Abstract
CXCR2 and its ligands have been shown to play an important role in tumor angiogenesis, therapy resistance and progression. In this study, we investigated whether CXCR2 ligands are responsible for the survival advantage and metastasis of drug-resistant cells and examined the underlying mechanism(s) doxorubicin or paclitaxel resistant mammary tumor cells. Our results demonstrated that drug-resistant Cl66 cells upregulated CXCR2 ligands but down regulated expression of CXCR2. We observed delayed tumor growth but increased metastasis in mice using these drug-resistant cells. Furthermore, we observed differential upregulation of stem cell and mesenchymal markers in the doxorubicin and paclitaxel-resistant tumor cells. Abrogation of the CXCR2 signaling axis using CXCR2 ligand neutralization resulted in significant inhibition of drug-resistant cell growth. Together, our data suggest chemotherapy-specific differential regulation of CXCR2 ligands, stem cell-like and mesenchymal phenotypes, and enhanced metastasis in drug-resistant cells and targeting CXCR2 signaling may help circumvent therapy resistance in breast cancer.
Keywords: Chemokines, Therapy resistance, CXCR2, stem cell
1. Introduction
Breast cancer is the most frequently diagnosed cancer among women in the United States, representing more than one quarter (29%) of the total estimated new cancer cases in 2015 [1, 2]. However, the primary tumor itself is not the major cause of mortality in breast cancer patients. Resistance to conventional therapies and metastasis-driven mortality leads to a daunting difference in the relative survival of patients suffering from localized cancer in comparison with advanced stage disease [3]. Currently, the management of advanced stage breast cancer is of outmost concern and necessitates the development of strategies that can be used in combination with already existing therapies to treat primary tumor.
Recent findings in cancer have implicated the role of chemokines in therapy resistance and metastasis [4-6]. Chemokines, a subtype of cytokines, chemoattract various cell types. Chemokines have been classified into four main subfamilies: CXC, CC, CX3C and XC. In normal tissues, chemokines are involved in the inflammatory responses [5, 7], CXC chemokines are involved in the recruitment of immune cells, specifically neutrophils, to the site of inflammation and subsequent regeneration of the damaged tissue [8, 9]. However, during tumorigenesis, cancer cells are known to up regulate chemokines, but surprisingly instead of suppressing the oncogenic activity of cancer cells and help in tumor progression [10, 11]. We recently reported increased expression of CXCR2 ligands in the supernatants of chemotherapy treated breast cancer cells [6]. Our observations were supported by clinical studies that showed elevated serum levels of CXCL6 and CXCL8 in patients with metastatic breast cancer [12, 13]. Similarly, levels of CXCL1 and CXCL8 have been reported to increase after chemotherapeutic treatment [6, 14, 15]. We furthermore observed that attenuation of the CXCR2 signaling axis sensitized mammary tumor cells towards chemotherapy both in vitro and in vivo [6]. In addition, earlier reports have demonstrated the role of NF-kB in chemotherapy resistance and CXCR2 signaling [16, 17].
Based on accumulating evidence and our previous findings, we hypothesized that expression of CXCR2 ligands may play a critical role in the selection of aggressive mammary tumor cells with increased resistance to chemotherapy and higher metastatic potential than primary tumor cells. To elucidate the mechanism(s) of therapy resistance of breast cancer cells, we developed Cl66 murine mammary tumor cell lines resistant to doxorubicin or paclitaxel, commonly used chemotherapy drugs for breast cancer treatment. Using these cell lines, we evaluated the effect of CXCR2 signaling on various mechanisms responsible for mammary tumor cell aggressiveness and growth. Our results demonstrated that doxorubicin- and paclitaxel- resistant Cl66 cells had increased expression of CXCR2 ligands but downregulation of CXCR2 receptor. Furthermore, abrogation of the CXCR2 signaling axis decreased cell growth of doxorubicin- and paclitaxel- resistant Cl66 cells.
2. Material and methods
2.1. Cell culture
Two murine mammary adenocarcinoma cell lines Cl66 and 4T1 (6-thioguanine resistant cell line) [18, 19] and two doxorubicin or paclitaxel drug-resistant cell lines derived from Cl66 (Cl66-Dox and Cl66-Pac respectively) were used in this study. Doxorubicin (Cl66-Dox) and paclitaxel (Cl66-Pac) resistant cells were derived from Cl66, parent murine mammary tumor cell lines through continues selection of the cells in increasing drug concentrations. Cl66-Dox was maintained at 500 nM concentration of doxorubicin whereas Cl66-Pac cells were maintained at 400 nM concentration of paclitaxel for all the experiments. All the cell lines were maintained in Dulbecco's Modified Eagle Media (DMEM) (Mediatech, Hendon, VA) with 5% newborn calf serum (Sigma-Aldrich) or 5% fetal bovine serum (FBS), 1% vitamins, 1% L-glutamine and 0.08% gentamycin (Invitrogen, Carlsbad, CA). Cells were treated with different doses of doxorubicin, paclitaxel or CXCR2 antagonist.
2.2. mRNA expression analysis
Gene expression analyses were performed using quantitative RT-PCR [20]. In brief, cDNA was synthesized from 5 μg total RNA using SuperScript™ II Reverse Transcriptase (Invitrogen) and oligo(dT) primer. 2μl of first strand cDNA (1:10 dilution) was amplified using the specific primer sequences as listed in Table 1. Amplified products were resolved using a 1.5 % agarose gel containing ethidium bromide and were analyzed using an Alpha Imager gel documentation system (AlphaInnotech, San Leandro, CA). For real time quantitative RT-PCR 1ul of the undiluted cDNA products were amplified per reaction in duplicate with SYBR green master mix (Roche, Indianapolis IN) and primer mix at 10 mM concentration for each gene in a Bio-Rad iCycler (Bio-Rad, Hercules, CA). Real time PCR products were quantitated using the software Gene Expression Macro™ Version 1.1 © 2004 Bio-Rad Laboratories. The mRNA levels of glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene were used to normalize expression.
