Abstract
In nature, macroscopic excitation waves1,2 are found in a diverse range of settings including chemical reactions, metal rust, yeast, amoeba and the heart and brain. In the case of living biological tissue, the spatiotemporal patterns formed by these excitation waves are different in healthy and diseased states2,3. Current electrical and pharmacological methods for wave modulation lack the spatiotemporal precision needed to control these patterns. Optical methods have the potential to overcome these limitations, but to date have only been demonstrated in simple systems, such as the Belousov–Zhabotinsky chemical reaction4. Here, we combine dye-free optical imaging with optogenetic actuation to achieve dynamic control of cardiac excitation waves. Illumination with patterned light is demonstrated to optically control the direction, speed and spiral chirality of such waves in cardiac tissue. This all-optical approach offers a new experimental platform for the study and control of pattern formation in complex biological excitable systems.
Heart cells form a dense, well-coupled excitable medium that displays macroscopic propagating waves of activity with characteristic space scales that are orders of magnitude larger than the cells themselves. Wave dynamics and the resultant spatiotemporal patterns are central to the heart’s function. Planar waves, emanating from a central pacemaking source, act to synchronize contraction during the normal heart beat. However, aberrant re-entrant waves, with their characteristic spiral morphology, re-excite the tissue rapidly and underlie potentially deadly tachycardias and fibrillation.
Optical mapping with voltage-sensitive dyes3 has helped confirm the existence of planar and spiral waves in cardiac tissue. Beyond this observation, however, a comparable means for the manipulation of excitation waves is lacking. The ability to control wave shape, direction and velocity can provide mechanistic insights into wave dynamics and help elucidate the basis of a range of excitable tissue disorders. Wave control can also offer new research targets. For example, because spatiotemporal variations in the conduction velocity are potent triggers of re-entrant waves5, dynamic control of cardiac wave velocity can shed light on spiral wave initiation, as well as on wave stability as a function of tissue size6 and spatial heterogeneity7. From a theoretical point of view, an optical approach for perturbing excitation waves would have the spatiotemporal precision to control wave dynamics. Perturbation of excitation waves by light has been demonstrated in photosensitive versions of the Belousov–Zhabotinsky (BZ) chemical reaction4 and to some degree in light-sensitive Dictyostelium8 amoeba colonies. However, existing tools for the manipulation of excitation waves in living mammalian tissue are very limited. In the heart, waves can be initiated and terminated crudely by electrical or pharmacological means that lack spatial and/or temporal precision, and more recently, optical pacing has been demonstrated9, where excitation is triggered by sharp local temperature gradients (Supplementary Table 1). None of these approaches has been shown to provide fine wave control.
Optogenetics, the inscription of light sensitivity in mammalian tissues through the genetic expression of microbial opsins10,11, holds promise to enable fine spatiotemporal targeting of excitation waves. The ability to optically address specific cells and cell types has been leveraged in the neurosciences to track and control neural circuits12,13, but its use in the cardiac field is only in its infancy14, with early reports showing its utility in optical initiation15,16 and termination of cardiac electrical activity17,18. Fine wave control will benefit from the combined power of optical imaging and spectrally compatible high-resolution optogenetic perturbation techniques14,19. However, to date, optogenetics has exclusively focused on perturbing cell-level properties, while precise control of macroscopic waves has not been demonstrated in neural or cardiac preparations.
