Abstract
Protein nanopores such as α-hemolysin and MspA can potentially be used to sequence long strands of DNA quickly and at low cost. In order to provide high-speed sequencing, large arrays of nanopores are required that allow the nanopores to individually addressed, but current nanopore sequencing methods rely on ionic current measurements and such methods are likely to prove difficult to scale up. Here, we show that, by optically encoding the ionic flux through protein nanopores, the discrimination of nucleic acid sequences and the detection of sequence-specific nucleic acid binding events can be parallelized. We make optical recordings at a density of ~104 nanopores per mm2 in a single droplet interface bilayer. Nanopore blockades can discriminate between DNAs with sub-pA equivalent resolution, and specific miRNA sequences can be identified by differences in unzipping kinetics. By creating an array of 2500 bilayers with a micro-patterned hydrogel chip, we are also able to load different samples into specific bilayers, suitable for high-throughput nanopore recording.
Rapid advances in next-generation DNA sequencing make it possible to sequence a human genome in a matter of days for less than $10001. Fast human genome sequencing has initiated radical changes in clinical diagnosis, personalized medicine, and the study of genetic diseases2, 3. However, in addition to low-cost, for many of these benefits to be realised, sequencing needs to be significantly faster.
One of the most promising 3rd generation sequencing methods is nanopore sequencing. It offers advantages being inherently label-free, whilst realizing long read lengths, single-molecule resolution, low cost, high speed and portability4, 5, 6. Nanopore sequencing using ionic current recording in planar bilayers, utilizing enzyme ratcheting of the DNA7, 8, 9, has been developed by Oxford Nanopore Technologies10 and in academic laboratories11, 12, 13. Oxford Nanopore’s portable nanopore sequencer, with about 500 active pores per chip, is now available at test sites in a worldwide access program14.
Based on reported nanopore sequencing speeds (~28 ms median duration per nucleotide)12, an array of ~106 nanopores would be needed for 3×109 bases to be called with 10x coverage within (an average of) 15 min (Supplementary Text 1). This parallelization could conceivably be achieved by scaling current electrical recording methods to measure the ionic flux through each nanopore. However, since readouts from each nanopore must be separately addressed, it is difficult to see how this significant increase in scale might be achieved without sacrifices in device complexity, size and cost.
Optical recording of nanopore currents
In this paper we build on our earlier work in which nanopore ionic currents are converted into an optical signal15. By using Total Internal Reflection Fluorescence (TIRF) microscopy in Droplet Interface Bilayers (DIBs)16 (Supplementary Methods 2), we are able to image protein pores in a lipid bilayer with single-molecule resolution, and thereby detect pore blockades in many nanopores in parallel. Recent reports have used this method to identify blockades in solid state nanopores caused by DNA17, 18. Here we demonstrate the potential of this method to achieve the necessary increase in throughput required for high-speed nucleic acid detection with sequence specificity and eventually nanopore sequencing.
We monitor the fluorescence signal from the indicator dye Fluo-8 (Supplementary Materials) arising from Ca2+ flux through a nanopore. A ‘plume’ of Ca2+ appears as a bright spot at the location of each pore in the bilayer (Figure 1a). This provides an optical analogue of Single Channel Recording (oSCR). Similar methods have been pioneered by several labs as a means to observe Ca2+ flux through ion channels19, 20, 21.
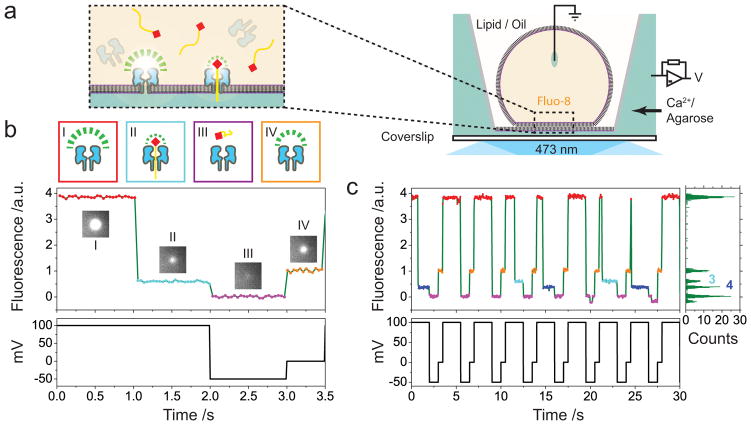
Figure 1. Optical detection of DNA by αHL in a DIB.