Table 1.
Primer sequences used in this study.
| Gene | Primers Sequence | Annealing temperature |
|---|---|---|
| mCXCR2 | Forward 5’- CACCGATGTCTACCTGCTGA -3’ Reverse 5’- CACAGGGTTGAGCCAAAAGT -3’ |
60°C |
| mCXCL1 | Forward 5’-TCGCTTCTCTGTGCAGCGCT-3’ Reverse 5’- GTGGTTGACACTTAGTGGTCT C-3’ |
55°C |
| mCXCL2 | Forward 5’-AGTGAACTGCGCTGTCAATG-3’ Reverse 5’-TTCAGGGTCAAGGCAAACTT-3’ |
57°C |
| mCXCL3 | Forward 5’-GCAAGTCCAGCTGAGCCGGGA-3’ Reverse 5’-GACACCGTTGGGATGGATCGCTTT-3’ |
68°C |
| mCXCL5 | Forward 5’-ATGGCGCCGCTGGCATTTCT-3’ Reverse 5’-CGCAGCTCCGTTGCGGCTAT-3’ |
68°C |
| mCXCL7 | Forward 5’-TCGTCCTGCACCAGGGCCTG-3’ Reverse 5’-AAGGGGAGCCAGCGCAACAA-3’ |
68°C |
| mGAPDH | Forward 5’- AGCCTCGTCCCGTAGACAAAA -3’ Reverse 5’- GATGACAAGCTTCCCATTCTCG -3’ |
59°C |
| mE-cadherin | Forward 5’-AGCCATTGCCAAGTACATCC-3’ Reverse 5’-AAAGACCGGCTGGGTAAACT-3’ |
60°C |
| mVimentin | Forward 5’-GCACTAACGAGTCCCTGGAG-3’ Reverse 5’-TGACGAGCCATCTCTTCCTT-3’ |
60°C |
| mRae1 | Forward 5’-CCCCAGTATCACCCAGCTTACAT-3’ Reverse 5’-CCCTCCTCTGGCCTCTCCTT-3’ |
60°C |
| mMult1 | Forward 5’-AGCTCATGTTGCACTGGAAA-3’ Reverse 5’-CATCCAAGAGAGGTGGTGGT-3’ |
55°C |
| mALDH1 | Forward 5’-GCTAGCTACAATGGAGGCACTCA-3’ Reverse 5’-GCAGCCTCCTAAATCCGACA-3’ |
55°C |
| mOct-4 | Forward 5’-GCATTCAAACTGAGGCACCA-3’ Reverse 5’-AGCTTCTTTCCCCATCCCA-3’ |
55°C |
| mSca-1 | Forward 5’-TCAGTCCTCCTGCAGACCTT-3’ Reverse 5’-ACTCCCACCTTGGAGCTTCT-3’ |
55°C |
| mABCB1 | Forward 5-GCTGCTGGTCCCATCTTCCAA-3’ Reverse 5’-TCACGACCTCACGTGTCTCT-3’ |
2.3. Cell proliferation assay
Tumor cells were plated in triplicate at a density of 5×103 cells/well in a 96 well plate and incubated for 24 hrs at 37°C. Cells were treated with different concentrations of doxorubicin or paclitaxel for 72 hrs. After 72 hrs, supernatants were collected from each well and fresh media was added to the cells along with 30μL of 5mg/ml MTT (3-(4, 5-Dimethylthiazol-2yl)-2, 5-diphenyltetrazolium bromide) (MP Biomedicals) and incubated for 2 hrs. After incubation, the media was aspirated and the precipitates were dissolved in 100 μL of DMSO (Fisher Scientific) with shaking. Absorbance was read at 570 nm on an ELx800 Bio-Tek plate reader. Percent inhibition was calculated using the following formula [100-(Absorbance of treatment / Absorbance of control group) ×100].
2.4. Enzyme-linked immunosorbant assay (ELISA)
Cell-free supernatants were collected from cells treated with different concentrations of doxorubicin or paclitaxel for 72 hrs of treatment. ELISA plates were coated with 100 μl per well of primary monoclonal antibody (2 μg/ml rat anti-mouse CXCL1/KC monoclonal, R&D Systems Inc) diluted in PBS (pH=7.4) and incubated overnight at 4°C. The next day, plates were washed and blocked with 300 μl of blocking buffer (as per manufacturer's protocol) for 1 hr. Standards (recombinant protein) and samples were added 100 μl/well in duplicate. After incubation, plates were washed and incubated with biotinylated secondary antibody 100 μl/well (0.2 μg/ml goat anti-mouse KC, R&D Systems Inc). After washing strepavidin-horseradish peroxidase (1:20000) was added and 3,3’,5,5’-tetramethylbenzidine substrate (100 μl/well) was used. Reactions were stopped using 0.15 mM sulphuric acid and plates were read at 450 nm using an ELx800 (Bio-Tek) plate reader. Concentrations were normalized to absorbance from the MTT assay described earlier.
