Abstract
Biodegradable conjugated polymer nanoparticles (CPNs) were prepared for high mitochondria targeting in live cancer cells. The degradable CPNs are nontoxic and specifically localized to mitochondria of live tumor cells through macropinocytosis followed by intracellular degradation and trafficking.
Conjugated polymer nanoparticles (CPNs) are intrinsic fluorescent soft materials fabricated by self-assembly of non-aqueous soluble conjugated polymers (CPs) in an aqueous solution.1, 2 Owing to their excellent photophysical (i.e., high brightness and photostability) and biophysical properties (i.e., high cellular entry and nontoxicity), CPNs have recently gained much attention for microscopic live cell/tissue imaging,3–5 biological sensing,6, 7 and nucleic acid delivery.8, 9 By introducing non-conjugated degradable linkers along the conjugated backbones, we have synthesized biodegradable CPs exhibiting similar fluorescent properties of fully conjugated CPs.10 The backbone modification with flexible linker affects the self-assembly properties of CPs, resulting in significantly changed cellular entry pathways.11
Despite successful cellular applications, the high molecular weights of CPs are not ideal for labelling or delivery to intracellular organelles due to inefficient diffusion and trafficking ability. For organelles with additional membranes such as nucleus and mitochondria, the necessary penetration of the organelle membranes further decreases the labeling and delivery efficiency. For the first time, here we present a strategy to achieve efficient mitochondria labeling of live cells using intracellular degradation of CPNs. We have designed biodegradable CPNs by self-assembly of disulfide containing CPs. Upon endocytosis, CPNs were disassembled and degraded to low molecular weight, fluorescent conjugated oligomers (COs) that were efficiently trafficked to mitochondria (Fig. 1). The results support that biodegradable CPNs are promising materials for labeling and therapeutic delivery to mitochondria, which is an important therapeutic target given that mitochondria dysfunction is related to various diseases such as cancer, obesity, diabetes, and neurodegenerative disorders.12
Fig. 1.
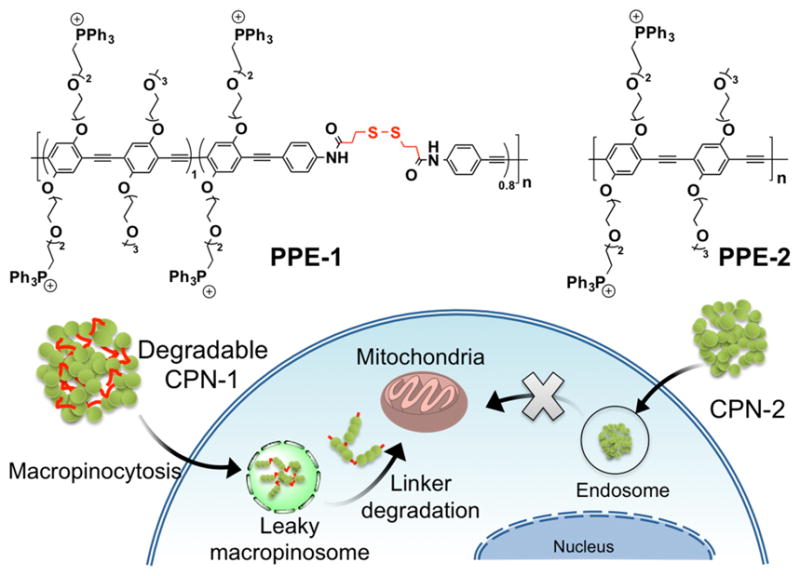
Chemical structures of PPEs with (PPE-1) and without (PPE-2) biodegradable linkers in the backbone and a schematic illustration of cellular entry and mitochondria localization of biodegradable CPNs. CPN-1 and CPN-2 were fabricated with the corresponding PPEs via self-assembly in water.
