Abstract
Noncoding RNAs (ncRNAs) comprise an important class of regulatory molecules that mediate a vast array of biological processes. This broad functional capacity has also facilitated the design of artificial ncRNAs with novel functions. To further investigate and harness these capabilities, we developed CRISPR-Display (“CRISP-Disp”), a targeted localization method that uses Sp. Cas9 to deploy large RNA cargos to DNA loci. We demonstrate that exogenous RNA domains can be functionally appended onto the CRISPR scaffold at multiple insertion points, allowing the construction of Cas9 complexes with protein-binding cassettes, artificial aptamers, pools of random sequences, and RNAs up to 4.8 kilobases in length, including natural lncRNAs. Unlike most existing CRISPR methods, CRISP-Disp allows simultaneous multiplexing of distinct functions at multiple targets, limited only by the number of available functional RNA motifs. We anticipate that this technology will provide a powerful method with which to ectopically localize functional RNAs and ribonucleoprotein (RNP) complexes at specified genomic loci.
Introduction
Noncoding RNAs (ncRNAs) are central components of diverse and fundamental processes in all kingdoms of life1. In eukaryotes, many well-established ncRNAs, and a number of newly identified mammalian long ncRNAs, are thought to help initiate or maintain regulatory processes within the nucleus1–3. However, mechanistic dissection of these putative nuclear regulators in vivo is often technically limited. For example, established knock-in and knockout strategies4, 5 lack the throughput required for high-resolution structure/function analysis, and cannot easily separate roles performed by an RNA transcript from those performed by a functional DNA element or by a cryptically encoded peptide5, 6.
Therefore, an experimental method that post-transcriptionally relocates a ncRNA transcript to an ectopic site would be an important tool for the study of natural ncRNA function. In addition, this method could provide a powerful engine for synthetic biology. Many RNA domains–both natural and artificial–have been adapted as components in synthetic regulators, reporters and scaffolds7–13. Targeting such RNA devices to specific DNA loci would enable a wide range of novel synthetic biological methods.
Towards these goals, we aimed to develop a general ncRNA ectopic localization system. We reasoned that this could be achieved using an artificial protein “conduit” that is programmed to bind a ncRNA and target it to specific DNA loci. (Supplementary Fig. 1a). A potentially powerful source for such a conduit is the S. pyogenes Cas9 nuclease (Sp. Cas9), an extremely high-affinity, programmable DNA-binding protein isolated from a type II CRISPR-associated system14–16. In a multitude of CRISPR-based biotechnology applications17–22, the guide that programs Cas9 DNA-targeting is presented in a so-called sgRNA, wherein the two natural Cas9 RNA cofactors (crRNA and tracrRNA)14, 15 are fused via an engineered loop (Supplementary Fig. 1b). Yet, despite recent work dissecting the determinants of Cas9 RNA recognition23–26, it remained unclear if and where large, structured RNA domains could be implanted within CRISPR complexes while maintaining RNA-directed localization.
Here, we demonstrate that a nuclease-deficient Sp. dCas9 mutant, “dCas9” (Refs. 17, 27–29)) can be co-opted to deploy a large RNA cargo to targeted DNA loci by directly linking that cargo to the sgRNA. We term this strategy, in which RNA domains are displayed on dCas9, CRISPR-Display, or “CRISP-Disp.” With the appropriate expression system and insertion point, CRISP-Disp does not appear inherently limited by the size or sequence composition of its RNA cargo. This allows dCas9 complexes to be functionalized with structured RNA domains, natural lncRNAs several kb in length, artificial RNA modules and pools of random sequences. Furthermore, these RNA-based functions can be simultaneously multiplexed using a shared pool of dCas9. This work provides initial insights into the general utility of CRISP-Disp for both the study of natural ncRNAs and the construction of novel RNA-based devices.
Results
Adapting CRISPR-Cas9 as an RNA Display Device
To assess potential ncRNA localization methods in vivo, we first implemented a highly sensitive reporter system in HEK293FT cells. In this system, the efficacy of an ectopically targeted regulator (protein or RNP) is reported by luciferase and fluorescent protein expression. RNA-targeting strategies were surveyed using both transiently transfected and stably integrated reporters (Supplementary Fig. 1c–f, Supplementary Table 1)27, 30.
In our initial strategy, the RNA-deploying “conduits” targeted to these reporters were based upon transcription activator-like effectors (TALEs)30. However, we were unable to coax TALES into recruiting ncRNAs to a chromatin-integrated locus (Supplementary Figs. 2–3). We ascribed this problem to the separable DNA- and RNA-binding activities of our conduit, which could be independently saturated without forming DNA•TALE•RNA ternary complexes (Supplementary Data).
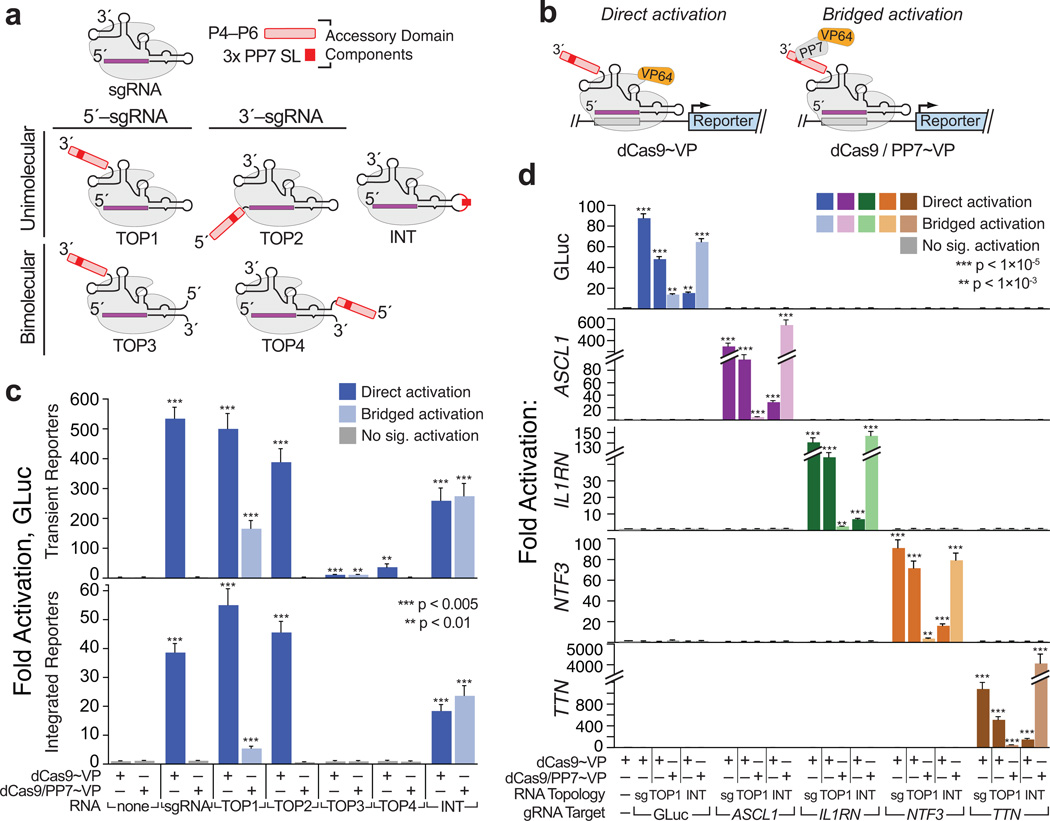
To circumvent this issue, we turned to the S. pyogenes CRISPR-(d)Cas9 system, which intrinsically couples its DNA- and RNA-binding activities (Supplementary Fig. 1b–f)15, 16. However, it was unclear a priori where insertions within the dCas9-bound RNAs would be tolerated, and how large they could be. To examine this, we devised five model cofactor RNAs (TOP1–4; INT) in which structured, 81–250 nt “accessory domains” were appended to the termini of, or internally within the sgRNA or tracrRNA15. Each accessory domain carried a cassette of stem-loops recognized by the PP7 phage coat protein31 (Fig. 1a; Supplementary Fig. 4a–b). At 357 nt, the largest of these constructs adds three-fold more sequence than does the longest modified sgRNA previously reported20, 25, 26.
Figure 1. Large, structured RNA domains can be functionally appended onto the sgRNA scaffold at multiple points.
(a) Design of “TOP” topology constructs. Accessory RNA domains are detailed in (Supplementary Fig. 4a–b); expression constructs in (Supplementary Fig. 1c). (b) Schematics summarizing direct activation (left) and bridged activation (right) assays. (c) Luciferase reporter assays of the five topology constructs. (d) Targeting minimal (“sg”) and expanded (“TOP1” and “INT”) sgRNAs to endogenous loci. ASCL1, IL1RN, NTF3 and TTN. were each targeted using mixed pools of four gRNAs28, 29, 33 (Supplementary Table 2). GLuc activation was measured by luciferase assays; endogenous gene activation was measured using qRT-PCR (Supplementary Table 3). Values are means ± standard deviation, (n=3) Luciferase assays; (n=4) qRT-PCR. Student’s one-tailed t-test relative to cells expressing dCas9~VP alone.
