Abstract
For this study, we sought to identify pre‐mRNA processing events modulated by changes in extracellular pH, inorganic phosphate, and antifungal drugs. We examined genes with at least four putative introns whose transcriptional level responded to these effectors. We showed that the intron retention levels of genes encoding asparagine synthetase 2, C6‐zinc finger regulator (fluffy), and a farnesyltransferase respond to amphotericin B, ketoconazole, and other effectors. In general, the assayed antifungals promoted the disruption of the structural domains of these proteins probably leading to their inactivation, which emphasize the complexity of the metabolic modulation exerted by antifungal signaling.
Keywords: alternative splicing, amphotericin B, asparagine synthetase, intron retention, ketoconazole, Neurospora crassa, Pi regulation
Abbreviations
- MAPK
mitogen‐activated protein kinase
- nuc‐1
transcriptional activator which controls phosphate acquisition
- nuc‐2
component of Pi‐regulated signal transduction pathway
- Pi
phosphate
- preg and pgov
regulatory genes which control phosphate acquisition
- qRT‐PCR
quantitative real‐time PCR
- RNAseq
next‐generation RNA sequencing technologies
- SRA
Sequence Read Archive
- TCA
tricarboxylic acid
Alternative splicing of pre‐mRNA transcripts is a regulated and complex molecular process that dramatically increases proteome diversity and fulfills important post‐transcriptional regulatory functions 1, 2, 3, 4, 5, 6, 7. There are at least five types of alternative splicing in eukaryotic microorganisms that are represented by alternate 5′ or 3′ splice sites, exon skipping, intron retention, and mutually exclusive events 2, 8, 9, 10. In filamentous fungi, the splicing of numerous genes is modulated in response to nutrient signaling. These include, for example, the genes hex‐1 of Neurospora crassa and pacC of Aspergillus nidulans, which respond to extracellular phosphate changes and ambient pH, respectively 11, 12, 13.
The filamentous fungus N. crassa is a heterothallic, free‐living, eukaryotic microorganism that is widespread in nature and is clearly visible due to its intense pink color. The importance of N. crassa in biochemical and molecular genetics research has been long recognized, making it an excellent model system for examining molecular responses to current signals in eukaryotic microorganisms 14, 15. Ambient signals are first detected by sensors, which then initiate the transmission of intracellular information via transduction pathways to a target 16, 17, 18, 19, 20. Changes in ambient temperature, light, pH, nutrients, and the presence of fungicides are among the extensive repertoire of fungal stressors that are detectable.
Phosphate (Pi) is a crucial ingredient in the synthesis of nucleic acids and the flow of genetic information 21, 22. In N. crassa, the molecular response to Pi deprivation consists of a highly conserved and hierarchical relationship among at least five genes—nuc‐2, preg, pgov, mak‐2, and nuc‐1—whose functioning allows for the activation of the transcriptional regulator NUC‐1 and its translocation into the nucleus. This translocation, in turn, leads to the transcriptional activation of several genes, including the Pi‐repressible phosphatases 15, 19, 23, 24, 25, 26, 27, 28, 29. Previously published microarray experiments using a mak‐2 knockout strain (Δmak‐2) confirmed that the mak‐2 gene, which encodes a mitogen‐activated protein kinase (MAPK), is an important component of the response to extracellular Pi changes in N. crassa 19. These experiments revealed many differentially expressed genes that are involved in various physiological processes related to Pi transport, metabolism, and regulation, as well as to post‐translational modification of proteins and the MAPK MAK‐2 signaling pathway. Among them are some genes that are not apparently related to Pi scavenging, such as the genes encoding asparagine synthetase 2 (KEGG: NCU04303) 19, 30, 31, 32, C6‐zinc finger regulator (fluffy, KEGG: NCU08726) 33, 34, 35, and a farnesyltransferase (KEGG: NCU05999) 36, 37. Several genes that are repressed in the wild‐type strain when grown in the presence of high Pi, such as nuc‐2, are repressed in the Δmak‐2 mutant strain when grown in either low‐ or high‐Pi media. In an extended model of the Pi‐signaling network, we proposed that the MAK‐2 and NUC‐2 proteins are functional in N. crassa that has been cultured under conditions of limited Pi, but are nonfunctional under sufficient Pi conditions, leading to a reduction in the transcription of Pi‐repressible genes. Here, we show that intron retention by the genes coding for asparagine synthetase 2 (asn‐2), C6‐zinc finger regulator (fluffy), and a farnesyltransferase (ram‐1) occurs in response to extracellular pH and Pi changes and the presence of antifungal drugs.
Materials and methods
Strains, culture conditions, disk diffusion assay, and cDNA synthesis
The N. crassa wild‐type St.L.74‐OR23‐1VA (FGSC No 2489) (control strain), and the Δmak‐2 mutant strain (FGSC No 11482) of N. crassa used in this study were obtained from the Fungal Genetics Stock Center, Kansas State University, Manhattan, Kansas 38 (www.fgsc.net). The Δmak‐2 mutant strain was generated by the Neurospora Functional Genomics Project Strains (www.fgsc.net). The strains were maintained on slants of Vogel's medium (1.5% agar, 2% sucrose).
The in vitro susceptibilities of two N. crassa strains (St.L.74A and Δmak‐2) to the antifungal drugs ketoconazole, amphotericin B, nystatin, and terbinafine were evaluated using a disk diffusion assay at pH 5.4 39, 40, 41. After spreading aliquots of 200 μL conidial suspension containing about 2 × 106 cells·mL−1 of each strain on the agar plate surface, filter paper disks (5‐mm diameter) containing different quantities of each antifungal drug were placed on the center of the plates. All fungi were tested with three biological replicates. Plates were incubated for 72 h at 30 °C. Inhibitor zones for the experiments were measured after visible fungal lawns had covered the control plates. These were plotted against the antifungal concentration to determine the standard deviation for the inhibition zone (< 10% for all experiments) and to define the antifungal concentrations used in gene expression assays.
For gene expression assays, conidia (approximately 106 cells·mL−1) were germinated for 5 h and 16 h at 30 °C in an orbital shaker (200 rpm) in low‐ (100 μm) or high‐Pi (10 mm) medium. The medium was adjusted to either pH 5.4 or 8.0 and was supplemented with 44 mm sucrose as the carbon source. Cells were grown in the presence or absence of amphotericin B (200 μg·mL−1) or ketoconazole (1 mg·mL−1) and prepared as previously described 42, 43.
Total RNA was extracted from approximately 100 mg of frozen mycelia using the Illustra RNAspin mini isolation kit (GE Healthcare, Little Chalfont, UK) and treated with RNAse‐free DNAse I (Invitrogen, Carlsbad, CA, USA). Purified RNA (1 μg) from each strain was reverse‐transcribed into cDNA with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) according to the manufacturer's instructions. The absence of genomic DNA in RNA preparations was confirmed by RT‐PCR using a primer set targeting an intron‐flanking region in the N. crassa actin gene.