Biodegradable poly(p-phenyleneethynylene)s (PPEs) were synthesized by polymerizing monomers including a disulfide-containing monomer (see ESI). Disulfide bonds are reducible under a high concentration of intracellular glutathione (~0.5–10 mM).13 To obtain the fluorescent properties comparable to a fully conjugated PPE, the amount of disulfide monomer in the PPE backbone was maintained less than 50%. To improve mitochondria targeting function, triphenylphosphonium (TPP) salts were introduced by reacting the PPEs containing bromine side chains with triphenylphosphine (see ESI). TPP is a well-known mitochondria-targeting small molecule and has been used for modifying small molecules, nanoparticles, and liposomes.14, 15 Proton and phosphorus nuclear magnetic resonance (NMR) spectroscopy confirmed the quantitative introduction of TPP salts (see ESI). The resulting PPE-1 (Mn = 9,300) and -2 (Mn = 16,000) were soluble in organic solvents such as dimethylsulfoxide (DMSO) and methanol. The ratio of repeating units of PPE-1 was calculated as ~1.0:0.8 using proton NMR integration values, indicating that the average length of COs along PPE-1 backbone is around 5 repeating units (i.e., pentamer) or slightly larger.
CPNs were prepared by diluting TPP-modified PPEs in DMSO with an excess amount of water (i.e., final DMSO concentration of less than 1 %).16 Using fully conjugated PPEs, non-degradable CPNs were also fabricated to support the concept that the degradability of CPNs is crucial for high intracellular organelle targeting. The photophysical and physical properties of both CPNs were somewhat similar. Both CPNs exhibit broad emission spectra centered on ~470 nm. Using nanoparticle tracking analysis, the mean hydrodynamic diameters of CPN-1 and -2 were determined as 139.9 ± 3.0 and 152.7 ± 6.7 nm, respectively (see ESI). The difference in the average size is believed to associate with the size distribution of CPNs. Indeed, both CPNs exhibit very similar mode diameters (i.e., the diameter of the major population) of 92–93 nm. Zeta potentials of CPN-1 and -2 were +24 and +29 mV, respectively. Attempts to take transmission electron microscopic (TEM) images were not successful as CPNs heavily aggregated on a copper grid during sample preparations.
The metabolic activities of human cervical carcinoma (HeLa) cells incubated with biodegradable CPN-1 and non-degradable CPN-2, respectively, for overnight were monitored to evaluate the toxicity of CPNs. Interestingly, while degradable CPN-1 exhibits no viability inhibition up to 40 μM, non-degradable CPN-2 exhibits substantially high toxicity starting from 10 μM (see ESI). To test whether the molecular weights of PPEs are associated with the toxicity, we synthesized a low molecular weight (Mn = 6,960) non-degradable PPE (PPE-3) by breaking the monomer stoichiometric balance in polymerization. The CPN fabricated with low molecular weight PPE (i.e., CPN-3) exhibits very similar toxicity to that of CPN-2, suggesting that the toxicity of CPN-2 is not related to the molecular weight of the corresponding PPE. Although further systematic investigations are necessary to better understand the backbone structure and the cellular toxicity relationship, we speculate that the degraded oligomers containing thiols may contribute to the lower toxicity by reducing the level of reactive oxygen species.17
CPNs were taken up by live cells and found in the cytosolic compartment (Fig. 2). Unlike CPN-2, which exhibits a characteristic punctuated pattern, the fluorescent image of CPN-1 was somewhat diffused along with some granular patterns (Fig. 2). The diffused pattern is indicative of disassembly of CPNs followed by degradation to COs via disulfide degradation under high intracellular glutathione concentration. Non-degraded CPNs were seen as punctuated dots. Nonetheless, the microscopic images are clearly different between biodegradable CPN-1 and non-degradable CPN-2. We ruled out the possibility that pre-degradation of CPN-1 in extracellular environment followed by diffusion or uptake of COs by cells, because disulfide reduction at the extracellular environment will be very inefficient due to lower glutathione concentration (i.e., one to three orders of magnitude lower).13
Fig. 2.