We subjected these RNA chimeras to two variations of our CRISPR transcription activator assay (Fig. 1b). In “direct activation” assays, they were coexpressed with dCas9 fused to the VP64 transcription activator (dCas~VP). In this format, reporter gene activation indicates that the sgRNA variant competently binds and targets dCas9. In “bridged activation” assays, constructs were coexpressed with dCas9 and PP7~VP, a chimera of PP7 and VP64. Bridged activation should only occur if the accessory domain remains intact in the mature dCas9 complexes.
Using transient reporters, we observed direct activation with all five topologies (Fig. 1c, top). However, while the activities of TOP1, TOP2 and INT were reduced less than twofold from that of the minimal sgRNA, TOP3 and TOP4 were ~9–30-fold less proficient. Critically, bridged activation was only observed with TOP1, TOP3 and INT, indicating that these constructs alone retained functional accessory domains in mature dCas9 complexes. These results were corroborated by FACS (Supplementary Fig. 4e) and qualitatively paralleled using integrated reporters (Fig. 1c, bottom). We attributed the lack of TOP2 bridged activation to partial degradation of that construct’s accessory domain, as indicated using RNA Immunoprecipitation (RIP) and qRT–PCR (Supplementary Fig. 4f)20, 25, 32.
Targeting long RNAs to endogenous genomic loci
To illustrate the general applicability of CRISP-Disp, we tested whether results from our reporter system could be recapitulated at endogenous loci. We generated pools of sgRNA, TOP1 and INT constructs targeting the human ASCL1, IL1RN, NTF3 and TTN promoters28, 29, 33 (Supplementary Table 2), and surveyed direct and bridged activation of these genes by qRT–PCR. As predicted, activation of each locus paralleled the results obtained using our GLuc reporter, demonstrating CRISP-Disp enables deployment of large RNA domains to genomic loci (Fig. 1d).
To confirm that dCas9 targeting fidelity is not perturbed by the addition of the accessory RNA domains, we performed mRNA seq from reporter cells expressing dCas9~VP and GLuc-targeting sgRNA, TOP1 and INT constructs. As predicted, changes in global gene expression induced by each RNA construct–a proxy for dCas9 off targeting–were essentially indistinguishable, indicating that TOP1- and INT-like accessory domains do not significantly alter dCas9 fidelity (Supplementary Fig. 5).
CRISP-Disp with “artificial lncRNA” scaffolds
We next sought to engineer CRISP-Disp for use with even longer ncRNAs, which required replacing the conventional RNA Polymerase III (Pol III) promoter used above with an RNA Polymerase II (Pol II) promoter and terminator. We therefore surveyed a variety of standard and noncanonical Pol II expression systems for the ability to generate nuclear-localized CRISP-Disp RNAs de novo (Supplementary Fig. 6–7; Supplementary Discussion). Although such Pol II transcripts were generally less effective than their Pol III-driven counterparts19, expression from several different backbones partially restored their activity (Supplementary Fig. 6b). Surprisingly, one such backbone (“CMV/3´Box”) also enabled bridged activation with TOP2, which was unviable under Pol III expression (Supplementary Fig. 6c–e).
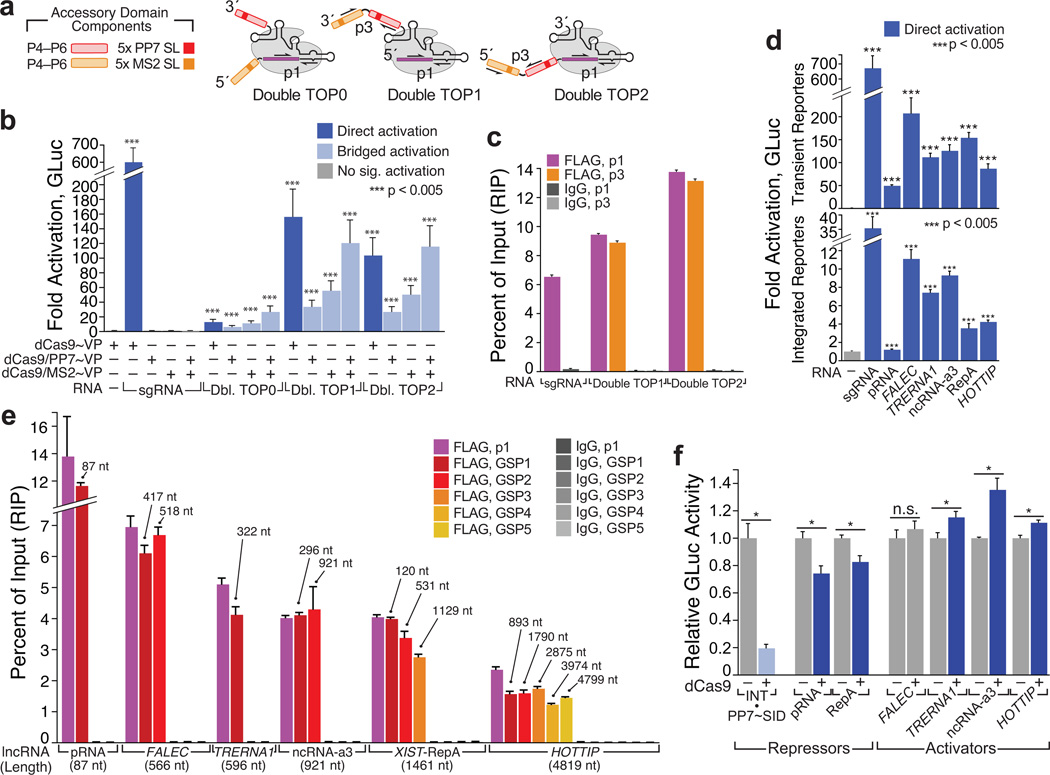
Next, we next attempted to build dCas9 complexes with model RNAs approaching the size of natural lncRNAs. We expanded our CMV/3´-Box TOP1 and TOP2 constructs with a second P4–P6 domain, bearing stem-loops recognized by the MS2 phage coat protein31, to generate a series of “artificial lncRNA” scaffolds with ~650 nt accessory domains (“Double TOP0–1” Fig. 2a, and Supplementary Fig. 4a–d). Although all three of these constructs induced measurable direct activation, Double TOP1–2 were more proficient, with activities nearly rivaling those of their single-domain counterparts (Fig. 2b). Moreover, all three constructs exhibited significant bridged activation. In transient reporter assays, activity monotonically increased upon coexpression with PP7~VP, MS2~VP and both activator proteins, indicating that each RNA construct retained both P4–P6 domains in mature dCas9 complexes (Fig. 2b). A qualitatively similar trend was observed with Double TOP1 using integrated reporters, despite the assay’s limited dynamic range (Supplementary Fig. 8). We furthermore confirmed the integrity of the Double TOP1–2 accessory domains by dCas9 RIP: for each construct, we obtained essentially stoichiometric yields of the sgRNA core and double P4–P6 accessory domains (Fig. 2c).
Figure 2. CRISPR-Display with artificial and natural lncRNAs.
(a) Design of “Double TOP” artificial lncRNA constructs. Accessory domains are detailed in (Supplementary Fig. 4a–d). The sgRNA core “p1” and domain-spanning “p3” qPCR primer pairs are indicated (Supplementary Table 3). (b) Direct and bridged activation assays using Double TOP (“Dbl TOP”) constructs, expressed from the CMV/3´Box backbone (Supplementary Fig. 6a). Transient reporter assays are shown. (c) RIP/qRT–PCR of dCas9•Double TOP1 and dCas9•Double TOP2. (d) sgRNAs appended with a battery of natural lncRNA domains (Supplementary Table 4) form functional complexes with dCas9~VP. Direct activation assays are shown. The minimal “pRNA” stem-loop34 was displayed internally; all other domains were appended on the sgRNA 3´ terminus. RNAs were expressed using the CMV/MASC system (Supplementary Fig. 6a).(e) LncRNA accessory domains remain intact in CRISP-Disp complexes. Immunopurified RNA was analyzed using qPCR primers targeting the sgRNA core (p1) and intervals along the each lncRNA domain (GSP1–GSP5, Supplementary Table 3). Above each primer set, the maximum distance from the sgRNA core is indicated. (f) Transient reporter assays with CRISP-Disp lncRNA constructs. Values are normalized relative to those of control cells expressing each lncRNA alone. For comparison, bridged repression with U6-driven INT, complexed with dCas9 and PP7~SID (Ref.39) is shown (left, light blue). Values are means ± standard deviation, (n=3) Luciferase assays; (n=4) qPCR. Student’s one-tailed t-test relative to cells expressing (b, d) dCas9~VP alone or (f) lncRNA alone. *, p < 0.05.
CRISP-Disp with natural lncRNA domains
A potentially powerful application of CRISP-Disp would be the ectopic localization of natural lncRNAs post-transcriptionally, since reconstitution of lncRNA activity at an ectopic site is unattainable by existing methods5, 6. To demonstrate the plausibility of this approach, we first established that natural long ncRNAs could be incorporated into CRISP-Disp complexes. We generated Pol II-driven TOP1- and INT-like constructs appended with human lncRNA domains: the repressive NoRC-binding pRNA stem-loop34 and Xist A-repeat (“RepA”) domains4, three enhancer-transcribed RNAs (eRNAs, Ref.35) and the 4799 nt transcription activator HOTTIP36 (Supplementary Table 4).