2.5. Tumor growth and metastasis
Female BALB/c or nude mice (6-8 weeks old) were purchased from the National Cancer Institute (Bethesda, MD). In accordance with approval from the American Association of Laboratory Animal Care and current regulations and standards of the US department of Agriculture, Department of Health and Human Services and National Institute of Health, the mice used for this study were maintained in facilities under specific pathogen-free conditions. All procedures performed in mice were in accordance with institutional guidelines and approved by the University of Nebraska Medical Center Institutional Animal Care and Use Committee (IACUC). Cl66-parent, Cl66-Dox or Cl66-Pac cells (50,000 in 50 μl of HBSS) were injected orthotopically in mammary fat pad (MFP) of BALB/c or nude female mice to study tumor growth and metastasis. Tumor growth was measured twice a week. Tumor volume was calculated using the formula π/6 × (smaller diameter)2 × (larger diameter). Primary tumors recovered from mice were fixed in formalin, embedded in paraffin and processed for histopathological evaluation and immunohistochemistry. Lungs were isolated and fixed in Bouin's fixative for monitoring lung metastases macro- and microscopically.
2.6. Immunofluorescence and immunohistochemistry
For immunofluroscence, 1000-5000 cells were seeded on chamber slides overnight and were washed twice the next day with PBS (pH=7.4). The slides were fixed with 4% paraformaldehyde for 20 mins followed by washing with PBS. After blocking, cells were incubated overnight at 4°C with following the primary antibodies: goat anti-vimentin, goat anti-CXCR2 (generous gift from Dr. Robert M Strieter, School of Medicine, University of Virginia, Charlottesville, VA), mouse anti-E-cadherin, mouse anti-P-cadherin, mouse anti-N-cadherin or mouse anti- β-catenin (generous gift from Dr. Keith Johnson, University of Nebraska Medical Center, Omaha, NE). The details of the respective dilutions and florescence-tagged secondary antibodies used in this study are mentioned in Table 2.
Table 2.
IHC and IF Antibodies dilutions with experimental conditions
| Antibody | Host | Source | Cat# | Blocking | 1 Ab Dilution | 2 Ab Dilution |
|---|---|---|---|---|---|---|
| CD31 | Rabbit | Abcam | ab28364 | 30min 5% goat serum | 1:100 | 1:500 Anti-Rabbit in goat |
| KI67 | Mouse | Origene | UM800033 | 30mins,10% Horse serum | 1:200 | 1:500 anti-mouse in horse |
| CXCR2 | Goat | Dr. Strieter's lab | NA | 30min 10% horse serum | 1:1000 | 1:250 anti-goat Cy3 in donkey |
| Vimentin | Goat | Dr. Keith Johnson | SC-7557 | 30min 10% horse serum | 1:200 | 1:250 anti-goat Cy3 in donkey |
| E-cadherin | Mouse | Dr. Keith Johnson | NA | 30min 5% goat serum | 1:5 | 1:400 Cy3 anti-mouse in goat |
| N-cadherin | Mouse | Dr. Keith Johnson | NA | 30min 5% goat serum | undiluted | 1:400 Cy3 anti-mouse in goat |
| P-cadherin | Mouse | Dr. Keith Johnson | NA | 30min 5% goat serum | 1:5 | 1:400 Cy3 anti-mouse in goat |
| β-catenin | Mouse | Dr. Keith Johnson | NA | 30min 5% goat serum | undiluted | 1:400 Cy3 anti-mouse in goat |
2.7. ALDEFLUOR assay
Aldefluor levels of the parent and resistant cells were determined using an ALDEFLUOR™ assay kit (Stem Cell technologies). Cells were resuspended in media and cell number and viability were determined by Trypan blue exclusion. Cells (1×105) were resuspended in 1ml of the assay buffer. 5μl of the activated ALDEFLUOR™ reagent was added to each ‘test’ tube and 5μl of the ALDEFLUOR™ DEAB reagent was added to the control tube for each cell type. All the test and control tubes were incubated for 30 mins at 37°C. Following incubation, tubes were centrifuged for 5 min at 250 g; after removal of the supernatants cell pellets were resuspended in 0.5 ml of the assay buffer and kept at 4°C until analysis. Results were analyzed as per the manufacture's protocol.
2.8. Statistical analysis
Analysis of in vitro data was performed using Kruskal-Wallis One Way Analysis of Variance on ranks with Tukey test for multiple comparison and Mann Whitney U – test. Analysis of in vivo assays was performed using the Mann-Whitney U-test and paired t-test using Sigma plot 11. All the values are expressed as mean ± SEM. p ≤ 0.05 was considered statistically significant.