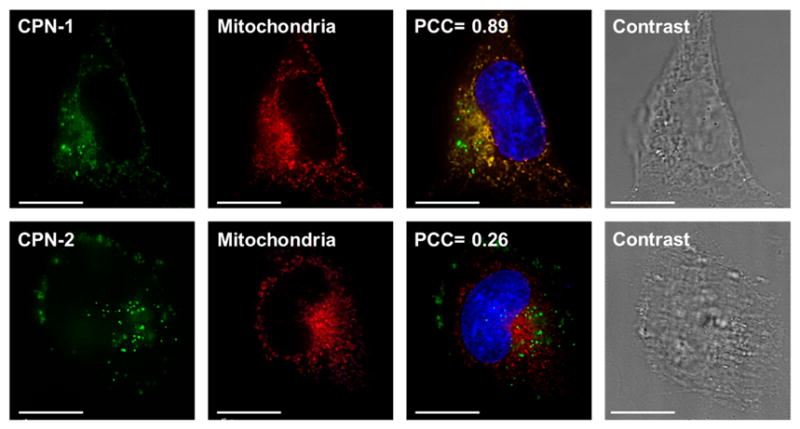
Microscopic images of HeLa cells incubated with CPNs for 18 h followed by mitochondria (red) and nuclear (blue) staining. The scale bar is 20 μm.
The PPE stock concentration in DMSO is influential for CPN formation and thus for fluorescent microscopic imaging. The higher stock concentration, the more punctuated spots observed in the microscopic images (see ESI). As polymer concentration increases, the chances of interpolymer aggregation via π-π staking in a poor solvent increase. The punctuated spots are indicative of aggregated nondegraded CPNs. Attempts to degrade CPNs under high reducing conditions were not conclusive, assuming that the strength of interpolymer interactions in an aqueous environment are strong. Partial backbone degradation to induce de-aggregation of CPNs was speculated from slight blue shifts on both absorption and emission wavelengths upon treating CPNs with excess amount of glutatihione (see ESI).
For analyzing subcellular localizations of CPNs, various organelles (i.e., nucleus, Golgi, endosome, and mitochondria) were post-stained using commercially available dyes. The Pearson’s Correlation Coefficient (PCC) was used to quantitatively evaluate co-localization.18 PCC scores of 0 and 1 correspond to uncorrelated and perfectly linearly correlated colors, respectively. Mean PCC scores from three independent images of an entire cell were selected and analyzed to increase the analysis objectivity. CPN-1 exhibited high mitochondrial co-localization (PCC ~0.8), while minimal endosome (~0.3) and Golgi apparatus (~0.2) co-localizations were observed. Meanwhile, CPN-2 showed high endosomal co-localization (~0.7) with minimal mitochondrial and Golgi apparatus co-localization. Non-degradable CPN-3, which was fabricated with low molecular weight PPE-3, also exhibits punctuated staining pattern with low mitochondria localization (~0.4), supporting that the degradability of CPNs is an important factor increasing mitochondria trafficking (see ESI).
The presence of punctuated spots in microscopic images and inefficient degradation of CPN-1 in a highly reducing environment imply that CPNs are hydrophobic despite of the charged TPP groups. To increase the hydrophilicity, CPNs were further complexed with an anionic linear polysaccharide, hyaluronic acid (HA). When the same biodegradable CPN-1 was complexed with HA (CPN-1/HA) followed by incubation with HeLa cells for overnight, the mitochondria imaging pattern was improved substantially with reduced punctuated spots (Fig. 3). As shown in Fig. 3, CPN-1/HA exhibits characteristic mitochondrial granular patterns and high PCC scores, while no changes were observed from CPN-2/HA complex. Non-degradable CPN exhibit punctuated fluorescent image patterns regardless of HA complexation.