Each construct induced significant direct activation, indicating that each formed functional dCas9-targeting complexes (Fig. 2d). This was further supported by RIP-qPCR: although overall dCas9 complexation appeared to decline monotonically with increasing sgRNA~lncRNA length, in all cases nearly quantitative yields of intact lncRNA domains were recovered, relative to the corresponding sgRNA core (Fig. 2e). We propose that the observed minor variation in construct integrity may be contingent on several factors, including length and RNA structure25.
We next tested if the lncRNA components of CRISP-Disp complexes could themselves regulate our reporter. Encouragingly, most of the lncRNA constructs repressed or activated GLuc expression as would be predicted from existing studies4, 34–36: pRNA and RepA diminished GLuc expression, while two eRNAs (TRERNA1, ncRNA-a3) and HOTTIP induced moderate activation (Fig. 2f). Although these effects were quite modest, they may have been limited by technical aspects of our assay, or by absence of particular factors in HEK293T cells. Regardless, these mild initial results demonstrate the plausibility of larger-scale lncRNA functional studies with CRISP-Disp.
CRISP-Disp with a diverse array of RNA species
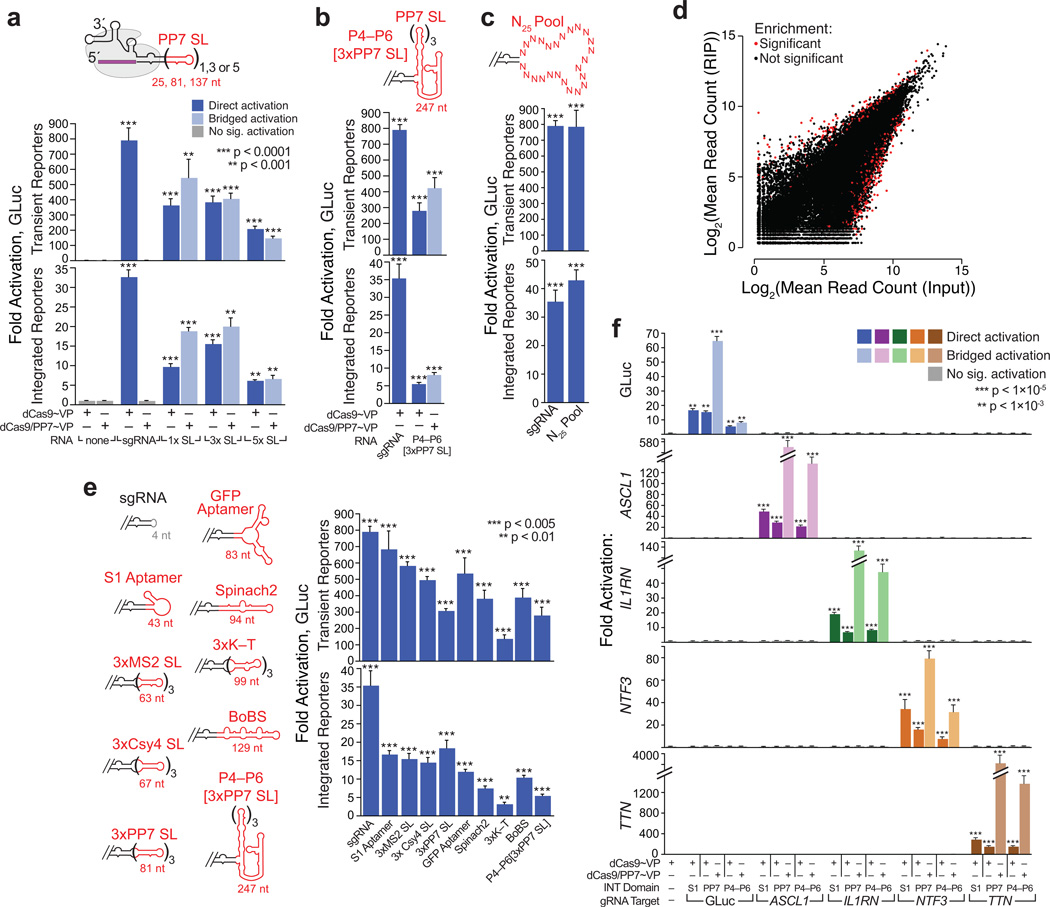
We were particularly intrigued by the proficiency of U6-driven INT (Fig. 1a,c), and reasoned that the sgRNA “engineered loop” might provide a universal insertion point for exogenous RNA domains. To explore this possibility, we first examined the influence of internal insert size. Encouragingly, INT-like constructs bearing 25–137 nt cassettes of PP7 stem-loops each induced robust activation in all assay formats (Fig. 3a), as did a construct bearing a ~250 nt domain equivalent to those of TOP1–4 (Fig. 3b; Supplementary Table 5). Hence, dCas9 can easily accommodate even large, structurally discontinuous inserts within the sgRNA core.
Figure 3. CRISPR-Display with a compendium of structurally diverse RNA domains.
(a) INT insert size has a modest effect on CRISP-Disp efficacy. “x SL” refers to the number of internal PP7 stem-loops. (b) Functional INT inserts can be large and structurally discontinuous with the sgRNA core. Accessory domain is detailed in (Supplementary Fig. 4a–b). ***, p < 0.005 (c) Assembly of functional CRISP-Disp INT complexes is independent of the inserted sequence. Direct activation assays with a pool of ~1.2×106 unique INT–N25 variants (Supplementary Fig. 9). ***, p < 0.005 (d) dCas9 binds to nearly all expressed INT–N25Pool variants. See also (Supplementary Fig. 10) (e) Assembling functional CRISP-Disp complexes bearing an assortment of natural and artificial RNA domains. Left: Cartoons depicting INT constructs. Insert lengths are indicated in red; details in (Supplementary Table 5). BoBS, “Bunch of Baby Spinach,” (Supplementary Fig. 11). Right: direct activation activities, sorted by construct length (f) Targeting INT-like constructs bearing an RNA device (S1 aptamer, “S1”) and a large domain (P4–P6[3xPP7], “P4–P6”) to endogenous loci. Constructs were targeted to ASCL1, IL1RN, NTF3 and TTN, as in (Fig. 1d). Data for the original INT (“PP7”) are included for comparison. Values are means ± standard deviation, (n=3) Luciferase assays; (n=4) qPCR. Student’s one-tailed t-test relative to cells expressing dCas9~VP alone. RNA constructs were expressed from the human U6 promoter.
To examine the influence of internal insert sequence, we synthesized a pool of ~1.2×106 unique sgRNA variants displaying internal cassettes of 25 random nucleotides (Supplementary Fig. 9). In aggregate, the activity of this INT-N25Pool rivaled that of the minimal sgRNA (Fig. 3c), implying that many of the variants formed productive CRISP-Disp complexes. To confirm this, we immunoprecipitated dCas9•INT-N25Pool complexes and analyzed the copurified sgRNA sequences by deep sequencing (RIP-Seq, Fig. 3d, Supplementary Fig. 10). Fewer than 0.01% and 0.02% of the sequence variants were significantly enriched or de-enriched, respectively; motif analysis of these variants revealed no apparent sequence constraints influencing sgRNA•dCas9 complexation. Although this pool represents a small (~1.1×10−9), sampling of the total 25mer sequence space, we extrapolate that CRISP-Disp is not intrinsically limited by the sequence of an internal insert.
To explore the potential utility of INT-like inserts, we next generated a series of INT-like constructs displaying an array of functional RNA domains, comprising natural protein-binding motifs31, 37, 38 and artificial aptamers that bind proteins7, 8 and small molecules13 (Supplementary Fig. 11 and Supplementary Table 5). Satisfyingly, all constructs were viable CRISP-Disp substrates (Fig. 3e). Variation in construct efficacy did not appear to result from limiting RNA expression levels (Supplementary Fig. 12), implying that design of a high-efficiency CRISP-Disp construct for a given motif may require some structural or sequence optimization.
Next, we used INT-like constructs to deploy structured RNA domains to endogenous loci. Constructs bearing the S1 streptavidin aptamer, a potentially useful RNA device7, and P4–P6[3xPP7], the largest INT-like construct tested, each activated ASCL1, IL1RN, NTF3, and TTN28, 29, 33 as predicted (Fig. 3f). Furthermore, INT-P4–P6[3xPP7] did not appear to perturb dCas9 targeting fidelity (Supplementary Fig. 5). We therefore infer that INT-like constructs can display RNA large RNA domains to specific loci genome-wide.