RNA‐seq quantification of intron retention events
Publicly available RNAseq data (next‐generation RNA sequencing technologies) from the Sequence Read Archive at the NCBI database (http://www.ncbi.nlm.nih.gov/sra) was used to quantify intron retention events in N. crassa. Seven RNAseq experiments for N. crassa wild‐type strains grown on glucose (GEO: GSM1238604 and GSM1238606), xylose (GEO: GSM1238609), arabinose (GEO: GSM1238602 and GSM1238597), sucrose (GEO: GSM899613), and cellulose (Avicel, GEO: GSM899607) were used. For intron retention quantification, SRA files were downloaded from the database and converted to fastq format using the SRA toolkit. Reads were then aligned to fasta files containing sequences of 50 nt representing each exon–intron boundary for the genes of interest (KEGG: NCU04303, NCU08726, and NCU05999) using Bowtie 44, 45. The number of reads mapped at each boundary was used to estimate the prevalence of intron retention events in each gene.
Qualitative and quantitative real‐time PCR (qRT‐PCR)
For qualitative expression analysis, primer pairs that yield PCR products surrounding each intron sequence (Table 1) were used to amplify the transcription products of the genes coding for asparagine synthetase 2 (KEGG: NCU04303), C6‐zinc finger regulator (KEGG: NCU08726), and a farnesyltransferase (KEGG: NCU05999). For each PCR reaction, we used approximately 100 ng of cDNA and 10 pmol of each oligonucleotide. Thermocycler conditions were 95 °C for 2 min, followed by 35 cycles at 95 °C for 30 s with an annealing temperature of 60 °C for 1 min. Only relevant results are shown in the figures.
Table 1.
Primers used in RT‐PCR and/or qRT‐PCR experiments
| Gene product name (gene name) | IDa | Primer sequences (5′‐3′) | Amplicon length (bp) | Amplicon spliced (bp) | Intron analyzed | Experiments |
|---|---|---|---|---|---|---|
| Asparagine synthetase 2 (asn‐2) | NCU04303 | |||||
| F: | ATCCGTCACCGTGGTCCTGAT | 188 | 49 | Intron 3 | RT‐PCR | |
| R: | TGTTGTGGCAAGTGACGCTGC | |||||
| F: | CAACCTTTCGCCCAACC | 63 | – | Intron 3 | qRT‐PCR | |
| R: | TTGACTCGATTGACACTTTTAT | |||||
| F: | GCGGCAGCGTCACTTG | 142 | 57 | Intron 4 | RT‐PCR | |
| R: | CCGACGATACTGAGACGCT | |||||
| F: | TGCGGATAAGACGATGGTTG | 182 | 112 | Intron 5 | RT‐PCR | |
| R: | TGCCAAGCGTCTTGTTTCTA | |||||
| C6‐zinc finger (fluffy) | NCU08726 | |||||
| F: | GCCAAGACAACACCTAACACCGAA | 299 | 149 | Intron 1 | RT‐PCR | |
| R: | AGATGGTCCTTGGTACGCCATTTC | |||||
| CaaX farnesyltransferase beta subunit Ram1 (ram‐1) | NCU05999 | |||||
| F: | CAACCCAACAACGAGGCCCATGGC | 503 | 313 | Intron 3 | RT‐PCR | |
| R: | CGACTATAGAGACTCTCCGGCGCA | |||||
| Actin | NCU04173 | |||||
| F: | GTATGTGCAAGGCCGGTTTCG | 305 | 109 | Introns 3/4 |
RT‐PCR qRT‐PCR |
|
| R: | TCCTTCTGGCCCATACCGATCATG | |||||
| Tubulin alpha‐1 | NCU09132 | |||||
| F: | CCTCGTCTTCCACTCCTTCGG | 445 | 190 | Intron 4 |
RT‐PCR qRT‐PCR |
|
| R: | ACTGTGTTCGAGGGTGCTGTG |
Gene accession number at the N. crassa genome database at the Broad Institute.
Quantitative real‐time PCR amplifications of NCU04303 (asparagine synthetase 2) were performed with the StepOne Plus Real‐Time PCR system (Applied Biosystems) using the oligonucleotides in Table 1. The qPCR experiments were performed in a 12.5‐μL reaction containing 6.25‐μL of the SYBR Green PCR Master Mix (Applied Biosystems), 50 ηg of cDNA, and 1 μL of each primer. The PCR protocol included an initial denaturation step at 50 °C for 2 min and at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. A dissociation curve was generated at the end of each PCR cycle to verify the amplification of a single product. Relative transcript quantities were calculated using the ΔΔC t method 11, 46 with N. crassa actin and β‐tubulin as the internal reference genes. Analysis was performed using the onestep software v2.2 (Applied Biosystems, Foster City, CA, USA). The control strain grown in high‐Pi conditions, pH 5.4 for 5 h and in the absence of antifungal was used as a reference for calculating relative gene expression. Statistical analysis was performed using one‐way ANOVA followed by the Tukey's ad hoc test using graphpad prism v5.1 software (La Jolla, CA, USA).
Results and Discussion
Previously, genome‐wide differential transcription profiling using a Δmak‐2 knockout strain of N. crassa grown under phosphate shortage revealed 912 unique genes that were found to be differentially expressed when compared to the expression in the control strain (S.t.L.74A) 19. Functional annotation of those differentially expressed genes using the Gene Ontology 47, 48 and N. crassa Genome Database 49, 50 showed that these genes were primarily involved in various physiological processes related to phosphate transport, metabolism, and regulation; posttranscriptional modification of proteins; and MAPK signaling pathways. In order to identify alternative splicing events in the transcriptional processing of these 912 genes we preselected 13 of them, which met the criteria of having at least four putative introns and being responsive to changes in ambient Pi or a mak‐2 − background 19. Qualitative expression analyses by RT‐PCR identified three candidate genes coding for asparagine synthetase 2 (asn‐2) (KEGG: NCU04303), C‐6‐zinc finger (fluffy) (KEGG: NCU08726), and CaaX farnesyl transferase beta subunit Ram1 (ram‐1) (KEGG: NCU05999) for further study (Table 1).
Assays assessing the in vitro susceptibility of N. crassa to some antifungal drugs showed that the Δmak‐2 knockout strain is more susceptible to ketoconazole and terbinafine than the wild‐type strain. In contrast, the Δmak‐2 strain is more tolerant to amphotericin B but has the same tolerance to nystatin (Fig. 1). These results suggest that MAK‐2 affects the inhibitory mechanism and the sensing of these antifungal drugs. Ketoconazole and amphotericin B were therefore selected for our intron retention assays because they presented opposite effects in a comparison of the wild‐type strain with the Δmak‐2 strain. Furthermore, the control strain is not affected by terbinafine at the concentrations used (Fig. 1).