Fig. 3.
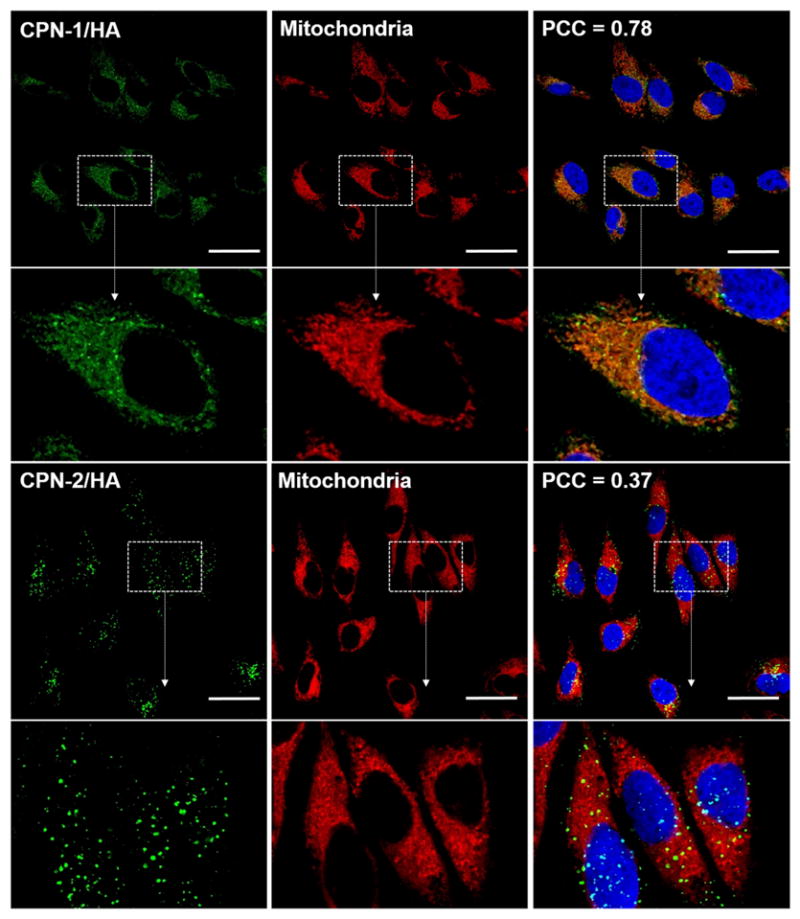
Confocal microscopic images of HeLa cells incubated with CPN/HA (green) for 18 h followed by mitochondria (red) and nucleus (blue) staining. The scale bar is 20 μm.
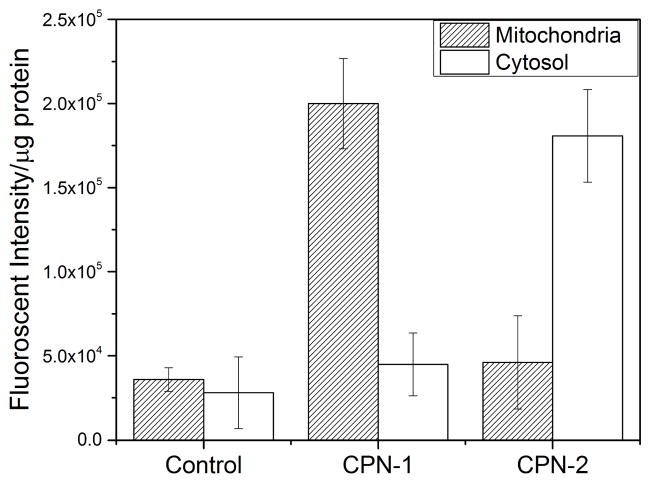
To further confirm that CPN-1 indeed trafficked to mitochondria, live cells incubated with CPNs were treated with a commercial mitochondria isolation kit followed by centrifugation to isolate a mitochondria fraction. Fluorescence spectroscopic analysis of the mitochondrial fraction of live HeLa cells clearly supports the high mitochondrial localization of CPN-1. As shown in Fig. 4, fluorescent signals were exclusively observed from the mitochondria fraction of live HeLa cells incubated with CPN-1. CPN-2 exhibited high cytosolic fluorescent intensity compared to the mitochondria fraction, supporting the data from microscopic image analysis.