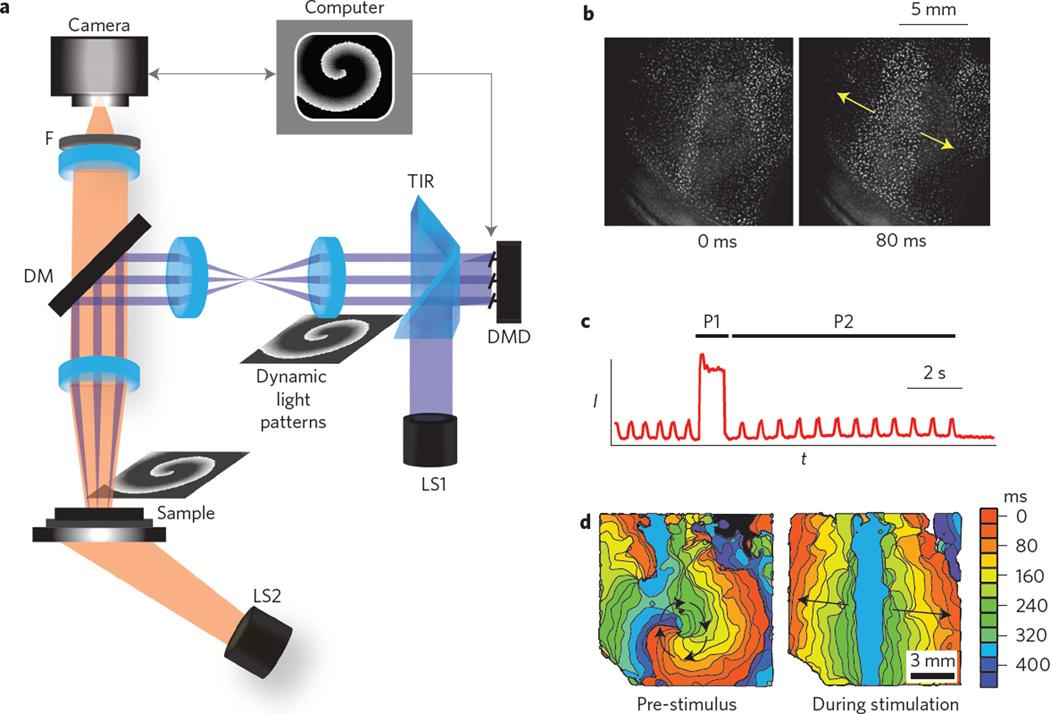
In this Letter, we demonstrate control of wave propagation and pattern formation in a biological excitable medium by combining dye-free macroscopic optical imaging of excitation with high-resolution dynamic optogenetic perturbation. Cardiac monolayers of coupled primary cardiomyocytes, known to support classic travelling excitation waves20,21, were genetically modified to uniformly express channelrhodopsins, adding optical responsiveness without otherwise altering innate functionality22. We used dynamic illumination patterns for optogenetic wave control in the cardiomyocyte monolayers. A user-generated sequence of binary images was uploaded to a digital micromirror device (DMD). Patterned excitation light was generated by reflecting collimated light from a blue light-emitting diode (LED) using a total-internal-reflection (TIR) prism to the DMD during image playback. Light from mirrors in the ‘on’ position was steered by a dichroic mirror and re-imaged on the sample by a ×1, 0.25 numerical aperture (NA) objective for optical stimulation (Fig. 1a). Macroscopic excitation–contraction waves were monitored over a large field of view using a dye-free (non-fluorescent) optical imaging technique. Interference patterns generated by the interaction of light from an off-axis, partially coherent LED source with the sample (Fig. 1a) allowed for direct visualization of wave activity during optogenetic stimulation (Fig. 1b; Supplementary Figs 4 and 5). Recorded data could be used to generate activity versus time plots (Fig. 1c) and activation maps (Fig. 1d), highlighting travelling wavefronts. The advantages of the imaging modality used here and in other dye-free implementations23,24 (Supplementary Table 1) include the use of low light levels, low-NA optics and the possibility for non-invasive long-term monitoring. In addition, the imaging modality is easily integrated with optogenetic stimulation and with other fluorescence-based imaging techniques (Supplementary Fig. 4) because any visible wavelength can be used to capture wave activity. The utility of the developed all-optical system is illustrated here by three proof-of-concept examples of the optical control of the wave properties: direction, speed and chirality.
Figure 1. All-optical system for control of wave dynamics in biological media.
a, Experimental set-up, including an actuation light source (LS1: 10WLED, 460 nm), a total-internal-reflection prism (TIR) and a computer-controlled digital micromirror device (DMD). Generated light patterns are projected via lenses and a dichroic mirror (DM, 510 nm) to the biological sample. A second light source (LS2: white LED, bandpass filtered at 580 ± 20 nm) provides oblique trans-illumination for dye-free imaging onto a scientific complementary metal-oxide semiconductor (sCMOS) camera through an objective lens (×1, 0.25 NA) and a long-pass emission filter (F, >580 nm). b, Example of minimally filtered images in response to optical line stimulation in cardiac monolayers. c,d, Intensity (I) versus time (t) trace from a single pixel (c) and activation maps (d) showing ongoing spontaneous activity (a spiral) pre-stimulus, terminated by a strong global optical stimulation (P1) and followed by periodic optical stimulation (P2) by a line stimulus. For b–d, see also Supplementary Movies 1 and 2.