CRISP-Disp enables concomitant deployment of multiple functionalities
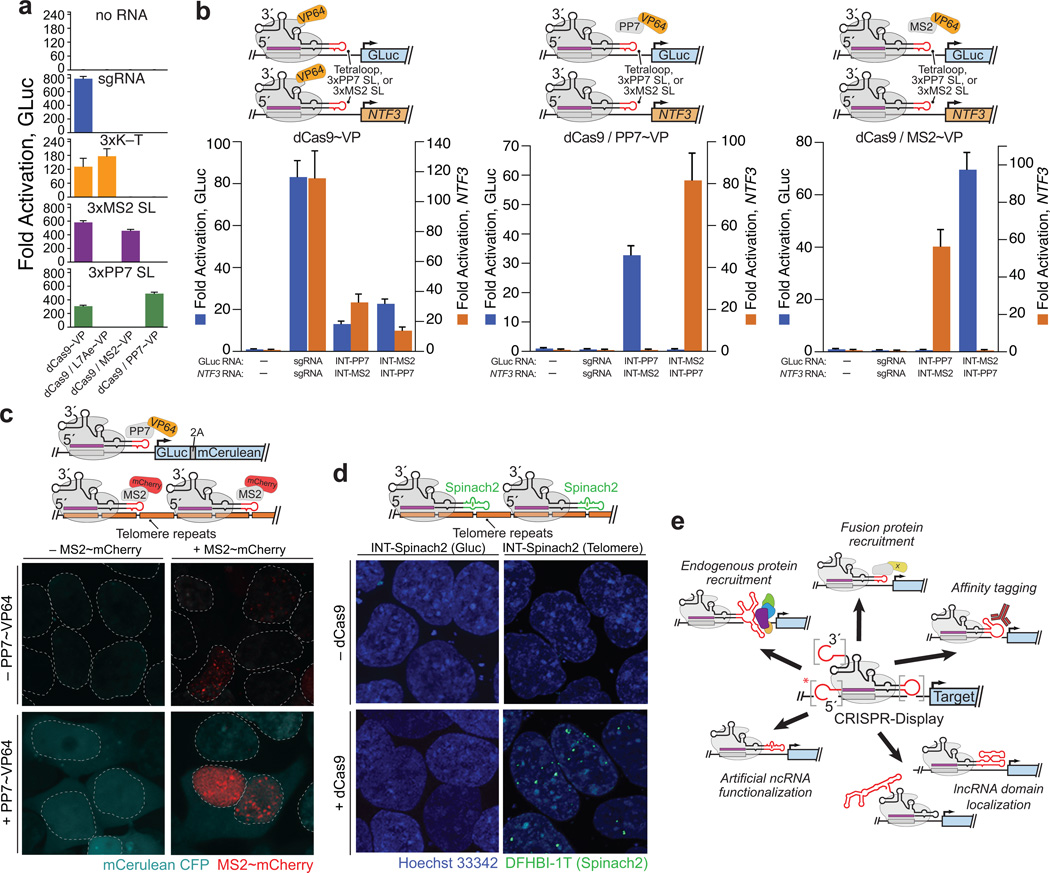
One potential advantage of CRISP-Disp is that it enables disparate functions to be simultaneously performed at diverse loci in the same cell, using a single toolset. This modularity could be achieved using a set of orthogonal RNA-binding proteins: each protein would be fused to a unique functional domain and targeted by sgRNAs bearing its cognate RNA motif25. To demonstrate the plausibility of this scheme, we first established a preliminary set of such RNA-binding proteins (A. fulgidus L7Ae, MS2 and PP7)31, 37 for display on dCas9. As predicted, when fused to VP64, each protein was only competent to induce bridged reporter activation when coexpressed with an sgRNA bearing its cognate RNA motif (Fig. 4a).
Figure 4. CRISPR-Display expands the functional repertoire of CRISPR-based methods.
(a–c) CRISP-Disp enables modular, simultaneous control of multiple functions using a shared pool of dCas9. (a) Bridged activation assays using an orthogonal set of RNA-binding proteins. K–T: a “kink-turn” motif, recognized by L7Ae (Supplementary Table 5)37; (b) Simultaneous binding and transcription activation of distinct genomic loci. Indicated sgRNAs and INT derivatives (“INT-PP7,” “INT-MS2”) were coexpressed in direct and bridged activation assays. Left: direct activation. Middle: bridged activation with PP7~VP. Right: bridged activation with MS2~VP. (c) Simultaneous activation and imaging of distinct genomic loci. Top: schematic of the experimental design. 2A: a 2A “self-cleaving” peptide. Middle and Bottom: Integrated GLuc reporter cells, activated and imaged using the indicated constructs. GLuc- and telomere-targeting INT derivatives were coexpressed in all experiments. Dotted lines denote nuclear membranes. (d) CRISPR-Display of novel RNA-based functions. Top: schematic of the experimental design. Middle and Bottom: aptamer-based imaging of DNA loci using the artificial fluorescent aptamer “Spinach2” (Ref. 13). (e) Novel, multiplexable functions made possible by CRISPR-Display. Accessory domains on the sgRNA 5´ end (asterisk) are not tolerated by most expression systems (Supplementary Fig. 6). Values in (a, b) are means ± standard deviation. (n=3) Luciferase assays; (n=4) qPCR. Confocal fluorescence images (c, d), at 63× magnification.
As a first demonstration of CRISP-Disp modularity, we bound dCas9 to multiple genomic targets, but activated only one (Fig. 4b). We coexpressed complementary pairs of GLuc- and NTF3-targeting INT-like sgRNA variants bearing PP7 and MS2 stem-loops (GLuc[3xPP7] with NTF3[3xMS2], or GLuc[3xMS2] with NTF3[3xPP7]) in integrated reporter cells. In the presence of dCas9~VP, we observed robust activation of both target genes, regardless of the sgRNA pair used (Fig. 4b, left), indicating that dCas9 had bound both loci under all conditions. However, in the presence of dCas9 and PP7~VP, only the gene targeted by sgRNAs bearing PP7 stem-loops was activated; the converse was observed upon coexpression of dCas9 and MS2~VP (Fig. 4b, middle and right). Hence, CRISP-Disp enables modular control of gene expression, as has been recently demonstrated with TOP1-like constructs25.
As a second demonstration of CRISP-Disp modularity, we sought to simultaneously activate one locus and image another (Fig. 4c). To accomplish this, we implemented a “bridged CRISPR-imaging” approach, in which targeted loci (here, telomeres18) are visualized using a ternary complex comprising dCas9, MS2~mCherry, and INT-like [3xMS2] constructs (Supplementary Fig. 13a). Using bridged telomere imaging in HEK293FT cells, we observed numerous (8–55; average of 26.6, in ~97 mCherry+ cells) fluorescent nuclear foci, which required expression of dCas9 and the proper cognate sgRNA (Supplementary Fig. 13b–c). Next, to simultaneously activate one locus and image another, we employed our integrated reporter cells, performing bridged activation (targeted by INT) at the GLuc reporter, and bridged imaging at telomeres (Fig. 4c, top). As predicted, each function could be individually or concomitantly controlled by expression with PP7~VP64, MS2~mCherry or both (Fig. 4c, bottom).
CRISP-Disp of autonomous RNA domains
Another potential advantage of CRISP-Disp is that it allows autonomously functional RNA domains, such as ribozymes, aptamers and regulatory devices7–13, to be targeted to individual loci. As a preliminary illustration of this approach, we targeted “Spinach2,” an artificial aptamer that binds to and induces fluorescence in a cell-permeable dye13, to telomeres18. When we coexpressed a Spinach2-appended telomere-targeting sgRNA with dCas9 and treated cells with the Spinach2 ligand DFHBI-1T (Ref. 13), we observed numerous (10–20; average of 12, in ~20% of cells) nuclear fluorescent foci (Fig. 4d and Supplementary Figure 14). Critically, no fluorescent foci were observed in the absence of dCas9, or when Spinach2 was targeted to the Gluc reporter. Although the Spinach2 signal was less robust than that observed using conventional CRISPR-imaging18, or with our own bridged imaging approach, this experiment represent an important proof-of-principle, demonstrating that artificial RNA domains can be harnessed to imbue dCas9 with novel properties.
Discussion
In the short time since its initial characterization14, 15, Sp. CRISPR-Cas9 has already been coopted for a host of exquisitely powerful genome modification and regulatory technologies17–22. We envision that CRISPR-Display, which limits the function dCas9 to DNA-targeting and “outsources” all other roles to RNA-based domains, will provide the basis for an even wider array of methods (Fig. 4e).
CRISPR-Display’s ability to ectopically localize diverse RNA cargos to targeted DNA loci has several implications. First, CRISP-Disp allows implementation of cargo-specific functions at each of its target sites, using a common toolkit. In theory, the breadth of distinct functions accessible in a single CRISP-Disp experiment is limited only by the number of orthogonal RNA domains or RNA/binding-protein pairs available (as in Fig. 4a–c). For example one might simultaneously activate and repress transcription, epigenetically mark histones and DNA, induce double-stranded breaks and image discrete sets of target loci. Our preliminary experiments, which targeted discrete–though unrelated–genomic loci for binding, activation and imaging (Fig. 4a–c) provide a small glimpse into the potential power this approach holds.
Second, since CRISP-Disp is not intrinsically limited by RNA length (Figs. 2, 3a–b), it may provide a method for the locus-targeted reconstitution of natural regulatory RNAs, which could dramatically advance the study of lncRNA mechanism1–6. For example, CRISP-DISP could be used to bring a long noncoding RNA domain to a target locus, and assess if the RNA molecule alone is functional when decoupled from the act of its transcription. The preliminary data presented here (Fig. 2d–f) demonstrate the plausibility of such experiments. However, more thorough, bona fide lncRNA reconstitution will likely require consideration of many variables (e.g., the chromatin state at the targeted locus), and may depend on the particular lncRNA under consideration..