(a) Schematic of a single DIB. A 60x TIRF objective is used both for illumination and imaging. A voltage protocol is applied using Ag/AgCl electrodes present in the agarose substrate and in the droplet. The insert shows a cartoon of the detection process. (b) A representative fluorescent trace from a single DNA blockade cycle. Streptavidin tethered ssDNA, which is an anion, is driven into the pore at +100 mV. Simultaneously, the cationic Ca2+ flows oppositely to the DNA flow and binds with Fluo-8 to be fluorescent. (I). Streptavidin (red squares)-tethered ssDNA (yellow line) is driven into the pore, partially blocking the Ca2+ flux (II). At −50 mV, the trapped ssDNA is released (III). The fluorescence at −50 mV diminishes due to the near reversal of the Ca2+ flux at negative potentials. Then the applied bias is returned to 0 mV (IV) and the cycle repeats. The fluorescence at 0 mV comes from the diffusion of Ca2+ from the agarose substrate. The trace amplitude is normalized so that the mean intensities of (III) and (IV) are 0 and 1. The normalized fluorescence amplitude of (II) identifies the captured DNA. (c) A sequence of nanopore blockades with a mixture of two types of DNA (X5, cyan, histogram level 3; C40, blue, histogram level 4). An additional lower fluorescence level at −50 mV is occasionally populated due to gating of the αHL pore at the negative potential.
We previously reported DNA discrimination with single-base resolution by measuring the ionic flux through an α-hemolysin (αHL) nanopore. The DNA is immobilized by tethering to streptavidin, mimicking a step in the translocation produced by a processive enzyme22, 23. A first step to evaluate parallel oSCR is to perform similar measurements to calibrate our method and determine the equivalent-current and time resolutions that are possible. Given the similar sensitivities to changes in residual current reported for tethered22 and enzyme-ratcheted DNA11,12, we can use tethered DNA to help evaluate the viability of oSCR for nanopore sequencing.
Under an applied potential of +100 mV, each αHL pore in a DIB appears as a bright fluorescent spot. By controlling the hydration of the agarose substrate under the DIB, we are able to restrict nanopore diffusion to less than 20 nm during the recording time (~30 min). The capture of a streptavidin-tethered DNA results in an immediate decrease in fluorescence intensity that is specific to the type of DNA present in the pore (Figure 1b, c). Flipping the applied potential to −50 mV releases the streptavidin-DNA from the pore. After tracking (Supplementary Video 2) and quantifying the fluorescence from individual pores, the signal is normalized to the (unblocked) pore fluorescence at 0 mV and −50 mV (Supplementary Methods 3, Supplementary Video 1).
Resolution of optical nanopore recording
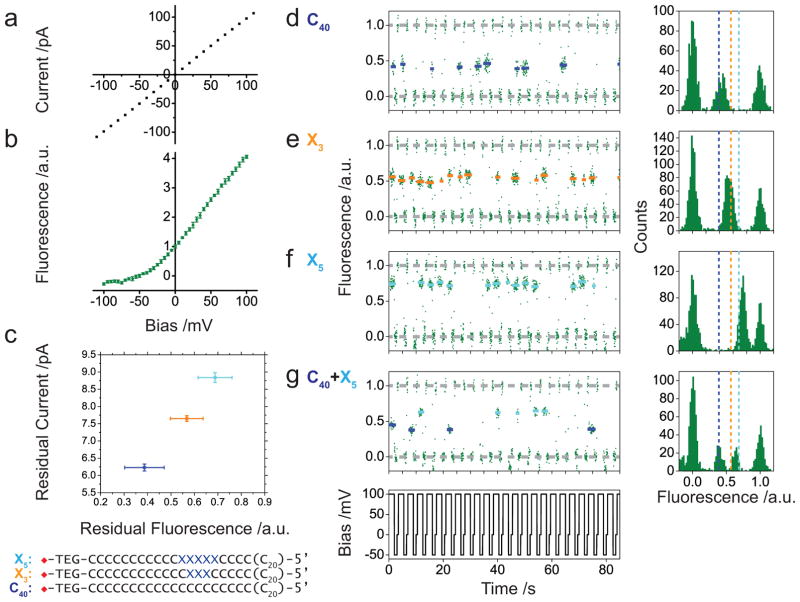
To determine our resolution, we examine the relation between applied potential, fluorescence, and equivalent currents using different homopolymeric DNAs. The current-voltage (I/V) relation in a BLM (Figure 2a, Supplementary Figure 3) can be compared to the equivalent fluorescence-voltage (F/V) response in a DIB (Figure 2b, Supplementary Figure 4). The F/V response shows an expected asymmetry, as Fluo-8 and Ca2+ are present on opposite sides of the bilayer. To measure the amplitude resolution of our method, a set of streptavidin-tethered poly-C ssDNA with increasing lengths of an abasic insert are used (C40, X3 and X5, Supplementary Table 1). There is an approximately linear relationship between the residual current (Supplementary Table 4) and the residual fluorescence (Figure 2c, Supplementary Table 2) with a fitted slope of 8.7 pA per normalized fluorescence unit. From this we can estimate that the standard error in the mean signal from a particular blocking level in our fluorescence measurements is equivalent to an error of 0.6 pA. From this plot we can conclude our optical measurement is only approximately 6 times less sensitive than our corresponding patch clamp measurements (Supplementary Methods 8). This estimate is further supported by examining the distribution of fluorescence amplitudes from an ‘all-points’ histogram for ~10 blocking events (Figure 2d–f). Each type of DNA appears as a single peak at a specific location in the histogram. There is sufficient resolution to resolve a mixture of two types of DNA that are separated by around one pA in their residual currents (Figure 2g). The minimum separation achieved using αHL WT in this work is promising, but insufficient for resolving the four nucleotides required for nanopore sequencing.