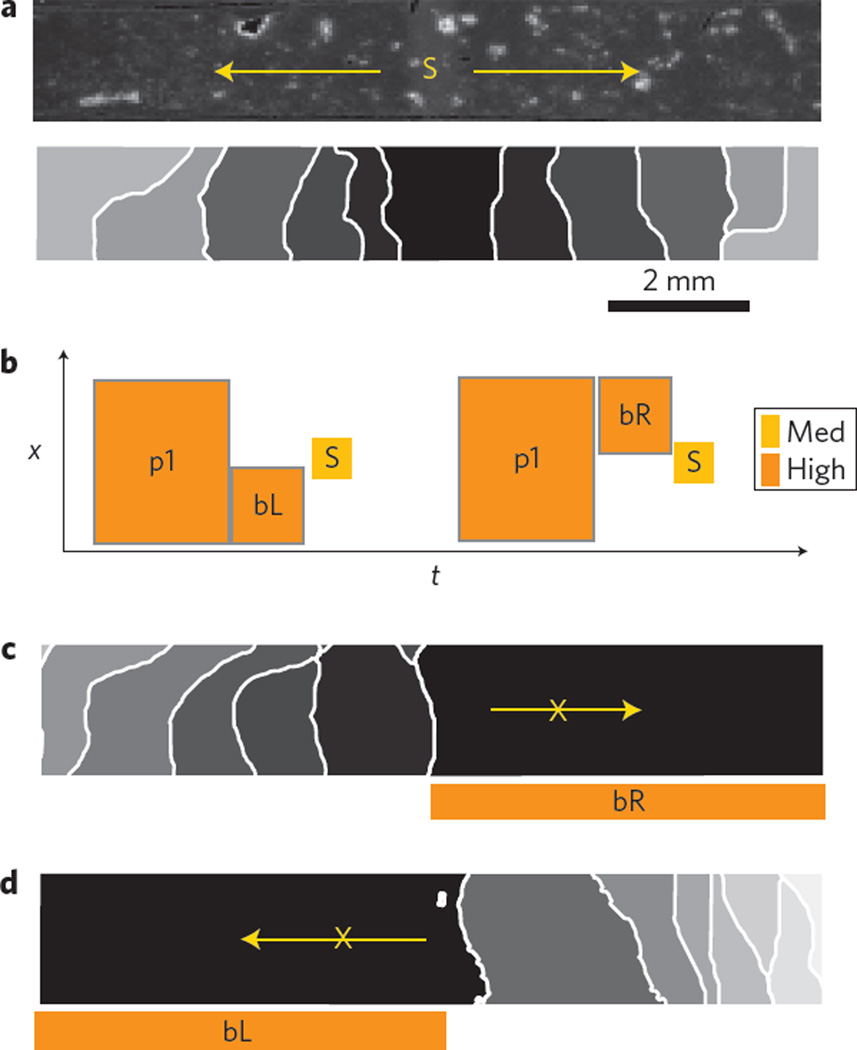
In homogeneous cardiac tissue, a stimulus triggers uniform wave propagation in all directions. However, certain conditions can temporarily block the wave in a particular direction. Such a unidirectional block enables the wave to curl and circle back on itself, leading to re-entry and lethal cardiac arrhythmias5,25. Experimentally, this unidirectional block can be recreated by generating a wavefront that interacts with the wake of a pre-existing wave, but reliable results are difficult to obtain with conventional electrical stimulation. Here, we use light to generate a unidirectional block of wave propagation in a genetically modified cardiac monolayer without the need for interaction with a pre-existing wavefront. Optogenetic stimulation can hold the affected cells in a depolarized (non-excitable) state, and timed release from this state marks the start of a refractory period, after which the cells are excitable again. Figure 2 illustrates that timed, space-defined removal of light, after a global depolarizing optical clamp, results in the control of tissue refractoriness and selection of the exact time and location of the unidirectional block upon new stimulation. Bidirectional propagation is triggered in Fig. 2b, and right or left unidirectional blocks are demonstrated in Fig. 2c,d using asymmetric light release.
Figure 2. Optical control of cardiac wave direction.
a, Sample and applied light stimulus S, inducing bidirectional propagation. b–d, Schematic representation (b) of the applied light protocol in space and time (x–t) with pre-conditioning stimuli p1 and blocking stimuli bL and bR to set tissue refractoriness before stimulus S, resulting in a right-side (c) or a left-side (d) unidirectional block. Here p1 is 350 ms, bR and bL are 50 ms and S is 10 ms. Irradiance levels are high (1,200 W m−2) and medium (700 W m−2). Activation maps show isochrones at 100 ms spacing.