Figure 1.
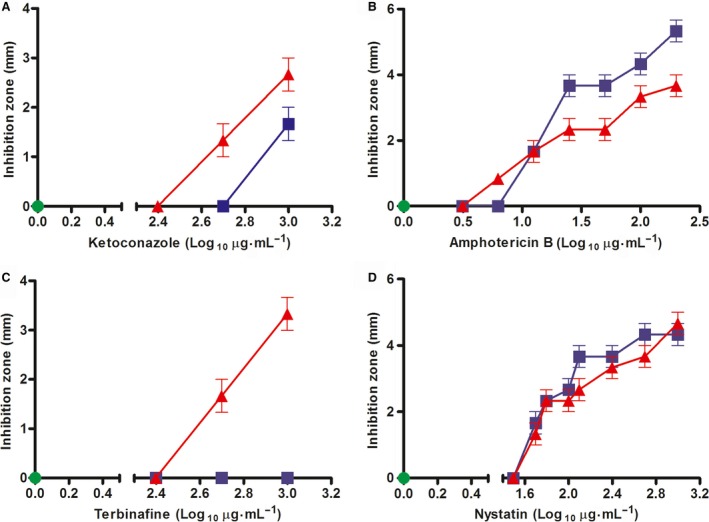
In vitro susceptibility of two N. crassa strains exposed to antifungal drugs at pH 5.4. The inhibition zones, measured in mm, were plotted against antifungal concentrations. The St.L.74A and Δmak‐2 strains were incubated in the absence of antifungal (control) (green circles). Strains were also incubated with ketoconazole (A), amphotericin B (B), terbinafine (C), and nystatin (D). Blue squares and red triangles represent the inhibition zones observed for the St.L.74A and Δmak‐2 strains, respectively.
Prediction of putative intron retention in these three genes using publicly available RNAseq data showed that different introns in the same gene might undergo differential splicing depending on the experimental conditions (Fig. 2). Our experimental confirmation (Figs 3, 4, 5, 6) of the retention of some of these predicted introns is useful since it allows the identification of alternative splicing events by using the next‐generation RNA sequencing technologies.
Figure 2.
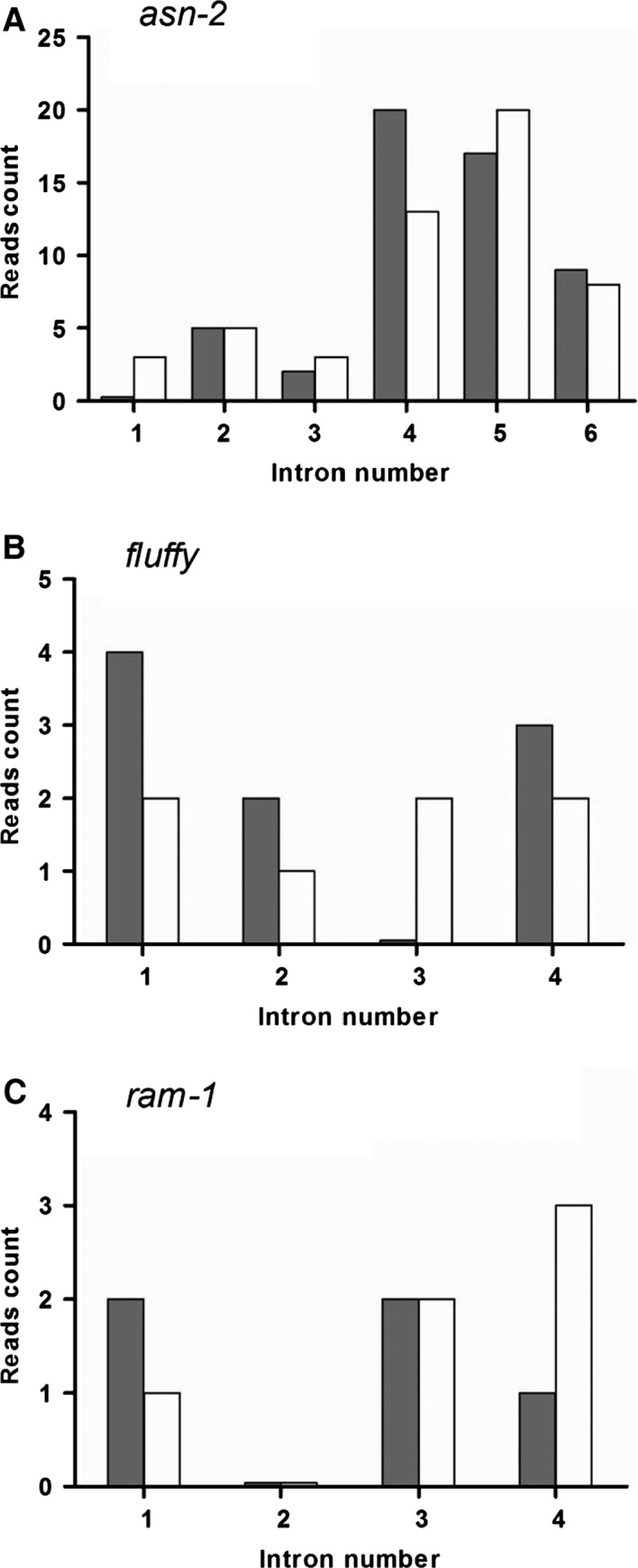
Intron retention evaluated by RNAseq in N. crassa. The Sequence Read Archive at the NCBI database (http://www.ncbi.nlm.nih.gov/sra) was used to quantify intron retention. Reads were then aligned to fasta files containing sequences of 50 nt representing each exon‐intron boundary for the genes of interest. The number of reads mapping at each boundary was used to estimate the prevalence of intron retention in each gene analyzed (5′‐ gray; 3′‐ white). (A) Asparagine synthetase 2 (asn‐2) (NCU04303); (B) C6‐zinc finger regulator (fluffy) (NCU08726); (C) farnesyltransferase (ram1) (NCU05999).
Figure 3.
