting feature to point out is that PI clustering seems to be a general mechanism for either multivalent or monophosphate PIs 19. The precise regulatory role of PI clustering in trafficking and signal transduction has still to be established, but it certainly opens up exciting perspectives in the field. For instance, PI clustering might orchestrate the different steps in carrier biogenesis. Also, the ability of cellular receptors to engage PI clustering might determine their sorting to the appropriate compartment. The physiological implication of PI clustering in living organisms has yet to be established. Recent studies have already shown that the oligomerization of Sec14-nodulin proteins controls the localization of PI(4,5)P 2 and signaling landscape in polarized membrane morphogenesis in Arabidopsis thaliana root hairs 82, 83. Despite the role of PIs in many cellular processes, certain PI isoforms and functions have often been elusive due to the lack of detection or labeling strategies, which is typically limited to the use of PI-binding motifs with all of the associated side effects. The development of novel experimental strategies capable of detecting the intrinsic dynamics of PIs or of exploiting the recently developed sub-100nm life cell imaging techniques 84 will be key to unraveling the regulatory role of PI clustering.
Acknowledgments
We thank Dr. Stefano Vanni (Institut de Pharmacologie Moléculaire et Cellulaire, UMR 7275, France) for kindly performing and providing the numerical simulations shown in Figure 2.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Peter Mayinger, Division of Nephrology & Hypertension and Department of Cell & Developmental Biology, Oregon Health & Science University, Portland, OR, USA
Volker Haucke, Leibniz-Institut für Molekulare Pharmakologie (FMP), Berlin, Germany
Vytas A Bankaitis, Department of Molecular and Cellular Medicine, Texas A&M Health Science Center, College Station, TX, USA
Tamás Balla, Molecular Signal Transduction NICHD, National Institutes of Health, Bethesda, MD, USA
Funding Statement
This work was supported by grants from the Agence Nationale de la Recherche (ANR) (ANR-13-BSV2-0004-01) and the ERC (MYODYN, # 339847).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 4 approved]
References
- 1. Di Paolo G, De Camilli P: Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443(7112):651–7. 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- 2. Moser von Filseck J, Čopič A, Delfosse V, et al. : INTRACELLULAR TRANSPORT. Phosphatidylserine transport by ORP/Osh proteins is driven by phosphatidylinositol 4-phosphate. Science. 2015;349(6246):432–6. 10.1126/science.aab1346 [DOI] [PubMed] [Google Scholar]
- 3. Balla T: Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev. 2013;93(3):1019–137. 10.1152/physrev.00028.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 4. Berridge MJ, Irvine RF: Inositol phosphates and cell signalling. Nature. 1989;341(6239):197–205. 10.1038/341197a0 [DOI] [PubMed] [Google Scholar]
- 5. De Matteis MA, Godi A: PI-loting membrane traffic. Nat Cell Biol. 2004;6(6):487–92. 10.1038/ncb0604-487 [DOI] [PubMed] [Google Scholar]
- 6. Kutateladze TG: Translation of the phosphoinositide code by PI effectors. Nat Chem Biol. 2010;6(7):507–13. 10.1038/nchembio.390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. van Meer G, Voelker DR, Feigenson GW: Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9(2):112–24. 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Koldsø H, Shorthouse D, Hélie J, et al. : Lipid clustering correlates with membrane curvature as revealed by molecular simulations of complex lipid bilayers. PLoS Comput Biol. 2014;10(10):e1003911. 10.1371/journal.pcbi.1003911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Krauss M, Haucke V: Phosphoinositide-metabolizing enzymes at the interface between membrane traffic and cell signalling. EMBO Rep. 2007;8(3):241–6. 10.1038/sj.embor.7400919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Laux T, Fukami K, Thelen M, et al. : GAP43, MARCKS, and CAP23 modulate PI(4,5)P 2 at plasmalemmal rafts, and regulate cell cortex actin dynamics through a common mechanism. J Cell Biol. 2000;149(7):1455–72. 10.1083/jcb.149.7.1455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McLaughlin S, Murray D: Plasma membrane phosphoinositide organization by protein electrostatics. Nature. 2005;438(7068):605–11. 10.1038/nature04398 [DOI] [PubMed] [Google Scholar]