Fig. 4.
Fluorescence analysis of mitochondrial and cytosolic fractions of HeLa cells incubated with CPNs. CPN-1 was found at the mitochondrial fraction, while CPN-2 was found mainly in the cytosolic fraction. The error bar represents ±standard deviation (n = 3).
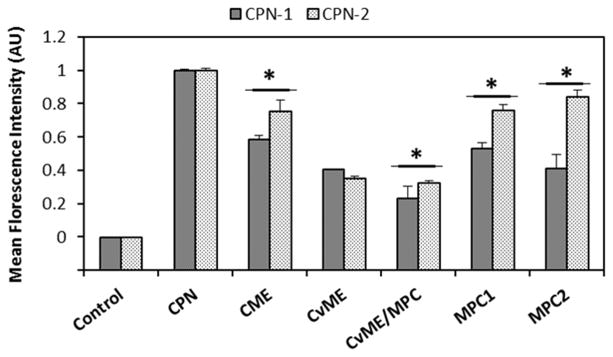
This dramatic subcellular localization difference between CPNs with the same side chains (i.e., TPP) implies that self-assembly structures of CPNs possibly influence cellular entry routes and the intracellular fates. We previously found that CPs with flexible linkers tend to form aggregates with high interpolymer interactions compared with CPs without the flexibility.19 Because of the flexible disulfide containing units, the surface properties of CPN-1 will be different and influential for cellular interactions and subsequent cellular entry. To test endocytosis pathways, live HeLa cells pretreated with pharmacological inhibitors of various endocytosis pathways were incubated with CPNs for 2 h and mean fluorescence intensity of live cells was monitored by flow cytometry.20 As shown in Fig. 5, CPNs use various endocytosis pathways to enter HeLa cell, determined by the reduced uptake of CPNs under the inhibitors tested. In addition to caveolae-mediated endocytosis (CvME), uptake of CPN-1 via macropinocytosis (MPC) was also noticeably reduced. Although further mechanistic and trafficking studies are needed, we speculate that the high mitochondria localization of CPN-1 is closely related to a series of cellular processes of MPC internalization of CPN-1, better escape of macropinosomes, degradation of disulfide bonds to generate low molecular weight COs, and increased diffusion for trafficking to mitochondria.
Fig. 5.
Endocytosis inhibition study under pharmacological inhibitors treatments. Mean fluorescence intensity of HeLa cell (Control), CPNs only (CPN), chlorpromazine (24 μM, CME), genistein (210 μM, CvME), methyl-β-cyclodextrin (1000 μM, CvME/MPC), LY294002 (120 μM, MPC1), and cytochalasin D (0.04 μM, MPC2) were measured using flow cytometry. Error bars represent standard deviation. *p < 0.05
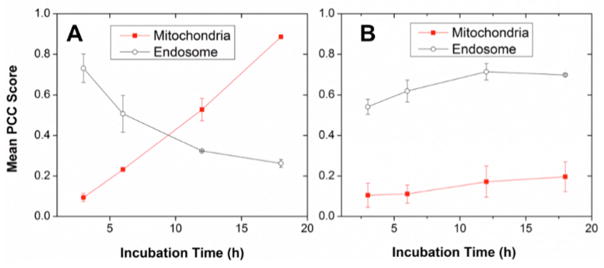
To determine whether the observed CPN-1 mitochondrial specificity is related to the degradation kinetics, we measured endosome and mitochondrial co-localization as a function of incubation time. Subcellular localization was monitored by fluorescent microscopic imaging, with CPNs incubated for 3, 6, 12, and 18 h in live HeLa cells. PCC scores between CPNs and mitochondrial or endosomal markers were measured and averaged from three independent trials (Fig. 6). CPN-1 exhibited a time-dependent mitochondrial and endosomal co-localization. As incubation time increased, CPN-1 localized more strongly toward the mitochondria, with almost exclusive mitochondrial localization at 18 h incubation. CPN-2, meanwhile, did not show time-dependent co-localization, exhibiting no significant change in mitochondrial or endosomal localization as incubation time increased. Although CPN-2 has high concentration of mitochondria targeting TPPs, the non-degradability, large molecular weight, and poor endosome escaping and diffusion efficiency limit mitochondria targeting. The time-dependency of CPN-1 on co-localization supports that intracellular degradation of biodegradable linkers play a significant role in mitochondrial trafficking of TPP functionalized COs.