3. Results
3.1. Increased expression of CXCR2 ligands in drug-resistant cells
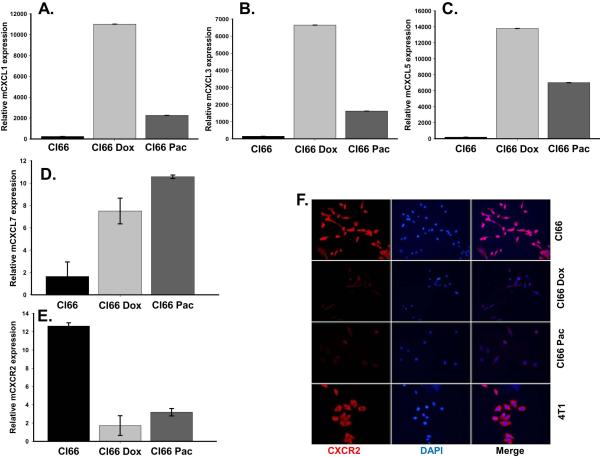
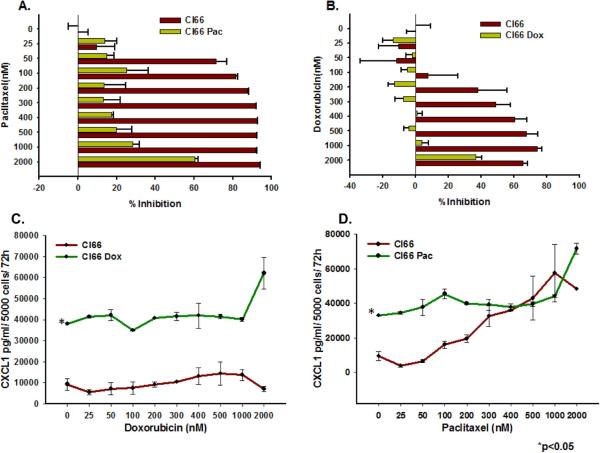
We observed higher expression of the CXCR2 ligands CXCL1, CXCL3, CXCL5 and CXCL7 in drug-resistant Cl66-Dox and Cl66-Pac cells in comparison with parent Cl66 cells at the mRNA level (Figure 1A-D.) Similarly, the CXCL1 protein level was also elevated in drug-resistant cells (Figure 2B). However, in contrast to the ligands, the expression of the receptor CXCR2 was downregulated in resistant cells (Figure 1E, F). Moreover, the drug-resistant cells were insensitive to increased drug concentrations (Figure 2A, B). There was a concomitant increase in CXCL1 expression in resistant cells when treated with increasing doses of chemotherapy (Figure 2C, D). Similar observations were made using a drug-resistant cells derived from 4T1 cells (data not shown), which were used as positive control in this study. As we observed higher expression of CXCR2 ligands by resistant cells, we evaluated the effect of inhibiting CXCR2 signaling with respect to drug resistance. In order to study CXCR2 inhibition, we treated drug-resistant and parent Cl66 cells with CXCR2 antagonists. Treatment with CXCR2 antagonist inhibited the growth of drug-resistant cells but not of parent cells (data not shown).
Figure 1. Chemotherapeutic drugs up-regulate CXCR2 ligands in doxorubicin- and paclitaxel-resistant Cl66 cells.
A-D) Expression of CXCR2 ligands CXCL1, CXCL3, CXCL5 and CXCL7 in Cl66, Cl66-Dox and Cl66-Pac cells. E) Real time PCR showing expression of CXCR2 receptor in parent and drug-resistant Cl66 cells. F) Immunofluorescence showing protein level of CXCR2 in Cl66, Cl66-Dox, Cl66-Pac cells and 4T1 cells.
Figure 2. Doxorubicin- and paclitaxel-resistant Cl66 cells are insensitive to doxorubicin and paclitaxel treatment and express higher amounts of CXCL1.
A, B) Graph showing comparison of cell viability among parent and drug-resistant cells at different drug concentrations using MTT assay. C, D) Graphs showing CXCL1 protein expression basally as well as with increasing drug treatment concentrations in Cl66-Dox and Cl66-Pac cells in comparison with the parent Cl66 cells by ELISA.
3.2. Drug-resistant cells showed delayed tumor growth and higher metastatic potential
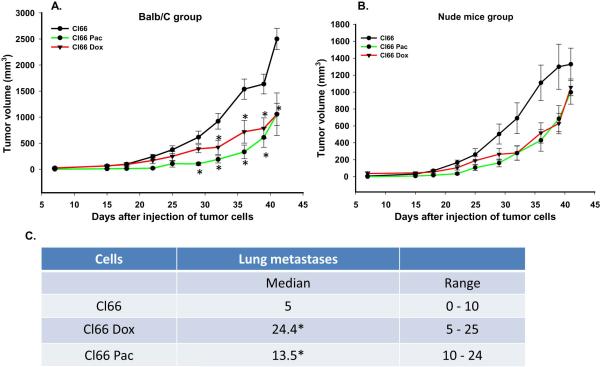
Next, we evaluated the behavior of drug-resistant Cl66 cells in vitro and in vivo. In vitro growth was examined and we found no difference in vitro growth among the parent, Cl66 Pac or C66 Dox cells (data not shown). In order to evaluate the in vivo tumor growth, we injected parent or drug-resistant Cl66 cells in the MFP of 8-week old BALB/c mice and monitored tumor growth. Mice injected with drug-resistant cells showed a delay in tumor appearance and slower tumor growth as compared to mice injected with the parent Cl66 cells (Figure 3A). To determine whether immunogenic responses elicited by resistant cells was responsible for delay in the tumor growth, we repeated these experiments using nude mice. We observed a similar trend for tumor growth in nude mice (Figure 3B). We analyzed the lungs of the tumor bearing mice and observed significantly higher metastatic lung nodules in BALB/c tumor bearing mice injected with resistant tumors as compared to mice injected with parent Cl66 cells Figure 3C). We also observed a higher serum concentration of CXCL1 (KC) in drug-resistant tumor bearing mice (data not shown).
Figure 3. Mice injected with drug-resistant Cl66 cells demonstrate slow tumor growth and higher metastases.
A, B) Tumor growth kinetics in BALB/c (A) and nude mice (B) for Cl66, Cl66-Dox and Cl66-Pac (n=5) injected cells. C) Table showing the number of metastatic lung nodules in BALB/c mice bearing tumors formed by Cl66, Cl66-Dox and Cl66-Pac cells.
3.3. Drug-resistant tumors show higher aggressiveness and angiogenesis
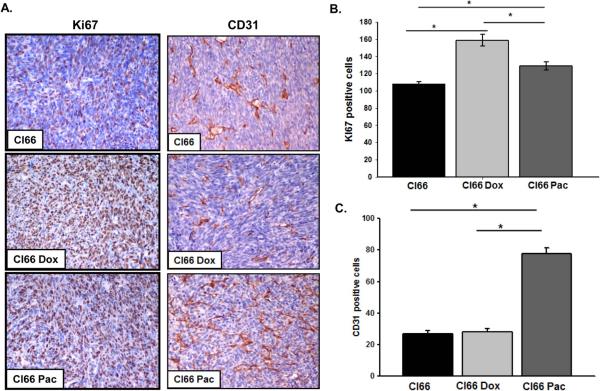
Next, in order to determine the in situ proliferation in tumors formed by parent or drug-resistant Cl66 cells in mice, we investigated for the expression of Ki67 using immunohistochemistry. We observed that tumors formed by drug-resistant Cl66 cells had higher expression of Ki67 in comparison with tumors formed by the parent Cl66 cells (p ≤ 0.001) (Figure 4A, B). Tumors formed by Cl66-Dox cells had significantly higher expression of Ki67 than tumor formed by Cl66-Pac cells (p = 0.002) or parent Cl66 cells (p ≤ 0.001) (Figure 4A, B).
Figure 4. Tumors formed by drug-resistant cells (Cl66-Dox and Cl66 Pac) show higher proliferation and angiogenesis in comparison with tumors formed by parent Cl66 cells.
A) Immunohistochemical analysis of Ki67 and CD31 in tumors formed by Cl66, Cl66-Dox and Cl66-Pac cells in BALB/c mice. B) Quantification of Ki67 in tumors. Five different areas positive for Ki67 were counted for each tumor section and mean ± SE were plotted. Cl66 vs Cl66-Dox p < 0.001, Cl66 vs Cl66-Pac p < 0.001 and Cl66-Pac vs Cl66-Dox p = 0.002. C) Quantification of CD31 in tumors. Five different areas positive for CD31 were counted for each tumor section and mean ± SE were plotted. Cl66 vs Cl66-Pac and Cl66-Dox vs Cl66-Pac p < 0.001.
As CXCR2 signaling has been associated with tumor angiogenesis, we evaluated the extent of angiogenesis in tumors formed by drug-resistant and parent cells. Immunohistochemical analysis of tumors formed by drug-resistant and parent cells revealed a significant increase in CD31 staining in tumors formed by Cl66-Pac in comparison with tumor formed by Cl66-Dox or parent Cl66 cells (p ≤ 0.001) (Figure 4A, C).
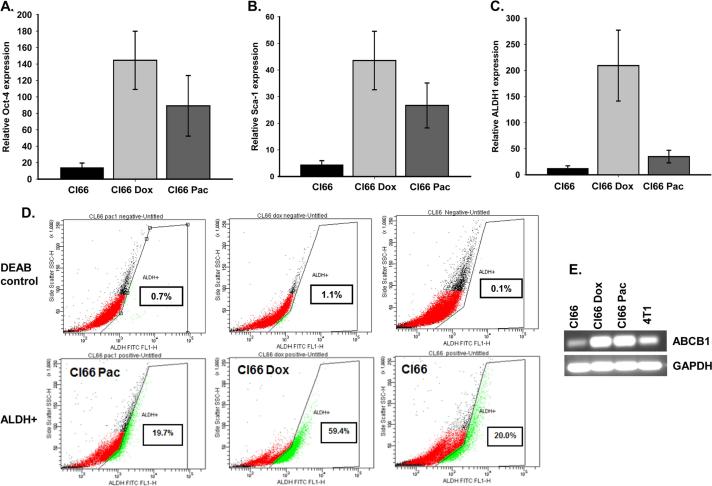
3.4. Drug-Resistant tumors expressed high levels of stem cell markers
It has been shown that selection of drug-resistant cancer cells enriches the cancer stem cell population [21, 22]. We evaluated the expression of different stem cell markers in tumors formed by resistant or parent Cl66 cells by RT-PCR. We observed higher expression of Oct-4, Sca-1, and ALDH1 stem cell markers in tumors formed by resistant cells (Figure 5A-C). To further evaluate whether drug-resistant cells have stem cell characteristics, we examined ALDH1 expression in resistant and parent cells growing in culture using RT-PCR. We observed an increase in expression of ALDH1 at the mRNA level in resistant cells in comparison with the parent Cl66 cells; Cl66-Dox cells expressed higher levels than Cl66-Pac cells (Figure 5D). We analyzed ALDH expression using an ALDEFLOUR enzymatic assays, we observed similar results. The ALDEFLOUR assay showed higher ALDH expression in Cl66-Dox cells in comparison with Cl66-Pac or parent Cl66 cells (Figure 5D). We also evaluated the expression of the commonly up-regulated multiple drug-resistant pump ABCB1 in breast cancer cells. Our results demonstrate that drug-resistant cells, Cl66-Dox and Cl66-Pac up-regulated ABCB1 as compared to parent Cl66 cells (Figure 5E).
Figure 5. Tumors formed by drug-resistant cells (Cl66-Dox and Cl66 Pax) show higher expression levels of stem cell markers in comparison with tumor formed by parent Cl66 cells.
A) Relative expression of Oct4, (B) Sca1 and (C) ALDH1 in tumors formed by parent Cl66 and drug-resistant Cl66-Dox and Cl66-Pac cells by real time PCR. D) ALDH1 expression using ALDH1 enzymatic assay in parent Cl66 and drug-resistant Cl66-Dox and Cl66-Pac cells growing in vitro. Diethylaminobenzaldehyde (DEAB) an inhibitor of ALDEFLOUR was used as a negative control for each cell line. E. Expression of multi-drug resistance pump gene ABCB1 in Cl66 parent and drug-resistant Cl66-Dox and Cl66-Pac cells by RT-PCR.