Third, CRISP-Disp expands the scope of Cas9-based methods by making available the broad functional repertoire of artificial RNA devices. While our initial illustration of this principal focused on imaging (Fig. 4d), other applications built from aptamers, ribozymes, sensors, processors and scaffolds7–13 are possible. For example, CRISP-Disp with RNA scaffolds might allow enzymatic activities to be uniquely targeted to discrete subnuclear sites12; RNA processors might enable complex regulatory functions to be gated in response to external stimuli or small molecules9–11, 19.
Finally, CRISP-Disp may provide a platform for the isolation of novel functional RNA domains. That the sgRNA scaffold can accommodate an expansive breadth of internally inserted sequences (Fig. 3c,d), suggests that it could be used as the backbone for SELEX. Such selections might yield, for example, aptamers that sequester or recruit endogenous protein complexes to target loci, potentially allowing host proteins to be re-targeted genome-wide.
The present studies establish a preliminary framework for the implementation of CRISPR-Display, including “best practices” for construct design (Supplementary Data). Moreover, the proof-of-principle experiments presented here hint to the larger scope of novel applications CRISPR-Display enables (Fig. 4e). We anticipate that this application list is, at best, preliminary.
ONLINE METHODS
Plasmid synthesis
Mammalian expression and reporter constructs were generated using standard restriction enzyme-based and ligation-independent cloning methods. Components were acquired as follows: The T7 promoter-targeting TALE30 was the generous gift of Feng Zhang (Broad Institute). Gaussia and Cypridina luciferases were derived from pGLuc-Basic and pCLuc-Basic, respectively (New England Biolabs). dCas9 (S. pyogenes D10A/H841A Cas9) was isolated from Addgene plasmid 47754, the EF1a promoter from Addgene plasmid 11154, mCerulean from Addgene plasmid 23244, Venus from Addgene 15753 and the human Ubiquitin C promoter (hUBCPro) used to drive expression of L7Ae~VP, PP7~VP (Supplementary Fig. 2a, bottom) and MS2~mCherry (Supplementary Fig. 13a, right) from Addgene plasmid 17627. All other components were synthesized de novo from small synthetic oligonucleotides or from gBlocks (Integrated DNA Technologies).
The backbone for Lentiviral reporter constructs was derived from pLenti6.3/TO/V5-DEST (Life Technologies), from which the Tet-reponsive promoter and Gateway cloning sites were removed. The backbone for the T7 TALE and MS2~VP constructs was derived from pcDNA3.1(+) (Life Technologies) in which the Neomycin expression cassette was removed. All other constructs were cloned into pNEB193 (New England Biolabs).
L7Ae, MS2 and PP7 were codon-optimized for expression in human cells and synthesized as gBlocks (Integrated DNA Technologies). The PP7 construct consists of two tandem copies of the non-aggregating ΔFG mutant31 joined by a flexible seven amino acid linker with the sequence GSTSGSG (Supplementary Fig. 2a, bottom). Similarly, the MS2 construct consists of two tandem copies of the non-aggregating V75E/A81G mutant40 joined by the same linker. L7Ae was designed according to a published sequence37.
INT-like constructs (Figs. 3,4 and Supplementary Table 5) were cloned as follows. We first cloned an INT general-purpose cloning vector, “sgINTgpc,” containing the following pertinent sequence:
GATCTAGATACGACTCACTATGTTTAAGAGCTATGCTGCGAATACGAGAAGTCTTCTTTTTTGA AGACAATCGTATTCGCAGCATAGCAAGTTTAAATAAGGCTAGTCCGTTATCAACTTGAAAAAGT GGCACCGAGTCGGTGCTTTTTTT
…Wherein italicized nucleotides denote the GLuc-targeting protospacer sequence (Supplementary Table 2), underlined nucleotides denote an extended sgRNA stem1 (Ref. 18) and bold nucleotides denote two outward-facing BbsI restriction sites. This cassette is under expression of a human U6 promoter (not shown). Inserts cloned into this backbone had the general format: 5´–CGAG–[Insert]–CTCGT–3´, wherein underlined nucleotides denote the sticky ends used for cloning; the additional C following the insert restores base-pairing at the end of stem1. These inserts were generated by PCR and restriction digestion with BbsI, or by annealing synthetic, 5´-phosphorylated oligonucleotides (following the protocol used for the N25 pool, below). Inserts were ligated into BbsI-digested, gel-purified sgINTgpc using the Quick Ligation Kit (New England Biolabs).
All sgRNAs and derivatives were initially cloned bearing a GLuc-targeting protospacer (Supplementary Table 2). ASCL1-, IL1RN-, NTF3-, TTN- and telomere-targeting constructs (Figs 1d, 3f, 4b–d; Supplementary Table 2) were derived from these parental constructs using an inverse-PCR method, using a forward primer that anneals downstream of the protospacer and a reverse primer that anneals to the 3´-end of the U6 promoter. Namely, PCR products were amplified with primers of the general format:
Forward: TAGTAGAAGACAAXXXXXXXXXXXXXGTTTAAGAGCTATGCTGCGAATACG Reverse: TAGTAGAAGACAAYYYYYYYYYYYYGGTGTTTCGTCCTTTCCAC
…Wherein bold nucleotides denote BbsI restriction sites; X’s denote nucleotides 9–21 of the new protospacer sequence; Y’s denote the reverse complement of nucleotides 1–9 of the new protospacer; underlined nucleotides are reverse complementary to one another. PCR products were purified using the QIAgen PCR cleanup kit, digested with BbsI and DpnI, purified again and quantified by UV-vis spectroscopy. Products (25 ng, in 11 µL final) were self-ligated using the Quick Ligation Kit (New England Biolabs).
All plasmid sequences were confirmed by Sanger sequencing (GeneWiz) prior to use.
Cloning the N25 Pool
Pool oligonucleotides (Integrated DNA Technologies) were as follows:
5´–[P]–CGAGNNNNNNNNNNNNNNNNNNNNNNNNNC–3´ 5´–[P]–ACGAGNNNNNNNNNNNNNNNNNNNNNNNNN–3´
…Wherein 5´–[P] denotes a 5´ Phosphate, and N denotes an equimolar mixture of all four nucleotides. Oligonucleotides were each resuspended in annealing buffer (10 mM Tris, pH 7.0, 50 mM NaCl) to 100 µM. 10 µL of each oligo were mixed in a 0.2 mL PCR tube; this mixture was heated to 95°C for 10 minutes and slowly annealed to 25°C over the course of two hours in a thermocycler. The reaction was snap-cooled on ice and diluted 100-fold with ice-cold annealing buffer. 1 µL of this diluted duplex mix was ligated into 25 ng of BbsI-cut sgINTgpc, in 12 µL final volume, using the Quick Ligation Kit (New England Biolabs). The entire reaction was transformed into 120 µL of XL10-Gold ultracompetent cells (Agilent), plated onto 12 LB Ampicillin plates and grown overnight at 37°C. Seven bacterial colonies were picked for Sanger sequencing (Supplementary Fig. 9), and the remainder were pooled by scraping the plates into 100 mL of liquid LB(Amp). Bacteria were pelleted by ultracentrifugation, and the plasmid pool was harvested in a single plasmid maxi-prep (QIAgen) (Supplementary Figs. 9–10).
Cell Culture, stable and transient transgene expression
HEK 293FT cells (ATCC) were maintained on gelatinized plates in high glucose Dulbecco’s modified Eagle’s medium (DMEM, Gibco), supplemented with 10% FBS, 1× penicillin/streptomycin and 2 mM L–Glutamine (Gibco). Cells were grown at 37°C and 5% CO2 in a humidified incubator.
Lentiviral particles were generated using standard second generation packaging plasmids, in 293T cells. Integrated reporter cells were generated as follows: 250,000 HEK 293FT cells were plated per well of a gelatinized six-well dish and incubated overnight. Growth media was thereafter removed; cells were washed once in warmed PBS, and supplied with 1.7 mL fresh warmed media supplemented with 200 µL CLuc reporter lentivirus and 8 µg/mL polybrene. After 24 hr this process was repeated with a second dose of CLuc virus. Cells were subsequently passaged onto 10 cm gelatinized plates and selected with 2 µg/mL puromycin. CLuc reporter cells were then plated onto gelatinized six-well dishes and transduced with GLuc reporter lentivirus following the same transduction protocol. GLuc-transduced cells were not selected with hygromycin prior to use. A lentiviral variant of our EF1α-dCas9 construct (Supplementary Fig. 1c) was also used for aptamer-based imaging (Fig. 4d) following the same transduction protocol, without antibiotic selection. To enrich for cells that expressed low levels of dCas9 (Ref. 18), we transiently transfected GLuc reporter, U6-INT and PP7~VP plasmids, as in analytical luciferase assays (see below), and collected GLuc+ cells by FACS.