Figure 2. Amplitude resolution of oSCR for DNA identification.
(a) Current – Voltage response from a single αHL nanopore in a BLM (cis: 1.32 M KCl, 8.8 mM HEPES, pH: 7.0; trans: 0.66 M CaCl2, 8.8 mM HEPES, pH: 7.0). (b) The equivalent optical Fluorescence – Voltage relation for a single αHL in a DIB. (c) Comparison of the response from optical and electrical recording for three different DNAs. Error bars for the residual fluorescence represent the standard deviation of 120 events (Supplementary Table 2). Error bars for the residual current represent the full width at half maximum (FWHM) of the peak fitting (Supplementary Table 4, Supplementary Figure 7). (d–g), Normalized fluorescence traces (Supplementary Methods 5) for different types of streptavidin-tethered ssDNA (d, C40; e, X3; f, X5; g, C40+X5). The fluorescence intensity is normalized so that the amplitude is 0 at −50 mV and 1 at 0 mV when the pore is open. Each blockade is fitted to the mean value of the corresponding data points and overlaid with colour-coded bars. All-points histograms are displayed on the right of each trace. The centre of the colour-coded dashed lines is assigned according to the mean values for each type of DNA blockade as shown in Figure 2c. C40 and X5 are distinguished in g by a 50% threshold between the mean amplitudes corresponding to the two states. The total DNA concentration in the droplet for d–f: 267 nM. For g: 133.5 nM each for C40 and X5.
Base calling with MspA
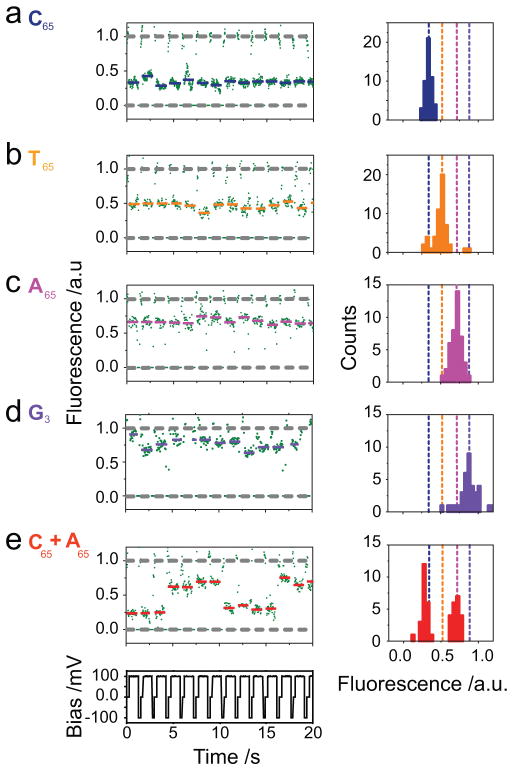
Like α-hemolysin, engineered Mycobacterium smegmatis porin A (MspA) has been shown to distinguish all four DNA bases24 and in addition shows improved separation between ionic current blockade levels12, 25. Following this previous work, we sought to exploit MspA with our method. Ionic fluxes through the MspA pore are blocked by a length of around four nucleotides at a given time13, 25. Using the M2 MspA (Supplementary Materials) pore26, 65-mer homopolymers (C65, T65, A65 or G3 in background of A65) (Supplementary Table 1) can be distinguished (Figure 3a–d and Supplementary Video 3). Event histograms show discrete amplitudes corresponding to the four DNAs (Supplementary Table 3). This pattern of current blocks is corroborated by equivalent measurements in BLMs under the same conditions (Supplementary Table 5 and Supplementary Figure 8). Mixing C65 and A65 results in two classes of blockades appearing at separate and distinguishable levels in the fluorescence trace (Figure 3e).
Figure 3. Optical discrimination of four nucleotides using the MspA M2 nanopore.
When streptavidin-tethered biotinylated DNA oligonucleotides block a pore, the ion flux is restricted and we observe a reversible stepwise change in the fluorescence intensity. (a–d), Normalized optical traces (Supplementary Methods 6) for the following 65-mers (Supplementary Table 1): C65 (5′-Biotin-CCCCCCCCCCCC-CCC-C35-CCTGTCTCCCTGCCG-3′), T65 (5′-Biotin-TTTTTTTTTTTT-TTT-T35-CCTGTCTCCCTGCCG-3′), A65 (5′-Biotin-AAAAAAAAAAAA-AAA-A35-CCTGTCTCCCTGCCG-3′) or G3 in background of A65 (5′-Biotin-AAAAAAAAAAAA-GGG-A35-CCTGTCTCCCTGCCG-3′). The blockades are fitted to the mean value of the corresponding data points and represented with a coloured bar. Event histograms displaying the mean amplitudes are on the right of the traces for each DNA oligonucleotide (Supplementary Table 3). (e), Normalized optical trace and corresponding event histogram for a mixture of C65 and A65.