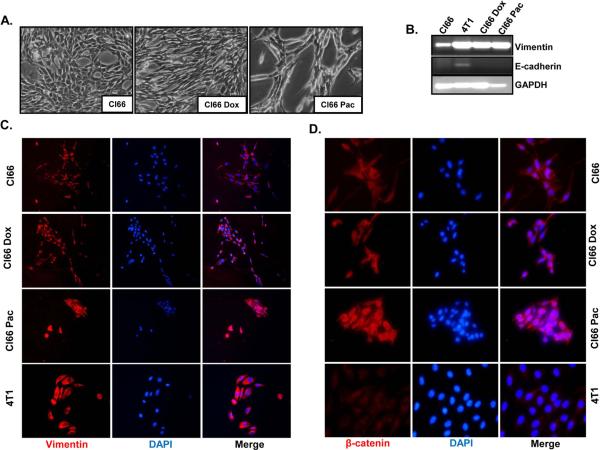
3.5. Drug-resistant cells showed epithelial to mesenchymal transition (EMT)
We observed a difference in morphology between drug-resistant and parent Cl66 cells (Figure 6A). Drug-resistant Cl66 cells appeared more elongated than parent cells. In order to investigate the reason for the observed morphological differences in these cells, we evaluated the expression of different EMT markers in drug-resistant and parent cells using RT-PCR and immunofluorescence (Figure 6A-D). We observed higher vimentin levels in drug-resistant cells at the mRNA level (Figure 6B) and we confirmed our observation using immunofluorescence (Figure 6C). Furthermore, we observed higher β-catenin levels (Figure 6D) and lower E-cadherin (data not shown) in drug-resistant cells in comparison with parent Cl66 cells.
Figure 6. Drug-resistant Cl66 cells show transition from an epithelial to mesenchymal phenotype.
A) Photographic images showing in vitro cell morphology of parent Cl66, Cl66-Dox and Cl66-Pac cells. B) Expression of vimentin and E-cadherin in Cl66 parent and drug-resistant Cl66-Dox and Cl66-Pac cells by RT-PCR. Vimentin (C) and (D) β-catenin protein expression in parent Cl66, Cl66-Dox and Cl66-Pac cells examined by immunofluorescence. Blue color represents DAPI whereas red represents cy3 for vimentin (C) and β-catenin (D).
4. Discussion
In this study, we report upregulation of CXCR2 ligands, stem cell-like and mesenchymal phenotypes as well as enhanced metastasis in mammary tumor cells selected with different chemotherapeutic agents and demonstrate the role of CXCR2 signaling axis following chemotherapy in maintaining the cancer stem cell-like phenotype and resistance. We used doxorubicin- and paclitaxel- resistant Cl66 cell lines derived from parent Cl66 murine mammary tumor cells. We observed that drug-resistant cells express higher levels of CXCR2 ligands, but lower levels of CXCR2 suggesting a feedback loop in the tumor cells which causes down regulation of the receptor when the ligands are upregulated. It has also been reported that CXCR2 signaling is important during senescence [23, 24]. The diminished expression of this receptor in resistant cells may suggest a mechanism adapted by these cells to overcome senescence.
In our in vivo studies, we observed that the tumors in mice injected with drug-resistant Cl66 cells grew slower than tumors in the mice injected with parent Cl66 cells. Surprisingly, the end point analysis of the tumor from mice injected with drug-resistant Cl66 cells revealed higher angiogenesis and proliferation with a higher incidence of metastasis than mice injected with parent Cl66 cells. These results suggest that the resistant cells may require support from the non-resistant or parent cells to support their growth, particularly during the initial stages of tumor development. Although, we observed delayed tumor growth, the resistant cells were able to survive, and being more inherently aggressive than the parent cells, they formed more metastases. Our findings are very similar to a recent publication on overexpression of human pituitary tumor transforming gene 1 in breast cancer [25]. Another possibility is that the resistant cells were removed by NK cells during earlier phases of tumor development. Higher expression of NKG2D ligands by resistant cells opens up the likelihood of attracting NK cells which ultimately helps in the killing of cancer cells [26]. Resistant cells grew slower in both BALB/c and nude mice and the only immune cells which are common between BALB/c and nude mice is NK cells. This is supported by a recent publication that showed decarbazine treatment upregulated NKG2D ligands to attract NK cells leading to killing of cancer cells [26].
We reported that drug-resistant Cl66 cells express stem cell markers. It has been documented that selecting MCF7 breast cancer cells against andriamycin enriches a population of CD44+ and CD24−/low cells [21, 22]. To investigate the possibility of cancer stem cell-like cells in our drug-resistant population, we performed an in vitro ALDEFLOUR assay with drug-resistant or parent Cl66 cells. Cl66-Dox cells had a higher percentage of ALDH positive cells than Cl66-Pac or parent Cl66 cells. This observation was in accordance with what we observed in vivo. Thus, the expression of cancer stem cell markers by resistant cells brings up the possibility of enrichment of cancer stem cells in the drug-resistant cell population which were quiescent during the initial stages of tumor development. Studies involving breast CSCss show that cells expressing CSC markers are resistant to radiation. Resistance has been seen in both in vitro and in vivo models of breast cancer stem cell response to chemo or radiation therapy [27, 28]. This resistance is further increased after ectopic expression of Wnt or β-catenin which are involved in developmental pathways and self-renewal of normal stem cells [29, 30]. These studies suggest that therapy activates the pathways involved in self-renewal of normal stem cells in cancer cells or increases the quiescent stem cell population or triggers dying cells to send signals to cancer stem cells to proliferate. Studies suggest that breast tumors may be initiated and maintained by these cells which possess stem cell-like characteristics [31]. These tumor cells are resistant to therapy and are able to metastasize to distant organs. We observed significantly enhanced metastases in tumors established from drug resistant cell lines. Furthermore, the emergence of cells expressing stem cell markers after neo-adjuvant therapy in cancer patients and in clinical trials also supports the in vitro and in vivo findings [6].
Furthermore, we observed a difference in morphology and enhanced EMT characteristics in Cl66-Pac cells in comparison with parent Cl66 cells. These differences were not observed in Cl66-Dox cells. These findings suggest that the type of drug used for selecting resistant cells will determine the behavior of cells in vivo and in vitro conditions. This also suggests that the tumor microenvironment plays a crucial role in controlling the behavior of these cells in our in vivo analysis.
Lastly, we reported that abrogating CXCR2 signaling inhibits in vitro cell growth of resistant cells. One of the downstream targets of CXCR2 signaling is NF-κB which has been reported to control various signaling pathways especially those involved during proliferation and survival of cancer cells. It has been shown that chemotherapeutic drugs upregulate NF-κB [17, 32] and higher expression of CXCR2 ligands by resistant cells raises the possibility that resistant cells may be using this pathway to their benefit to evade immunosurveillance. These findings also emphasize that cancer cells adapt various pathways to outwit chemotherapy. The CXCR2 signaling pathway is not only involved during primary tumor growth, but is also important to the process of breast cancer metastasis. Whether there is a direct or indirect relationship between CXCR2 signaling and stem cell or EMT markers needs further investigation.
In conclusion, the present study highlights the importance of CXCR2 signaling in breast cancer therapy resistance and suggests that targeting this signaling may help reduce the problem of resistance in cancer. Furthermore, the availability of CXCR2 antagonists for the treatment of asthma and other chronic obstructive pulmonary diseases (COPD) with minimal side effects [33] points to the fact that these s could be easily utilized for breast cancer treatment.
Highlights.
Differential regulation of CXCR2 ligands and stem cell-like and mesenchymal phenotypes depends on chemotherapy used.
Drug-resistant cell lines formed smaller primary tumors but were highly metastatic.
Targeting CXCR2 signaling may help circumvent therapy resistance in breast cancer.
Acknowledgements
This work was supported in part by Susan G. Komen for the Cure grant KG090860, U54CA163120 and Cancer Center Support Grant (P30CA036727) from National Cancer Institute, National Institutes of Health. Bhawna Sharma was supported through University of Nebraska Medical Center Graduate Student Fellowship/Assistantship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors declare no conflict of interest.
Reference List
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: A Cancer Journal for Clinicians. 2013 doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA: A Cancer Journal for Clinicians. 2014 doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Sharma B, Singh R. Emerging candidates in breast cancer stem cell maintenance, therapy resistance and relapse. Journal of Carcinogenesis. 2011;10:36–36. doi: 10.4103/1477-3163.91119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Acharyya S, Oskarsson T, Vanharanta S, Malladi S, Kim J, Morris P.-á., Manova-Todorova K, Leversha M, Hogg N, Seshan V.-á., Norton L, Brogi E, Massagu+¬ J. A CXCL1 Paracrine Network Links Cancer Chemoresistance and Metastasis. Cell. 2012;150:165–178. doi: 10.1016/j.cell.2012.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nannuru KC, Sharma B, Varney ML, Singh RK. Role of chemokine receptor CXCR2 expression in mammary tumor growth, angiogenesis and metastasis. J Carcinog. 2011;10:40. doi: 10.4103/1477-3163.92308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma B, Nawandar DM, Nannuru KC, Varney ML, Singh RK. Targeting CXCR2 Enhances Chemotherapeutic Response, Inhibits Mammary Tumor Growth, Angiogenesis, and Lung Metastasis. Molecular Cancer Therapeutics. 2013;12:799–808. doi: 10.1158/1535-7163.MCT-12-0529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Crusz SM, Balkwill FR. Inflammation and cancer: advances and new agents. Nat Rev Clin Oncol. 2015;12:584–596. doi: 10.1038/nrclinonc.2015.105. [DOI] [PubMed] [Google Scholar]
- 8.Luster AD. Chemokines--chemotactic cytokines that mediate inflammation. N.Engl.J Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 9.Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annu.Rev.Immunol. 2000;18:217–42. 217–242. doi: 10.1146/annurev.immunol.18.1.217. [DOI] [PubMed] [Google Scholar]
- 10.Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- 11.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002 Dec 19-26;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benoy IH, Salgado R, Van Dam P, Geboers K, Van Marck E, Scharpe S, Vermeulen PB, Dirix LY. Increased Serum Interleukin-8 in Patients with Early and Metastatic Breast Cancer Correlates with Early Dissemination and Survival. Clinical Cancer Research. 2004;10:7157–7162. doi: 10.1158/1078-0432.CCR-04-0812. [DOI] [PubMed] [Google Scholar]
- 13.Batchu RB, Moreno AM, Szmania S, Gupta SK, Zhan F, Rosen N, Kozlowski M, Spencer T, Spagnoli GC, Shaughnessy J, Barlogie B, Tricot G, Van Rhee F. High-Level Expression of Cancer/Testis Antigen NY-ESO-1 and Human Granulocyte-Macrophage Colony-Stimulating Factor in Dendritic Cells with a Bicistronic Retroviral Vector. Hum.Gene Ther. 2003;14:1333–1345. doi: 10.1089/104303403322319417. [DOI] [PubMed] [Google Scholar]
- 14.De Larco JE, Wuertz BRK, Rosner KA, Erickson SA, Gamache DE, Manivel JC, Furcht LT. A Potential Role for Interleukin-8 in the Metastatic Phenotype of Breast Carcinoma Cells. American Journal of Pathology. 2001;158:639–646. doi: 10.1016/S0002-9440(10)64005-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lev DC, Ruiz M, Mills L, McGary EC, Price JE, Bar-Eli M. Dacarbazine causes transcriptional up-regulation of interleukin 8 and vascular endothelial growth factor in melanoma cells: a possible escape mechanism from chemotherapy. Mol Cancer Ther. 2003;2:753–763. [PubMed] [Google Scholar]
- 16.Cusack JC, Liu R, Baldwin AS. NF- kappa B and chemoresistance: potentiation of cancer drugs via inhibition of NF- kappa B. Drug Resist. Updat. 1999;2:271–273. doi: 10.1054/drup.1999.0094. [DOI] [PubMed] [Google Scholar]
- 17.Huang S, DeGuzman A, Bucana CD, Fidler IJ. Level of interleukin-8 expression by metastatic human melanoma cells directly correlates with constitutive NF-kappaB activity. Cytokines Cell Mol.Ther. 2000;6:9–17. doi: 10.1080/13684730050515868. [DOI] [PubMed] [Google Scholar]
- 18.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52:1399–1405. [PubMed] [Google Scholar]
- 19.Wilson TJ, Nannuru KC, Singh RK. Cathepsin G Recruits Osteoclast Precursors via Proteolytic Activation of Protease-Activated Receptor-1. Cancer Research. 2009;69:3188–3195. doi: 10.1158/0008-5472.CAN-08-1956. [DOI] [PubMed] [Google Scholar]
- 20.Li A, Dubey S, Varney ML, Dave BJ, Singh RK. IL-8 directly enhanced endothelial cell survival, proliferation, and matrix metalloproteinases production and regulated angiogenesis. J Immunol. 2003;170:3369–3376. doi: 10.4049/jimmunol.170.6.3369. [DOI] [PubMed] [Google Scholar]
- 21.Calcagno AM, Ambudkar SV. Molecular mechanisms of drug resistance in single-step and multi-step drug-selected cancer cells. Methods in molecular biology (Clifton, N.J.) 2010;596:77–93. doi: 10.1007/978-1-60761-416-6_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Calcagno AM, Salcido CD, Gillet JP, Wu CP, Fostel JM, Mumau MD, Gottesman MM, Varticovski L, Ambudkar SV. Prolonged drug selection of breast cancer cells and enrichment of cancer stem cell characteristics. Journal of the National Cancer Institute. 2010;102:1637–1652. doi: 10.1093/jnci/djq361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Acosta JC, O'Loghlen A, Banito A, Guijarro MV, Augert A, Raguz S, Fumagalli M, Da Costa M, Brown C, Popov N, Takatsu Y, Melamed J, Adda di Fagagna F, Bernard D, Hernando E, Gil J.s. Chemokine Signaling via the CXCR2 Receptor Reinforces Senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- 24.Cichowski K, Hahn WC. Unexpected Pieces to the Senescence Puzzle. Cell. 2008;133:958–961. doi: 10.1016/j.cell.2008.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruan JW, Liao YC, Lua I, Li MH, Hsu CY, Chen JH. Human pituitary tumor-transforming gene 1 overexpression reinforces oncogene-induced senescence through CXCR2/p21 signaling in breast cancer cells. Breast cancer research : BCR. 2012;14:R106. doi: 10.1186/bcr3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hervieu A, Rebe C, Vegran F, Chalmin F, Bruchard M, Vabres P, Apetoh L, Ghiringhelli F, Mignot G. Dacarbazine-Mediated Upregulation of NKG2D Ligands on Tumor Cells Activates NK and CD8 T Cells and Restrains Melanoma Growth. J Invest Dermatol. 2013;133:499–508. doi: 10.1038/jid.2012.273. [DOI] [PubMed] [Google Scholar]
- 27.Hambardzumyan D, Squatrito M, Holland EC. Radiation resistance and stem-like cells in brain tumors. Cancer Cell. 2006;10:454–456. doi: 10.1016/j.ccr.2006.11.008. [DOI] [PubMed] [Google Scholar]
- 28.Shafee N, Smith CR, Wei S, Kim Y, Mills GB, Hortobagyi GN, Stanbridge EJ, Lee EY. Cancer stem cells contribute to cisplatin resistance in Brca1/p53-mediated mouse mammary tumors. Cancer Res. 2008;68:3243–3250. doi: 10.1158/0008-5472.CAN-07-5480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Phillips TM, McBride WH, Pajonk F. The response of CD24(−/low)/CD44+ breast cancer-initiating cells to radiation. Journal of the National Cancer Institute. 2006;98:1777–1785. doi: 10.1093/jnci/djj495. [DOI] [PubMed] [Google Scholar]
- 30.Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM. WNT/beta-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci U S A. 2007;104:618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Korkaya H, Wicha MS. Selective targeting of cancer stem cells: a new concept in cancer therapeutics. BioDrugs : clinical immunotherapeutics, biopharmaceuticals and gene therapy. 2007;21:299–310. doi: 10.2165/00063030-200721050-00002. [DOI] [PubMed] [Google Scholar]
- 32.Brea-Calvo G, Siendones E, Sanchez-Alcazar JA, de Cabo R, Navas P. Cell survival from chemotherapy depends on NF-kappaB transcriptional up-regulation of coenzyme Q biosynthesis. PloS one. 2009;4:e5301. doi: 10.1371/journal.pone.0005301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nair P, Gaga M, Zervas E, Alagha K, Hargreave FE, O'Byrne PM, Stryszak P, Gann L, Sadeh J, Chanez P, o.b.o.t.s. investigators Safety and efficacy of a CXCR2 antagonist in patients with severe asthma and sputum neutrophils: a randomized, placebo-controlled clinical trial. Clinical & Experimental Allergy. 2012;42:1097–1103. doi: 10.1111/j.1365-2222.2012.04014.x. [DOI] [PubMed] [Google Scholar]