Transient transfections were per performed using Lipofectamine 2000 (Life Technologies), following the manufacturer’s protocol. For luciferase assays, 125,000 cells in 0.6 mL media were plated per well of gelatinized 12-well dishes and incubated overnight. Transfection mixes contained 33 ng of each luciferase reporter plasmid (where appropriate), 59 ng of dCas9 or dCas9~VP plasmid, 66 ng of PP7~VP, L7Ae~VP or MS2~VP (where appropriate), 11.6 ng of U6–driven or 542 ng of Pol II-driven sgRNA variants. For experiments using TOP3 and TOP4 (Fig. 1a,c, Supplementary Fig. 6b), 11.6 ng of a separate U6-driven gRNA plasmid was also included. For FACS, (Supplementary Figs. 1e, 2b, 4e and 6d) transfection mixes also contained 10 ng of an mCherry cotransfection control. In all cases, the total transfected plasmid mass was brought to 750 ng per well using pNEB193 (New England Biolabs) in 18 µL final volume, with 2.25 µL Lipofectamine 2000.
For RNA immunoprecipitation (RIP) and RIP-Seq experiments, 2.1 million cells in 10 mL growth media were plated onto gelatinized 10 cm dishes and grown overnight. Transfection mixes were as described above, but all masses and volumes were scaled 15.7–fold to account for the increase in growth area and cell number. RIP transfection mixes included each luciferase reporter to independently monitor CRISP-Disp function.
To test CRISP-Disp function at endogenous loci (Figs. 1d, 3f), cells were plated in gelatinized 12-well dishes as in standard luciferase assays. Transfection mixes were similar to those described28, and contained 500 ng dCas9 or dCas9~VP plasmid, 500 ng GLuc-Targeting sgRNA construct or 500 ng of a mix containing equal masses (125 ng each) of four ASCL1-, IL1RN-, NTF3- or TTN-targeting constructs28, 29, 33 (Supplementary Table 2). Where appropriate, 556 ng of PP7~VP plasmid was also included. All mixes were brought to 1556 ng per well using pNEB193, in 38 uL final volume, with 4.7 µL Lipofectamine 2000.
For multiplexing experiments (Fig 4b) cells were plated in gelatinized 12-well dishes as above. Transfection mixes contained 250 ng dCas9 or dCas9~VP, 250 ng GLuc-targeting sgRNA variant, 250 ng of a mix containing equal masses (62.5 ng each) of four NTF3-targeting constructs (Supplementary Table 2), and 278 ng of PP7~VP or MS2~VP, where appropriate. In all cases the total transfected mass was brought to 1028 ng using pNEB193, in 30 µL volume, with 3.1 µL Lipofectamine 2000.
In bridged imaging experiments (Fig. 4c, Supplementary Fig. 13), 80,000 cells in 1 mL growth media were plated per well of untreated Nunc Lab-Tek glass two-chamber slides (Thermo Scientific). Twenty-four hours thereafter, growth media was changed, and cells were transfected with 440 ng dCas9 and 235 ng of each modified sgRNA. Where appropriate, 440 ng of PP7~VP64 and/or 100 ng of MS2~mCherry were included. The total transfected mass was brought to 1500 ng with pNEB193, in 11.4 µL, with 4.5 µL Lipofectamine 2000, according to the manufacturer’s protocol. For aptamer-based imaging (Fig. 4d), 80,000 dCas9-transduced (“+dCas9,” see above) or untransduced (“–dCas9”) cells in 1 mL growth media were plated per well of Nunc Lab-Tek glass two-chamber slides that had been treated as follows. Wells were coated with 100 µg/mL poly-L-lysine (Millipore) overnight at 4°C. The next day, wells were washed twice with ddH2O, UV sterilized for five minutes in a biosafety cabinet, coated with 100 µg/mL rat collagen-I (Corning) and 50 µg/mL laminin (Life Technologies) for two hours at 37°C, and dried prior to plating cells. Transfections were performed 24 hours thereafter, with 600 ng (telomere- or GLuc-targeting) INT-spinach2 construct, 600 ng of pNEB193 and 4.5 ng of an mCherry cotransfection control, in a total volume of 11.4 µL, with 3.8 µL Lipofectamine 2000, according to the manufacturer’s protocol. All live-cell imaging experiments were performed 48–72 hours post-transfection (see below).
Luciferase and FACS Assays
Luciferase assays were performed using the BioLux Gaussia and Cypridina Luciferase Assay kits (New England Biolabs), following the manufacturer’s protocols. Growth media (200 µL) was harvested three days after transfection and, if not used immediately, was stored in the dark at 4°C in parafilm-sealed 96-well dishes. 20 µL of each experimental sample was manually pipetted into black-walled 96-well plates (Corning) and assayed using a FLUOstar OPTIMA Luminometer equipped with automatic injectors (BMG Labtech). Gaussia and Cypridina assays were performed in parallel. For each, a single empirically determined gain was applied to all samples within an experimental series. Each sample was injected with 50 µL of luciferase assay buffer and mixed for two seconds prior to data acquisition. Signal was integrated over 20 seconds using an open (unfiltered) top-down optic.
For each sample, experimental raw luciferase signals were background-subtracted, and the ratio of Luciferase values, (GLuc/CLuc), was calculated. Biological replicates (at least three per experiment) were used to calculate a mean value, <GLuc/Cluc>. Fold activation was then calculated relative to a control sample in which dCas9~VP was expressed in the absence of an sgRNA construct:
Statistical significance testing likewise used this dCas9~VP control as the basis of comparison.
For FACS assays, cells were propagated and transfected in gelatinized 12-well dishes, as described for luciferase assays, and analyzed three days after transfection. Cells were harvested by trypsinization, quenched by the addition of chilled growth media, diluted threefold in chilled staining media (Hank’s Balanced Salt Solution (HBSS, Gibco), supplemented with 2% Donor Bovine Serum (DBS, Atlanta Biologicals)), and pelleted at 200 g in a swinging bucket rotor. Cells were resuspended in chilled staining media and analyzed on a BD LSR II Flow Cytometer (BD Sciences), equipped with HcRed, CFP and YFP filters. Voltages, compensations and gates were empirically determined using unstained and single color controls, according to standard methods. 100,000 mCherry+ cells were recorded from each sample.
RNA Immunoprecipitation (RIP)
Cells were propagated on gelatinized 10-centimeter dishes, transfected as described above, and harvested three days after transfection. Thereafter, RIP was performed essentially as described previously41. Growth media was aspirated, and cells were washed twice with 10 mL room temperature PBS (Gibco). Cells were crosslinked by incubation in 0.1% (v/v) formaldehyde in PBS for 10 minutes at room temperature, under very gentle agitation. Crosslinking was quenched by the addition of Glycine to 133 mM and gentle agitation for an additional five minutes at room temperature, after which the liquid phase was aspirated. Crosslinked cells were washed twice with room temperature PBS, harvested by scraping, allotted into samples of 1×107 cells (typically three samples per 10 cm dish), and pelleted at 200 g in a swinging bucket rotor. PBS was aspirated and cell pellets were flash-frozen in liquid nitrogen and stored at −80°C until use.
Cell pellets were thawed on ice, gently resuspended into 1 mL of ice-cold RIPA(+) buffer (standard RIPA supplemented with 0.1 U/µL RNAseOUT (Life Technologies), 1× EDTA-free Proteinase Inhibitor Cocktail (Thermo Scientific) and 0.5 mM DTT), and lysed for 10 minutes at 4°C with end-over-end agitation. Samples were then sheared using a Branson Digital Sonifier 250 (Emerson Industrial Automation) at 10% amplitude for three 30-second intervals (0.7 seconds on + 1.3 seconds off), with 30-second resting steps between intervals. Samples were held in ice-cold metal thermal blocks throughout sonication. Sheared samples were then clarified by ultracentrifugation and diluted with 1 mL each of ice-cold Native Lysis Buffer(+) (25 mM Tris, pH 7.4, 150 mM KCl, 5 mM EDTA, 0.5% (v/v) NP-40, supplemented with inhibitors and DTT, as above), filtered through a 0.45 µm syringe-mounted filter, and flash-frozen in liquid nitrogen before use.
Clarified lysates were thawed on ice and pre-cleared by incubation with buffer-equilibrated magnetic Protein G beads (Life Technologies) for 30 minutes at 4°C, with end-over-end rotation. 100 µL aliquots were removed and frozen, to serve as “input” normalization controls. Cleared lysates corresponding to 5×106 cells were then incubated with 6 µg rabbit anti-FLAG (SIGMA) or Rabbit normal IgG (Cell Signaling Technology), for two hours at 4°C with end-over-end rotation. Buffer-equilibrated magnetic Protein G beads were then added and the samples were again rotated end-over-end for one hour at 4°C. Beads were collected and twice washed with Native Lysis Buffer(+) for 10 minutes at 4°C, with end-over-end rotation. Immunoprecipitated RNA was thereafter isolated as described below.
RNA Isolation, Quantitative RT–PCR and mRNA Seq
Whole cell RNA (Figs. 1d, 3f, 4b and Supplementary Figs. 5 and 12) and RNA from subcellular fractions (Supplementary Fig. 7) were isolated by extraction with Trizol and Trizol-LS Reagent (Life Technologies), respectively, following the manufacturer’s protocols. RNA was precipitated with isopropanol using GlycoBlue (Life Technologies) as a carrier, and subsequently purified using RNEasy spin columns (QIAgen), following the manufacturer’s “RNA Cleanup” protocol, with on-column DNase treatment.