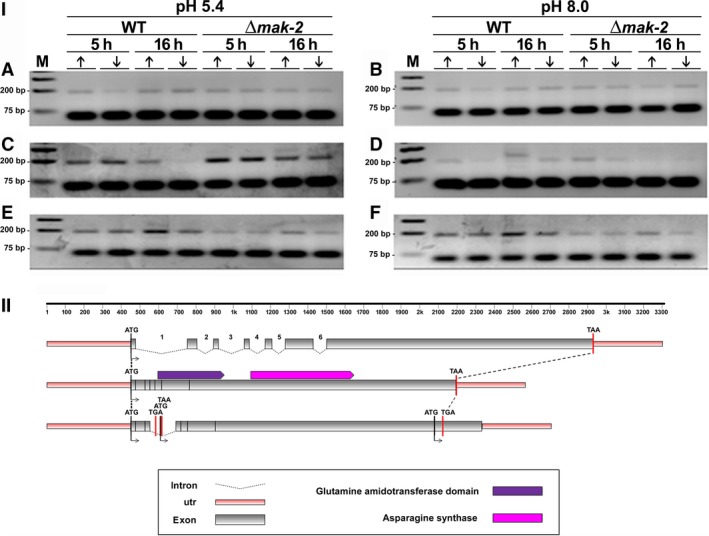
(I) Retention of intron‐3 visualized by RT‐PCR during pre‐mRNA processing of the asn‐2 gene in N. crassa. Strains Δmak‐2 mutant and St.L.74A were incubated for 5 h and 16 h in high‐ (10 mm) (↑) or low‐Pi (100 μm) (↓) at pH 5.4 and pH 8.0 in the absence of antifungal (control) (A, B), with amphotericin B (C, D), and with ketoconazole (E, F). (M) Molecular weight ladder. Sizes expected for the amplified fragments were 49 bp and 188 bp for nonretention or retention of the intron, respectively. (II) Schematic overview of intron‐3 retention, as compared to the genomic DNA and mRNA organization of the asn‐2 gene of N. crassa.
Figure 4.
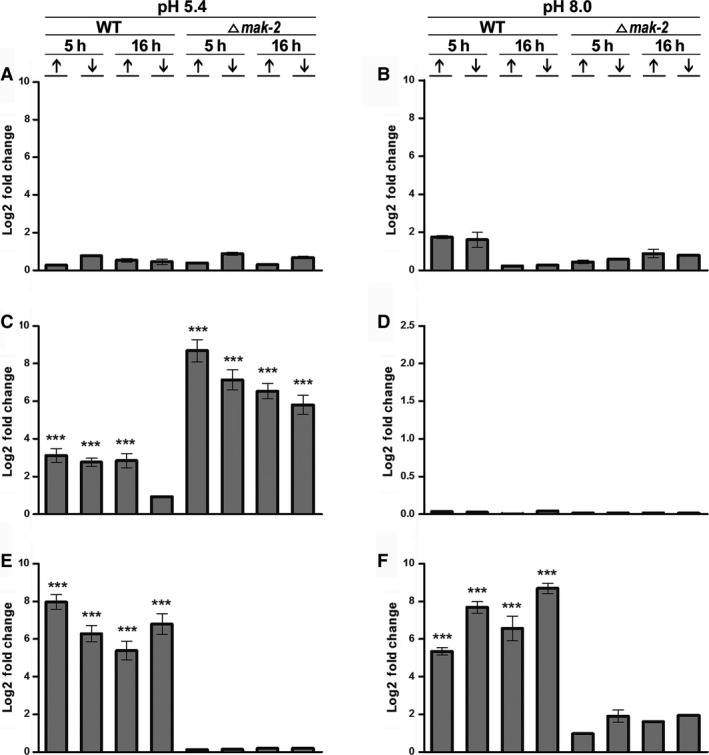
Real‐time PCR (qRT‐PCR) validation of intron‐3 transcript levels in the asn‐2 gene in N. crassa. Strains Δmak‐2 mutant and St.L.74A were incubated for 5 h and 16 h in high‐ (10 mm) (↑) or low‐Pi (100 μm) (↓), at pH 5.4 and pH 8.0, in the absence of antifungal (control) (A, B), with amphotericin B (C, D), and with ketoconazole (E, F). qRT‐PCR data are representative of the average values ± standard deviation (SD) obtained from three independent experiments. Statistically significant values are indicated by asterisks: Tukey's ad hoc test, ***P < 001.
Figure 5.
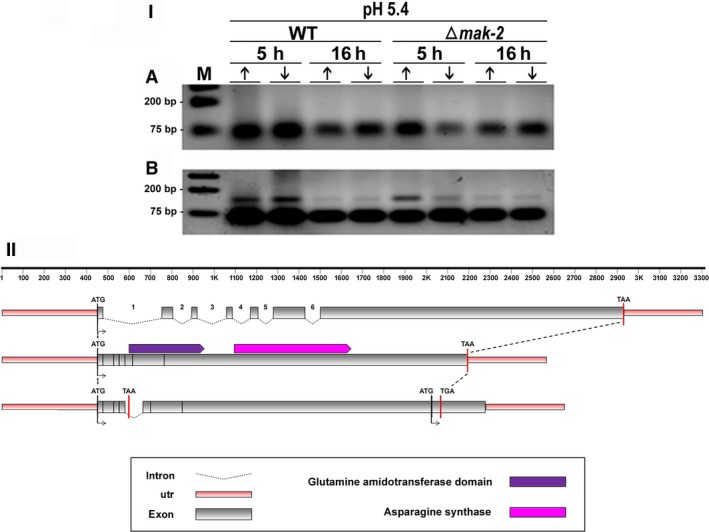
(I) Retention of intron‐4 visualized by RT‐PCR during pre‐mRNA processing of the asn‐2 gene in N. crassa. Strains Δmak‐2 mutant and St.L.74A were incubated for 5 h and 16 h in high‐ (10 mm) (↑) or low‐Pi (100 μm) (↓) at pH 5.4 in the absence (A) and the presence of ketoconazole (B). (M) Molecular weight ladder. Sizes expected for the amplified fragments are 57 and 142 bp for nonretention or retention of the intron, respectively. (II) Schematic overview of intron‐4 retention, as compared to the genomic DNA and mRNA organization of the asn‐2 gene of N. crassa.
Figure 6.