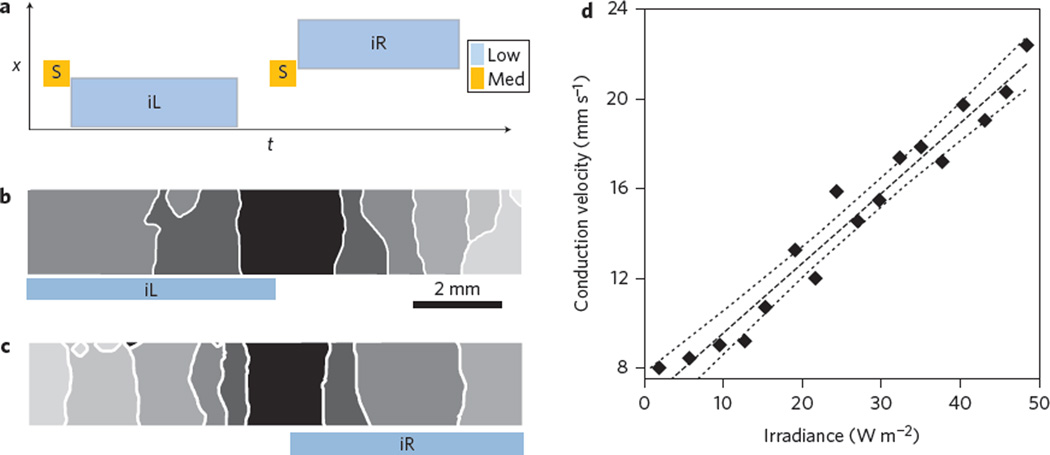
Wave conduction velocity plays a key role in excitable media dynamics as it directly influences the spatial scale that can accommodate a re-entrant circuit6. Experimentally, only crude pharmacological tools exist to influence cardiac conduction velocity (largely defined by cell–cell coupling)26. Here, we report that low light application can speed up propagation, as dosed subthreshold depolarization brings cells closer to the threshold for excitation and yields shorter activation times. The conduction velocity can be increased in user-defined regions of the tissue by applying low light ahead of a triggered wave (Fig. 3). The conduction velocity is increased first to the left (Fig. 3b) and then to the right (Fig. 3c) without affecting the propagation properties in un-illuminated regions of the tissue. We were able to precisely control the conduction velocity by varying the light levels. A linear velocity increase was demonstrated over a wide range of light intensities corresponding to conduction velocities ranging from 10 to 30 mm s−1 (Fig. 3d; Supplementary Fig. 6).
Figure 3. Optical control of cardiac wave conduction velocity.
a, Schematic of applied optical stimulation protocol in space (x) and time (t). b,c, Activation maps of controlled left-side (b) and right-side (c) increase of conduction velocity by light, indicated by the larger spacing of the isochrones. d, Relationship between conduction velocity and irradiance, with linear regression best fit (dashed line) and 95% confidence intervals (dotted lines). Results from nine independent experiments are shown in Supplementary Fig. 6. iR and iL are 500 ms. Isochrones are 100 ms apart. Irradiance levels for the stimulating pulse (‘Med’ in a) are 700 W m−2, with low irradiance varying between 0 and 80 W m−2 applied as shown in b and c. For b and c, see also Supplementary Movie 3.