miRNA detection
These measurements exploit differences in residual ionic flux to distinguish different species. However, if sufficiently dissimilar, mean event durations can also be used to discriminate different species. One important application that exploits event durations is the detection of specific microRNAs (miRNA)27. miRNAs are short non-coding RNA fragments that play an important role in the post-transcriptional regulation of gene expression28, 29. Because of this role, they are also important disease markers. When hybridized with a pre-designed DNA probe and subjected to an electrical potential in a nanopore, miRNAs can be forced to unzip and are subsequently translocated through the pore. The unzipping kinetics, are controlled by the strength of hybridization, and therefore can be used to reveal miRNA identity27.
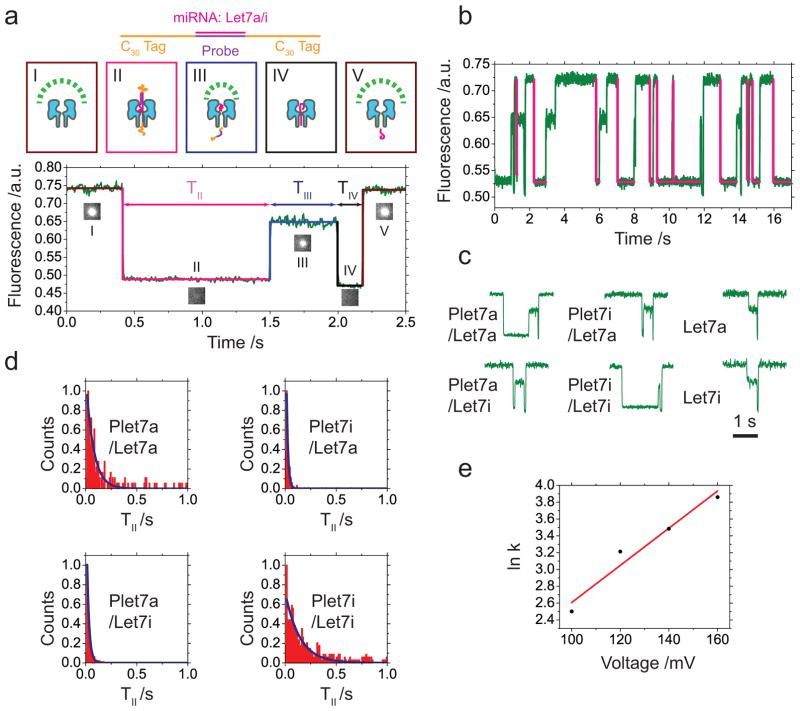
As a proof of concept, we designed DNA probes (Plet7a and Plet7i) complementary to the let-7 (Let7a and Let7i) miRNA (Supplementary Table 1) that are distinguished by a 4 base difference in sequence (Figure 4a). Let-7 is a direct regulator of RAS expression in human cells that is expressed at higher levels in more differentiated tumors30. Combinations of miRNA with DNA probes are thermally annealed (Supplementary Methods 1) before pipetting a droplet containing one type of probe/miRNA to form a DIB (Figure 1a). Under a constant potential (+160 mV), fluorescent spots derived from each αHL pore blink spontaneously (Supplementary Video 4), corresponding to miRNA unzipping and translocation events. A typical unzipping event includes three characteristic blockade levels (Figure 4a, b). Combinations of fully complementary probe and miRNA sequence generate long event durations, those with base mis-matches show short events, and controls missing the DNA probe show no unzipping events (Figure 4c, Supplementary Figure 10, Supplementary Table 6). Event durations show an exponential distribution (Figure 4d) and reflect the strength of hybridization between the miRNA and the probe. The fitted rate constant (Supplementary Text 2) for the unzipping process (Supplementary Table 7) determines the miRNA identity (Figure 4d). The voltage dependence of the rate constant for a weakly hybridizing combination (Plet7a/Let7i) shows an expected exponential relationship with the applied potential (Figure 4e, Supplementary Text 2). At low potentials, strongly hybridized combinations (Plet7i/Let7i) show blockade (TII) durations that are too long to measure. (Supplementary Figure 11).
Figure 4. Detection of miRNA sequences by oSCR based on unzipping event duration.
(a) A representative miRNA unzipping event. miRNA (Let7a or Let7i) hybridizes with the DNA probe (Plet7a or Plet7i). Poly-C30 ssDNA tags on both ends of the probe are designed to enable pore-threading and initiate unzipping. At +160 mV, an open nanopore (I) shows a decrease in fluorescence when the hybridized complex is captured and subsequently unzipped (II). Following unzipping, when the DNA probe has translocated through the pore the miRNA remains in the vestibule (III). The miRNA then translocates (IV) and the pore re-opens (V). (b) A series of miRNA (Plet7a/Let7a) unzipping events at +160 mV. Magenta fitting lines (Supplementary Figure 9) highlight capture/unzipping (II) events. (c) Different probe/miRNA combinations show different capture/unzipping times (II). Matched miRNA and probe generate long events. miRNA without probe shows no capture/unzipping events (II). (d) Histograms of capture/unzipping event (II) lifetimes for all the probe/miRNA combinations fit with exponentials. The fitted rate constant for unzipping reflects the hybridization strength (Supplementary Text 2). (e) Dependence of the unzipping rate constant on applied potential for Plet7a/Let7i (Supplementary Figure 11).
Optical recording from nanopores in a bilayer array
Multiple fluorescence traces (>100) from active nanopores can be analysed simultaneously at a density of 104 pores per mm2 (Supplementary Figure 5, 17, Supplementary Video 3, 4), even for our limited (150 μm × 150 μm) field of view (Figure 5a, b). Although it is possible to utilize the super-resolved locations of individual nanopores, a more conservative estimate of the maximum measurement density can be made by observing adjacent pores, which can easily be resolved when separated by around 3 μm (Supplementary Figure 6). This corresponds to an effective measurement density of 105 pores per mm2 (Supplementary Text 1). Therefore, our measurements suggest that oSCR can be used to parallelize the detection of specific nucleic acids for both DNA sequencing and miRNA screening.
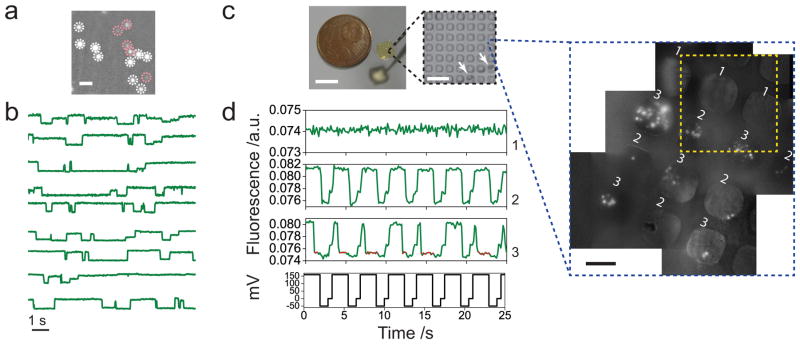
Figure 5. High throughput and multi-sample oSCR.
(a) A frame containing multiple fluorescence spots representing open (white dashed circles) and blocked nanopores (pink dashed circles, DNA: streptavidin tethered C40). Scale bar: 10 μm. (b) Parallel recordings of nine fluorescence traces simultaneously extracted from the same field of view. The fluorescence traces show spontaneous amplitude transitions due to consecutive miRNA (Plet7a/Let7i) capture/unzipping events (Supplementary Video 4). oSCR results in a and b are both recorded in DIBs. (c) A Fluo-8 containing hydrogel chip with cast pillar-array layer, scale bar: 4 mm (Supplementary Figure 13). The image inset in a black dashed square shows separation of the formed bilayers from the unformed. After HHBa formation (Supplementary Video 6), the pillar array is lifted slowly to separate the bilayers. Boundary lines delimiting the area containing formed bilayers are visualized (white arrows). Scale bar: 140 μm. Inset in blue dashed square: multiple fluorescence images have been stitched together to show an expanded view of an area of the chip containing various biological samples (1. −αHL, -DNA; 2. +αHL, -DNA; 3. +αHL, +DNA). Yellow dashed square: a single frame accommodates four bilayers at a time. Scale bar: 40 μm. (d) Parallel single-molecule nanopore activity from a full frame HHBa recording. The fluorescence traces are recorded simultaneously from nanopores in different bilayers of the array (see c, yellow dashed square). The unspotted HHB shows constant fluorescence (Trace 1). The HHB loaded with αHL shows fluorescence changes synchronized with the voltage protocol (Trace 2). DNA (C40) blockades (red points on the trace) are only detected in Trace 3.
However, these high throughput measurements from a single DIB are limited to a single sample of pores plus analytes in any bilayer. The area of an individual DIB is also limited to ~1 mm2 due to the increased likelihood of bilayer rupture for large area DIBs at potentials more than 100 mV. Therefore, parallelization of, not only the nanopores, but also of the bilayers would significantly improve the utility of oSCR. In principle, a parallel array of droplets equipped with an array of inserted electrodes could be used. However, this approach presents significant technical challenges for electrode fabrication, alignment and insertion. We have circumvented this problem by creating bilayer arrays between an agarose substrate and a micro-patterned agarose chip.
To demonstrate this concept, a Fluo-8 containing hydrogel chip (Figure 5c) is fabricated by using agarose cast from a lithographic master (Supplementary Figure 13). An array of micro-pillars (50 μm × 50 μm) at a density of 200 bilayers per mm2 (Figure 5c, Supplementary Figure 16) can accommodate different analytes and pores. Samples are loaded into the array with a pulled glass capillary driven by a spotting robot (Supplementary Figure 15). After loading, the chip is flipped and incubated in the lipid/oil solution for 10 min. Then, when the chip surface is brought into contact with the hydrogel-coated substrate coverslip, bilayers form spontaneously (Figure 5c black dashed inset, Supplementary Video 6) to create a Hydrogel-Hydrogel Bilayer array (HHBa).
Following bilayer formation, individual inserted nanopores (αHL in this case) can be resolved as fluorescent spots in each bilayer. Nanopores can be selectively loaded into individual bilayers of the array (Supplementary Figure 15) and oSCR for multiple different samples of pores or analytes can be recorded simultaneously in the same field of view (Figure 5c blue dashed inset). oSCR with different combinations of αHL and streptavidin-ssDNA in a HHBa is shown in Figure 5d. As the HHBa forms slightly further away from the glass coverslip surface compared to oSCR with individual DIBs, our illumination intensity is reduced, and we observe a reduced signal-to-noise ratio compared to an individual DIB.
Limitations
These methods are not without limitations. The electron-multiplying CCD used in these experiments limits our temporal resolution to approximately 2 ms, which although too slow for many ion channels, is suitable for measuring current blockades relevant for nanopore sequencing based on enzyme ratcheting. Given the relatively large photon fluxes we detect, the time resolution could be improved by replacing this camera with a less-sensitive, but faster (sCMOS) detector.
In the present configuration, the maximum period for data acquisition is limited by accumulation of Ca2+ in the droplet to around 30 minutes. As Ca2+ moves into the droplet, the background fluorescence increases. A 30-minute maximum duration places little restriction on most conceivable nanopore measurements, particularly if the goal is high-speed high throughput detection. Furthermore, in the present set up, the total area we can image with our camera of 150 × 150 μm. This gives a maximum theoretical throughput of approximately 3000 pores31. We are investigating lens-free32 and macroscopic TIRF imaging33 as potential avenues for improvement.
With our current sensitivity, oSCR would require around a 36-fold decrease in sequencing speed from a single nanopore, as compared to direct electrical detection of nanopore currents. However, this reduction is already more than compensated by the parallelization enabled by oSCR, notwithstanding any future improvements in optical detection sensitivity.
Improvements in HHBa device alignment and geometry with more mechanically flexible hydrogel materials should readily improve the signal:noise we report through greater control of the position of the bilayer relative to the underlying substrate. The simplicity of our design would also be compatible with future portable high throughput devices requiring integrated modules for microfluidics, voltage generation, LED illumination and on-chip imaging.
Conclusions
An optical rather than an electrical readout has enabled significant gains in the parallelization of ionic flux recording through nanopores, whilst retaining amplitude resolution comparable to that achieved with conventional single-channel electrical recording (~1pA) with no requirements for biomolecule labelling. We have used this amplitude resolution to demonstrate single base resolution using oSCR with a MspA nanopore; successfully combining this method with enzyme-ratcheting of the DNA would enable parallel DNA sequencing.
Scaling DIBs beyond a single bilayer also enables easy loading of different nanopores and different analytes into separate bilayers. We use straightforward, low-cost fabrication methods and simple materials to create a miniaturized (5 mm × 5 mm) disposable device. Loading different types of nanopore into different bilayers in the array has the potential to improve the base-calling ability of nanopore sequencing.
Overall, this ~1000-fold improvement in measurement density over current methods has the potential to overcome current barriers in nanopore-based sequencing and sensing. Comparing to current state of the art electrical measurement in nanopores, we estimate that with these potential improvements in technology, optical recording from an array of nanopore should produce sequencing signals with a rate of 106 bases mm−2 s−1, which could in principle produce a human genome sequence in as little as 15 minutes.
Methods
DIB formation
Protein nanopores (1.2 nM) and analytes (streptavidin-tethered ssDNA [267 nM for αHL and 2 μM for MspA] or miRNA [267 nM]) are placed in a 200 – 350 nL droplet (1.32 M KCl, 8.8 mM HEPES, 0.4 mM EDTA, pH 7.0 [αHL] / 8.0 [MspA], 40 μM Fluo-8), which is incubated in 3 mM 1,2-diphytanoyl-sn-glycero-3-phosphocholine (DPhPC) in hexadecane (Supplementary Materials) to form a lipid monolayer coating. The droplet is transferred by pipetting onto a cover slip in the measurement chamber (Figure 1a, Supplementary Figure 1). The cover slip has been spin-coated (3000 rpm, 30 s) with a thin layer (~200 nm) of agarose (0.66 M CaCl2, 8.8 mM HEPES, pH 7.0 [αHL] / 8.0 [MspA]) and subsequently incubated with 3 mM DPhPC in hexadecane. Upon contact with the monolayer on the agarose, the lipid-coated droplet spontaneously forms a DIB. A ground electrode (Ag/AgCl), is inserted into the droplet, with a corresponding active electrode (Ag/AgCl) in the substrate agarose. Voltage protocols are applied with a patch clamp amplifier (Axopatch 200B, Molecular Devices). Nanopores present in the droplet spontaneously insert into the DIB and the ion flux is detected both electrically (Supplementary Figure 2) and optically (Figure 1).
TIRF Microscopy
TIRF measurements are performed with a Nikon Eclipse Ti microscope equipped with a 60x oil immersion objective (Plan Apo TIRF, Nikon). The fluorescence is excited by a 473 nm Argon ion laser (Shanghai Dream Laser Technologies) and imaged with an electron-multiplying CCD camera (Ixon3, Andor). In the TIRF recording, the full field of view is 150 μm × 150 μm. Parameters such as the exposure time, EM gain and the binning size are optimized to achieve the best S/N ratio for each recording. The highest recording rate that has been tested is 2 ms per frame.
Data Analysis
The fluorescence images are recorded in .sif format (Andor Solis). Fluorescence traces are extracted from the image series by tracking and Gaussian fitting for each individual fluorescent spot. For DNA identification, the trace is further normalized according to the reference fluorescence levels (Supplementary Methods 3 and 4).
Hydrogel-Hydrogel Bilayer Array
The PDMS moulds for the hydrogel chips are fabricated by standard photolithography and soft lithographic methods (Supplementary Methods 6,7). The hydrogel chip is fabricated by filling a PDMS mould with buffer-containing (1.32 M KCl, 8.8 mM HEPES, 0.4 mM EDTA, pH 7.0) molten agarose (3% v/v, agarose for routine use) followed by desication, gel casting and peeling off. The cast hydrogel chip is infused with Fluo-8 (40 μM) and immersed in hexadecane before spot loading (Supplementary Figure 13). Following loading (Supplementary Figure 15, Supplementary Video 5), the chip is flipped and placed on an electrode ring (Supplementary Figure 14) both for electrical connection and chip manipulation. Due to the larger chip size (5 mm × 5 mm) compared with a droplet (less than 1 mm diameter), a lipid/oil chamber with an enlarged opening is designed for HHBa formation and measurement (Supplementary Figure 12). The HHBa forms spontaneously when the chip is annealed with the substrate agarose in a solution of 3 mM DPhPC in hexadecane (Supplementary Video 6). Voltage protocols are applied with a patch clamp amplifier (Axopatch 200B, Molecular Devices). Nanopores are monitored with TIRF microscopy (Supplementary Figure 16) as described above. A 30 ms exposure time is used for recording. Four bilayers can be imaged simultaneously (Supplementary Video 7).
Supplementary Material
Acknowledgments
The authors thank Dirk Aarts for clean room access, Tanuj Sapra for the clean room training and Ellina Mikhailova for the protein preparation. This work is supported by funding from the US National Human Genome Research Institute (NHGRI) “$1000 Genome” research grant “R01 HG003709” and Oxford Nanopore Technologies Ltd.
Footnotes
Author Contributions
S.H., M.R.R., O.C., H.B. & M.I.W. designed the experiments. S.H., M.R.R., O.C. performed the experiments. S.H. wrote the data analysis package program for the measurements with αHL and performed the related data analysis. M.I.W. designed the data analysis methodology for the measurements with MspA. M.R.R. performed the MspA related data analysis. S.H. designed and wrote the spotting robot program for the hydrogel array. S.H., M.R.R., O.C., H.B. & M.W. wrote the paper.
References
- 1.Mardis ER. Anticipating the $1,000 genome. Genome Biology. 2006;7:12. doi: 10.1186/gb-2006-7-7-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bayley H. Sequencing single molecules of DNA. Current Opinion in Chemical Biology. 2006;10:628–637. doi: 10.1016/j.cbpa.2006.10.040. [DOI] [PubMed] [Google Scholar]
- 3.Xuan JK, Yu Y, Qing T, Guo L, Shi LM. Next-generation sequencing in the clinic: Promises and challenges. Cancer Letters. 2013;340:284–295. doi: 10.1016/j.canlet.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Branton D, et al. The potential and challenges of nanopore sequencing. Nat Biotechnol. 2008;26:1146–1153. doi: 10.1038/nbt.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Venkatesan BM, Bashir R. Nanopore sensors for nucleic acid analysis. Nature Nanotech. 2011;6:615–624. doi: 10.1038/nnano.2011.129. [DOI] [PubMed] [Google Scholar]
- 6.Huang S. Nanopore-based sensing devices and applications to genome sequencing: a brief history and the missing pieces. Chinese Science Bulletin. 2014;59:4918–4928. [Google Scholar]
- 7.Cockroft SL, Chu J, Amorin M, Ghadiri MR. A single-molecule nanopore device detects DNA polymerase activity with single-nucleotide resolution. J Am Chem Soc. 2008;130:818–820. doi: 10.1021/ja077082c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu J, Gonzalez-Lopez M, Cockroft SL, Amorin M, Ghadiri MR. Real-Time Monitoring of DNA Polymerase Function and Stepwise Single-Nucleotide DNA Strand Translocation through a Protein Nanopore. Angew Chem Int Ed. 2010;49:10106–10109. doi: 10.1002/anie.201005460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lieberman KR, et al. Processive Replication of Single DNA Molecules in a Nanopore Catalyzed by phi29 DNA Polymerase. J Am Chem Soc. 2010;132:17961–17972. doi: 10.1021/ja1087612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pennisi E. DNA Sequencers Still Waiting for the Nanopore Revolution. Science. 2014;343:829–830. doi: 10.1126/science.343.6173.829. [DOI] [PubMed] [Google Scholar]