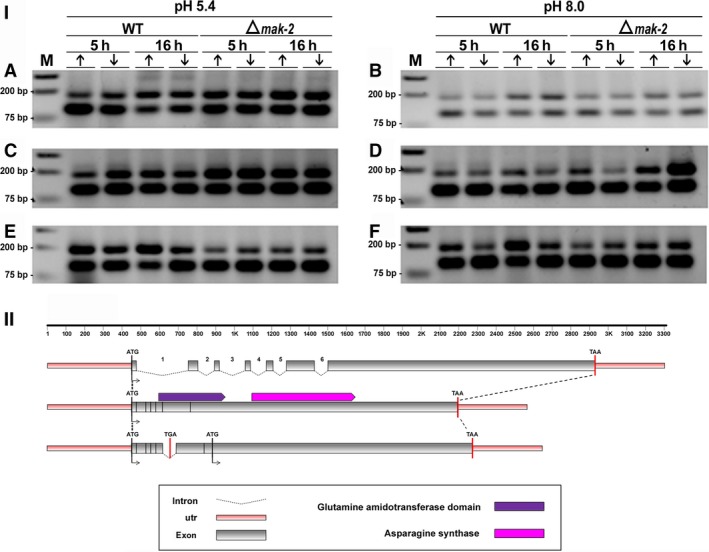
(I) Retention of intron‐5 visualized by RT‐PCR during pre‐mRNA processing of the asn‐2 gene in N. crassa. Strains Δmak‐2 mutant and St.L.74A were incubated for 5 h and 16 h in high‐ (10 mm) (↑) or low‐Pi (100 μm) (↓) at pH 5.4 and pH 8.0 in the absence of antifungal (A, B), with amphotericin B (C, D), and with ketoconazole (E, F). (M) Molecular weight ladder. Sizes expected for the amplified fragments are 112 and 182 bp for nonretention or retention of the intron, respectively. (II) Schematic overview of intron‐5 retention, as compared to the genomic DNA and mRNA organization of the asn‐2 gene of N. crassa.
Genes and proteins
Asparagine synthetase 2
Asparagine is biosynthesized in eukaryotes from l‐aspartate using glutamine as the amino group donor, which is an enzymatic reaction driven by the hydrolysis of ATP 30. Asparagine synthetase 2, encoded by the asn‐2 gene, catalyzes this metabolic pathway. However, in some organisms such as Escherichia coli, a second metabolic pathway utilizes either ammonia or glutamine as the amino group donor 30. Little is known about the roles of asparagine, glutamate, glutamine, and aspartate, aside from being important sources for the de novo biosynthesis of nucleotides and various amino acids. However, asparagine may act as a metabolic regulator of cellular adaptation to glutamine depletion likely by supporting the availability of TCA cycle intermediates and the supply of reduced nitrogen needed to maintain the synthesis of nonessential amino acids. Additionally, glutamine derivatives have been proposed to be an essential component of glutamine‐dependent cell survival in mammals. Thus, asparagine synthetase‐2 may play a regulatory role in the intricate metabolism of nitrogen and carbon in eukaryotes 32. In N. crassa, transcription of the asn‐2 gene is up‐regulated in cultures with abundant Pi, a physiological condition where the transcriptional regulator NUC‐1 is nonfunctional. Thus, the asn‐2 gene is not Pi‐repressible. We also provided conclusive evidence that transcription of Pi‐repressible genes is down‐regulated in the Δmak‐2 strain, irrespective of the supply of Pi, whereas, for some non‐Pi‐repressible genes, this effect is not observed 19. Interestingly, another asn gene, coding for an asparagine synthetase domain‐containing protein 1, here renamed asn‐1 (KEGG: 07300.7), is also not Pi‐repressible 19.
Pre‐mRNA processing of the asn‐2 gene of N. crassa (KEGG: NCU04303) apparently responds to different effectors, depending on the intron being considered (Figs 3, 4, 5, 6). Six putative introns were identified in the asn‐2 gene and were referred to as introns 1– 6, based on their 5′ to 3′ locations (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html). The retention of intron‐3 is weak at both pH 5.4 and pH 8.0 in the absence of antifungal drugs (Fig. 3IA,B), which is in agreement with the RNA‐seq analysis (Fig. 2). In the presence of amphotericin B, however, retention increases at pH 5.4 but not at pH 8.0 (Fig. 3IC,D). In the presence of ketoconazole, retention of intron‐3 increases at pH 5.4 and pH 8.0 in the 74A strain, but not in the ∆mak‐2 strain (Fig. 3IE,F). These observations were confirmed by qRT‐PCR analysis (Fig. 4), which indicated that these antifungal drugs indeed affect the transcriptional process in N. crassa. Schematic representation of intron‐3 retention, as compared to the genomic DNA and mRNA organization of the asn‐2 gene of N. crassa, showed disruption of the asparagine synthase and glutamine amidotransferase domains of the protein leading to a probable inactivation of the catalytic activities of this enzyme. It is also observed a nucleotide long sequence, probably not translated (Fig. 3II).
Intron‐4 was consistently removed from the asn‐2 transcripts in the absence of antifungal drugs at both pH 5.4 and 8.0. Interestingly, intron‐4 was retained in both 74A, and Δmak‐2 strains cultured for 5h in the presence of ketoconazole at pH 5.4, irrespective of current Pi changes (Fig. 5I) but was spliced out at pH 8.0 (not shown) in both wild‐type and Δmak‐2 strains. Our prediction of intron‐4 retention (Fig. 2) in the absence of antifungal drugs was not confirmed by our data (Fig. 5I). However, retention of this intron may be affected by the different composition of the culture medium used for the RNAseq experiments, as occurred in the antifungal treatment (Fig. 5I). Schematic representation of intron‐4 retention, as compared to the genomic DNA and mRNA organization of the asn‐2 gene of N. crassa, showed disruption of the asparagine synthase and glutamine amidotransferase domains of the protein leading to a probable inactivation of both catalytic activities of this enzyme. It also showed a nucleotide long sequence which was probably not translated (Fig. 5II).
The retention of intron‐5 in the asn‐2 transcripts was observed in all culture conditions tested, irrespective of the absence or presence of antifungal, levels of extracellular Pi, strains assayed, or incubation time (Fig. 6I), confirming our predicted results. However, it is clear that retention of intron‐5 is lower at pH 8.0 in the absence of antifungal compared to pH 5.4 (Fig. 6I). Schematic representation of intron‐5 retention, as compared to the genomic DNA and mRNA organization of the asn‐2 gene of N. crassa (Fig. 6I), showed disruption of the glutamine amidotransferase domain of the protein leading to a probable inactivation of the catalytic activity of this enzyme. However, retention of intron‐5 led to a nucleotide long sequence in which the sequence of the asparagine synthase domain seems conserved. This sequence defines a putative protein possibly capable of retaining this enzymatic activity (Fig. 6II).
C6‐zinc finger regulator (fluffy) (KEGG: NCU08726)
The fluffy gene of N. crassa (NCU08726) is a transcription factor that directly regulates two of the five genes involved in cellular conidiation 33, 34, 35. Four putative introns were identified in the fluffy gene and were referred to as introns 1–4, in 5′ to 3′ order of appearance. We correctly predicted the retention of intron‐1 in the RNAseq experiments at pH 8.0 in the absence of antifungal drugs. Interestingly, retention was not observed in any other culture conditions assayed (Fig. 7I, and results not shown). The schematic representation of intron‐1 retention, as compared to the genomic DNA and mRNA organization of the fluffy gene of N. crassa, showed disruption of its DNA‐binding domain leading to a predicted inactive protein. Also, retention of intron‐1 led to a nucleotide long sequence that defines a putative protein in which the zinc‐finger domain is absent (Fig. 7II).
