Abstract
Cell therapy regimens are frequently compromised by low-efficiency cell homing to therapeutic niches. Improvements in this regard would enhance effectiveness of clinically-applicable cell therapy. The major regulators of tissue-specific cellular migration are chemokines and therefore selection of therapeutic cellular populations for appropriate chemokine receptor expression would enhance tissue-homing competence. A number of practical considerations preclude the use of antibodies in this context and alternative approaches are required. Here we demonstrate that appropriately labelled chemokines are at least as effective in detecting their cognate receptors as commercially available antibodies. We also demonstrate the utility of biotinylated-chemokines as cell-sorting reagents. Specifically we demonstrate, in the context of CCR7 (essential for lymph node-homing of leukocytes), the ability of biotinylated-CCL19 with magnetic-bead sorting to enrich for CCR7-expressing cells. The sorted cells demonstrate improved CCR7 responsiveness and lymph node-homing capability and the sorting is effective for both T cells and dendritic cells. Importantly the ability of chemokines to detect CCR7, and sort for CCR7 positivity, crosses species being effective on murine and human cells. This novel approach to cell sorting is therefore inexpensive, versatile and applicable to numerous cell-therapy contexts. We propose that this represents a significant technological advance with important therapeutic implications.
Introduction
Leukocyte migration is regulated by chemokines which possess a characteristic conserved cysteine motif(1-3) and interact with G-protein coupled receptors(4). To date, approximately 45 chemokines, and 18 receptors, have been identified. Chemokine biology can be simplified by defining them as being inflammatory, or homeostatic, according to the contexts in which they function(2, 5). Thus inflammatory chemokines control leukocyte migration to inflamed sites throughout the body whereas homeostatic chemokines regulate basal leukocyte trafficking to specific tissues and tissue compartments. It is now clear that cells carry ‘address-codes’ specifying tissue-specific migratory capacity and key components of cellular ‘address-codes’ are receptors for homeostatic chemokines(6). Notably, this aspect of regulation of cellular migration is rarely considered in current cell therapy regimens, which frequently require tissue-specific targeting of therapeutic cells for effective clinical outcome. Accordingly, cells used for therapy invariably display heterogeneous expression of appropriate homing chemokine receptors which contributes to the inefficient migration of these cells to their therapeutic niche(7-10).
The best example of chemokine-dependent tissue-specific migration, and one of importance to cellular therapy, is the role for CCL19 and CCL21, and their cognate receptor CCR7, in specifying cell migration to lymph nodes (LNs)(11-14). Thus antigen presenting cells, such as dendritic cells (DCs), following antigen encounter at infected/inflamed sites, upregulate CCR7 which supports their migration to LNs(15-18). Naive and central memory T cells also express CCR7 which specifically marks a population requiring transit to LNs for effector function. The importance of CCR7 for LN migration of DCs and T cells is supported by numerous studies with CCR7-deficient mice(13). Thus CCR7 is essential for cell migration to LNs and the development of adaptive immune responses and tolerance. In cellular therapies, therapeutic DCs and T cells typically display varied CCR7 expression levels(19). As a result, much of the lack of success of DC, and T cell, immunotherapy has been attributed to poor cellular homing to LNs compounded by possible tolerance induction by immature CCR7− DCs(20, 21). A number of approaches have been developed to try to overcome this including direct intra-lymphatic injection of DCs, intranodal injection(22), adenoviral over-expression of CCR7(23) and ‘trogocytosis’(24). Each of these approaches has disadvantages and is of limited clinical use. It is clear that new insights are required to improve therapeutic cell homing in these, and other, cell therapy contexts.
We present a novel approach to this problem involving the use of biotinylated chemokines to enrich for cells bearing their cognate receptors. Such technology is important given the dearth of high-quality antibodies to many chemokine receptors, along with the expense, and other considerations, associated with production of antibodies for clinical cell sorting. The approach described has numerous other advantages including the ability to chemically synthesise biotinylated chemokines to complete purity at relatively low cost and (given the conservation of chemokines) the ability to use biotinylated human chemokines in both human and veterinary clinical and experimental contexts. Specifically, we demonstrate the ability of biotinylated CCL19 (bCCL19) to detect, and enrich for, CCR7-expressing T cells and DCs. We further demonstrate that bCCL19-sorted T cells and DCs are fully functional, displaying heightened responses to CCR7 ligands, and that the DCs have enhanced LN-homing capacity. They are therefore likely to represent improved cellular products for immunotherapy. The requirement for specific chemokine receptors in other tissue-specific cellular therapy contexts means that the approaches described will be of broad clinical applicability. Overall, we conclude that the use of chemokines as novel cell sorting agents is simple, inexpensive, versatile and ideally suited to clinical development.
Materials and Methods
Human cell culture
Human buffy coats were obtained from Scottish National Blood Transfusion Service (approved by Glasgow NHS Trust-East Ethics Committee). PBMC were isolated using Ficoll-paque gradient (GE Healthcare). Short-term polyclonal cultures of human T cells were generated exactly as previously described(25). CD14+ monocytes were isolated using magnetic bead separation (Miltenyi Biotec), as per manufacturer’s instructions. Immature DCs were generated as described(26) and matured by re-plating 0.5×106 cells/ml in complete medium with addition of 20μg/ml Polyinosinic-polycytidylic and 1μg/ml prostaglandin E2 (both from Sigma-Aldrich) for 48 hours(27).
HEK cells were cultured and transfected with D6 and CCR7 as described(28-30), and binding and internalisation assays performed as previously reported(31-33).
Mice
C57Bl/6 D6-deficient mice and WT C57Bl/6 mice with Ly5.2, Ly5.1 and Ly5.1/5.2 heterozygous expression were maintained in the Glasgow University Biological Services Facility under specific pathogen-free conditions. All experiments were conducted under the auspices of a UK Home Office Licence and approved by the University of Glasgow Ethical Review committee.
Generation of bone marrow-derived DC (BM-DC)
Mouse bone marrow (BM) cells were flushed from femurs and tibias and plated at 106 cells/ml in RPMI (Invitrogen) supplemented with 10% fetal calf serum (PAA, UK), L-glutamine (2mM), penicillin (100U/mL), streptomycin (100μg/mL)(Sigma) and 20ng/ml recombinant murine GM-CSF (Peprotech) for 7 days. Non-adherent cells were re-plated and medium supplemented on days 2 and 4. On day 7, cells were re-plated at 106 cells/ml and matured with 100ng/ml ultra-pure LPS (Source Bioscience) and 50ng/ml recombinant murine TNF-α (Peprotech) for 24 hours.
Antibodies and flow cytometry
Antibodies used are listed in Table 1. Dead cell exclusion was performed using DRAQ7 (Biostatus). Flow cytometric data were acquired using the MACSQuant Flow cytometer and analysed using MACSQuantify™ version 2.4 software (Miltenyi Biotec).
Table 1. Antibodies and suppliers.
| Antibody | Clone number | Supplier |
|---|---|---|
| anti-human CD27-PECy5.5 | Clone 0323 | eBioscience |
| anti-human CD83-APC | Clone HB15e | eBioscience |
| anti-mouse CD45.2-APC | Clone 104 | eBioscience |
| anti-mouse CD11c-Pacific Blue | Clone N418 | eBioscience |
| anti-mouse CXCR4-PE | Clone 2B11 | eBioscience |
| murine IgG2b-PE isotype control | Clone eBMG2b | eBioscience |
| murine IgG2aκ-PE isotype control | Clone eBM2a | eBioscience |
| anti-human CD4-VioBlue | Clone VIT4 | Miltenyi Biotec |
| anti-human CD8-VioBlue | Clone BW135/80 | Miltenyi Biotec |
| anti-human CD45RA-APC | Clone T6D11 | Miltenyi Biotec |
| anti-human CD62L-FITC | Clone 145/15 | Miltenyi Biotec |
| anti-mouse CD45.2-VioBlue | Clone 104-2 | Miltenyi Biotec |
| anti-biotin-PE | Clone Bio3-18E7 | Miltenyi Biotec |
| anti-human CCR7-PE, | Clone 150503 | R&D Systems |
| anti-human CCR7-APC | Clone 150503 | R&D Systems |
| anti-human CCR4-PE | Clone 205410 | R&D Systems |
| anti-mouse CD45.1-Brilliant Violet | Clone A20 | BioLegend |
| anti-mouse CD45.1-APC | Clone A20 | BioLegend |
| anti-mouse CD45.2-APC | Clone 104 | BioLegend |
| anti-mouse F4/80-alexaFluor488 | Clone BM8 | BioLegend |
| anti-mouse MHC Class II-Brilliant Violet 421 | Clone M5/114.15.2 | BioLegend |
| anti-mouse CD40-FITC | Clone 3/23 | BioLegend |
| anti-mouse CD86-PECy7 | Clone GL1 | BioLegend |
| anti-mouse CCR4-PE | Clone 2G12 | BioLegend |
| Armenian hamster IgG-PE | Clone HTK888 | BioLegend |
| isotype control |
Labelled chemokine generation
As chemokines are relatively small proteins (65-80 amino acids) they can be generated by total chemical synthesis and site-specifically labelled during this process. For all the biotinylated chemokine proteins used in this study, the biotin moieties were added to a Lysine residue introduced at the extreme Carboxy-terminus of the proteins. All chemokine synthesis was performed by Almac Sciences, Scotland (Edinburgh, Scotland).
Chemokine uptake and competition assay
Cells were re-suspended in buffer and 100μL/well aliquoted into round-bottom 96-well plates. CCL17-SAPE (CCL17-Streptavidin-PE) or alexa-fluor647-labelled CCL22 were added to appropriate wells at a final concentration of 600ng/mL. For the competition assay, 20-fold excess unlabelled-CCL22 was added to wells containing CCL17-SAPE. For surface staining/chemokine uptake assays, cells were incubated at either 4°C or 37°C for 30 or 60 minutes. For competition assay, cells were incubated at 37°C for 60 minutes. To prevent adherence of cells, samples were agitated every 20 minutes by gentle pipetting. Cells were washed and labelled with 1μL DRAQ7 dead cell discriminator prior to flow cytometric analysis.
Cell staining using biotinylated CCL19
Both murine and human cell staining was performed in PEB buffer (0.5% media-grade bovine serum albumin (Millipore) and 2mM EDTA (Ambion) in PBS (Invitrogen)). Single biotinylated CCL19 (bCCL19), CCL17 (bCCL17), CXCL12 (bCXCL12), double-biotinylated CCL19, native CCL22 and alexa-fluor647-labelled CCL22 were generated by chemical synthesis as described above (Almac Sciences), and used to label target cells. Initial experiments were conducted using bCCL19. This was either added directly to cells which were then washed and bCCL19 detected with PE-conjugated streptavidin (SAPE, Invitrogen, final concentration 10μg/mL) or bCCL19 was detected using PE-labelled anti-biotin antibody, or was precomplexed to SAPE (10μg/mL) by incubating the indicated concentrations at 4°C for 30 minutes. For all other cell staining using biotinylated chemokines, including uptake, the chemokine was precomplexed with SAPE at 4°C for 30 minutes. Cells were re-suspended at 106 cells/test in a volume of 100μL. Single-cell suspensions of thymocytes for CCL17-SAPE experiments were obtained from D6-deficient mice by passing the thymus through a 100μm filter. Murine bone marrow cells for CXCL12-SAPE staining were obtained by flushing bone marrow from the femur and tibia of mice and performing red cell lysis (Miltenyi Biotec). Human and mouse DCs were pre-incubated with human or mouse Fc receptor-block (Miltenyi Biotec), respectively, for 5 minutes prior to antibody staining. Antibody staining for cell surface lineage markers was performed at 4°C for 10 minutes. In all experiments cells were then washed with PEB and labelled with 1 μL DRAQ7 dead cell discriminator prior to flow cytometric analysis.
Cell sorting
Cells were washed and resuspended in PEB at a maximal concentration of 107 cells/100μl. Cells were incubated at 4°C with indicated concentrations of bCCL19 conjugated to SAPE (final concentration 50μg/ml SAPE), for 30 minutes (unless otherwise stated). Murine BM-DCs were Fc receptor-blocked prior to addition of bCCL19-SAPE as before. Cells were washed with PEB, stained with anti-PE microbeads (Miltenyi Biotec as per manufacturer’s instructions) and isolated using MACS-LS columns at room temperature in ice-cold buffer.
CFSE dilution assay
T cell proliferation was measured as previously described(34) with minor modifications. Cells were resuspended in PBS and incubated with 1μM CFSE (final concentration) for 10 minutes at 37°C, before quenching with complete medium on ice for 10 minutes. Cells were washed twice in complete medium before replating with either 20U/mL IL-2 alone or 20U/mL IL-2 plus T cell activation/expansion beads (Miltenyi Biotec, as per manufacturer’s instructions), for up to 5 days, and fed using complete medium plus IL-2 (20U/mL) on day 3. Cells were plated in 6 replicate wells, and proliferation measured daily from a fresh well using the MACSQuant Analyser and MACSQuantify™ version 2.4 software.
In vitro transwell migration assay
For Transwell assays (3μm-pore Transwell plates; Fisher Scientific), cells were washed and incubated for 1 hour at 37°C in chemotaxis buffer (RPMI/0.5% BSA). Chemotaxis buffer (600μl) was added to the bottom of each well and incubated at 37°C for 10 minutes before being aspirated and replaced with dilutions of unlabelled CCL19 (0, 10, 100 and 500ng/mL; Miltenyi Biotec) in chemotaxis buffer, and the Transwell membranes inserted into each well (10 minutes, 37°C). T cell suspensions (100μl containing 5×105 cells) were added to the inserts in each well and incubated for 3 hours at 37°C. The inserts were then removed and the lower chamber aspirated and the cells labelled with DRAQ7 and counted using the MACSQuant Analyser and MACSQuantify™ version 2.4 software. Data were expressed as a Migration Index, calculated as [individual count for each replicate]/[average background migration].
In vivo cell migration
Both Ly5.1 and Ly5.2 matured BM-DCs were stained with bCCL19-SAPE complex as for sorting, but only Ly5.1 cells were enriched into a >95% CCR7+ population. Equal numbers (7×105) of sorted Ly5.1 and unsorted Ly5.2 cells were mixed 1:1 in PBS/0.5% BSA, and injected into the right footpad of Ly5.1/Ly5.2 heterozygous mice. At 48 hours post-injection, draining popliteal LNs were removed and dissociated in HBSS with Blendzyme Liberase DL (0.84 Wünsch Units/ml as per manufacturer’s instructions, Roche Ltd). LN cells were passed through a 40μm filter, stained with Ly5.1 and Ly5.2 antibodies and analysed by flow cytometry. Single Ly5.1+ and Ly5.2+ expressing DCs were detected against the background level of endogenous Ly5.1+/Ly5.2+ DC. A reciprocal experiment was also performed, with Ly5.1 cells left unsorted and Ly5.2 enriched for CCR7+ cells.
Statistical analysis
Statistical analysis used GraphPad Prism version 5.03 software, with specific tests detailed in the results section. P values < 0.05 were considered statistically significant.
Results
Biotinylated chemokines as chemokine receptor detection reagents
In initial experiments we examined the ability of biotinylated CCL17 (binds to CCR4 and D6(35)) and biotinylated CXCL12 (binds to CXCR4 and CXCR7(36)), in comparison with commercially available antibodies, to detect their cognate receptors on primary cells. As shown (Figure 1A), in comparison to isotype controls or anti-human CCR4 antibodies, anti-mouse CCR4 antibodies detected CCR4 expression in murine thymocytes (11.99%). Importantly, streptavidin-PE (SAPE)-conjugated CCL17 detected CCR4 at levels equivalent to those detected by the antibody. CCR4 detection by biotinylated CCL17 was optimal at 37°C (16.23%) but still detectable at 4°C (8.24%) (Figures 1B and 1C). The thymocytes used were from D6-deficient mice. Therefore the only receptor which CCL17 is capable of interacting with is CCR4. These data demonstrate that SAPE-conjugated biotinylated CCL17 can detect CCR4 on primary murine thymocytes.
Figure 1. Labelled Chemokines as receptor detection agents.
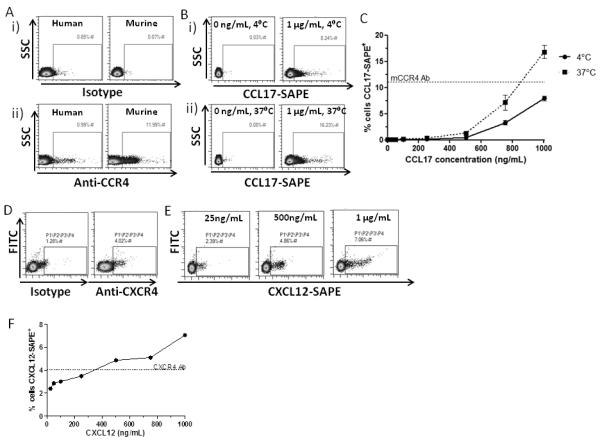
(A) CCR4 expression on thymocytes. Thymocytes from D6KO mice were labelled with either (i) relevant isotype controls or (ii) anti-human or anti-mouse CCR4 antibodies. (B) CCR4 detection was enhanced at 37°C compared to 4°C. (C) Incubating the cells with an increasing concentration of CCL17-SAPE complex showed that, at 37°C, the chemokine was able to label CCR4+ cells as well as, if not better than, species-specific antibody. n = 3, mean ± SEM. (D) CXCR4 expression on bone marrow cells. Cells flushed from murine bone marrow were stained with either anti-CXCR4 antibody or isotype to compare levels of CXCR4 detection. (E) CXCR4 detection at 37°C increased with increasing concentration of CXCL12-SAPE. (F) At concentrations above 500ng/mL, CXCL12-SAPE gives superior staining of CXCR4+ cells than the commercially available antibody.
CXCR4 is involved in stem cell migration and function. Anti-murine CXCR4 antibodies detected CXCR4 expression on approximately 4% of total bone marrow cells (Figure 1D). As shown (Figure 1E), SAPE-conjugated biotinylated CXCL12 was at least as good as, and potentially superior to, the antibody used at detecting CXCR4 in total murine bone marrow cells (7.06% versus 4.02%) (Figures 1E and 1F).
Finally we examined ability of labelled chemokines to detect receptors on heterologous transfectants. D6 binds both CCL17 and CCL22(29, 32, 33). As shown (Supplementary Figure 1A) SAPE-conjugated biotinylated CCL17, as well as AlexaFluor-labelled CCL22, were both able to detect identical levels of D6 positivity (76%) in transfected HEK cells at a relatively low concentration of chemokine (<100ng/ml; Supplementary Figure 1B) and the two chemokines were able to cross compete on this receptor (Supplemental Figure 1C).
Thus chemokines have utility as versatile, receptor-specific, detection agents for use in flow cytometric detection of chemokine receptors on primary cells and heterologous transfectants. In this regard, labelled chemokines are at least as effective as the commercially available antibodies used.
Biotinylated CCL19 detects CCR7 on T cells
To determined the value of biotinylated chemokines for detecting CCR7-expressing cells, which are of significant cellular therapeutic interest, we generated biotinylated human CCL19 (bCCL19). To test the ability of bCCL19 to functionally interact with CCR7, we measured its binding to, and internalisation by, CCR7-expressing HEK cells. As shown (Figure 2Ai), bCCL19 pre-complexed with SAPE (bCCL19-SAPE) displayed minimal non-specific binding, and uptake, by untransfected HEK but did bind to, and was internalised by, CCR7-transfected HEK cells. This was competed out by unlabelled human CCL19 (Figure 2Aii). bCCL19 was internalised into cytoplasmic vesicles in CCR7-expressing HEK cells (but not untransfected HEK cells) following receptor interaction (Figure 2Aiii). These results confirm that bCCL19 is active in terms of CCR7 binding and internalisation.
Figure 2. bCCL19 can detect CCR7 on T cells using flow cytometry.
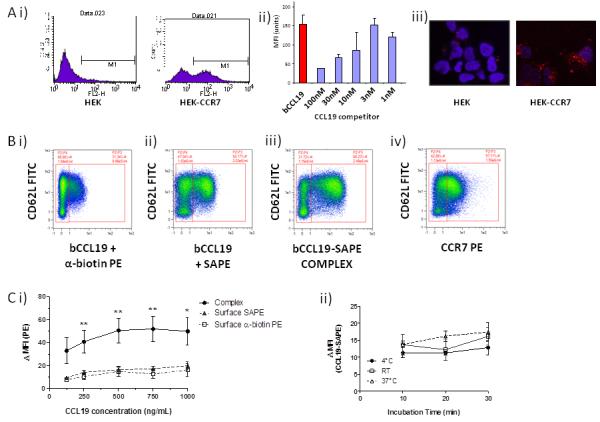
(A) i) Representative flow cytometric data showing the binding of the SAPE tagged bCCL19 to HEK cells transfected with CCR7 (HEK-CCR7) but not to untransfected HEK cells (HEK). ii) The binding of bCCL19 to HEK-CCR7 cells is competable with unlabelled CCL19 (concentrations in nM). iii) confocal imaging demonstrating the uptake of PE-tagged bCCL19 into intracellular vesicles in HEK-CCR7, but not HEK, cells. All of the above experiments in were performed at 37°C. (B) Representative dot-plots of T cells stained with (i) bCCL19 followed by antibiotin-PE antibody; (ii) bCCL19 followed by SAPE; (iii) pre-complexed bCCL19-SAPE; (iv) CCR7-PE antibody. Plots taken from one donor with 250ng/mL bCCL19 used for staining, with gating to indicate positive and negative populations. (C) Optimisation of staining protocol using human T cells. (i) T cells were stained using either pre-complexed bCCL19-SAPE (circles), or with bCCL19 alone, followed by surface staining with either SAPE (triangles) or anti-biotin PE (squares) (n = 4 different donors. Data represents mean ± SEM, one-way ANOVA [125] p = 0.1; [250] **p = 0.0056; [500] **p = 0.0076; [750] **p = 0.0059; [1000] *p = 0.025). Data are expressed as ΔMFI, which was calculated as [MFI of CCR7/CD62L-positive population]/[MFI of CCR7/CD62L-negative population]. (ii) T cells were incubated with 250 ng/mL CCL19-SAPE complex at 3 different temperatures for increasing lengths of time to determine effects on staining as described above (n = 2 different donors).
To investigate the usefulness of bCCL19 as a flow cytometric marker for CCR7 on leukocytes, we stained human T cells of mixed CCR7 positivity, with bCCL19 in combination with various secondary detection agents. Controls showing no non-specific binding of secondary detection agents to leukocytes are presented in Supplemental Figure 2. As shown in Figure 2Bi, whilst bCCL19 and PE-labelled anti-biotin antibody resulted in modest CCR7 staining on T cells (31.34%), bCCL19 staining followed by SAPE-based detection enhanced staining (52.17%) (Figure 2Bii) suggesting that streptavidin-based tetramerisation improves CCR7 detection by bCCL19. This was further enhanced (to 66.23%) by pre-complexing bCCL19 and SAPE prior to addition to T cells (Figure 2Biii). Flow cytometric staining using the bCCL19-SAPE complex was consistently 10-fold stronger than that seen with a commercially available PE-labelled anti-CCR7 antibody (Figure 2Biv). Notably the percentage of T cells detected as being CCR7+ was similar suggesting that bCCL19, and anti-CCR7, were identifying the same cell population.
To optimise the staining protocol (Figure 2C) we performed dose-response studies, which confirm that pre-complexing gave the strongest staining which was maximal at 250-500ng/ml bCCL19 (Figure 2Ci). Importantly, neither incubation time (10-30 minutes) nor temperature (4°C or 37°C) had a significant impact on CCR7 staining with bCCL19-SAPE (Figure 2Cii). Therefore these flow cytometric data indicate that SAPE-conjugated bCCL19 represents an efficient reagent for CCR7 detection on human T cells.
bCCL19 can be used to sort CCR7+ T cells
We next investigated the possibility that bCCL19-SAPE tetramers, with anti-PE magnetic microbeads, could be used to sort CCR7+ T cells. To this end, we sorted T cells using 250ng/ml SAPE-conjugated bCCL19, according to the protocol outlined in Figure 3A. As shown (Figure 3Bi and ii), this protocol enriched for CCR7 positivity from 36% in the pre-sort population to 98.76% in the sorted population. In addition, yields of CCR7+ T cells were consistently around 50%. We also generated double biotin-labelled CCL19 to determine whether enhanced streptavidin binding might increase cell yield after sorting. As shown (Figure 3Biii), this made no significant difference to the yield of CCR7+ cells obtained. Data from numerous bCCL19-based sorts for CCR7+ demonstrate the consistency and completeness achieved with this sorting approach (Figure 3C).
Figure 3. bCCL19 can be used to sort for CCR7+ Human T cells.
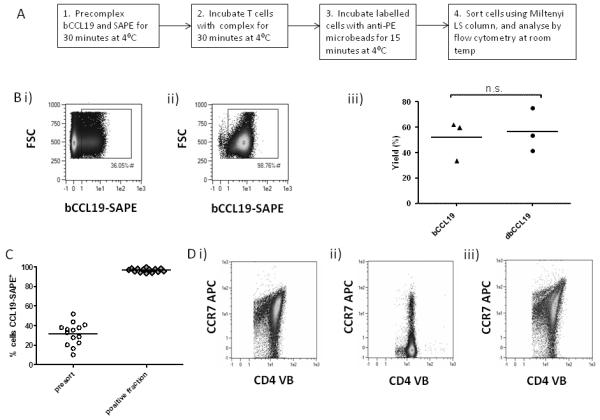
(A) Sorting method used for T cells. (B) T cells, following 11 days of in vitro culture, were stained with 250ng/mL bCCL19 conjugated to SAPE, and magnetically sorted using anti-PE microbeads. (i) The pre-sort sample. (ii) Sorted cells. (iii) Yield of cells using single (sCCL19) and double (dCCL19) biotinylated CCL19. p = 0.568, 2-tailed students T-test. (C) Percentage of cells labelled with bCCL19-SAPE before (presort) and after (positive fraction) sorting. (n = 14). (D) Receptor recycling in T cells after incubation with bCCL19. PBMC-derived T cells were incubated with 100ng/ml bCCL19 in duplicate. (i) T cells stained for CD4 and CCR7 prior to incubation with bCCL19. (ii) One aliquot of PBMC was counterstained with anti-CD4-VioBlue and anti-CCR7-APC mAb after incubation with bCCL19. (iii) The remaining bCCL19-bound cells were then warmed to 37°C for 60 minutes, washed in cold PEB and stained with anti-CD4 and anti-CCR7 as before.
Chemokines binding to their receptors induce receptor internalisation and it is possible that ligand-based sorting for CCR7+ cells may induce a CCR7− phenotype in the cells, impairing their subsequent in vivo function. However, whilst bCCL19 did indeed induce receptor internalisation (Figure 3Di and ii) as indicated by a significant drop in cell surface CCR7 (detected using anti-CCR7 antibody) on T cells, levels were fully restored through recycling following incubation of the sorted cells in culture medium for one hour at 37°C (Figure 3Diii). Thus bCCL19-SAPE is capable of highly efficient sorting of CCR7+ T cells from a mixed population.
bCCL19 sorted T cells are predominantly of memory and naive phenotypes
As shown (Figure 4A) bCCL19 enriched CD4+ T cells from the mixed population to 85.85% CCR7-positivity. In keeping with CCR7 expression, the enriched population had a largely naive (bCCL19+CD62L+CD27+CD45RAhi) or central memory (bCCL19+CD62L+CD27+CD45RAlo) phenotype with the relative representation of CCR7+ naive and central memory cells being similar to the pre-sort population (Figure 4Ci). For CD8+ T cells (Figure 4B), bCCL19 enriched for more minor populations (13.44%) again displaying naive and central memory phenotypes. In contrast to CD4+ cells, sorted CD8+ T cells displayed a significantly increased proportion of naive cells compared with the pre-sort samples (Figure 4Cii). The relative percentages of CD4+ and CD8+ T cells were maintained post-sort suggesting that the overall spread of CCR7 expression in the pre-sort populations is similar (Figure 4Ciii)
Figure 4. Human T cell sort phenotypes and in vitro migration.
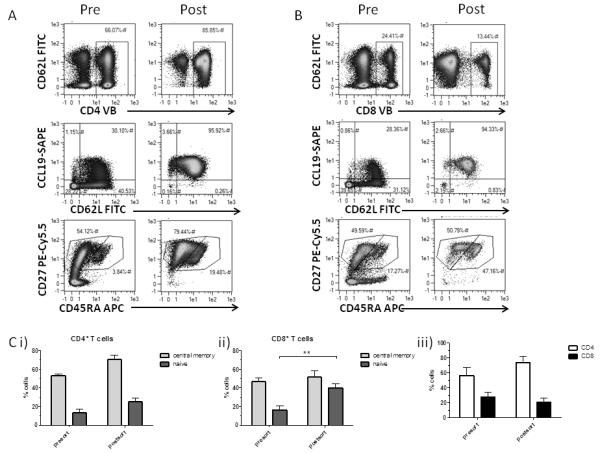
(A) Phenotyping of bCCL19-sorted CD4+ T cells, using CD27, CD45RA, CD62L antibodies and CCL19-SAPE. (B) Phenotyping of bCCL19-sorted CD8+ T cells, using CD27, CD45RA, CD62L antibodies and bCCL19-SAPE. For both (A) and (B), presort samples are on the left and post-sort samples on the right. (C) (i & ii) Proportion of central memory to naive phenotypes in CD4 (i) and CD8 (ii) pre- and post-sort cells; (iii) Proportion of CD4 to CD8 cells in pre- and post-sort samples. (results are mean ± SEM of 6 or 7 experiments). All results were analysed using two-tailed paired student’s T-test (**p=0.038)
bCCL19-SAPE-sorted T cells are functional
To confirm functionality of sorted T cells, we compared their proliferative capacity, to that of unsorted T cells, in response to exposure to activation/expansion beads (see Materials and Methods section for details) which are designed to mimic antigen presentation. Proliferation was measured using a CFSE dilution assay. Simple incubation of T cells with bCCL19-SAPE did not effect the proliferative status of either i) CD4+ or ii) CD8+ cells (compared 1 and 2 in Figure 5A). In contrast, bCCL19-purified CCR7+ cells displayed markedly higher proliferation with both CD4+ and CD8+ cells undergoing up to five cell divisions over 5 days (compare 1 and 2 with 3). Thus bCCL19-sorted T cells are highly proliferative compared to the unsorted population and this is likely to be a reflection of their enhanced ability to respond to antigen presentation, or equivalent stimuli, such as is represented by the use of activation/expansion beads. To demonstrate that bCCL19-sorted T cells display enhanced functional CCR7 responses, we examined their responses in migration assays. Following bCCL19-based sorting the cells (unsorted, bCCL19− and bCCL19+) were allowed to rest overnight to restore surface CCR7 levels and then examined for migratory responses to increasing CCL19 concentrations using Transwell migration assays. As shown (Figure 5B), bCCL19-sorted cells displayed significantly enhanced chemotaxis to CCL19 compared to either unsorted T cells or the bCCL19− population. Though not reaching significance, the bCCL19− fraction consistently displayed reduced chemotaxis toward CCL19 compared to bCCL19 stained but unsorted cells. These data demonstrate that bCCL19-sorted T cells are fully functional and display enhanced CCR7-dependent chemotaxis.
Figure 5. bCCL19-SAPE sorted T cells are functional.
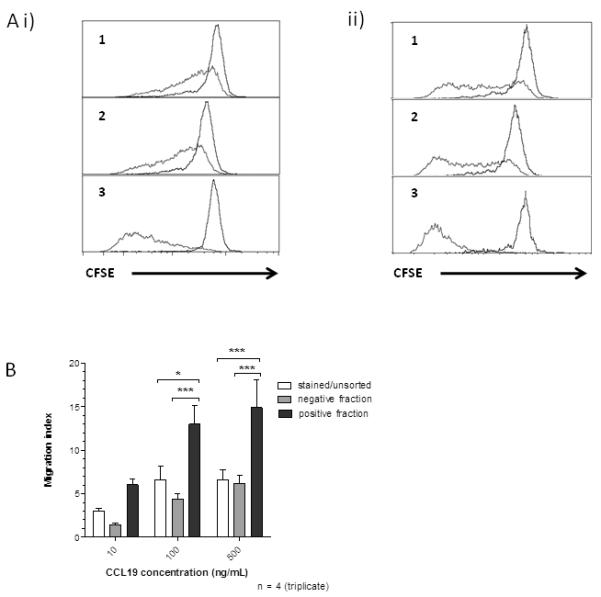
(A) Measure of proliferation of T cells using CFSE dilution assays. T cells were sorted and immediately labelled with CFSE. They were then placed in culture with IL-2 alone (20 U/mL; controls - black lines), or with activation beads plus IL-2 (red lines) and left for five days following which CFSE intensity was evaluated using flow cytometry. (i) CD4+ T cells; (ii) CD8+ T cells. (1) unsorted cells not exposed to bCCL19-SAPE; (2) unsorted cells incubated with bCCL19-SAPE for 30 minutes and (3) positive fraction from sorts. Plots are representative data from one of 4 separate experiments. (B) In vitro migration of sorted T cells. Migration experiments were conducted using unsorted cells that had been exposed to the bCCL19-SAPE complex (stained/unsorted - white bars); the negative fraction from the bCCL19-SAPE sorts (grey bars); and the highly purified bCCL19-SAPE+ fraction from the sorts (black bars). All results are mean ± SEM, n=4, 2-way ANOVA with Bonferroni post-hoc test. * p < 0.05, *** p < 0.001)
Dendritic cells can be stained and sorted using bCCL19
To examine the ability of bCCL19 to detect CCR7 on cells other than T cells, we generated immature and mature (CCR7-expressing) human DCs. As shown (Figure 6A), whilst immature DCs displayed low level bCCL19 staining (0.15%) indicative of low CCR7 positivity (Figure 6Ai), matured DCs displayed markedly enhanced (38.06%), although variable, CCR7 positivity (Figure 6Aii). bCCL19 staining correlated with CD83 expression, indicating that bCCL19 is specifically detecting CCR7 on mature DCs (Figure 6Aii). Data from multiple experiments revealed the consistent ability of bCCL19 to stain for CCR7 on immature and mature DCs (Figure 6Aiii). These data demonstrate that bCCL19 can detect CCR7 on monocyte-derived DCs.
Figure 6. Dendritic cell staining and sorting with bCCL19-SAPE.
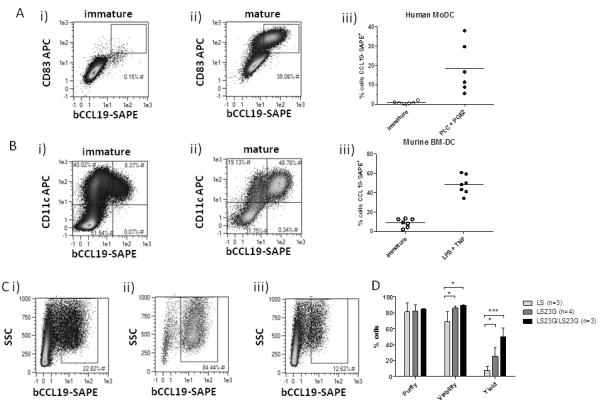
(A) Immature DCs were grown from CD14+ fractions of buffy coats and matured with PolyI:C/PGE2 for 48 hours. Cells were stained as in Figure 1. (i) Immature DCs. (ii) Mature DCs. (iii) Maturation of human monocyte-derived DC (MoDC) with PolyI:C and PGE2 increases the percentage of cells staining positive with bCCL19-SAPE (n=6). bCCL19-SAPE+ gates were set using SAPE controls where <1% of cells showed positivity for SAPE. (B) bCCL19-SAPE staining of murine BMDCs. Immature DCs were grown from murine BM in the presence of GM-CSF and matured with LPS and TNF-α for 24 hours. Cells were stained using 250ng/mL bCCL19 conjugated to SAPE. (i) Immature DCs (ii) DCs matured with LPS/TNF-α. (iii) Percentage of cells staining positive with bCCL19 (n = 7). Gates were set as in (A). (C) Initial sorting of DCs. The percentage of cells staining positive with bCCL19-SAPE is increased from the presort sample (i) to the post-sort sort fraction (ii) leaving behind a negative sort fraction depleted for bCCL19+ cells (iii). (D) Quantitative data from comparative DC sorts. Sorting was initially performed with a single LS column. The sort was then performed using LS columns fitted with 23G needles (LS23G, dark grey bars), to slow the flow of cells through the column. After the first sort, the negative fraction of the sample was passed through a second LS23G (LS23G/LS23G, black bars), to increase the yield of positive cells from the sort. Statistics were calculated using 2-way repeated measures ANOVA with Bonferroni post-test. n=3-4, *p<0.05, ***p<0.001.
Next we examined whether bCCL19 was capable of sorting CCR7+ DCs. As we wished to examine the impact of bCCL19-based CCR7 sorting on in vivo migration competence of DCs, we present data on sorting of murine BM-DCs. As shown (Figure 6B), similar to human monocyte-derived DCs, CCL19 staining was able to distinguish between immature (Figure 6Bi) and mature (Figure 6Bii) BM-DCs on the basis of CCR7 positivity. In repeat experiments (Figure 6Biii), whilst immature BM-DCs were predominantly CCR7−, mature DCs displayed approximately 50% CCR7 positivity. Next we examined the ability of bCCL19 to enrich for CCR7+ cells from mixed populations of CCR7+ and CCR7− mature DCs. As shown (Figure 6C), whilst the overall percentage of CCR7 positivity in the pre-sort sample was around 23% (Figure 6Ci), bCCL19-based sorting enriched for CCR7+ cells (to 84.44%) following magnetic bead selection (Figure 6Cii) leaving behind a population depleted in CCR7+ cells (12.62%) (Figure 6Ciii). In multiple experiments, we observed a high-level of purity and viability of the sorted DC population. However, initial yields were <20% but could be improved by attaching a 23G needle to the outlet of the sorting column, thus reducing flow rate, and enhancing yields to >20% (Figure 6D). Yields were further enhanced to >50% (Figure 6D) by passing the flow-through from the initial cell enrichment through a second sorting column (again with a 23G needle attached). Thus, in addition to T cells, bCCL19 can be used to detect CCR7+ DCs and enrich for these cells using magnetic bead technology to generate clinically usable yields.
bCCL19-sorted dendritic cells display enhanced LN migration competence
As CCR7 mediates DC migration to LNs from peripheral tissues, we next examined bCCL19-sorted DCs for their LN-homing potential. The experiment outlined in Figure 7A used DCs generated from Ly5.1 and Ly5.2 mouse BM which were enriched for CCR7 expression using bCCL19 (Ly5.1), or left unsorted (Ly5.2). Equal numbers of the DCs were mixed and adoptively transferred into the right footpad of recipient heterozygous Ly5.1/Ly5.2 mice. Experiments using reciprocal Ly5.2 sorted/Ly5.1 unsorted DCs were also carried out. As shown (Figure 7Bi), only Ly5.1/Ly5.2 heterozygous cells were detected in contralateral popliteal LNs however, substantial numbers of Ly5.1+ and Ly5.2+ cells were detectable in the popliteal LN draining the injection site, against a background of Ly5.1/Ly5.2 heterozygous cells (Figure 7Bii). As shown in two separate experiments involving reciprocal use of Ly5.1 and Ly5.2 DCs (Figure 7Ci and ii), it was observed that significantly more bCCL19-sorted DCs migrated to the draining LN than unsorted, but bCCL19-labelled, cells. The marked differences in the numbers of unsorted and sorted DCs reaching the LNs in Ci and Cii are likely to be peculiarities of the individual experiments and not a consequence of Ly5.1 or Ly5.2 contribution to DC migratory kinetics. These data confirm the functional competence of bCCL19-sorted DCs and highlight bCCL19-based sorting as a novel strategy for the isolation of DCs with enhanced LN-homing competence.
Figure 7. In vivo homing of CCL19 sorted cells.
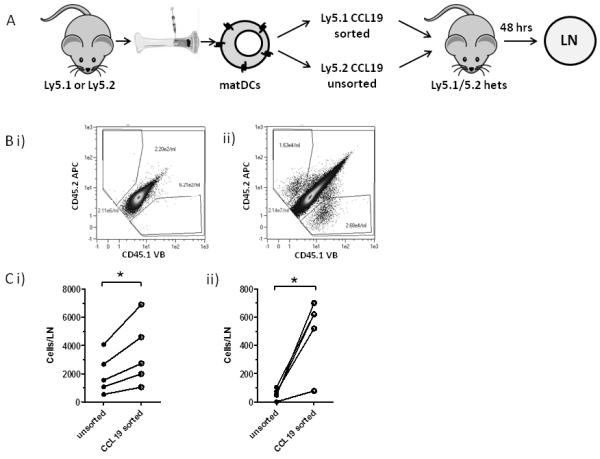
(A) DCs were grown from BM of Ly5.1 and Ly5.2 mice. The Ly5.2 DCs were exposed to the bCCL19-SAPE complex but left unsorted, while the Ly5.1 DCs were sorted using the bCCL19-SAPE complex and anti-PE microbeads (Miltenyi). The sorted and unsorted cells were mixed at a ratio of 1:1 (7 × 105 cells of each cell type) in PBS + 0.5% BSA, and injected into the rear right footpads of Ly5.1/Ly5.2 heterozygous mice. After 48 h the draining popliteal LN, and the contralateral popliteal LN, were removed, the cells isolated, and stained with anti-CD45.1 and anti-CD45.2 antibodies. Single-stained cells were counted using the MACSQuant flow cytometer and results expressed as cells per LN. The reciprocal experiment where Ly5.1 cells were unsorted and Ly5.2 cells were sorted was also performed. (B) Representative flow cytometry plots of draining popliteal LN from the experiment. (i) A contralateral popliteal LN. (ii) A draining popliteal LN from the sorted Ly5.1 experiment. (C) Number of transferred cells per LN: bCCL19-sorted cells (open circles) and unsorted cells (closed circles) were mixed at a ratio of 1:1 and transferred into recipient footpads.(i) Ly5.1 sorted vs Ly5.2 unsorted, (ii) Ly5.2 sorted and Ly5.1 unsorted. All results are mean ± SEM, and statistical significance was determined using Two-tailed paired students T-test. *p<0.05.
Discussion
The ability to preselect therapeutic cell populations, on the basis of chemokine receptor expression, would improve their ability to home to specific therapeutic niches and lead to marked enhancements in therapeutic efficacy. This is clear in the context of DC migration to LNs for immune-initiating therapeutic function(13, 16), but is also relevant to selection of cells for homing to BM(37), skin(38), gut(39), or indeed any site for which chemokine homing receptors are known. Unfortunately the relative lack of high-quality antibodies to chemokine receptors means that simple immune-based selection for chemokine receptor bearing cells is not always possible. In addition, antibodies are expensive and invariably species-specific. Here we describe a novel use of chemokines as versatile reagents capable of detecting, and sorting for, specific chemokine receptor-bearing cells. The advantages of chemokines over antibodies in this context are: (i) they are relatively inexpensive to produce in high-quantity and high purity by chemical synthesis; (ii) they can cross species and are amenable to detection of orthologous receptors in multiple species; (iii) they are adaptable for GMP-grade therapeutic use. Specifically we show that bCCL19 can detect CCR7 on T cells and DCs sort these cells on this basis. The enriched CCR7+ T cells and DCs are fully functional and, as would be anticipated, bCCL19-enriched DCs display enhanced LN-migration competence. This has implications for DC therapy and suggests that pre-selection of cells using bCCL19 would provide a ‘fitter’ cell product with enhanced LN-migration competence. In addition, it would deplete immature DCs within the therapeutic population that may otherwise induce tolerance to therapeutic antigens(20, 21). With regards the therapeutic use of T cells, both ‘natural’(40) and artificially-transduced Ag-specific T cell therapies(41) utilise CCR7+ naïve and central memory T cells. These cells possess the ability to re-populate both central and effector immunity niches leading to elaboration of full immune responses upon adoptive transfer. Conversely, CCR7− effector clones could not fully repopulate immune responses against CMV(40). We have shown that bCCL19 makes an excellent sorting agent for isolating fully functional naive and central memory T cells. Thus isolating T cells in this manner has direct applicability to developing clinical protocols where CCR7+ T cells will be applied.
The ability to produce biotinylated chemokines through chemical synthesis means that chemokines relevant in other cell homing contexts can, in principle, be utilised in cell sorting applications. One example of this is the potential use of biotinylated-CXCL12 for detecting and isolating BM-homing haemopoietic stem cell populations(37, 42-45). We have demonstrated the utility of biotinylated CXCL12 for CXCR4 detection on BM cells. To attempt to provide an initial assessment of clinical utility, we tried using biotinylated CXCL12 to measure stem cell mobilisation to peripheral blood following G-CSF administration in mice. However, in this context the biotinylated CXCL12 could not detect CXCR4 on the mobilised cells perhaps as a result of the reported downregulation of CXCR4 upon G-CSF mobilisation of stem cells (46). Importantly, this does not preclude clinical utility in other contexts. All secondary reagents involved in the use of biotinylated chemokines for flow cytometry and cell sorting are commercially available and adaptable for immediate use with any biotinylated chemokine. In addition, all secondary reagents used for cell sorting in this study have been developed for GMP-grade use. Accordingly, the use of biotinylated chemokines as cell sorting agents in a range of clinical contexts is feasible using currently available reagents and technologies.
In summary we provide evidence demonstrating a novel use for chemokines as reagents for identifying, and isolating, cells bearing their cognate receptors. In this context, biotinylated chemokines have important therapeutic potential and are also likely to prove useful in a variety of experimental and veterinary contexts.
Supplementary Material
Acknowledgments
Funding sources: This study was supported by grants from the Medical Research Council and the Scottish Chief Scientist’s Office.
References
- 1.Rot A, von Andrian UH. Chemokines in innate and adaptive host defense: basic chemokinese grammar for immune cells. Annu Rev Immunol. 2004;22:891–928. doi: 10.1146/annurev.immunol.22.012703.104543. [DOI] [PubMed] [Google Scholar]
- 2.Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity. 2000;12:121–127. doi: 10.1016/s1074-7613(00)80165-x. [DOI] [PubMed] [Google Scholar]
- 3.Zlotnik A, Yoshie O, Nomiyama H. The chemokine and chemokine receptor superfamilies and their molecular evolution. Genome biology. 2006;7:243. doi: 10.1186/gb-2006-7-12-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murphy PM, Baggiolini M, Charo IF, Hebert CA, Horuk R, Matsushima K, Miller LH, Oppenheim JJ, Power CA. International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol Rev. 2000;52:145–176. [PubMed] [Google Scholar]
- 5.Mantovani A. The chemokine system: redundancy for robust outputs. Immunol Today. 1999;20:254–257. doi: 10.1016/s0167-5699(99)01469-3. [DOI] [PubMed] [Google Scholar]
- 6.Parsonage G, Filer AD, Haworth O, Nash GB, Rainger GE, Salmon M, Buckley CD. A stromal address code defined by fibroblasts. Trends Immunol. 2005;26:150–156. doi: 10.1016/j.it.2004.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chavakis E, Koyanagi M, Dimmeler S. Enhancing the Outcome of Cell Therapy for Cardiac Repair Progress From Bench to Bedside and Back. Circulation. 2010;121:325–335. doi: 10.1161/CIRCULATIONAHA.109.901405. [DOI] [PubMed] [Google Scholar]
- 8.Kang SK, Shin IS, Ko MS, Jo JY, Ra JC. Journey of mesenchymal stem cells for homing: strategies to enhance efficacy and safety of stem cell therapy. Stem cells international. 2012;2012:342968–342968. doi: 10.1155/2012/342968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ziadloo A, Burks SR, Gold EM, Lewis BK, Chaudhry A, Merino MJ, Frenkel V, Frank JA. Enhanced Homing Permeability and Retention of Bone Marrow Stromal Cells by Noninvasive Pulsed Focused Ultrasound. Stem Cells. 2012;30:1216–1227. doi: 10.1002/stem.1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garzon-Muvdi T, Quinones-Hinojosa A. Neural Stem Cell Niches and Homing: Recruitment and Integration into Functional Tissues. Ilar Journal. 2010;51:3–23. doi: 10.1093/ilar.51.1.3. [DOI] [PubMed] [Google Scholar]
- 11.Bromley SK, Thomas SY, Luster AD. Chemokine receptor CCR7 guides T cell exit from peripheral tissues and entry into afferent lymphatics. Nat Immunol. 2005;6:895–901. doi: 10.1038/ni1240. [DOI] [PubMed] [Google Scholar]
- 12.Debes GF, Arnold CN, Young AJ, Krautwald S, Lipp M, Hay JB, Butcher EC. Chemokine receptor CCR7 required for T lymphocyte exit from peripheral tissues. Nat Immunol. 2005;6:889–894. doi: 10.1038/ni1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8:362–371. doi: 10.1038/nri2297. [DOI] [PubMed] [Google Scholar]
- 14.Forster R, Schubel A, Breitfeld D, Kremmer E, Renner-Muller I, Wolf E, Lipp M. CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell. 1999;99:23–33. doi: 10.1016/s0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 15.Braun A, Worbs T, Moschovakis GL, Halle S, Hoffmann K, Bolter J, Munk A, Forster R. Afferent lymph-derived T cells and DCs use different chemokine receptor CCR7-dependent routes for entry into the lymph node and intranodal migration. Nature Immunology. 2011;12:879–U878. doi: 10.1038/ni.2085. [DOI] [PubMed] [Google Scholar]
- 16.Forster R, Braun A, Worbs T. Lymph node homing of T cells and dendritic cells via afferent lymphatics. Trends in Immunology. 2012;33:271–280. doi: 10.1016/j.it.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 17.Sallusto F, Schaerli P, Loetscher P, Schaniel C, Lenig D, Mackay CR, Qin S, Lanzavecchia A. Rapid and coordinated switch in chemokine receptor expression during dendritic cell maturation. Eur J Immunol. 1998;28:2760–2769. doi: 10.1002/(SICI)1521-4141(199809)28:09<2760::AID-IMMU2760>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 18.Sozzani S, Allavena P, D’Amico G, Luini W, Bianchi G, Kataura M, Imai T, Yoshie O, Bonecchi R, Mantovani A. Differential regulation of chemokine receptors during dendritic cell maturation: a model for their trafficking properties. J Immunol. 1998;161:1083–1086. [PubMed] [Google Scholar]
- 19.Ritter U, Wiede F, Mielenz D, Kiafard Z, Zwirner J, Korner H. Analysis of the CCR7 expression on murine bone marrow-derived and spleen dendritic cells. J Leukoc Biol. 2004;76:472–476. doi: 10.1189/jlb.0104037. [DOI] [PubMed] [Google Scholar]
- 20.Hilkens CMU, Isaacs JD, Thomson AW. Development of Dendritic Cell-Based Immunotherapy for Autoimmunity. International Reviews of Immunology. 2010;29:156–183. doi: 10.3109/08830180903281193. [DOI] [PubMed] [Google Scholar]
- 21.Jonuleit H, Giesecke-Tuettenberg A, Tuting T, Thurner-Schuler B, Stuge TB, Paragnik L, Kandemir A, Lee PP, Schuler G, Knop J, Enk AH. A comparison of two types of dendritic cell as adjuvants for the induction of melanoma-specific T-cell responses in humans following intranodal injection. International Journal of Cancer. 2001;93:243–251. doi: 10.1002/ijc.1323. [DOI] [PubMed] [Google Scholar]
- 22.Lesterhuis WJ, de Vries IJM, Schreibelt G, Lambeck AJA, Aarntzen EHJG, Jacobs JFM, Scharenborg NM, van de Rakt MWMM, de Boer AJ, Croockewit S, van Rossum MM, Mus R, Oyen WJG, Boerman OC, Lucas S, Adema GJ, Punt CJA, Figdor CG. Route of Administration Modulates the Induction of Dendritic Cell Vaccine-Induced Antigen-Specific T Cells in Advanced Melanoma Patients. Clinical Cancer Research. 2011;17:5725–5735. doi: 10.1158/1078-0432.CCR-11-1261. [DOI] [PubMed] [Google Scholar]
- 23.Okada N, Mori N, Koretomo R, Okada Y, Nakayama T, Yoshie O, Mizuguchi H, Hayakawa T, Nakagawa S, Mayumi T, Fujita T, Yamamoto A. Augmentation of the migratory ability of DC-based vaccine into regional lymph nodes by efficient CCR7 gene transduction. Gene therapy. 2005;12:129–139. doi: 10.1038/sj.gt.3302358. [DOI] [PubMed] [Google Scholar]
- 24.Somanchi SS, Somanchi A, Cooper LJN, Lee DA. Engineering lymph node homing of ex vivo-expanded human natural killer cells via trogocytosis of the chemokine receptor CCR7. Blood. 2012;119:5164–5172. doi: 10.1182/blood-2011-11-389924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campbell JDM, Cook G, Robertson SE, Fraser A, Boyd KS, Gracie JA, Franklin IM. Suppression of IL-2-induced T cell proliferation and phosphorylation of STAT3 and STAT5 by tumor-derived TGF beta is reversed by IL-15. Journal of Immunology. 2001;167:553–561. doi: 10.4049/jimmunol.167.1.553. [DOI] [PubMed] [Google Scholar]
- 26.Campbell JD, Piechaczek C, Winkels G, Schwamborn E, Micheli D, Hennemann S, Schmitz J. Isolation and generation of clinical-grade dendritic cells using the CliniMACS system. Methods in molecular medicine. 2005;109:55–70. doi: 10.1385/1-59259-862-5:055. [DOI] [PubMed] [Google Scholar]
- 27.Scandella E, Men Y, Gillessen S, Forster R, Groettrup M. Prostaglandin E2 is a key factor for CCR7 surface expression and migration of monocyte-derived denctritic cells. Blood. 2002;100:1354–1361. doi: 10.1182/blood-2001-11-0017. [DOI] [PubMed] [Google Scholar]
- 28.McKimmie CS, Singh MD, Hewit K, Lopez-Franco O, Le Brocq M, Rose-John S, Lee KM, Baker AH, Wheat R, Blackbourn DJ, Nibbs RJB, Graham GJ. An analysis of the function and expression of D6 on lymphatic endothelial cells. Blood. 2013 doi: 10.1182/blood-2012-04-425314. [DOI] [PubMed] [Google Scholar]
- 29.Nibbs RJ, Wylie SM, Pragnell IB, Graham GJ. Cloning and characterization of a novel murine beta chemokine receptor, D6. Comparison to three other related macrophage inflammatory protein-1alpha receptors, CCR-1, CCR-3, and CCR-5. J Biol Chem. 1997;272:12495–12504. doi: 10.1074/jbc.272.19.12495. [DOI] [PubMed] [Google Scholar]
- 30.Nibbs RJ, Wylie SM, Yang J, Landau NR, Graham GJ. Cloning and characterization of a novel promiscuous human beta-chemokine receptor D6. J Biol Chem. 1997;272:32078–32083. doi: 10.1074/jbc.272.51.32078. [DOI] [PubMed] [Google Scholar]
- 31.Comerford I, Milasta S, Morrow V, Milligan G, Nibbs R. The chemokine receptor CCX-CKR mediates effective scavenging of CCL19 in vitro. Eur J Immunol. 2006;36:1904–1916. doi: 10.1002/eji.200535716. [DOI] [PubMed] [Google Scholar]
- 32.Hansell CAH, Schiering C, Kinstrie R, Ford L, Bordon Y, McInnes IB, Goodyear CS, Nibbs RJB. Universal expression and dual function of the atypical chemokine receptor D6 on innate-like B cells in mice. Blood. 2011;117:5413–5424. doi: 10.1182/blood-2010-11-317115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weber M,E, Blair CV, Simpson M, O’Hara PE, Blackburn A, Rot G, Graham J, Nibbs RJ. The chemokine receptor D6 constitutively traffics to and from the cell surface to internalize and degrade chemokines. Mol Biol Cell. 2004;15:2492–2508. doi: 10.1091/mbc.E03-09-0634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hasbold J, Gett AV, Deenick E, Avery D, Jun J, Hodgkin PD. Quantitative analysis of lymphocyte differentiation and proliferation in vitro using carboxyfluorescein diacetate succinimidyl ester. Immunology and cell biology. 1999;77:516–522. doi: 10.1046/j.1440-1711.1999.00874.x. [DOI] [PubMed] [Google Scholar]
- 35.Graham GJ. D6 and the atypical chemokine receptor family: novel regulators of immune and inflammatory processes. Eur J Immunol. 2009;39:342–351. doi: 10.1002/eji.200838858. [DOI] [PubMed] [Google Scholar]
- 36.Sierro F, Biben C, Martinez-Munoz L, Mellado M, Ransohoff RM, Li M, Woehl B, Leung H, Groom J, Batten M, Harvey RP, Martinez AC, Mackay CR, Mackay F. Disrupted cardiac development but normal hematopoiesis in mice deficient in the second CXCL12/SDF-1 receptor, CXCR7. Proc Natl Acad Sci U S A. 2007;104:14759–14764. doi: 10.1073/pnas.0702229104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lapidot T, Dar A, Kollet O. How do stem cells find their way home? Blood. 2005;106:1901–1910. doi: 10.1182/blood-2005-04-1417. [DOI] [PubMed] [Google Scholar]
- 38.Homey B, Alenius H, Muller A, Soto H, Bowman EP, Yuan W, McEvoy L, Lauerma AI, Assmann T, Bunemann E, Lehto M, Wolff H, Yen D, Marxhausen H, To W, Sedgwick J, Ruzicka T, Lehmann P, Zlotnik A. CCL27-CCR10 interactions regulate T cell-mediated skin inflammation. Nat Med. 2002;8:157–165. doi: 10.1038/nm0202-157. [DOI] [PubMed] [Google Scholar]
- 39.Pabst O, Ohl L, Wendland M, Wurbel MA, Kremmer E, Malissen B, Forster R. Chemokine receptor CCR9 contributes to the localization of plasma cells to the small intestine. Journal of Experimental Medicine. 2004;199:411–416. doi: 10.1084/jem.20030996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Berger C, Jensen MC, Lansdorp PM, Gough M, Elliott C, Riddell SR. Adoptive transfer of effector CD8(+) T cells derived from central memory cells establishes persistent T cell memory in primates. Journal of Clinical Investigation. 2008;118:294–305. doi: 10.1172/JCI32103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hillerdal V, Nilsson B, Carlsson B, Eriksson F, Essand M. T cells engineered with a T cell receptor against the prostate antigen TARP specifically kill HLA-A2(+) prostate and breast cancer cells. P Natl Acad Sci USA. 2012;109:15877–15881. doi: 10.1073/pnas.1209042109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lapidot T, Petit I. Current understanding of stem cell mobilization: the roles of chemokines, proteolytic enzymes, adhesion molecules, cytokines, and stromal cells. Exp Hematol. 2002;30:973–981. doi: 10.1016/s0301-472x(02)00883-4. [DOI] [PubMed] [Google Scholar]
- 43.Nilsson SK, Simmons PJ, Bertoncello I. Hemopoietic stem cell engraftment. Exp Hematol. 2006;34:123–129. doi: 10.1016/j.exphem.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Whetton AD, Graham GJ. Homing and mobilization in the stem cell niche. Trends Cell Biol. 1999;9:233–238. doi: 10.1016/s0962-8924(99)01559-7. [DOI] [PubMed] [Google Scholar]
- 45.Winkler IG, Levesque JP. Mechanisms of hematopoietic stem cell mobilization: when innate immunity assails the cells that make blood and bone. Exp Hematol. 2006;34:996–1009. doi: 10.1016/j.exphem.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Petit I, Szyper-Kravitz M, Nagler A, Lahav M, Peled A, Habler L, Ponomaryov T, Taichman RS, Arenzana-Seisdedos F, Fujii N, Sandbank J, Zipori D, Lapidot T. G-CSF induces stem cell mobilization by decreasing bone marrow SDF-1 and up-regulating CXCR4. Nat Immunol. 2002;3:687–694. doi: 10.1038/ni813. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