- 12. van den Bogaart G, Meyenberg K, Risselada HJ, et al. : Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479(7374):552–5. 10.1038/nature10545 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Honigmann A, van den Bogaart G, Iraheta E, et al. : Phosphatidylinositol 4,5-bisphosphate clusters act as molecular beacons for vesicle recruitment. Nat Struct Mol Biol. 2013;20(6):679–86. 10.1038/nsmb.2570 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Bogdanov M, Dowhan W, Vitrac H: Lipids and topological rules governing membrane protein assembly. Biochim Biophys Acta. 2014;1843(8):1475–88. 10.1016/j.bbamcr.2013.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li L, Shi X, Guo X, et al. : Ionic protein-lipid interaction at the plasma membrane: what can the charge do? Trends Biochem Sci. 2014;39(3):130–40. 10.1016/j.tibs.2014.01.002 [DOI] [PubMed] [Google Scholar]
- 16. Carvalho K, Ramos L, Roy C, et al. : Giant unilamellar vesicles containing phosphatidylinositol(4,5)bisphosphate: characterization and functionality. Biophys J. 2008;95(9):4348–60. 10.1529/biophysj.107.126912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhao H, Michelot A, Koskela EV, et al. : Membrane-sculpting BAR domains generate stable lipid microdomains. Cell Rep. 2013;4(6):1213–23. 10.1016/j.celrep.2013.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 18. Saarikangas J, Zhao H, Pykäläinen A, et al. : Molecular mechanisms of membrane deformation by I-BAR domain proteins. Curr Biol. 2009;19(2):95–107. 10.1016/j.cub.2008.12.029 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Picas L, Viaud J, Schauer K, et al. : BIN1/M-Amphiphysin2 induces clustering of phosphoinositides to recruit its downstream partner dynamin. Nat Commun. 2014;5: 5647. 10.1038/ncomms6647 [DOI] [PubMed] [Google Scholar]
- 20. Ellenbroek WG, Wang YH, Christian DA, et al. : Divalent cation-dependent formation of electrostatic PIP 2 clusters in lipid monolayers. Biophys J. 2011;101(9):2178–84. 10.1016/j.bpj.2011.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khelashvili G, Weinstein H, Harries D: Protein diffusion on charged membranes: a dynamic mean-field model describes time evolution and lipid reorganization. Biophys J. 2008;94(7):2580–97. 10.1529/biophysj.107.120667 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 22. Golebiewska U, Gambhir A, Hangyás-Mihályné G, et al. : Membrane-bound basic peptides sequester multivalent (PIP 2), but not monovalent (PS), acidic lipids. Biophys J. 2006;91(2):588–99. 10.1529/biophysj.106.081562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Domanov YA, Aimon S, Toombes GE, et al. : Mobility in geometrically confined membranes. Proc Natl Acad Sci U S A. 2011;108(31):12605–10. 10.1073/pnas.1102646108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Peter BJ, Kent HM, Mills IG, et al. : BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303(5657):495–9. 10.1126/science.1092586 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 25. Cullen PJ: Endosomal sorting and signalling: an emerging role for sorting nexins. Nat Rev Mol Cell Biol. 2008;9(7):574–82. 10.1038/nrm2427 [DOI] [PubMed] [Google Scholar]
- 26. Heo WD, Inoue T, Park WS, et al. : PI(3,4,5)P 3 and PI(4,5)P 2 lipids target proteins with polybasic clusters to the plasma membrane. Science. 2006;314(5804):1458–61. 10.1126/science.1134389 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 27. Arumugam S, Petrov EP, Schwille P: Cytoskeletal pinning controls phase separation in multicomponent lipid membranes. Biophys J. 2015;108(5):1104–13. 10.1016/j.bpj.2014.12.050 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Honigmann A, Sadeghi S, Keller J, et al. : A lipid bound actin meshwork organizes liquid phase separation in model membranes. eLife. 2014;3:e01671. 10.7554/eLife.01671 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 29. Liu AP, Fletcher DA: Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophys J. 2006;91(11):4064–70. 10.1529/biophysj.106.090852 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Kusumi A, Nakada C, Ritchie K, et al. : Paradigm shift of the plasma membrane concept from the two-dimensional continuum fluid to the partitioned fluid: high-speed single-molecule tracking of membrane molecules. Annu Rev Biophys Biomol Struct. 2005;34:351–78. 10.1146/annurev.biophys.34.040204.144637 [DOI] [PubMed] [Google Scholar]