RNA from RIP and RIP-Seq experiments (Figs. 2c,e, 3d and Supplementary Figs. 2d, 4f, and 6e) was isolated as follows. Following RIP (see above), protein G beads were suspended in 56 µL nuclease-free water, and processed alongside input samples (56 µL; 5.6% of the total). All samples were brought to 100 µL with 3× Reverse Crosslinking Buffer (final concentrations: 1× PBS, 2% N-Lauroyl Sarcosine, 10 mM EDTA, 5 mM DTT, 0.4 U/µL RNaseOUT and 2 mg/mL proteinase K (Ambion)). Formaldehyde crosslinks were reversed by incubation in a thermocycler at 42°C for one hour, and then 55°C for one hour. RNA was thereafter purified using four volumes (400 µL) Agencourt RNAClean XP Beads (Beckman Coulter), following the manufacturer’s protocol, and eluted into 30 µL nuclease-free water. Residual DNA was removed by treatment with 5 U RNase-free DNAs (RQ1, Promega) in 50 µL, following the manufacturer’s protocol. RNA was subsequently purified using four volumes (200 µL) Agencourt RNAClean XP beads, eluted into 20 µL nuclease-free water, and stored at −20°C until use.
cDNA was synthesized using SuperScript III Reverse Transcriptase (Life Technologies), according to the manufacturer’s protocol, priming from anchored oligo-dT21, random hexamers (Life Technologies) or a gene specific primer (Integrated DNA Technologies), where appropriate. Target RNA abundance was quantified by qRT-PCR on a 7900HT Fast Real-Time PCR System (Applied Biosystems), using Rox-normalized FastStart Universal SYBR Green Master Mix (Roche) and gene-specific primers (Supplementary Table 3), in quadruplicate. Non-reverse-transcribed RNA was used as a negative control. “Clipped” data were processed using Realtime PCR Miner42, to calculate CT and efficiency values (Supplementary Table 3). Bulk gene expression measurements (Figs. 1c, 3f, 4b and Supplementary Fig. 12) were normalized to a GAPDH internal control (Supplementary Table 3); RIP measurements were normalized to input RNA levels. In subcellular fractionation experiments (Supplementary Fig. 7), the yield of RNA in each compartment was quantified relative to the unfractionated input level, as in RIP experiments. Data analysis was performed using standard methods.
For global gene expression analysis (Supplementary Fig. 5), Poly(A)+ mRNA seq libraries were prepared using the TruSeq RNA sample preparation kit, v2 (Illumina) as described41. Indexed libraries were pooled and subjected to 50 cycles of paired end sequencing, followed by 25 cycles of indexing, on two lanes of an Illumina HiSeq 2500 (FAS Center for Systems Biology, Harvard). For characterization of gene expression, sequencing reads were mapped to a custom gene set comprising UCSC known human genes (hg19), appended with dCas9, GLuc, CLuc and sgRNA constructs, using TopHat2 with default options43. Differential analysis of gene expression was assessed using Cuffdiff2 with default options44. Genes plotted in (Supplementary Fig. 5) were restricted to the top 75% of expressed genes, based on FPKM values.
Error Propagation and Reproducibility
For Luciferase and qRT-PCR assays, experimental uncertainties were propagated as described previously45. Given S, the sum or difference of values A, B, uncertainly was calculated using the formula:
…wherein σA and σB are the measurement errors of A and B, respectively. For P, the product or quotient of values A and B, uncertainty was calculated using the formula:
The uncertainty of other functions, F(x), was calculated using the first derivative approximation:
Sample sizes were determined in accordance with standard practices used in similar experiments in the literature; no sample-size estimates were performed to ensure adequate power to detect a prespecified effect size. Experiments were neither randomized nor blinded to experimental conditions. No samples were excluded from analysis.
Subcellular Fractionation
Cytoplasmic and nuclear fractions (Supplementary Fig. 7) were isolated as described46, 47. Briefly, cells were grown and transfected in gelatinized 10-cm dishes, as described for RIP experiments, above. Three days after transfection, cells were harvested by trypsinization, quenched with growth media, pelleted and washed thrice with ice-cold PBS. Cells were gently resuspended in five packed cell pellet volumes (“cv’s”) of ice-cold Cyto Extract Buffer(+) (20 mM Tris, pH 7.6, 0.1 mM EDTA, 2 mM MgCl2, supplemented with 0.5 U/µL RNAseOUT and 1× EDTA-free Proteinase Inhibitor Cocktail), and swollen by incubation at room temperature for two minutes, and on ice for ten minutes more. Cells were then lysed by addition of CHAPS to 0.6% final, gentle pipetting, and two passages through a syringe equipped with a 20G needle. Lysate was clarified by centrifugation at 500g in a tabletop microcentrifuge at 4°C; 70% of the resulting supernatant was retrieved as the cytoplasmic fraction. The pellet, corresponding to nuclei and cell debris, was washed twice by gentle resuspension into five cv’s of Nuclear Wash Buffer(+) (Cyto Extract Buffer, supplemented to 0.6% CHAPS and with inhibitors, as above), followed by centrifugation at 500g. Washed nuclei were gently resuspended into two cv’s of Nuclei Resuspension Buffer(+) (10 mM Tris, pH 7.5, 150 mM NaCl, 0.15% (v/v) NP–40, supplemented with inhibitors, as above) layered onto a cushion of five cv’s Sucrose Buffer(+) (10 mM Tris, pH 7.5, 150 mM NaCl, 24% (w/v) Sucrose, plus inhibitors), and pelleted at 14,000 rpm in a tabletop microcentrifuge at 4°C. The resulting pelleted nuclei were resuspended into two cv’s of ice-cold PBS and pelleted at 500g, before harvesting RNA. We confirmed the success of our fractionations by two methods: western blotting and qRT-PCR (Supplementary Fig. 7b–c). In western blots, aliquots of whole cell lysate, the cytoplasmic fraction and PBS-suspended nuclei were probed using antibodies against (α/β)-Tubulin and Fibrillarin (Cell Signaling Technology). For qPCR, extracted RNA (see above) was quantified using primers against the cytoplasmic ncRNA SNHG5 and the nuclear ncRNA XIST (Supplementary Table 3).
N25 RNA Library Preparation, Sequencing and Analysis
For the N25 RIP-Seq experiment (Fig. 3d), cell growth, transfection, RIP and RNA preparation were performed as described above, in triplicate. Seven deep sequencing libraries were prepared: one from the starting plasmid pool, three from replicates of the input RNA, and three from replicates of the immunoprecipitated RNA. The plasmid pool library was generated directly via PCR, using 5 ng of plasmid template in a 50 µL reaction, amplified through 19 cycles of PCR with Pfu Ultra II HS polymerase (Agilent), according to the manufacturer’s protocol. Gene-specific PCR primers that bracketed the N25 insertion site, appended with standard Illumina adapters and indexes, were used (Supplementary Table 6, and Supplementary Fig. 9). For each input and RIP library, 10 ng RNA was reverse-transcribed in 20 µL as described above, using a gene specific primer (Supplementary Table 6, and Supplementary Fig. 9). Each cDNA reaction was used in its entirety as PCR template, using the same primer design as was used for the plasmid pool, but with different Illumina indexes. The pools were amplified in 200 µL, through 26 cycles of PCR with Pfu Ultra II HS polymerase (Agilent), according to the manufacturer’s protocol. The resulting deep sequencing libraries were purified twice with 1.0 volume of Agencourt AMPure XP Beads (Beckman Coulter), according the manufacturer’s protocol, and eluted in EB Buffer (QIAgen). The plasmid pool library contained traces of high molecular weight contaminants (not shown) that were removed by “reverse selection:” the sample was treated with 0.65 volumes of AMPure XP Beads, and the unbound fraction was retained. The integrity and concentration of each final library was measured using a “DNA High Sensitivity” assay on an Agilent 2100 model Bioanalyzer (Supplementary Fig. 9).
Libraries were denatured in 50 mM NaOH, diluted in buffer HT1 (Illumina) and combined to yield a 20 pM pool, according to standard protocols. This pool was doped with TailorMix Indexed PhiX Control Library (SeqMatic), at a ratio of 7:3 N25Pool:PhiX, and sequenced on two lanes of an Illumina HiSeq 2500 (FAS Center for Systems Biology, Harvard) for 150 cycles, followed by 25 cycles of indexing.
Random insert sequences were extracted from raw sequencing reads by removing the constant sequences abutting each side of the insertion point. The number of occurrences of each random sequence within each individual sample was then tabulated. Sequence counts were used to calculate enrichment using DESeq2 (Ref.48).
Live Cell Imaging
Images in (Supplementary Fig. 1d) were collected on an Axio Observer D1 system (Zeiss), equipped with eYFP and eCFP filters.