Figure 7.
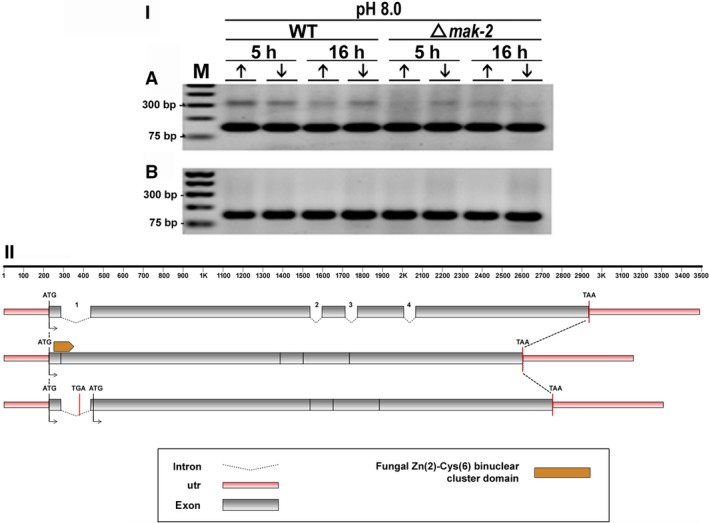
(I) Retention of intron‐1 visualized by RT‐PCR during pre‐mRNA processing of the fluffy gene in N. crassa. Strains Δmak‐2 mutant and St.L.74A were incubated for 5 h and 16 h in high‐ (10 mm) (↑) or low‐Pi (100 μm) (↓) at pH 8.0 in the absence (A) and presence (B) of ketoconazole. (M) Molecular weight ladder. Sizes expected for the amplified fragments are 149 and 299 bp for nonretention or retention of the intron, respectively. (II) Schematic overview of intron‐1 retention, as compared to the genomic DNA and mRNA organization of the fluffy gene of N. crassa.
CaaX farnesyltransferase beta subunit Ram1 (ram‐1) (KEGG: NCU05999)
The ram‐1 gene of N. crassa (NCU05999) codes for a farnesyltransferase, an enzyme that catalyzes the addition of farnesyl pyrophosphate to a cysteine of a G protein, resulting in a farnesylated protein and a diphosphate. G proteins play crucial roles in many signaling processes 7, 36, 37. Thus, the proper functioning of this farnesyltransferase is likely necessary for signal transduction in eukaryotic microorganisms. The catalytic domain of the prenyltransferase‐like activity of this enzyme is harbored in the N‐terminal domain, whereas the catalytic domain of the prenyltransferase and squalene oxidase activities is repeated over the protein body (http://www.broadinstitute.org/annotation/genome/neurospora/MultiHome.html). Four putative introns were identified in the ram‐1 gene. The retention of intron‐3 was observed only at pH 5.4 in both wild‐type and ∆mak‐2 strains cultured for 16 h and in the absence of antifungal drugs (Fig. 8I). The schematic representation of intron‐3 retention, as compared to the genomic DNA and mRNA organization of the ram‐1 gene of N. crassa, showed disruption of its catalytic repeat domain leading to a predicted inactive protein (Fig. 8II).
Figure 8.
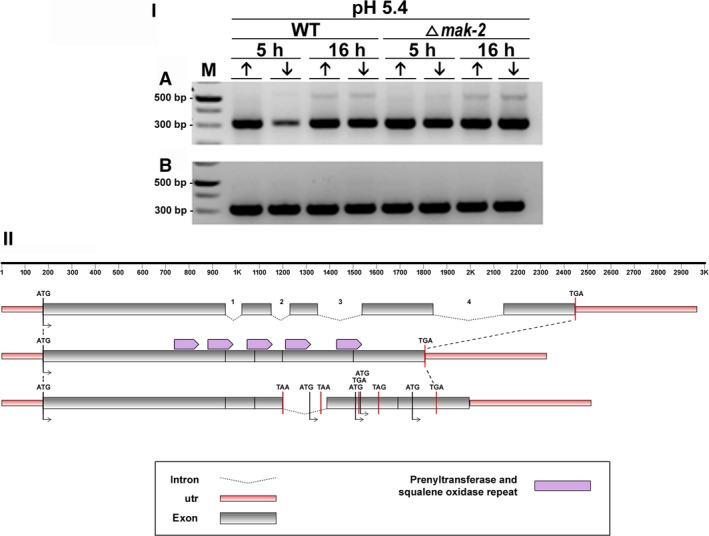
(I) Retention of intron‐3 visualized by RT‐PCR during pre‐mRNA processing of the gene coding for farnesyl transferase beta subunit Ram1 in N. crassa. Strains Δmak‐2 mutant and St.L.74A were incubated for 5 h and 16 h in high‐ (10 mm) (↑) or low‐Pi (100 μm) (↓) at pH 5.4 in the absence (A) and presence (B) of ketoconazole. (M) Molecular weight ladder. Sizes expected for the amplified fragments are 313 and 503 bp for nonretention or retention of the intron, respectively. (II) Schematic overview of intron‐3 retention, as compared to the genomic DNA and mRNA organization of the gene coding for a farnesyltransferase (beta subunit Ram1) of N. crassa.
Conclusions
Intron retention is one of the five top types of alternative splicing in eukaryotic microorganisms that are represented by alternate 5′ or 3′ splice sites. The splicing of numerous genes is modulated in response to nutrient signaling, including many cellular stressors. Here, we show that intron retention in the genes encoding asparagine synthetase 2 (asn‐2), C6‐zinc finger transcription factor (fluffy), and a farnesyltransferase (ram‐1) is modulated by antifungal drugs. In general, the assayed antifungal promoted the disruption of the structural domains of these proteins probably leading to their inactivation. Antifungal‐induced intron retention in some cases is dependent on MAPK protein MAK‐2. Furthermore, we showed that putative introns in the same gene can undergo differential splicing, and confirmed by RT‐PCR experiments the differential retention of introns predicted by RNAseq analyses. Intron retention in N. crassa, and probably in other fungi, is an adaptive genetic response to the toxic effect of antifungal. The antifungal transduction signaling pathway comprises, besides the regulation of mRNA processing in many genes, also the spliceosomal machinery, which may affect the whole fungal metabolism.
Our findings provide an overview of the broad spectrum of metabolic and cellular activities that can be affected by antifungals outside of the inhibition of enzymes of the ergosterol biosynthetic pathway. Thus, antifungals can affect many cellular processes through differential intron retention, leading to changes in gene transcription and the rate of transcript synthesis or degradation, thereby influencing recruitment of many cellular components such as proteins, nucleic acids, and by‐products. These results emphasize the complexity of the metabolic modulation exerted by antifungal signaling.