- 31. Kwik J, Boyle S, Fooksman D, et al. : Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proc Natl Acad Sci U S A. 2003;100(24):13964–9. 10.1073/pnas.2336102100 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Johnson CM, Chichili GR, Rodgers W: Compartmentalization of phosphatidylinositol 4,5-bisphosphate signaling evidenced using targeted phosphatases. J Biol Chem. 2008;283(44):29920–8. 10.1074/jbc.M805921200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lang T, Bruns D, Wenzel D, et al. : SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 2001;20(9):2202–13. 10.1093/emboj/20.9.2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Manneville JB, Casella JF, Ambroggio E, et al. : COPI coat assembly occurs on liquid-disordered domains and the associated membrane deformations are limited by membrane tension. Proc Natl Acad Sci U S A. 2008;105(44):16946–51. 10.1073/pnas.0807102105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vanni S, Hirose H, Barelli H, et al. : A sub-nanometre view of how membrane curvature and composition modulate lipid packing and protein recruitment. Nat Commun. 2014;5: 4916. 10.1038/ncomms5916 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 36. McMahon HT, Boucrot E: Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12(8):517–33. 10.1038/nrm3151 [DOI] [PubMed] [Google Scholar]
- 37. Prévost C, Zhao H, Manzi J, et al. : IRSp53 senses negative membrane curvature and phase separates along membrane tubules. Nat Commun. 2015;6: 8529. 10.1038/ncomms9529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Posor Y, Eichhorn-Gruenig M, Puchkov D, et al. : Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 2013;499(7457):233–7. 10.1038/nature12360 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 39. Roux A, Cuvelier D, Nassoy P, et al. : Role of curvature and phase transition in lipid sorting and fission of membrane tubules. EMBO J. 2005;24(8):1537–45. 10.1038/sj.emboj.7600631 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 40. Liu J, Sun Y, Drubin DG, et al. : The mechanochemistry of endocytosis. PLoS Biol. 2009;7(9):e1000204. 10.1371/journal.pbio.1000204 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 41. Henne WM, Kent HM, Ford MG, et al. : Structure and analysis of FCHo2 F-BAR domain: a dimerizing and membrane recruitment module that effects membrane curvature. Structure. 2007;15(7):839–52. 10.1016/j.str.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 42. Henne WM, Boucrot E, Meinecke M, et al. : FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 2010;328(5983):1281–4. 10.1126/science.1188462 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 43. Collins BM, McCoy AJ, Kent HM, et al. : Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109(4):523–35. 10.1016/S0092-8674(02)00735-3 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Höning S, Ricotta D, Krauss M, et al. : Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol Cell. 2005;18(5):519–31. 10.1016/j.molcel.2005.04.019 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Yamabhai M, Hoffman NG, Hardison NL, et al. : Intersectin, a novel adaptor protein with two Eps15 homology and five Src homology 3 domains. J Biol Chem. 1998;273(47):31401–7. 10.1074/jbc.273.47.31401 [DOI] [PubMed] [Google Scholar]
- 46. Hussain NK, Jenna S, Glogauer M, et al. : Endocytic protein intersectin-l regulates actin assembly via Cdc42 and N-WASP. Nat Cell Biol. 2001;3(10):927–32. 10.1038/ncb1001-927 [DOI] [PubMed] [Google Scholar]
- 47. Adams A, Thorn JM, Yamabhai M, et al. : Intersectin, an adaptor protein involved in clathrin-mediated endocytosis, activates mitogenic signaling pathways. J Biol Chem. 2000;275(35):27414–20. 10.1074/jbc.M004810200 [DOI] [PubMed] [Google Scholar]
- 48. Ford MG, Pearse BM, Higgins MK, et al. : Simultaneous binding of PtdIns(4,5)P 2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291(5506):1051–5. 10.1126/science.291.5506.1051 [DOI] [PubMed] [Google Scholar]