- 11.Cherf GM, et al. Automated forward and reverse ratcheting of DNA in a nanopore at 5-angstrom precision. Nat Biotechnol. 2012;30:344–348. doi: 10.1038/nbt.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manrao EA, et al. Reading DNA at single-nucleotide resolution with a mutant MspA nanopore and phi29 DNA polymerase. Nat Biotechnol. 2012;30:349–353. doi: 10.1038/nbt.2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Laszlo AH, et al. Decoding long nanopore sequencing reads of natural DNA. Nat Biotechnol. 2014;32:829–833. doi: 10.1038/nbt.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bayley H. Nanopore sequencing: from imagination to reality. Clinical Chemistry. 2015;61:25–31. doi: 10.1373/clinchem.2014.223016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heron AJ, Thompson JR, Cronin B, Bayley H, Wallace MI. Simultaneous Measurement of Ionic Current and Fluorescence from Single Protein Pores. J Am Chem Soc. 2009;131:1652–1653. doi: 10.1021/ja808128s. [DOI] [PubMed] [Google Scholar]
- 16.Bayley H, et al. Droplet interface bilayers. Molecular Biosystems. 2008;4:1191–1208. doi: 10.1039/b808893d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ivankin A, et al. Label-Free Optical Detection of Biomolecular Translocation through Nanopore Arrays. ACS Nano. 2014;8:10774–10781. doi: 10.1021/nn504551d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anderson BN, et al. Probing Solid-State Nanopores with Light for the Detection of Unlabeled Analytes. ACS Nano. 2014;8:11836–11845. doi: 10.1021/nn505545h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demuro A, et al. Calcium dysregulation and membrane disruption as a ubiquitous neurotoxic mechanism of soluble amyloid oligomers. Journal of Biological Chemistry. 2005;280:17294–17300. doi: 10.1074/jbc.M500997200. [DOI] [PubMed] [Google Scholar]
- 20.Zou H, Lifshitz LM, Tuft RA, Fogarty KE, Singer JJ. Visualization of Ca2+ entry through single stretch-activated cation channels. Proc Natl Acad Sci USA. 2002;99:6404–6409. doi: 10.1073/pnas.092654999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang SQ, Song LS, Lakatta EG, Cheng HP. Ca2+ signalling between single L-type Ca2+ channels and ryanodine receptors in heart cells. Nature. 2001;410:592–596. doi: 10.1038/35069083. [DOI] [PubMed] [Google Scholar]
- 22.Stoddart D, Heron AJ, Mikhailova E, Maglia G, Bayley H. Single-nucleotide discrimination in immobilized DNA oligonucleotides with a biological nanopore. Proc Natl Acad Sci USA. 2009;106:7702–7707. doi: 10.1073/pnas.0901054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wallace EVB, et al. Identification of epigenetic DNA modifications with a protein nanopore. Chemical Communications. 2010;46:8195–8197. doi: 10.1039/c0cc02864a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Derrington IM, et al. Nanopore DNA sequencing with MspA. Proc Natl Acad Sci USA. 2010;107:16060–16065. doi: 10.1073/pnas.1001831107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manrao EA, Derrington IM, Pavlenok M, Niederweis M, Gundlach JH. Nucleotide Discrimination with DNA Immobilized in the MspA Nanopore. Plos One. 2011;6:e25723. doi: 10.1371/journal.pone.0025723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Butler TZ, Pavlenok M, Derrington IM, Niederweis M, Gundlach JH. Single-molecule DNA detection with an engineered MspA protein nanopore. Prc Natl Acad Sci USA. 2008;105:20647–20652. doi: 10.1073/pnas.0807514106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Y, Zheng DL, Tan QL, Wang MX, Gu LQ. Nanopore-based detection of circulating microRNAs in lung cancer patients. Nature Nanotech. 2011;6:668–674. doi: 10.1038/nnano.2011.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 29.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 30.Johnson SM, et al. RAS is regulated by the let-7 MicroRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 31.Gross LCM, Castell OK, Wallace MI. Dynamic and Reversible Control of 2D Membrane Protein Concentration in a Droplet Interface Bilayer. Nano Lett. 2011;11:3324–3328. doi: 10.1021/nl201689v. [DOI] [PubMed] [Google Scholar]
- 32.Coskun AF, Su TW, Ozcan A. Wide field-of-view lens-free fluorescent imaging on a chip. Lab on a Chip. 2010;10:824–827. doi: 10.1039/b926561a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Castell OK, Berridge J, Wallace MI. Quantification of Membrane Protein Inhibition by Optical Ion Flux in a Droplet Interface Bilayer Array. Angew Chem Int Ed. 2012;51:3134–3138. doi: 10.1002/anie.201107343. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.