Author contributions
AR and NMM‐R conceived and designed the project, supervised the research study, and prepared the manuscript. NSM and PMS acquired the data. RS‐R performed the bioinformatics analyses. AR, NSM, PMS, RS‐R, and NMM‐R analyzed and interpreted the data.
Acknowledgements
This work was supported by grants from the Brazilian Funding Agencies: São Paulo Research Foundation (FAPESP), (Grant No 2014/03847‐7 and Postdoctoral Fellowship No. 2012/03689‐7), Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) (Grants No 305252/2013‐5 and 304222/2013‐5), Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação de Apoio ao Ensino, Pesquisa e Assistência (FAEPA). We thank S. H. Castrechini, C. A. Vieira, and M. Mazucato for technical support. We thank P. R. Sanches for his computational support.
References
- 1. Mannella CA, Collins RA, Green MR and Lambowitz AM (1979) Defective splicing of mitochondrial rRNA in cytochrome‐deficient nuclear mutants of Neurospora crassa . Proc Natl Acad Sci USA 76, 2635–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kempken F (2013) Alternative splicing in ascomycetes. Appl Microbiol Biotechnol 97, 4235–4241. [DOI] [PubMed] [Google Scholar]
- 3. Strandberg R, Tzelepis G, Johannesson H and Karlsson M (2013) Coexistence and expression profiles of two alternative splice variants of the pheromone receptor gene pre1 in Neurospora crassa . Arch Microbiol 195, 773–780. [DOI] [PubMed] [Google Scholar]
- 4. Geng C and Paukstelis PJ (2014) An in vitro peptide complementation assay for CYT‐18‐dependent group I intron splicing reveals a new role for the N‐terminus. Biochemistry 53, 1311–1319. [DOI] [PubMed] [Google Scholar]
- 5. Lamech LT, Mallam AL and Lambowitz AM (2014) Evolution of RNA‐protein interactions: non‐specific binding led to RNA splicing activity of fungal mitochondrial tyrosyl‐tRNA synthetases. PLoS Biol 12, e1002028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Yan X, Sablok G, Feng G, Ma J, Zhao H and Sun X (2015) nagnag: Identification and quantification of NAGNAG alternative splicing using RNA‐Seq data. FEBS Lett 589, 1766–1770. [DOI] [PubMed] [Google Scholar]
- 7. Subramani PA, Narala VR, Michael RD, Lomada D and Reddy MC (2015) Molecular docking and simulation of Curcumin with Geranylgeranyl Transferase1 (GGTase1) and Farnesyl Transferase (FTase). Bioinformation 11, 248–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chapman RE and Walter P (1997) Translational attenuation mediated by an mRNA intron. Curr Biol 7, 850–859. [DOI] [PubMed] [Google Scholar]
- 9. Kawahara T, Yanagi H, Yura T and Mori K (1997) Endoplasmic reticulum stress‐induced mRNA splicing permits synthesis of transcription factor Hac1p/Ern4p that activates the unfolded protein response. Mol Biol Cell 8, 1845–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mulder HJ and Nikolaev I (2009) HacA‐dependent transcriptional switch releases hacA mRNA from a translational block upon endoplasmic reticulum stress. Eukaryot Cell 8, 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leal J, Squina FM, Freitas JS, Silva EM, Ono CJ, Martinez‐Rossi NM and Rossi A (2009) A splice variant of the Neurospora crassa hex‐1 transcript, which encodes the major protein of the Woronin body, is modulated by extracellular phosphate and pH changes. FEBS Lett 583, 180–184. [DOI] [PubMed] [Google Scholar]
- 12. Trevisan GL, Oliveira EH, Peres NTA, Cruz AH, Martinez‐Rossi NM and Rossi A (2011) Transcription of Aspergillus nidulans pacC is modulated by alternative RNA splicing of palB . FEBS Lett 585, 3442–3445. [DOI] [PubMed] [Google Scholar]
- 13. Rossi A, Cruz AHC, Santos RS, Silva PM, Silva EM, Mendes NS and Martinez‐Rossi NM (2013) Ambient pH sensing in filamentous fungi: pitfalls in elucidating regulatory hierarchical signaling networks. IUBMB Life 65, 930–935. [DOI] [PubMed] [Google Scholar]
- 14. Beadle GW and Tatum EL (1941) Genetic control of biochemical reactions in Neurospora . Proc Natl Acad Sci USA 27, 499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Metzenberg RL and Chia W (1979) Genetic control of phosphorus assimilation in Neurospora crassa: dose‐dependent dominance and recessiveness in constitutive mutants. Genetics 93, 625–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bahn YS, Xue C, Idnurm A, Rutherford JC, Heitman J and Cardenas ME (2007) Sensing the environment: lessons from fungi. Nat Rev Microbiol 5, 57–69. [DOI] [PubMed] [Google Scholar]
- 17. Kholodenko BN and Birtwistle MR (2009) Four‐dimensional dynamics of MAPK information processing systems. Wiley Interdiscip Rev Syst Biol Med 1, 28–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Richthammer C, Enseleit M, Sanchez‐Leon E, Marz S, Heilig Y, Riquelme M and Seiler S (2012) RHO1 and RHO2 share partially overlapping functions in the regulation of cell wall integrity and hyphal polarity in Neurospora crassa . Mol Microbiol 85, 716–733. [DOI] [PubMed] [Google Scholar]
- 19. Gras DE, Persinoti GF, Peres NTA, Martinez‐Rossi NM, Tahira AC, Reis EM, Prade RA and Rossi A (2013) Transcriptional profiling of Neurospora crassa ∆mak‐2 reveals that mitogen‐activated protein kinase MAK‐2 participates in the phosphate signaling pathway. Fungal Genet Biol 60, 140–149. [DOI] [PubMed] [Google Scholar]
- 20. Nozawa SR, Ferreira‐Nozawa MS, Martinez‐Rossi NM and Rossi A (2003) The pH‐induced glycosylation of secreted phosphatases is mediated in Aspergillus nidulans by the regulatory gene pacC‐dependent pathway. Fungal Genet Biol 39, 286–295. [DOI] [PubMed] [Google Scholar]
- 21. Lenburg ME and O'Shea EK (1996) Signaling phosphate starvation. Trends Biochem Sci 21, 383–387. [PubMed] [Google Scholar]
- 22. Paytan A and McLaughlin K (2007) The oceanic phosphorus cycle. Chem Rev 107, 563–576. [DOI] [PubMed] [Google Scholar]
- 23. Kang S (1993) Functional domains of the transcriptional activator NUC‐1 in Neurospora crassa . Gene 130, 259–264. [DOI] [PubMed] [Google Scholar]