- 49. Stahelin RV, Long F, Peter BJ, et al. : Contrasting membrane interaction mechanisms of AP180 N-terminal homology (ANTH) and epsin N-terminal homology (ENTH) domains. J Biol Chem. 2003;278(31):28993–9. 10.1074/jbc.M302865200 [DOI] [PubMed] [Google Scholar]
- 50. Engqvist-Goldstein AE, Kessels MM, Chopra VS, et al. : An actin-binding protein of the Sla2/Huntingtin interacting protein 1 family is a novel component of clathrin-coated pits and vesicles. J Cell Biol. 1999;147(7):1503–18. 10.1083/jcb.147.7.1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ford MG, Mills IG, Peter BJ, et al. : Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419(6905):361–6. 10.1038/nature01020 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 52. Takei K, Slepnev VI, Haucke V, et al. : Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1(1):33–9. 10.1038/9004 [DOI] [PubMed] [Google Scholar]
- 53. Ringstad N, Gad H, Löw P, et al. : Endophilin/SH3p4 is required for the transition from early to late stages in clathrin-mediated synaptic vesicle endocytosis. Neuron. 1999;24(1):143–54. 10.1016/S0896-6273(00)80828-4 [DOI] [PubMed] [Google Scholar]
- 54. Qualmann B, Kelly RB: Syndapin isoforms participate in receptor-mediated endocytosis and actin organization. J Cell Biol. 2000;148(5):1047–62. 10.1083/jcb.148.5.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang Q, Navarro MV, Peng G, et al. : Molecular mechanism of membrane constriction and tubulation mediated by the F-BAR protein Pacsin/Syndapin. Proc Natl Acad Sci U S A. 2009;106(31):12700–5. 10.1073/pnas.0902974106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lundmark R, Carlsson SR: Sorting nexin 9 participates in clathrin-mediated endocytosis through interactions with the core components. J Biol Chem. 2003;278(47):46772–81. 10.1074/jbc.M307334200 [DOI] [PubMed] [Google Scholar]
- 57. Pylypenko O, Lundmark R, Rasmuson E, et al. : The PX-BAR membrane-remodeling unit of sorting nexin 9. EMBO J. 2007;26(22):4788–800. 10.1038/sj.emboj.7601889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Bashkirov PV, Akimov SA, Evseev AI, et al. : GTPase cycle of dynamin is coupled to membrane squeeze and release, leading to spontaneous fission. Cell. 2008;135(7):1276–86. 10.1016/j.cell.2008.11.028 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 59. Faelber K, Posor Y, Gao S, et al. : Crystal structure of nucleotide-free dynamin. Nature. 2011;477(7366):556–60. 10.1038/nature10369 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Klein DE, Lee A, Frank DW, et al. : The pleckstrin homology domains of dynamin isoforms require oligomerization for high affinity phosphoinositide binding. J Biol Chem. 1998;273(42):27725–33. 10.1074/jbc.273.42.27725 [DOI] [PubMed] [Google Scholar]
- 61. Mao Y, Balkin DM, Zoncu R, et al. : A PH domain within OCRL bridges clathrin-mediated membrane trafficking to phosphoinositide metabolism. EMBO J. 2009;28(13):1831–42. 10.1038/emboj.2009.155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Dho SE, French MB, Woods SA, et al. : Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J Biol Chem. 1999;274(46):33097–104. 10.1074/jbc.274.46.33097 [DOI] [PubMed] [Google Scholar]
- 63. Salcini AE, Confalonieri S, Doria M, et al. : Binding specificity and in vivo targets of the EH domain, a novel protein-protein interaction module. Genes Dev. 1997;11(17):2239–49. 10.1101/gad.11.17.2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mishra SK, Keyel PA, Hawryluk MJ, et al. : Disabled-2 exhibits the properties of a cargo-selective endocytic clathrin adaptor. EMBO J. 2002;21(18):4915–26. 10.1093/emboj/cdf487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Maurer ME, Cooper JA: The adaptor protein Dab2 sorts LDL receptors into coated pits independently of AP-2 and ARH. J Cell Sci. 2006;119(Pt 20):4235–46. 10.1242/jcs.03217 [DOI] [PubMed] [Google Scholar]
- 66. Yun M, Keshvara L, Park CG, et al. : Crystal structures of the Dab homology domains of mouse disabled 1 and 2. J Biol Chem. 2003;278(38):36572–81. 10.1074/jbc.M304384200 [DOI] [PubMed] [Google Scholar]