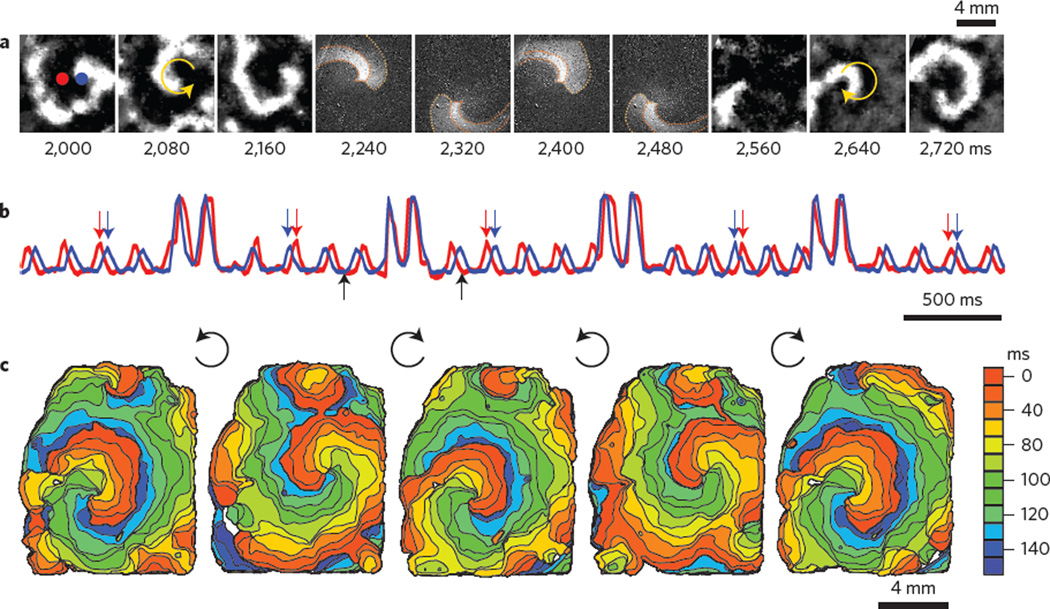
Finally, spiral waves are a prototypical example of self-organization in distributed excitable media1,2, and are observed in autocatalytic chemical reactions, yeast, amoeba colonies, heart, cortical and retinal preparations. Experimental control of spiral wave dynamics is an important tool for understanding pattern formation in excitable media. Recently, global optogenetic stimulation was used to abolish cardiac spiral waves18,27. Here, we demonstrate a finer level of optical control, exemplified by light-controlled reversal of cardiac spiral wave chirality. The spiral chirality (direction of rotation) is a fundamental property that affects how the wave interacts with other wavefronts as well as the underlying medium28,29. To our knowledge, the controlled modulation of spiral wave chirality has not been demonstrated experimentally in any excitable system, although theoretical ideas to do so have been proposed29. To reverse the chirality of an ongoing spiral wave, we transiently imposed a computer-generated (using a cellular automaton model) counter-rotating spiral wave of shaped light with a slightly higher frequency of rotation compared to the native spiral (Fig. 4a). Within one to two rotations, the imposed wave effectively overwrites the existing spiral. The shaped refractory gradient left after light removal perpetuates the spiral by allowing the light-triggered wave to persist and re-enter along the path of the imposed spiral. Interestingly, the spirals drift over a few rotations to a preferred location that is different for clockwise and anticlockwise spirals, and the latter are faster (18%), probably indicating asymmetry in the underlying tissue microstructures. The chirality control was robustly deployed repeatedly, resulting in the same outcome, independent of the phase of the ongoing spiral (Fig. 4b,c). Chirality reversal was repeated 16 times in three independent preparations.
Figure 4. Optical control of spiral wave chirality in cardiac monolayer.
a, Snapshots from an ongoing anticlockwise spiral wave (frames 2,000–2,160), an optically applied computer-generated clockwise spiral wave (frames 2,240–2,480) and the persisting spiral wave post-chirality reversal (frames 2,560–2,720). b, Activity traces from the red and blue pixels indicated in a, showing four light-controlled chirality reversals. Computer-generated spirals were imposed at random phases for less than two rotations, as seen in the four higher-intensity transients. Black arrows indicate the time period presented in a. Red and blue arrows indicate the switch of order of excitation at the chosen locations due to chirality reversal. c, Activation maps for the initial spiral wave and the four resultant spirals after each of the chirality reversals. See Supplementary Movies 4 and 5 for minimally processed and colourized data, respectively (for more details see Supplementary Fig. 3).
In summary, we have presented a new all-optical framework for the modulation of cardiac excitation waves, in ways that are impossible using conventional techniques. Our current implementation shows wavefront control in thin, genetically modified tissues, but the proposed approach is applicable to a wide range of new experimental targets, providing insights into pattern formation in the intact heart. Indeed, we demonstrate control of waves in cardiac tissue at a level normally associated with computer models or simpler experimental systems such as photosensitive chemical reactions4. Furthermore, because diverse excitable media display similar macroscopic dynamics, insights from the cardiac experiments described here will be relevant to a broad range of biological and chemical reaction–diffusion systems. The ability to precisely control light will enable new research on pattern formation in complex biological excitable media.
Methods
Biological excitable medium: cardiac monolayer cultures
Our experimental goal was to use optogenetic tools to modulate a biological excitable medium that can support a wide range of spatiotemporal patterns. We used an established experimental model of cardiac syncytium—a monolayer culture of primary neonatal rat ventricular myocytes. Cardiomyocytes were isolated from Sprague–Dawley rat pups. Cells were cultured in high serum conditions at moderate plating densities and imaged while in culture medium to generate isotropic preparations that spontaneously display complex waves20,30,31.
Hearts were isolated from neonatal Sprague–Dawley rat pups (P3–P4) according to Schedule 1 of the UK Home Office Animals Scientific Procedures Act (1986). The atria were dissected and discarded, and the ventricles were cut into small 0.1–0.2 mm2 blocks and subjected to enzymatic digestion in trypsin (3 h at 4 °C on a shaker, 1 mg ml−1, Worthington), followed by series of collagenase digestions (type IV, 1 mg ml−1, Sigma Aldrich). Following trypsinization, the tissue was rinsed with cold Hank’s buffered salt solution (HBSS, Sigma Aldrich) for 3 min. After the wash, 5 ml collagenase was added to the tissue and stirred in a shaker bath at 37 °C for 2 min. The first supernatant was discarded and the process was repeated. Following each collagenase treatment, the tissue was gently agitated with a wide-tip pipette. The supernatant containing myocytes was transferred to a conical tube containing 3 ml HBSS and stored on ice. This collagenase digestion step was repeated several times. The tubes were balanced using HBSS and centrifuged at 1,000 r.p.m. for 6–8 min. Following centrifugation, the supernatant was discarded and HBSS was added to the pellet and gently agitated. The cell suspension was then strained using a 0.22 µm sterile cell strainer. Centrifugation was repeated once again (1,000 r.p.m., 6 min). The supernatant was then discarded and culture medium was added. The isolated cells were pre-plated in an incubator (37 °C, 5% CO2) for an hour to allow fibroblasts to attach. The ventricular myocytes in the supernatant were then carefully removed from the dish and a cell count was performed using a haemocytometer and trypan blue. The myocytes were plated on 35 mm poly-lysine-coated Petri dishes (bio coat poly-d-lysine, 35 mm Petri plates, Corning) at a density of 700,000–1,000,000 cells (per 35 mm Petri dish) in culture medium (68% DMEM, 17% M199, 10% horse serum, 5% FBS and 1% penicillin/streptomycin, all from Sigma Aldrich).
Optogenetic modification of cardiomyocytes with AdChR2
Adenoviral vectors containing the transgene for hChR2(H134R)-eYFP were prepared in collaboration with the Stony Brook University Stem Cell Centre based on the expression cassette of the plasmid pcDNA3.1/hChR2(H134R)-eYFP (#20940; Addgene)22.
Cardiomyocytes were infected using a previously published method22 and an optimized dose of adenovirus (multiplicity of infection, MOI 15–25) at 37 °C for 2 h. The MOI was optimized during preliminary experiments based on desired transgene expression (by the eYFP reporter) and minimal cell death (by propidium iodide staining). At this MOI, >95% of the myocytes expressed ChR2 within 48 h, as confirmed by eYFP reporter visualization, without an increase in cell death compared with control22. Following infection, the culture medium was gently removed and fresh culture medium was added. The medium was initially changed after 12 h, and then replaced every 48 h. Functional measurements were performed on the samples from day 9 onwards.
Dye-free imaging
We developed a dye-free imaging system in which off-axis oblique illumination, a commonly used technique for enhancing contrast in microscopic samples, was used to obtain high-contrast high-resolution images of macroscopic wave propagation (field of view, 1–4 cm2). This technique relies on cellular excitation–contraction induced changes in the optical properties of the tissue. The imaging system uses a semi-coherent light source (a narrowband LED) that has a coherence length comparable to the axial thickness of the sample. An Olympus MVX10 macroscope and Andor Neo sCMOS camera (2,560 × 2,160 pixels, 6.5 µm pixels), fitted with a 580 nm longpass filter (Fig. 1a, ‘F’), was used to record macroscopic wave patterns from a sample illuminated by a white LED fitted with a bandpass filter (580 ± 20 nm, Fig. 1a, ‘LS2’). LEDs were supplied by Cairn Research and filters were supplied by Chroma Technology. Experiments were carried out in an Okolab (Indigo Scientific) stage incubation chamber controlled for heat (33–37 °C), CO2 (5%) and humidity. Samples were allowed to equilibrate in these conditions for 20 min before commencing recording. The dye-free imaging enabled re-examination of samples over multiple days, and their viability and consistent optical responsiveness was confirmed. Frame rates were varied between 25 and 100 fps depending on the observed wave velocity and the desired record duration.
Computer-controlled dynamic patterned-light stimulation
Dynamic space–time patterns (movies) for optical stimulation were generated using a computer-controlled DMD from Vialux. Both the DMD and the recording camera(s) were controlled using custom-written software in Java and Python. Sequences of binary images were downloaded to the Vialux DMD unit and projected onto the sample. Precise control of the light level was achieved by controlling the LED current and by simulating grey levels by rapidly toggling the DMD state. For example, the fine steps in light level for Fig. 3d were achieved by projecting patterned light every fifth frame with a duty cycle of 500 µs and increasing the LED current between 0 and 25% maximum intensity. In Fig. 4, sequences of spiral wave patterns were generated by simulating clockwise and anticlockwise rotating spiral waves in a cellular automaton excitable media (CA) model31. The speed of rotation of the projected spiral was controlled by varying the time between projected frames.
Supplementary Material
Acknowledgments
The authors thank T. Wilson, H. Bien and S. Aslam for discussions and technical assistance, and E. Mann for use of the DMD projector (Royal Society Grant RG110135). G.B. acknowledges support from the BHF Centre of Research Excellence, Oxford (RE/08/004). R.A.B.B. holds an EPSRC Developing Leaders Grant, a Goodger award, a Winston Churchill Fellowship and Paul Nurse Junior Research Fellowship (Linacre College, Oxford). J.T. acknowledges support from the Bakala Foundation. This work was supported by MR/K015877/1 (G.B.), NIH R01 HL111649 and NSF-Biophotonics grant 1511353 (E.E.), as well as a NYSTEM grant C026716 to the Stony Brook Stem Cell Centre.
Footnotes
Author contributions
G.B. and E.E. initiated the project and provided guidance. R.A.B.B. and G.B. performed the experiments. G.B. wrote the software to collect and analyse the data. C.M.A. and E.E. developed and provided biological materials and guidance on the optogenetic manipulations. R.A.B.B., A.C., J.T. and A.K. helped with data interpretation and figure preparation. G.B. and E.E. wrote the manuscript with contributions from all authors. All authors were involved in analysis of the results and revision of the manuscript.
Supplementary information is available in the online version of the paper.
Competing financial interests
The authors declare no competing financial interests.
References
- 1.Krinsky VI. Self-Organization: Autowaves and Structures Far From Equilibrium. Springer; 1984. [Google Scholar]
- 2.Winfree AT. The Geometry of Biological Time. Vol. 12. Springer Science & Business Media; 2001. [Google Scholar]
- 3.Davidenko J, Pertsov A, Salomonsz R, Baxter W, Jalife J. Stationary and drifting spiral waves of excitation in isolated cardiac muscle. Nature. 1992;355:349–351. doi: 10.1038/355349a0. [DOI] [PubMed] [Google Scholar]
- 4.Steinbock O, Müller SC. Light-controlled anchoring of meandering spiral waves. Phys. Rev. E. 1993;47:1506. doi: 10.1103/physreve.47.1506. [DOI] [PubMed] [Google Scholar]
- 5.Mines G. On circulating excitations in heart muscles and their possible relation to tachycardia and fibrillation. Trans. R. Soc. Canada. 1914:43–55. [Google Scholar]
- 6.Garrey WE. The nature of fibrillary contraction of the heart: its relation to tissue mass and form. Am. J. Physiol. 1914;33:397–414. [Google Scholar]
- 7.Shajahan TK, Nayak AR, Pandit R. Spiral-wave turbulence and its control in the presence of inhomogeneities in four mathematical models of cardiac tissue. PLoS ONE. 2009;4:e4738. doi: 10.1371/journal.pone.0004738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miura K, Siegert F. Light affects cAMP signaling and cell movement activity in Dictyostelium discoideum. Proc. Natl Acad. Sci. USA. 2000;97:2111–2116. doi: 10.1073/pnas.040554497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jenkins MW, et al. Optical pacing of the embryonic heart. Nature Photon. 2010;4:623–626. doi: 10.1038/nphoton.2010.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nature Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- 11.Nagel G, et al. Channelrhodopsin-2, a directly light-gated cation-selective membrane channel. Proc. Natl Acad. Sci. USA. 2003;100:13940–13945. doi: 10.1073/pnas.1936192100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yizhar O, Fenno LE, Davidson TJ, Mogri M, Deisseroth K. Optogenetics in neural systems. Neuron. 