Live fluorescence images (Fig. 4c–d, and Supplementary Figs. 13–14) were taken with an LSM 700 Inverted Confocal Microscope (Harvard Center for Biological Imaging), with an aperture setting of 1 A.U., using the DAPI filter for Hoechst 33342, the CFP filter for mCerulean, the mCherry filter for mCherry and the FITC filter for DFHBI-1T, where appropriate. In bridged imaging experiments (Fig. 4c; Supplementary Fig. 13), cells were imaged two days post-transfection, in their growth media. Images are max-merges of 37–47 Z-stacks, taken with a step size of 0.33 µm, at 63× magnification. For aptamer-based imaging (Fig. 4d and Supplementary Fig. 14), growth media was replaced with imaging media (Fluorobrite DMEM (Life Technologies), 25 mM HEPES, 5 mM MgSO4, 1 µg/ml Hoechst 33342 (Life Technologies), and 20 µM DFHBI-1T (Lucerna)) for 30 minutes at 37°C. Images in (Fig. 4c–d) are max-merges of 20–30 Z-stacks, taken with a step size 0.35 µm, at 63× magnification.
Supplementary Material
Acknowledgments
We thank L. Cong and F. Zhang for advice and reagents regarding TALE domains, Y. Sancak, C. Fulco, S. Donovan and M. Morse for their general technical assistance, G. Kenty for his help with luminometry, D. Hendrickson and D. Tenen for their RIP expertise, C. Gerhardinger and C. Daly for their support with deep sequencing, M. Tabebordbar, J. LaVecchio and S. Ionescu and M. Sauvageau for their assistance with FACS. We are grateful to all members of the Rinn lab for their thoughtful discussions and critiques. This work was supported by US National Institutes of Heath grants P01GM099117. D.M.S. is a former Howard Hughes Medical Institute fellow of the Jane Coffin Childs Memorial Fund for Medical Research; J.L.R. is the Alvin and Esta Star Associate Professor.
D.M.S. and J.L.R. have filed for patents concerning the design and expression of extended sgRNAs to expand the repertoire of dCas9 function.
Footnotes
Accession Codes
The Gene Expression Omnibus SuperSeries accession number for the data reported in this paper is GSE66756. This accession encompasses the RIP-Seq data in SubSeries GSE62305 and total mRNA sequencing in SubSeries GSE66755.
Author Contributions
D.M.S. designed and performed experiments; F.E.H. designed and performed microscopy experiments (Figs. 4c–d and Supplementary Figs. 13–14); S.T.Y. assisted with computational analysis (Fig. 3d and Supplementary Figs. 5, 10); J.L.R. directed research. D.M.S. and J.L.R. wrote the manuscript.
Competing financial interests
The authors declare competing financial interests.
References
- 1.Cech TR, Steitz JA. The noncoding RNA revolution-trashing old rules to forge new ones. Cell. 2014;157:77–94. doi: 10.1016/j.cell.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annual review of biochemistry. 2012;81:145–166. doi: 10.1146/annurev-biochem-051410-092902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ulitsky I, Bartel DP. lincRNAs: genomics, evolution, and mechanisms. Cell. 2013;154:26–46. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Minks J, Baldry SE, Yang C, Cotton AM, Brown CJ. XIST-induced silencing of flanking genes is achieved by additive action of repeat a monomers in human somatic cells. Epigenetics & chromatin. 2013;6:23. doi: 10.1186/1756-8935-6-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sauvageau M, et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. eLife. 2013;2:e01749. doi: 10.7554/eLife.01749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bassett AR, et al. Considerations when investigating lncRNA function in vivo. eLife. 2014;3:e03058. doi: 10.7554/eLife.03058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Walker SC, Good PD, Gipson TA, Engelke DR. The dual use of RNA aptamer sequences for affinity purification and localization studies of RNAs and RNA-protein complexes. Methods in molecular biology. 2011;714:423–444. doi: 10.1007/978-1-61779-005-8_26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tome JM, et al. Comprehensive analysis of RNA-protein interactions by high-throughput sequencing-RNA affinity profiling. Nature methods. 2014;11:683–688. doi: 10.1038/nmeth.2970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Auslander S, et al. A general design strategy for protein-responsive riboswitches in mammalian cells. Nat Meth. 2014;11:1154–1160. doi: 10.1038/nmeth.3136. [DOI] [PubMed] [Google Scholar]
- 10.Liang JC, Bloom RJ, Smolke CD. Engineering biological systems with synthetic RNA molecules. Molecular cell. 2011;43:915–926. doi: 10.1016/j.molcel.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carothers JM, Goler JA, Juminaga D, Keasling JD. Model-driven engineering of RNA devices to quantitatively program gene expression. Science. 2011;334:1716–1719. doi: 10.1126/science.1212209. [DOI] [PubMed] [Google Scholar]
- 12.Delebecque CJ, Lindner AB, Silver PA, Aldaye FA. Organization of intracellular reactions with rationally designed RNA assemblies. Science. 2011;333:470–474. doi: 10.1126/science.1206938. [DOI] [PubMed] [Google Scholar]
- 13.Song W, Strack RL, Svensen N, Jaffrey SR. Plug-and-play fluorophores extend the spectral properties of Spinach. Journal of the American Chemical Society. 2014;136:1198–1201. doi: 10.1021/ja410819x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garneau JE, et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 15.Jinek M, et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sternberg SH, Redding S, Jinek M, Greene EC, Doudna JA. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature. 2014;507:62–67. doi: 10.1038/nature13011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu PD, Lander ES, Zhang F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell. 2014;157:1262–1278. doi: 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nissim L, Perli SD, Fridkin A, Perez-Pinera P, Lu TK. Multiplexed and Programmable Regulation of Gene Networks with an Integrated RNA and CRISPR/Cas Toolkit in Human Cells. Molecular cell. 2014;54:698–710. doi: 10.1016/j.molcel.2014.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryan OW, et al. Selection of chromosomal DNA libraries using a multiplex CRISPR system. eLife. 2014;3 doi: 10.7554/eLife.03703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilbert LA, et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell. 2014 doi: 10.1016/j.cell.2014.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Citorik RJ, Mimee M, Lu TK. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nature biotechnology. 2014 doi: 10.1038/nbt.3011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Briner, Alexandra E, et al. Guide RNA Functional Modules Direct Cas9 Activity and Orthogonality. Molecular cell. 2014;56:333–339. doi: 10.1016/j.molcel.2014.09.019. [DOI] [PubMed] [Google Scholar]
- 24.Wright AV, et al. Rational design of a split-Cas9 enzyme complex. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:2984–2989. doi: 10.1073/pnas.1501698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zalatan JG, et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell. 2015;160:339–350. doi: 10.1016/j.cell.2014.11.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konermann S, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilbert LA, et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maeder ML, et al. CRISPR RNA-guided activation of endogenous human genes. Nature methods. 2013;10:977–979. doi: 10.1038/nmeth.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Pinera P, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nature methods. 2013;10:973–976. doi: 10.1038/nmeth.2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang F, et al. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nature biotechnology. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chao JA, Patskovsky Y, Almo SC, Singer RH. Structural basis for the coevolution of a viral RNA-protein complex. Nature structural & molecular biology. 2008;15:103–105. doi: 10.1038/nsmb1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chavez A, et al. Highly efficient Cas9-mediated transcriptional programming. Nature methods. 2015 doi: 10.1038/nmeth.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mayer C, Neubert M, Grummt I. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO reports. 2008;9:774–780. doi: 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orom UA, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang KC, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472:120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Saito H, et al. Synthetic translational regulation by an L7Ae-kink-turn RNP switch. Nature chemical biology. 2010;6:71–78. doi: 10.1038/nchembio.273. [DOI] [PubMed] [Google Scholar]
- 38.Sternberg SH, Haurwitz RE, Doudna JA. Mechanism of substrate selection by a highly specific CRISPR endoribonuclease. Rna. 2012;18:661–672. doi: 10.1261/rna.030882.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cong L, Zhou R, Kuo YC, Cunniff M, Zhang F. Comprehensive interrogation of natural TALE DNA-binding modules and transcriptional repressor domains. Nature communications. 2012;3:968. doi: 10.1038/ncomms1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.LeCuyer KA, Behlen LS, Uhlenbeck OC. Mutants of the bacteriophage MS2 coat protein that alter its cooperative binding to RNA. Biochemistry. 1995;34:10600–10606. doi: 10.1021/bi00033a035. [DOI] [PubMed] [Google Scholar]
- 41.Kelley DR, Hendrickson DG, Tenen D, Rinn JL. Transposable elements modulate human RNA abundance and splicing via specific RNA-protein interactions. Genome Biol. 2014;15:537. doi: 10.1186/s13059-014-0537-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. Journal of computational biology : a journal of computational molecular cell biology. 2005;12:1047–1064. doi: 10.1089/cmb.2005.12.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nature biotechnology. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shechner DM, Bartel DP. The structural basis of RNA-catalyzed RNA polymerization. Nature structural & molecular biology. 2011;18:1036–1042. doi: 10.1038/nsmb.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosner M, Hengstschlager M. Detection of cytoplasmic and nuclear functions of mTOR by fractionation. Methods in molecular biology. 2012;821:105–124. doi: 10.1007/978-1-61779-430-8_8. [DOI] [PubMed] [Google Scholar]
- 47.Bhatt DM, et al. Transcript dynamics of proinflammatory genes revealed by sequence analysis of subcellular RNA fractions. Cell. 2012;150:279–290. doi: 10.1016/j.cell.2012.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.