- 67. Vicinanza M, D'Angelo G, Di Campli A, et al. : Function and dysfunction of the PI system in membrane trafficking. EMBO J. 2008;27(19):2457–70. 10.1038/emboj.2008.169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Cruz-Garcia D, Ortega-Bellido M, Scarpa M, et al. : Recruitment of arfaptins to the trans-Golgi network by PI(4)P and their involvement in cargo export. EMBO J. 2013;32(12):1717–29. 10.1038/emboj.2013.116 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Wang J, Morita Y, Mazelova J, et al. : The Arf GAP ASAP1 provides a platform to regulate Arf4- and Rab11-Rab8-mediated ciliary receptor targeting. EMBO J. 2012;31(20):4057–71. 10.1038/emboj.2012.253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. van Weering JR, Sessions RB, Traer CJ, et al. : Molecular basis for SNX-BAR-mediated assembly of distinct endosomal sorting tubules. EMBO J. 2012;31(23):4466–80. 10.1038/emboj.2012.283 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 71. Bonifacino JS, Hurley JH: Retromer. Curr Opin Cell Biol. 2008;20(4):427–36. 10.1016/j.ceb.2008.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. van Weering JR, Verkade P, Cullen PJ: SNX-BAR-mediated endosome tubulation is co-ordinated with endosome maturation. Traffic. 2012;13(1):94–107. 10.1111/j.1600-0854.2011.01297.x [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 73. Arkhipov A, Shan Y, Das R, et al. : Architecture and membrane interactions of the EGF receptor. Cell. 2013;152(3):557–69. 10.1016/j.cell.2012.12.030 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Abd Halim KB, Koldsø H, Sansom MS: Interactions of the EGFR juxtamembrane domain with PIP 2-containing lipid bilayers: Insights from multiscale molecular dynamics simulations. Biochim Biophys Acta. 2015;1850(5):1017–25. 10.1016/j.bbagen.2014.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 75. Cochet C, Filhol O, Payrastre B, et al. : Interaction between the epidermal growth factor receptor and phosphoinositide kinases. J Biol Chem. 1991;266(1):637–44. [PubMed] [Google Scholar]
- 76. Michailidis IE, Rusinova R, Georgakopoulos A, et al. : Phosphatidylinositol-4,5-bisphosphate regulates epidermal growth factor receptor activation. Pflugers Arch. 2011;461(3):387–97. 10.1007/s00424-010-0904-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Endres NF, Das R, Smith AW, et al. : Conformational coupling across the plasma membrane in activation of the EGF receptor. Cell. 2013;152(3):543–56. 10.1016/j.cell.2012.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 78. Wang Y, Gao J, Guo X, et al. : Regulation of EGFR nanocluster formation by ionic protein-lipid interaction. Cell Res. 2014;24(8):959–76. 10.1038/cr.2014.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sun Y, Hedman AC, Tan X, et al. : Endosomal type Iγ PIP 5-kinase controls EGF receptor lysosomal sorting. Dev Cell. 2013;25(2):144–55. 10.1016/j.devcel.2013.03.010 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 80. Szentpetery Z, Várnai P, Balla T: Acute manipulation of Golgi phosphoinositides to assess their importance in cellular trafficking and signaling. Proc Natl Acad Sci U S A. 2010;107(18):8225–30. 10.1073/pnas.1000157107 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 81. Hammond GR, Fischer MJ, Anderson KE, et al. : PI4P and PI(4,5)P 2 are essential but independent lipid determinants of membrane identity. Science. 2012;337(6095):727–30. 10.1126/science.1222483 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 82. Ghosh R, de Campos MK, Huang J, et al. : Sec14-nodulin proteins and the patterning of phosphoinositide landmarks for developmental control of membrane morphogenesis. Mol Biol Cell. 2015;26(9):1764–81. 10.1091/mbc.E14-10-1475 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 83. Vincent P, Chua M, Nogue F, et al. : A Sec14p-nodulin domain phosphatidylinositol transfer protein polarizes membrane growth of Arabidopsis thaliana root hairs. J Cell Biol. 2005;168(5):801–12. 10.1083/jcb.200412074 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 84. Li D, Shao L, Chen BC, et al. : ADVANCED IMAGING. Extended-resolution structured illumination imaging of endocytic and cytoskeletal dynamics. Science. 2015;349(6251):aab3500. 10.1126/science.aab3500 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation