Abstract
Tumor suppressor gene, STK11, encodes for serine–threonine kinase, which has a critical role in regulating cell growth and apoptosis. Mutations of the same lead to the inactivation of STK11, which eventually causes different types of cancer. In this study, we focused on identifying those driver mutations through analyzing structural variations of mutants, viz., D194N, E199K, L160P, and Y49D. Native and the mutants were analyzed to determine their geometrical deviations such as root-mean-square deviation, root-mean-square fluctuation, radius of gyration, potential energy, and solvent-accessible surface area using conformational sampling technique. Additionally, the global minimized structure of native and mutants was further analyzed to compute their intramolecular interactions and distribution of secondary structure. Subsequently, simulated thermal denaturation and docking studies were performed to determine their structural variations, which in turn alter the formation of active complex that comprises STK11, STRAD, and MO25. The deleterious effect of the mutants would result in a comparative loss of enzyme function due to variations in their binding energy pertaining to spatial conformation and flexibility. Hence, the structural variations in binding energy exhibited by the mutants, viz., D194N, E199K, L160P, and Y49D, to that of the native, consequently lead to pathogenesis.
Keywords: cancer, conformational sampling, intramolecular interaction, driver mutation, docking
Introduction
Cancer is driven mostly by somatic mutations. Efficiently identifying the driver mutations from a vast majority of passenger mutations, which do not contribute to cancer, is a difficult task. The gene STK11 is a tumor suppressor gene and encodes for serine–threonine kinase, which has a critical role in regulating cell growth and apoptosis.1,2 Inactivation of this gene leads to development of cancer. Many mutations in STK11 are small deletions or point/missense mutations that are present in the STK11 catalytic kinase domain, and a few of them occur within the noncatalytic COOH- terminal, thereby resulting in STK11 protein reduction, loss, or inactivation.3,4 Mutations in the STK11 gene also cause Peutz–Jeghers syndrome, a condition characterized by the development of noncancerous growths called hamartomatous polyps in the gastrointestinal tract.5
The catalytic kinase domain (residues 43–347) of STK11 associates with the pseudokinase STe20-related adaptor (STRAD; residues 59–431) and full-length scaffolding mouse protein 25 (MO25) in a 1:1:1 heterotrimeric complex in the cell.6–10 In contrast to the majority of the protein kinases that are regulated by phosphorylation, STK11 is activated by binding to STRAD and MO25 through an unknown, phosphorylation-independent molecular mechanism. Although STK11 lacks phosphorylation of the activation loop, it adopts an active conformation. The αC-helix of STK11 is rotated into the canonical closed conformation, by forming the conserved salt bridge between Lys (78) and Glu (98). This active conformation of STK11 appears to be achieved through contributions of both STRAD and MO25. The C-terminallobe of STRAD interacts with both N- and C-terminal lobes of STK11 kinase domain. Mutations in STK11 can lead to its inactivation without affecting this complex assembly.6
Compared to our previous study,10 we have suggested a distinct computational approach to analyze the functional impacts of selected mutations of STK11 in pathogenesis. Molecular dynamics simulation protocol and thermal annealing process were used to compare the native and mutants, viz., D194N, E199K, L160P, and Y49D. Mutant D194N has been reported in lung cancer11; E199K, reported in large intestine cancer12; L160P, reported in cervical cancer13; and Y49D, reported in skin cancer.14 The computational method followed here might distinguish the driver mutations of cancerous genes from a vast number of passenger mutations.
Materials and Methods
Datasets
The protein sequence and variants of STK11 were obtained from the Swiss-Prot database15,16 available at http://www.expasy.ch/sprot/. The 3D Cartesian coordinates of the protein STK11 were obtained from Protein Data Bank (PDB Id: 2 WTK) for in silico mutation modeling and docking studies.17
Modeling missense mutation on protein structures and energy minimization
SWISSPDB viewer18 was used for performing mutant modeling on STK11, and NOMAD-Ref server was used for performing the energy minimization for 3D structures.19 GROMACS force field embedded in NOMAD-Ref was used for energy minimization, based on the steepest descent, conjugate gradient, and limited-memory Broyden-Fletcher-Goldfarb-Shanno methods. It creates a GROMACS topology using the GROMOS96 vacuum force field.20
Prediction of disease-causing mutations by artificial neural network predictor, NetDiseaseSNP, and validation by Catalog of Somatic Mutations in Cancer database
For the prediction of disease-causing mutations, we used the tool NetDiseaseSNP,21 a sequence conservation-based predictor of the pathogenicity of mutations, which exploits the predictive power of artificial neural networks. This method derives sequence conservation from position-specific scoring matix (PSSM), based on the alignment algorithm of sorting intolerant from tolerant (SIFT), which is complemented with the calculation of surface accessibility by the predictor Net-SurfP.22 This approach provides NetDiseaseSNP the potential to extract all relevant information directly from protein sequences. NetDiseaseSNP encodes the SIFT score (normalized probability) for the SNP amino acid in one input neuron. SIFT predicts the effects of all possible substitutions at each position in the protein sequence. This server is available at http://www.cbs.dtu.dk/services/NetDiseaseSNP/. The artificial neural networks of this predictor will generate an output value close to 1 if the combination of features describing that particular mutation suggests that it might be involved in disease, and close to 0 for neutral mutations.
The database Catalogue of Somatic Mutations in Cancer (COSMIC)23 is the largest and ample resource for exploring the impact of somatic mutations in human cancer. In order to gain a deep sense of knowledge on the key cancer genes, many appropriate literatures were identified for each gene and then subjected to manual curation. This manual curation allows this database to capture very high detail across mutation positions and disease descriptions. The variants were subjected to a COSMIC search to extract the information of primary tissue affected. The COSMIC dataset can be assumed to be enriched for cancer driver mutations when compared with large-scale somatic mutation discovery datasets, which were expected to contain a fair number of passenger mutations.23
Ensemble analyses through normal mode-based simulation
Conformation sampling approach was used to generate ensembles to expand the chances of identifying an energetic landscape that closely matched the input structures.24 The Normal Mode-based Simulation (NMSim) approach25 has been shown to be a computationally efficient alternative to molecular dynamics simulations for conformational sampling of proteins and performs three types of simulations, viz., unbiased exploration of conformational space, pathway generation by a targeted simulation, and radius of gyration (RoG)-guided simulation. This Web server implements a three-step approach for multiscale modeling of protein conformational changes. Initially, the protein structure is coarse-grained, followed by a rigid cluster normal mode analysis that provides low-frequency normal modes, and finally, these modes are used to extend the recently introduced idea of constrained geometric simulations by biasing backbone motions of the protein, whereas, side chain motions are biased toward favorable rotamer states (NMSim). This program is accessible through http://www.nmsim.de. The RoG-guided simulation type was used here to generate the ensembles of native and mutant structures. The parameters used for the rigid cluster decomposition were as follows: energy cutoff for hydrogen bonds (−1.0 kcal/mol), method for placing hydrophobic constraints (3), cutoff for including hydrophobic constraints (0.35 Å). The method chosen for the normal mode analysis was rigid cluster normal mode analysis, and the distance cutoff for interactions between C-alpha atoms was set to 10 Å. The parameters for the simulation were as follows: number of trajectories (1), number of simulation cycles (500), number of NMSim cycles (1), frequency of writing out conformations (1), side chain distortions (0.3), normal mode range (1–50), ROG mode (1), and step size (0.5 Å). The structural diversity of native and mutant ensembles, obtained through NMSim program, was evaluated by their root-mean-square deviation (RMSD) and root-mean-square fluctuation (RMSF). Geometrical and conformational deviations of native and mutant ensembles were studied through RMSD.26 RMSF was used to study the fluctuation of residues present in native and mutant structures, which represents their atomic mobility.27,28 RoG is indicative of the level of compaction in the structure29 and calculated using VEGA ZZ package,30 which implements trajectory visualization and numerous calculations. Packing defects and folding patterns of native and mutants were studied through solvent-accessible surface area (SASA),31 which was computed using VEGA ZZ package.30 Changes in SASA are caused due to changes in their tertiary structures, which in turn are affected by the folding patterns of native and mutants.32
Furthermore, native and mutant ensembles were analyzed for their energetic contributions through Bayesian Analysis Conformation Hunt (BACH) algorithm (http://bachserver.pd.infn.it/in) in which the all-atom energy score was computed based on 1091 parameters. The BACH score was also used to discriminate the global minima of native and mutant ensemble.33,34
Analysis of protein integrity by computing interactions, hydrogen bond analysis, simulated thermal denaturation, and secondary structure analysis
Protein stability relies on various strong and weak interactions.35 Protein interactions calculator (PIC) program,36 a structure-based algorithm was used to compute the interactions stabilizing native and mutant structures. The program was used to compute the disulfide bridges, interactions involving hydrophobic, ionic, aromatic–aromatic residues, aromatic–sulfur residues, and cation–π interactions, which contributed to the overall integrity of native and mutant structures. In addition, the hydrogen bond analysis was done, as they stabilize secondary structure elements.37 HB Plot (http://virtuadrug.com/products/hb-plot/index.html) is a tool for exploring the protein structure and function by describing the structure as a network of hydrogen bonding interaction. This Web server plots three classes of hydrogen bonding by color coding: type 1 – short (distance smaller than 2.5 Å between donor and acceptor), type 2 – intermediate (between 2.5 Å and 3.2 Å), and type 3 – long hydrogen bonds (greater than 3.2 Å).37,38 Besides, the native and mutant structures were subjected to simulated thermal denaturation through Proflex, embedded in Stone-Hinge program available at http://stonehinge.bmb.msu.edu/to describe the folding patterns.39 This Web server simulated incremental thermal denaturation of the structure, as the calculated temperature rises, and hydrogen bonds weaker than the current energy level were broken. Hydrophobic interactions were maintained throughout the process, as the strength of these interactions increased somewhat with a modest increase in temperature.40 Furthermore, secondary structure elements were analyzed through STRIDE program, which assigns the secondary structure based on combined use of hydrogen bond energy and statistically derived backbone torsional angle information.41 Changes in α helix, β sheets, and coils of native and mutants were compared and visualized, as structural deterioration might cost the functional properties of structures.
Analysis of interaction of native and mutant with STRAD
We subjected the native and mutants for docking studies by using PatchDock42,43 in order to understand their functional activity with the binding partner, STRAD. Patch-Dock is a geometry-based molecular docking algorithm, and it is aimed at finding docking transformations that yield good molecular shape complementarity.42 Such transformations, when applied, induce both wide interface areas and small amounts of steric clashes. A wide interface ensured that several matched local features of the docked molecules that have complementary characteristics were included. The PatchDock algorithm divides the Connolly dot surface representation43,44 of the molecules into concave, convex, and flat patches. Then, complementary patches were matched to generate candidate transformations. Each candidate transformation was further evaluated by a scoring function that considered both geometric fit and atomic desolvation energy.44 PatchDock is available at http://bioinfo3d.cs.tau.ac.il/PatchDock/. The 10 best solutions of PatchDock analysis were selected for further flexible refinement and rescoring analysis through fast interaction refinement in molecular docking (FireDock) algorithm.31,45 This method targets the problem of flexibility and scoring of solutions produced by fast rigid-body docking algorithms. The output provided a list of refined complexes, sorted by a binding energy function, and a 3D visualization for observing and comparing the refined complexes.45 FireDock is available at http://bioinfo3d.cs.tau.ac.il/FireDock/firedock.html. The docked complexes of native and mutants were analyzed for binding energies, followed by visualization of binding sites using PyMol (http://www.pymol.org).
To identify the binding residues between STK11 and STRAD, we submitted the native and mutants to the protein interface recognition server, solvent accessibility based protein–protein interface identification and recognition (SPPIDER).46 This server integrates enhanced relative solvent accessibility predictions with high-resolution structural data. This Web server is available at http://sppider.cchmc.org/.
Results and Discussion
Retrieval of STK11structure and mutant modeling
The native structure (2 WTK) was obtained from PDB database, followed by in silico mutation modeling to generate structures for D194N, E199K, L160P, and Y49D mutants, and subsequently, all the mutant structures were energy minimized.
Identification of disease-causing missense mutations and validation
The four mutants, viz., D194N, E199K, L160P, and Y49D, were identified as disease causing through the score of 0.97, 0.91, 0.94, and 0.95, respectively, obtained from Net-DiseaseSNP (Table 1). Since the output values were close to 1, all the four selected mutants were considered as disease causing. The mutant D194N changed its amino acid from polar negatively charged to polar neutral, E199K changed from acid to basic, Y49D changed from polar neutral to polar negatively charged, and the mutant L160P retained its basic property.
Table 1.
Scoring of mutants by using NetDiseaseSNP and total energy calculation.
| STK11 | NETDiseaseSNP SCORE | TOTAL ENERGY KJ/mol | PRIMARY TISSUE AFFECTED |
|---|---|---|---|
| Native | – | −16734.766 | – |
| D194N | 0.97 | −12314.974 | Lung |
| E199K | 0.91 | −12326.152 | Large intestine |
| L160P | 0.94 | −10595.924 | Cervix |
| Y49D | 0.95 | −13090.824 | Skin |
Note: “–” indicates Not applicable.
For the validation of this result, we derived the data of corresponding mutation from COSMIC and identified the primary tissue affected by each mutation. The primary tissues affected by the four mutants, viz., D194N, E199K, L160P, and Y49D, were lung, large intestine, cervix, and skin, respectively,11–14 as illustrated in Table 1. The CDS mutation for the above mutants were also obtained from COSMIC, and it was found to be 580G > A, 595G > A, 479T > C, and 145T > G correspondingly.
Ensemble analysis of native and mutant structures
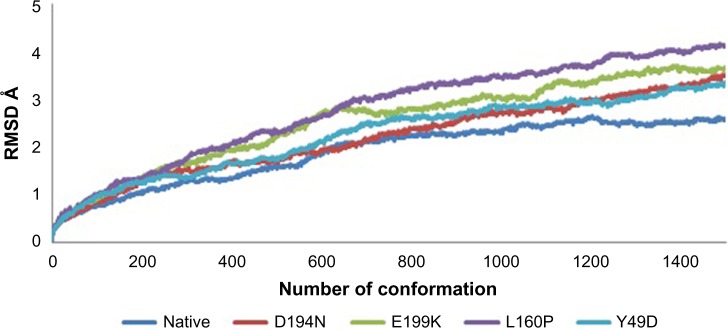
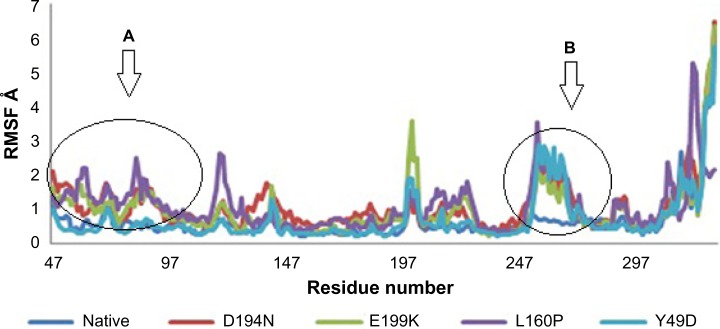
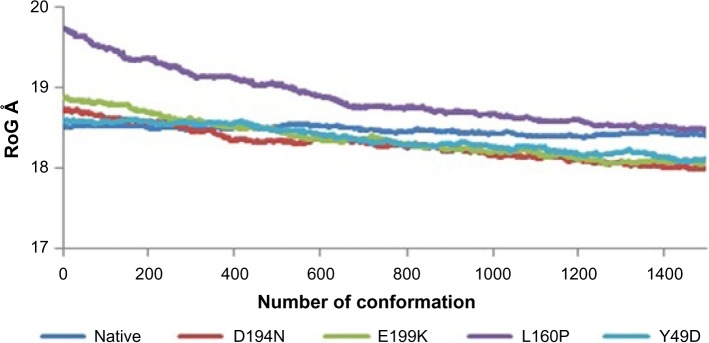
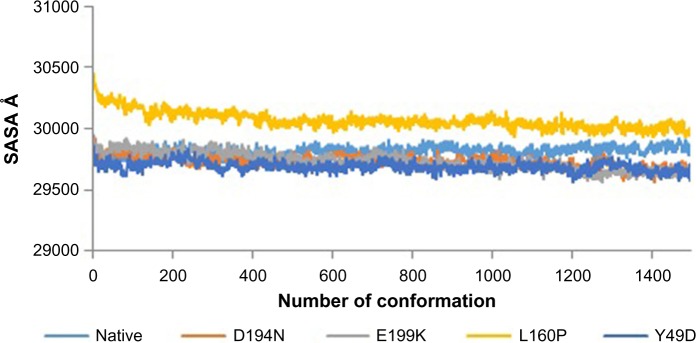
Divergence of the mutant structure from the native structure could be caused by substitutions, deletions, and insertion,47 and the deviation between the two structures could alter the functional activity48 with respect to binding efficiency of the binding partner, which was evaluated by their RMSD, RMSF, and RoG values. The stability of secondary structure elements and conformational changes were assessed by plotting these values obtained from trajectory. To analyze the deviations in conformational space and rigidity of the ensembles, RMSD plot of Cα atoms was computed for native and mutant (D194N, E199K, L160P, and Y49D) structures. The mean RMSD of native was found to be 1.9 Å. On the other hand, all the mutants possessed a higher mean value of RMSD as presented in Table 2 and Figure 1. These results suggested that mutant ensembles comprised structural deviations when compared with native. In addition, RMSF was studied to understand the overall flexibility of the residues of native and mutant. The mean RMSF of native was found to be 0.66 Å, whereas all the mutants possessed increased flexibility in their residues (Table 2 and Fig. 2). Increase in flexibility creates a large loss in enthalpy (weakened native contacts) that is unfavorable for the protein.49 The analysis of RMSF exposed that the residues 47–97 and 250–276 possessed a comparatively high flexibility change than the other residues in all the mutants. The region 47–97 had involved the5 binding residues, viz., Leu (67), Thr (71), Leu (72), Cys (73), and Arg (74)6 for the native–STRAD interaction. Furthermore, RoG analysis disclosed that the three mutants, viz., D194N, E199K, and Y49D, had a less RoG of 18.2918 Å, 18.3645 Å, and 18.3630 Å, respectively, when compared with native 18.4733 Å. Mutant L160P possessed higher value than the native, and it was found to be 18.8869 Å; this could have resulted because of the increase in the flexibility of this mutant (Table 2 and Fig. 3). This, in turn, showed the distinct compactness of mutant structures. Besides, SASA was analyzed, in which, the native exhibited 29,810.50 Å2 as an average SASA. Three mutants, viz., D194N, E199K, and Y49D, exhibited decreased SASA compared with native, and it was found to be 29,726.60 Å2, 29,730.12 Å2, and 29,686.71 Å2, respectively (Table 2 and Fig. 4), whereas, the mutant L160P showed an increased SASA of 30,070.69 Å2. This could have also resulted due to the high RMSF possessed by the mutant L160P, since there is correlation between the flexibility of the residue and SASA.50 As the flexibility increases, SASA will also increase, which in turn affects the stability of the protein unfavorably.
Table 2.
Trajectory analysis of native and mutants.
| STK11 | AVERAGE RMSD (Å) | AVERAGE RMSF (Å) | AVERAGE SURFACE AREA (Å) | AVERAGE RADIUS OF GYRATION (Å) |
|---|---|---|---|---|
| Native | 1.9 | 0.66 | 29810.50 | 18.47 |
| D194N | 2.18 | 1.05 | 29726.60 | 18.29 |
| E199K | 2.53 | 0.95 | 29730.12 | 18.36 |
| L160P | 2.79 | 1.10 | 30070.69 | 18.87 |
| Y49D | 2.24 | 0.70 | 29686.71 | 18.36 |
Figure 1.
RMSD variation of native and mutants.
Figure 2.
RMSF of native and mutants. (A) Highly fluctuated region (residues 47–97). (B) Highly fluctuated region (residues 250–276).
Figure 3.
RoG of native and mutants.
Figure 4.
SASA of native and mutants.
Analyzing structural events of global minima of native and mutants
Various types of intramolecular interactions,51 more specifically hydrogen bonds52 influence protein folding,40 which in turn play a major role in the structural property of a protein. The following analyses were carried out between native and mutants using global minimized structures. Hydrophobic, cation–π, and other interactions were contributed in stabilizing proteins.38 Therefore, intramolecular interactions for native and mutant structures were studied using the PIC program.36 Different interactions were used to study the variations in structural integrity of the structures, and the results were represented type wise in Table 3. From this result, it was noted that the native structure had a total of 999 interactions, while all the mutants had reduced number of interactions, among which L160P mutant had the lowest with 779 interactions. Reduction in the number of interactions in mutant structures might alter their structural integrity and folding patterns. Therefore, hydrogen bond analysis and dilution patterns were studied in detail to understand the folding patterns of native and mutant structures. Furthermore, energetic contributions of native and mutant structures were analyzed to explore the possible deviations, where the total energy possessed by the native was −16,734.766 kJ/mol, and that of the mutants, viz., D194N, E199K, L160P, and Y49D, were found to be −12,314.974 kJ/mol, −12,326.152 kJ/mol, −10,595.924 kJ/mol, and −13,090.824 kJ/mol, respectively (Table 1). This analysis clearly portrayed that all the four mutants showed increased total energy as compared with native STK11, thus showing alteration in the stability of mutants as compared with native.
Table 3.
Intra molecular interactions of native and mutants.
| STK11 | HI | M-M | M-S | S-S | II | AR-AR | AR-S | C-π | TOTAL NO. OF INTERACTIONS |
|---|---|---|---|---|---|---|---|---|---|
| Native | 257 | 390 | 180 | 127 | 29 | 7 | 4 | 5 | 999 |
| D194N | 257 | 384 | 177 | 112 | 30 | 7 | 5 | 3 | 975 |
| E199K | 258 | 381 | 177 | 110 | 28 | 7 | 6 | 2 | 969 |
| L160P | 244 | 319 | 103 | 68 | 29 | 7 | 4 | 5 | 779 |
| Y49D | 281 | 382 | 184 | 105 | 29 | 8 | 5 | 3 | 997 |
Abbreviations: HI, Hydrophobic Interactions; M-M, Main chain-Main chain hydrogen bond; M-S, Main chain- Side chain Hydrogen bond; S-S, Side chain-Side chain hydrogen bond; I-I, Ionic Interactions; Ar-Ar, Aromatic–Aromatic interaction; Ar-S, Aromatic–Sulphur interaction; C-π, Cation-π interaction.
Identifying the strength of hydrogen bonds, pattern of simulated thermal denaturation, and elements of secondary structure of native and mutants
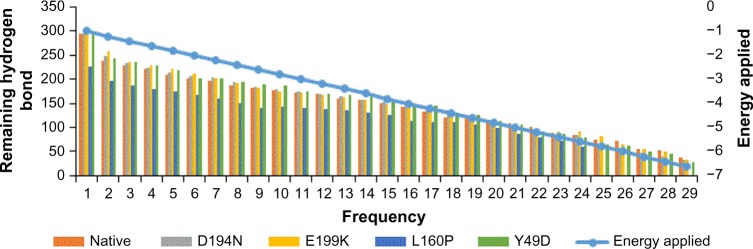
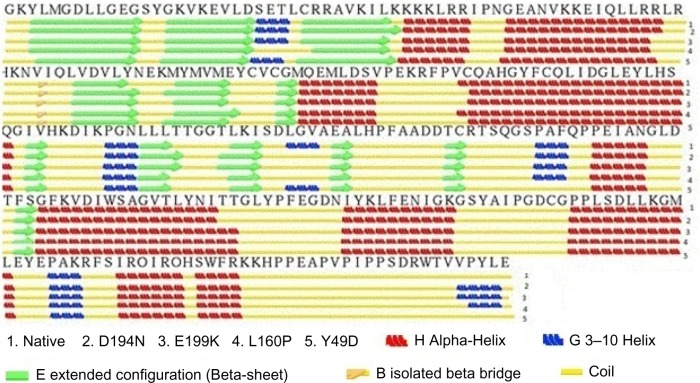
The pattern of hydrogen bond distribution affects secondary and tertiary structures of proteins.52 Besides, residues which are far apart in sequence are brought closer by protein folding, as aided by hydrogen bonds.53 Moreover, changes in folding patterns are known to affect the outline structures of proteins and arrangements of secondary structure elements.54 Therefore, native and mutant structures were subjected to hydrogen bond analysis and simulated thermal denaturation to analyze the number of hydrogen bonds present in the structure and check how hydrogen bonds were diluted on the application of various energy ranges. The total number of hydrogen bonds present in the native was 284, and most of them (282) reside in the intermediate range, and only 2 were in the long range. Of note, the three mutants, viz., D194N, E199K, and L160P, had less number of total hydrogen bonds of about 273, 283, and 225, respectively, than the native (Table 4). The intermediate range of hydrogen bonds was less in these three mutants, and also, more number of long-range hydrogen bonds was present, when compared with native. The mutant Y49D exhibited more number (292) of hydrogen bonds than the native. Of the 292 hydrogen bonds, about 13 were in the long range and 279 were in the intermediate range. This portrayed that the native possesses strong hydrogen bonds than the mutants, as the length of the hydrogen bonds correlates with the strength of the bond.37 Native and mutant structures exhibited different patterns of hydrogen bond dilution as noticeable from Table 5 and Figure 5. When the energy of −1 kJ/mol was applied to native and mutants, the remaining hydrogen bonds present in native was 276, whereas the mutants, viz., D194N, E199K, L160P, and Y49D, possessed 265, 274, 226, and 278 hydrogen bonds, respectively. For the energy of −5.8 kJ/mol, the mutant L160P showed complete dilution of hydrogen bonds, whereas the native and the three mutants, viz., D194N, E199K, and Y49D, had remaining hydrogen bonds of 74, 65, 81, and 64, respectively, which, in turn, suggested variations in their folding patterns. In addition, secondary structure elements of native and mutants were visualized and compared with the observed position level variations. It was noted that mutants showed variations in terms of secondary structure elements when compared with native (Fig. 6). These variations could have occurred due to the mutations that might rearrange the packed secondary structure54 of STK11. The position level variations in secondary structure elements of the mutants were as follows: mutant L160P comprised variations at positions 23–26 (three residues) where, helix was converted to coil. All the mutants showed variations from helix to coil at positions 170–173 (three residues), except Y49D, which showed variation from helix to coil at positions 193–196 (three residues). Three mutants, viz., E199K, L160P, and Y49D, showed variation from coil to helix at positions 203–204, while D194N and E199K showed variations from coil to helix at positions 281–284 (four residues) and L160P showed variation from coil to helix at positions 282–284 (three residues).
Table 4.
Hydrogen bond analysis of native and mutant STSTK11.
| STK11 | SHORT HB (<2.5 Å) | INTERMEDIATE HB (2.5 ≥3.2 Å) | LONG HB (>3.2 Å) | TOTAL HB |
|---|---|---|---|---|
| Native | 0 | 282 | 2 | 284 |
| D194N | 0 | 265 | 9 | 273 |
| E199K | 0 | 276 | 7 | 283 |
| L160P | 0 | 213 | 12 | 225 |
| Y49D | 0 | 279 | 13 | 292 |
Abbreviation: HB, Hydrogen Bond.
Table 5.
Simulated thermal denaturation of native and mutants.
| ENERGY (KJ/MOL) | REMAINING HYDROGEN BONDS | ||||
|---|---|---|---|---|---|
| NATIVE | D194N | E199K | L160P | Y49D | |
| −1 | 276 | 265 | 274 | 226 | 278 |
| −1.2 | 238 | 248 | 257 | 196 | 243 |
| −1.4 | 228 | 233 | 236 | 187 | 237 |
| −1.6 | 222 | 225 | 228 | 179 | 229 |
| −1.8 | 210 | 215 | 221 | 175 | 220 |
| −2 | 202 | 206 | 211 | 167 | 203 |
| −2.2 | 196 | 204 | 203 | 160 | 201 |
| −2.4 | 188 | 194 | 191 | 150 | 194 |
| −2.6 | 183 | 185 | 182 | 140 | 190 |
| −2.8 | 178 | 180 | 175 | 144 | 186 |
| −3 | 173 | 174 | 172 | 140 | 174 |
| −3.2 | 169 | 169 | 168 | 139 | 171 |
| −3.4 | 160 | 164 | 163 | 135 | 168 |
| −3.6 | 157 | 157 | 157 | 130 | 165 |
| −3.8 | 150 | 152 | 149 | 127 | 157 |
| −4 | 143 | 142 | 141 | 114 | 147 |
| −4.2 | 132 | 141 | 137 | 112 | 145 |
| −4.4 | 122 | 136 | 130 | 110 | 131 |
| −4.6 | 119 | 120 | 115 | 106 | 127 |
| −4.8 | 115 | 108 | 114 | 98 | 114 |
| −5 | 109 | 103 | 104 | 87 | 106 |
| −5.2 | 102 | 97 | 96 | 80 | 92 |
| −5.4 | 90 | 80 | 92 | 73 | 87 |
| −5.6 | 84 | 83 | 92 | 59 | 80 |
| −5.8 | 74 | 65 | 81 | 0 | 64 |
| −6 | 71 | 57 | 64 | 0 | 61 |
| −6.2 | 54 | 47 | 55 | 0 | 50 |
| −6.4 | 53 | 41 | 50 | 0 | 45 |
| −6.6 | 38 | 33 | 33 | 0 | 28 |
Figure 5.
Thermal denaturation of native and mutants with intervals of applied energy.
Figure 6.
Secondary structure analysis of native and mutants.
On the whole, the strength of hydrogen bonds, changes in folding patterns, and variations in secondary structure elements gave an idea about the structural changes in the mutant structures. These changes might alter the functional properties of mutant structures that could result in the pathogenesis of the disease, and therefore, functional properties were assessed through docking studies.55
Computation of binding affinity between native and mutants
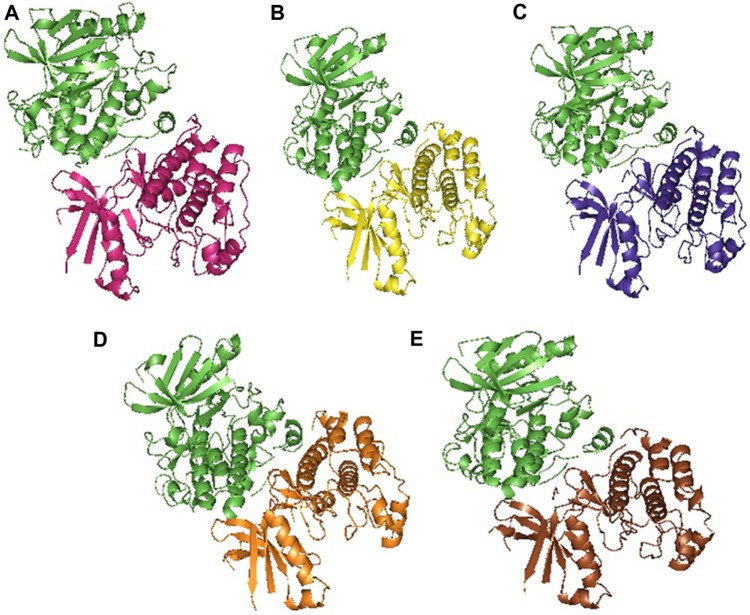
Native and mutant structures were analyzed for their binding affinity toward the partner STRAD. Docking was performed using the PatchDock between STRAD and the global minima of native and mutants of STK11 by specifying the receptor-binding sites such as Arg (74), Cys (73), Leu (72), Thr (71), and Thr (186) and the ligand-binding sites such as Leu (241), His (231), Phe (233), and Gln (251) to determine the binding efficiency in the form of the atomic contact energy (ACE). The 10 best solutions of PatchDock analysis were selected and further analyzed for flexible refinement and rescoring by FireDock. From the analysis, the ACE between STRAD and native STK11 was found to be −3.30 kJ/mol and all the mutants demonstrated low ACE when compared with the native (Table 6). Also, the energy contribution from hydrogen bonding, van der waals force of attraction, and repulsions to the complexes (Fig. 7) of all mutants were varying when compared with native as represented in Table 6. SPPIDER was used to calculate the contacts between the binding residues of native and mutant STK11 with STRAD, and the binding residues were Leu (67), Thr (71), Leu (72), Cys (73), Arg (74), Cys (151), Thr (186), Lys (191), Pro (315), Pro (317), Ile (318), Pro (320), Arg (323), Trp (324), Val (327), Leu (330), and Glu (331). Reduced ACE of mutants toward STRAD might be due to altered RMSF values of binding residues that were present in the highly fluctuated region. Changes in flexibility of these residues might have resulted in improper STRAD binding and eventual loss of functional activity of mutants, which could lead to cancer.
Table 6.
Energy contribution of ACE, attractive and repulsive Vdw and HB in the docked structure of native and mutants with STRAD.
| STK11 | ACE KJ/mol |
ATTRACTIVE Vdw KJ/mol |
REPULSIVE Vdw KJ/mol |
HB KJ/mol |
|---|---|---|---|---|
| Native | −3.30 | −16.50 | 6.57 | −2.99 |
| D194N | 3.76 | −25.34 | 7.84 | −2.00 |
| E199K | 4.21 | −17.99 | 6.25 | −2.61 |
| L160P | −2.29 | −27.24 | 8.28 | −1.92 |
| Y49D | −2.78 | −22.03 | 7.29 | −2.20 |
Abbreviations: ACE, Atomic Contact Energy; Vdw, Van der waals force; HB, Hydrogen Bond.
Figure 7.
Docked structures of (A) native (magenta) and STRAD (green), (B) D194N (yellow) and STRAD (green), (C) E199K (violet) and STRAD (green), (D) L160P (orange) and STRAD (green), (E) Y49D (brown) and STRAD (green).
Conclusion
Single-nucleotide polymorphism is the most common form of genetic variation,56 and missense mutations lead to substitution of amino acids, which eventually affected the function of STK11 and were often responsible for causing various types of cancer.11–14 Ensemble analysis of mutants, viz., D194N, E199K, L160P, and Y49D, substantiated considerable variations in terms of RMSD. RMSF of mutants showed variation, especially at the region between 47 and 97, where five binding residues were present, which suggested increased flexibility of residues as compared with native. RoG analysis throughout the ensembles substantiated that the mutants exhibited RoG in the range of 18.2918–18.8869 Å. The SASA analysis of mutants also showed variation from native, possessing SASA in the range of 29,726.60–30,070.69 Å. Furthermore, global minima of native and mutants were used for further analysis. Variations were observed between the native and mutant structures in terms of intramolecular interactions, number of hydrogen bonds, simulated thermal denaturation, and secondary structure elements. Therefore, it was obvious that mutants would have variations in affinity toward partner, which was analyzed through docking studies, where all four mutants had reduced ACE compared with native. This could have eventuated due to the change of flexibility in binding residues and changes in the secondary structure elements. Therefore, it was concluded that mutants, viz., D194N, E199K, L160P, and Y49D, have detrimental effects contributing to the pathogenesis of disease. The study gains interest since the deleterious effects that remained latent within the mutant structures were explored using conformational sampling method that could serve as an alternate to classical molecular dynamics and structural variations of these mutants, which were not reported yet, were explored and might shed more light onto the cancer research community.
Supplementary Material
Figure A. Diagrammatic representation of the computational approach to distinguish driver mutations.
Acknowledgments
The authors are grateful to VIT University for providing necessary facilities and infrastructure to carry out this study.
Footnotes
ACADEMIC EDITOR: J. T. Efird, Editor in Chief
PEER REVIEW: Six peer reviewers contributed to the peer review report. Reviewers’ reports totaled 2077 words, excluding any confidential comments to the academic editor.
FUNDING: Authors disclose no external funding sources.
COMPETING INTERESTS: Authors disclose no potential conflicts of interest.
Paper subject to independent expert blind peer review. All editorial decisions made by independent academic editor. Upon submission manuscript was subject to anti-plagiarism scanning. Prior to publication all authors have given signed confirmation of agreement to article publication and compliance with all applicable ethical and legal requirements, including the accuracy of author and contributor information, disclosure of competing interests and funding sources, compliance with ethical requirements relating to human and animal study participants, and compliance with any copyright requirements of third parties. This journal is a member of the Committee on Publication Ethics (COPE).
Author Contributions
Designed the computational analysis: ML. Analysed and contributed to the writing of the manuscript: DMP. Made critical revision and approved final version: RR. All authors reviewed and approved of the final manuscript.
REFERENCES
- 1.Alessi DR, Sakamoto K, Bayascas JR. LKB1-dependent signaling pathways. Annu Rev Biochem. 2006;75:137–63. doi: 10.1146/annurev.biochem.75.103004.142702. [DOI] [PubMed] [Google Scholar]
- 2.Karuman P, Gozani O, Odze RD, et al. The Peutz-Jegher gene product LKB1 is a mediator of p53-dependent cell death. Mol Cell. 2001;7(6):1307–19. doi: 10.1016/s1097-2765(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 3.Schumacher V, Vogel T, Leube B, et al. STK11 genotyping and cancer risk in Peutz-Jeghers syndrome. J Med Genet. 2005;42(5):428–35. doi: 10.1136/jmg.2004.026294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boudeau J, Sapkota G, Alessi DR. LKB1, a protein kinase regulating cell proliferation and polarity. FEBS Lett. 2003;546(1):159–65. doi: 10.1016/s0014-5793(03)00642-2. [DOI] [PubMed] [Google Scholar]
- 5.Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews. Seattle (WA): University of Washington; 1993. Available at http://www.ncbi.nlm.nih.gov/books/NBK1116/ [Google Scholar]
- 6.Zeqiraj E, Filippi BM, Deak M, Alessi DR, van Aalten DM. Structure of the LKB1-STRAD-MO25 complex reveals an allosteric mechanism of kinase activation. Science. 2009;326(5960):1707–11. doi: 10.1126/science.1178377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baas AF, Boudeau J, Sapkota GP, et al. Activation of the tumour suppressor kinase LKB1 by the STE20-like pseudokinase STRAD. EMBO J. 2003;22(12):3062–72. doi: 10.1093/emboj/cdg292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boudeau J, Scott JW, Resta N, et al. Analysis of the LKB1-STRAD-MO25 complex. J Cell Sci. 2004;117(pt 26):6365–75. doi: 10.1242/jcs.01571. [DOI] [PubMed] [Google Scholar]
- 9.Hawley SA, Boudeau J, Reid JL, et al. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2(4):28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lopus M, Rajasekaran R. Exploring the missense mutation impact on STK11 which cause Peutz-Jeghers syndrome by computational analysis. Int J ChemTech Res. 2014;6(2):1251–67. [Google Scholar]
- 11.Ji H, Ramsey MR, Hayes DN, et al. LKB1 modulates lung cancer differentiation and metastasis. Nature. 2007;448(7155):807–10. doi: 10.1038/nature06030. [DOI] [PubMed] [Google Scholar]
- 12.Dong SM, Kim KM, Kim SY, et al. Frequent somatic mutations in serine/threonine kinase 11/Peutz-Jeghers syndrome gene in left-sided colon cancer. Cancer Res. 1998;58(17):3787–90. [PubMed] [Google Scholar]
- 13.Kuragaki C, Enomoto T, Ueno Y, et al. Mutations in the STK11 gene characterize minimal deviation adenocarcinoma of the uterine cervix. Lab Invest. 2003;83(1):35–45. doi: 10.1097/01.lab.0000049821.16698.d0. [DOI] [PubMed] [Google Scholar]
- 14.Rowan A, Bataille V, MacKie R, et al. Somatic mutations in the Peutz-Jeghers (LKB1/STKII) gene in sporadic malignant melanomas. J Invest Dermatol. 1999;112(4):509–11. doi: 10.1046/j.1523-1747.1999.00551.x. [DOI] [PubMed] [Google Scholar]
- 15.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Res. 2000;28(1):45–8. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berman HM, Westbrook J, Feng Z, et al. The protein data bank. Nucleic Acids Res. 2000;28(1):235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–23. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 19.Lindahl E, Azuara C, Koehl P, Delarue M. NOMAD-Ref: visualization, deformation and refinement of macromolecular structures based on all-atom normal mode analysis. Nucleic Acids Res. 2006;34(Web Server):W52–6. doi: 10.1093/nar/gkl082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Delarue M, Dumas P. On the use of low-frequency normal modes to enforce collective movements in refining macromolecular structural models. Proc Natl Acad Sci U S A. 2004;101(18):6957–62. doi: 10.1073/pnas.0400301101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johansen MB, Izarzugaza JM, Brunak S, Petersen TN, Gupta R. Prediction of disease causing non-synonymous SNPs by the artificial neural network predictor NetDiseaseSNP. PLoS One. 2013;8(7):e68370. doi: 10.1371/journal.pone.0068370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Petersen B, Petersen TN, Andersen P, Nielsen M, Lundegaard C. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct Biol. 2009;9:51. doi: 10.1186/1472-6807-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forbes SA, Beare D, Gunasekaran P, et al. COSMIC: exploring the world’s knowledge of somatic mutations in human cancer. Nucleic Acids Res. 2015;43(Database issue):D805–11. doi: 10.1093/nar/gku1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kellogg EH, Leaver-Fay A, Baker D. Role of conformational sampling in computing mutation-induced changes in protein structure and stability. Proteins Struct Funct Bioinformatics. 2011;79(3):830–8. doi: 10.1002/prot.22921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kruger DM, Ahmed A, Gohlke H. NMSim web server: integrated approach for normal mode-based geometric simulations of biologically relevant conformational transitions in proteins. Nucleic Acids Res. 2012;40(W1):W310–6. doi: 10.1093/nar/gks478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Read RJ, Adams PD, Arendall WB, et al. A new generation of crystallographic validation tools for the protein data bank. Structure. 2011;19(10):1395–412. doi: 10.1016/j.str.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen J, Liang Z, Wang W, Yi C, Zhang S, Zhang Q. Revealing origin of decrease in potency of Darunavir and Amprenavir against HIV-2 relative to HIV-1 protease by molecular dynamics simulations. Sci Rep. 2014;4:6872. doi: 10.1038/srep06872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vendome J, Posy S, Jin X, et al. Molecular design principles underlying β-strand swapping in the adhesive dimerization of cadherins. Nat Struct Mol Biol. 2011;18(6):693–700. doi: 10.1038/nsmb.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobanov MI, Bogatyreva NS, Galzitskaia OV. Radius of gyration is indicator of compactness of protein structure. Mol Biol. 2008;42(4):701–6. [PubMed] [Google Scholar]
- 30.Pedretti A, Villa L, Vistoli G. VEGA–an open platform to develop chemo-bio-informatics applications, using plug-in architecture and script programming. J Comput Aided Mol Des. 2004;18(3):167–73. doi: 10.1023/b:jcam.0000035186.90683.f2. [DOI] [PubMed] [Google Scholar]
- 31.Mashiach E, Schneidman-Duhovny D, Andrusier N, Nussinov R, Wolfson HJ. FireDock: a web server for fast interaction refinement in molecular docking. Nucleic Acids Res. 2008;36(Web Server issue):W229–32. doi: 10.1093/nar/gkn186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Connolly ML. Solvent-accessible surfaces of proteins and nucleic acids. Science. 1983;221(4612):709–13. doi: 10.1126/science.6879170. [DOI] [PubMed] [Google Scholar]
- 33.Cossio P, Granata D, Laio A, Seno F, Trovato A. A simple and efficient statistical potential for scoring ensembles of protein structures. Sci Rep. 2012;2:351. [Google Scholar]
- 34.Trosset J-Y, Scheraga HA. Reaching the global minimum in docking simulations: a Monte Carlo energy minimization approach using Bezier splines. Proc Natl Acad Sci U S A. 1998;95(14):8011–5. doi: 10.1073/pnas.95.14.8011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogt G, Woell S, Argos P. Protein thermal stability, hydrogen bonds, and ion pairs. J Mol Biol. 1997;269(4):631–43. doi: 10.1006/jmbi.1997.1042. [DOI] [PubMed] [Google Scholar]
- 36.Tina KG, Bhadra R, Srinivasan N. PIC: protein interactions calculator. Nucleic Acids Res. 2007;35(Web Server):W473–6. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bikadi Z, Demko L, Hazai E. Functional and structural characterization of a protein based on analysis of its hydrogen bonding network by hydrogen bonding plot. Arch Biochem Biophys. 2007;461(2):225–34. doi: 10.1016/j.abb.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 38.McDonald IK, Thornton JM. Satisfying hydrogen bonding potential in proteins. J Mol Biol. 1994;238(5):777–93. doi: 10.1006/jmbi.1994.1334. [DOI] [PubMed] [Google Scholar]
- 39.Keating KS, Flores SC, Gerstein MB, Kuhn LA. StoneHinge: hinge prediction by network analysis of individual protein structures. Protein Sci. 2009;18(2):359–71. doi: 10.1002/pro.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seeliger D, Haas J, de Groot BL. Geometry-based sampling of conformational transitions in proteins. Structure. 2007;15(11):1482–92. doi: 10.1016/j.str.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 41.Heinig M, Frishman D. STRIDE: a web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 2004;32(Web Server issue):W500–2. doi: 10.1093/nar/gkh429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duhovny D, Nussinov R, Wolfson HJ. Algorithms in Bioinformatics. Springer; 2002. [Accessed October 3, 2014]. Efficient unbound docking of rigid molecules [Internet]; Available at http://link.springer.com/chapter/10.1007/3-540-45784-4_14. [Google Scholar]
- 43.Schneidman-Duhovny D, Inbar Y, Nussinov R, Wolfson HJ. PatchDock and SymmDock: servers for rigid and symmetric docking. Nucleic Acids Res. 2005;33(Web Server issue):W363–7. doi: 10.1093/nar/gki481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang C, Vasmatzis G, Cornette JL, DeLisi C. Determination of atomic desolvation energies from the structures of crystallized proteins. J Mol Biol. 1997;267(3):707–26. doi: 10.1006/jmbi.1996.0859. [DOI] [PubMed] [Google Scholar]
- 45.Andrusier N, Nussinov R, Wolfson HJ. FireDock: fast interaction refinement in molecular docking. Proteins. 2007;69(1):139–59. doi: 10.1002/prot.21495. [DOI] [PubMed] [Google Scholar]
- 46.Porollo A, Meller J. Prediction-based fingerprints of protein-protein interactions. Proteins. 2007;66(3):630–45. doi: 10.1002/prot.21248. [DOI] [PubMed] [Google Scholar]
- 47.Han JH, Kerrison N, Chothia C, Teichmann SA. Divergence of interdomain geometry in two-domain proteins. Structure. 2006;14(5):935–45. doi: 10.1016/j.str.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 48.Varfolomeev SD, Uporov IV, Fedorov EV. Bioinformatics and molecular mode-ling in chemical enzymology. Active sites of hydrolases. Biochemistry (Mosc) 2002;67(10):1099–108. doi: 10.1023/a:1020907122341. [DOI] [PubMed] [Google Scholar]
- 49.Verma D, Jacobs DJ, Livesay DR. Changes in lysozyme flexibility upon mutation are frequent, large and long-ranged. PLoS Comput Biol. 2012;8(3):e1002409. doi: 10.1371/journal.pcbi.1002409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H, Zhang T, Chen K, Shen S, Ruan J, Kurgan L. On the relation between residue flexibility and local solvent accessibility in proteins. Proteins. 2009;76(3):617–36. doi: 10.1002/prot.22375. [DOI] [PubMed] [Google Scholar]
- 51.Pace CN, Shirley BA, McNutt M, Gajiwala K. Forces contributing to the conformational stability of proteins. FASEB J. 1996;10(1):75–83. doi: 10.1096/fasebj.10.1.8566551. [DOI] [PubMed] [Google Scholar]
- 52.Hubbard RE, Kamran Haider M. eLS. John Wiley and Sons, Ltd; 2001. [Accessed May 6, 2015]. Hydrogen bonds in proteins: role and strength [Internet] Available at: http://onlinelibrary.wiley.com/doi/10.1002/9780470015902.a0003011.pub2/abstract. [Google Scholar]
- 53.Shirley BA. Protein Stability and Folding: Theory and Practice. Totowa, NJ: Humana Press; 1995. [Google Scholar]
- 54.Chothia C, Finkelstein AV. The classification and origins of protein folding patterns. Annu Rev Biochem. 1990;59(1):1007–35. doi: 10.1146/annurev.bi.59.070190.005043. [DOI] [PubMed] [Google Scholar]
- 55.Steiner T, Koellner G. Hydrogen bonds with π-acceptors in proteins: frequencies and role in stabilizing local 3D structures. J Mol Biol. 2001;305(3):535–57. doi: 10.1006/jmbi.2000.4301. [DOI] [PubMed] [Google Scholar]
- 56.1000 Genomes Project Consortium. Abecasis GR, Altshuler D, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure A. Diagrammatic representation of the computational approach to distinguish driver mutations.