2011;71:9–34. doi: 10.1016/j.neuron.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Lima SQ, Miesenböck G. Remote control of behavior through genetically targeted photostimulation of neurons. Cell. 2005;121:141–152. doi: 10.1016/j.cell.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Ambrosi CM, Klimas A, Yu J, Entcheva E. Cardiac applications of optogenetics. Prog. Biophys. Mol. Biol. 2014;115:294–304. doi: 10.1016/j.pbiomolbio.2014.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bruegmann T, et al. Optogenetic control of heart muscle in vitro and in vivo. Nature Methods. 2010;7:897–900. doi: 10.1038/nmeth.1512. [DOI] [PubMed] [Google Scholar]
- 16.Jia Z, et al. Stimulating cardiac muscle by light cardiac optogenetics by cell delivery. Circ. Arrhythmia Electrophysiol. 2011;4:753–760. doi: 10.1161/CIRCEP.111.964247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arrenberg AB, Stainier DYR, Baier H, Huisken J. Optogenetic control of cardiac function. Science. 2010;330:971–974. doi: 10.1126/science.1195929. [DOI] [PubMed] [Google Scholar]
- 18.Bingen BO, et al. Light-induced termination of spiral wave arrhythmias by optogenetic engineering of atrial cardiomyocytes. Cardiovasc. Res. 2014;104:194–205. doi: 10.1093/cvr/cvu179. [DOI] [PubMed] [Google Scholar]
- 19.Hochbaum DR, et al. All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins. Nature Methods. 2014;11:825–833. doi: 10.1038/nmeth.3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bub G, Glass L, Publicover NG, Shrier A. Bursting calcium rotors in cultured cardiac myocyte monolayers. Proc. Natl Acad. Sci. USA. 1998;95:10283–10287. doi: 10.1073/pnas.95.17.10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tung L, Zhang Y. Optical imaging of arrhythmias in tissue culture. J. Electrocardiol. 2006;39:S2–S6. doi: 10.1016/j.jelectrocard.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 22.Ambrosi CM, Entcheva E. Optogenetic control of cardiomyocytes via viral delivery. Methods Mol. Biol. 2014;1181:215–228. doi: 10.1007/978-1-4939-1047-2_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hwang S-M, Yea K-H, Lee KJ. Regular and alternant spiral waves of contractile motion on rat ventricle cell cultures. Phys. Rev. Lett. 2004;92:198103. doi: 10.1103/PhysRevLett.92.198103. [DOI] [PubMed] [Google Scholar]
- 24.Hwang S-M, Kim TY, Lee KJ. Complex-periodic spiral waves in confluent cardiac cell cultures induced by localized inhomogeneities. Proc. Natl Acad. Sci. USA. 2005;102:10363–10368. doi: 10.1073/pnas.0501539102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wiener N, Rosenblueth A. The mathematical formulation of the problem of conduction of impulses in a network of connected excitable elements, specifically in cardiac muscle. Arch. del Inst. Cardiol. México. 1946;16:205–265. [PubMed] [Google Scholar]
- 26.Jia Z, Bien H, Shiferaw Y, Entcheva E. Cardiac cellular coupling and the spread of early instabilities in intracellular Ca2+ Biophys. J. 2012;102:1294–1302. doi: 10.1016/j.bpj.2012.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ambrosi CM, Williams JC, Entcheva E. Optogenetic modulation of pacemaking, arrhythmia generation, and inhibition with sustained (non-pulsed) light. Circulation. 2014;130:A19856. [Google Scholar]
- 28.Quail T, Shrier A, Glass L. Spatial symmetry breaking determines spiral wave chirality. Phys. Rev. Lett. 2014;113:158101. doi: 10.1103/PhysRevLett.113.158101. [DOI] [PubMed] [Google Scholar]
- 29.Li B-W, Cai M-C, Zhang H, Panfilov AV, Dierckx H. Chiral selection and frequency response of spiral waves in reaction–diffusion systems under a chiral electric field. J. Chem. Phys. 2014;140:184901. doi: 10.1063/1.4874645. [DOI] [PubMed] [Google Scholar]
References
- 30.Bub G, Shrier A, Glass L. Spiral wave generation in heterogeneous excitable media. Phys. Rev. Lett. 2002;88:058101. doi: 10.1103/PhysRevLett.88.058101. [DOI] [PubMed] [Google Scholar]
- 31.Bub G, Shrier A, Glass L. Global organization of dynamics in oscillatory heterogeneous excitable media. Phys. Rev. Lett. 2005;94:028105. doi: 10.1103/PhysRevLett.94.028105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.