- 24. Kang S and Metzenberg RL (1993) Insertional mutagenesis in Neurospora crassa: cloning and molecular analysis of the preg+ gene controlling the activity of the transcriptional activator NUC‐1. Genetics 133, 193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Peleg Y, Addison R, Aramayo R and Metzenberg RL (1996) Translocation of Neurospora crassa transcription factor NUC‐1 into the nucleus is induced by phosphorus limitation. Fungal Genet Biol 20, 185–191. [DOI] [PubMed] [Google Scholar]
- 26. Robinson KA and Lopes JM (2000) Survey and summary: Saccharomyces cerevisiae basic helix‐loop‐helix proteins regulate diverse biological processes. Nucleic Acids Res 28, 1499–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Leal J, Squina FM, Martinez‐Rossi NM and Rossi A (2007) The transcription of the gene for iso‐orotate decarboxylase (IDCase), an enzyme of the thymidine salvage pathway, is downregulated in the preg c mutant strain of Neurospora crassa grown under phosphate starvation. Can J Microbiol 53, 1011–1015. [DOI] [PubMed] [Google Scholar]
- 28. Gras DE, Silveira HCS, Peres NTA, Sanches PR, Martinez‐Rossi NM and Rossi A (2009) Transcriptional changes in the nuc‐2A mutant strain of Neurospora crassa cultivated under conditions of phosphate shortage. Microbiol Res 164, 658–664. [DOI] [PubMed] [Google Scholar]
- 29. Gras DE, Silveira HCS, Martinez‐Rossi NM and Rossi A (2007) Identification of genes displaying differential expression in the nuc‐2 mutant strain of the mold Neurospora crassa grown under phosphate starvation. FEMS Microbiol Lett 269, 196–200. [DOI] [PubMed] [Google Scholar]
- 30. MacPhee KG, Nelson RE and Schuster SM (1983) Neurospora crassa mutants deficient in asparagine synthetase. J Bacteriol 156, 475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Seifi H, De Vleesschauwer D, Aziz A and Hofte M (2014) Modulating plant primary amino acid metabolism as a necrotrophic virulence strategy: the immune‐regulatory role of asparagine synthetase in Botrytis cinerea‐tomato interaction. Plant Signal Behav 9, e27995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang J, Fan J, Venneti S, Cross JR, Takagi T, Bhinder B, Djaballah H, Kanai M, Cheng EH, Judkins AR et al (2014) Asparagine plays a critical role in regulating cellular adaptation to glutamine depletion. Mol Cell 56, 205–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bailey LA and Ebbole DJ (1998) The fluffy gene of Neurospora crassa encodes a Gal4p‐type C6 zinc cluster protein required for conidial development. Genetics 148, 1813–1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rerngsamran P, Murphy MB, Doyle SA and Ebbole DJ (2005) Fluffy, the major regulator of conidiation in Neurospora crassa, directly activates a developmentally regulated hydrophobin gene. Mol Microbiol 56, 282–297. [DOI] [PubMed] [Google Scholar]
- 35. Olmedo M, Ruger‐Herreros C and Corrochano LM (2010) Regulation by blue light of the fluffy gene encoding a major regulator of conidiation in Neurospora crassa . Genetics 184, 651–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mende K, Homann V and Tudzynski B (1997) The geranylgeranyl diphosphate synthase gene of Gibberella fujikuroi: isolation and expression. Mol Gen Genet 255, 96–105. [DOI] [PubMed] [Google Scholar]
- 37. Quondam M, Barbato C, Pickford A, Helmer‐Citterich M and Macino G (1997) Homology modeling of Neurospora crassa geranylgeranyl pyrophosphate synthase: structural interpretation of mutant phenotypes. Protein Eng 10, 1047–1055. [DOI] [PubMed] [Google Scholar]
- 38. McCluskey K (2003) The Fungal Genetics Stock Center: from molds to molecules. Adv Appl Microbiol 52, 245–262. [DOI] [PubMed] [Google Scholar]
- 39. Noake T, Kuriyama T, White PL, Potts AJ, Lewis MA, Williams DW and Barnes RA (2007) Antifungal susceptibility of Candida species using the Clinical and Laboratory Standards Institute disk diffusion and broth microdilution methods. J Chemother 19, 283–287. [DOI] [PubMed] [Google Scholar]
- 40. Milici ME, Maida CM, Spreghini E, Ravazzolo B, Oliveri S, Scalise G and Barchiesi F (2007) Comparison between disk diffusion and microdilution methods for determining susceptibility of clinical fungal isolates to caspofungin. J Clin Microbiol 45, 3529–3533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Menezes EA, Vasconcelos Junior AA, Angelo MR, Cunha Mda C and Cunha FA (2013) Correlation between microdilution, Etest, and disk diffusion methods for antifungal susceptibility testing of fluconazole against Candida sp. blood isolates. Rev Soc Bras Med Trop 46, 106–107. [DOI] [PubMed] [Google Scholar]
- 42. Nyc JF, Kadner RJ and Crocken BJ (1966) A repressible alkaline phosphatase in Neurospora crassa . J Biol Chem 241, 1468–1472. [PubMed] [Google Scholar]
- 43. Rodrigues SA and Rossi A (1985) Effect of phosphate levels on the synthesis of acid phosphatases (EC 3.1.3.2) in Neurospora crassa . Genet Res 45, 239–249. [DOI] [PubMed] [Google Scholar]
- 44. Langmead B, Trapnell C, Pop M and Salzberg SL (2009) Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Henry VJ, Bandrowski AE, Pepin AS, Gonzalez BJ and Desfeux A (2014) OMICtools: an informative directory for multi‐omic data analysis. Database (Oxford) 2014, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Winer J, Jung CK, Shackel I and Williams PM (1999) Development and validation of real‐time quantitative reverse transcriptase‐polymerase chain reaction for monitoring gene expression in cardiac myocytes in vitro. Anal Biochem 270, 41–49. [DOI] [PubMed] [Google Scholar]
- 47. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25, 25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Vencio RZ, Koide T, Gomes SL and Pereira CA (2006) BayGO: bayesian analysis of ontology term enrichment in microarray data. BMC Bioinformatics 7, 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Galagan JE, Calvo SE, Borkovich KA, Selker EU, Read ND, Jaffe D, FitzHugh W, Ma LJ, Smirnov S, Purcell S (2003) The genome sequence of the filamentous fungus Neurospora crassa . Nature 422, 859–868. [DOI] [PubMed] [Google Scholar]
- 50. Wu C, Yang F, Smith KM, Peterson M, Dekhang R, Zhang Y, Zucker J, Bredeweg EL, Mallappa C, Zhou X (2014) Genome‐wide characterization of light‐regulated genes in Neurospora crassa . G3: Genes ‐ Genomes ‐ Genetics 4, 1731–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]