Fig. 6.
Time-course localizations of A) CPN-1 and B) CPN-2 on endosomes (open circles) and mitochondria (filled circles). Subcellular localization of CPN-1 changes from endosome to mitochondria, while CPN-2 remains constant regardless of incubation time. Error bars represent standard deviation from n = 3
In conclusion, we present a new strategy to achieve a unique mitochondrial-specific localization of CPs. We demonstrated that disulfide-containing CPNs exhibit active cellular internalization as polymer nanoparticles via a variety of endocytosis, and undergoes degradation to low molecular weight oligomers that were efficiently trafficked to target mitochondria. The concept we demonstrated here will lead to the development of novel CP-based materials for improved intracellular sensing, labeling, and potential therapeutic delivery.
Supplementary Material
Acknowledgments
J. H. M. acknowledges generous support from National Institute of Health (GM092778) and National Science Foundation (DMR1352317). M. T. was supported by the Dissertation Year Fellowship funded by the Florida International University Graduate School. E. M. was supported by NIH R25GM61347. S. L. acknowledges the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (NRF-2015R1A4A1041036).
Footnotes
Electronic Supplementary Information (ESI) available: synthesis and characterization of CPNs and details on the cellular studies. See DOI: 10.1039/x0xx00000x
Contributor Information
Sunwoo Lee, Email: sunwoo@chonnam.ac.kr.
Joong Ho Moon, Email: jmoon@fiu.edu.
Notes and references
- 1.Feng L, Zhu C, Yuan H, Liu L, Lv F, Wang S. Chemical Society Reviews. 2013;42:6620–6633. doi: 10.1039/c3cs60036j. [DOI] [PubMed] [Google Scholar]
- 2.Tuncel D, Demir HV. Nanoscale. 2010;2:484–494. doi: 10.1039/b9nr00374f. [DOI] [PubMed] [Google Scholar]
- 3.Rahim NAA, McDaniel W, Bardon K, Srinivasan S, Vickerman V, So PTC, Moon JH. Advanced Materials. 2009;21:3492–3496. [Google Scholar]
- 4.Wu C, Bull B, Szymanski C, Christensen K, McNeill J. Acs Nano. 2008;2:2415–2423. doi: 10.1021/nn800590n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moon JH, McDaniel W, MacLean P, Hancock LE. Angewandte Chemie-International Edition. 2007;46:8223–8225. doi: 10.1002/anie.200701991. [DOI] [PubMed] [Google Scholar]
- 6.Chen LH, McBranch DW, Wang HL, Helgeson R, Wudl F, Whitten DG. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:12287–12292. doi: 10.1073/pnas.96.22.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ho HA, Boissinot M, Bergeron MG, Corbeil G, Dore K, Boudreau D, Leclerc M. Angewandte Chemie-International Edition. 2002;41:1548–1551. doi: 10.1002/1521-3773(20020503)41:9<1548::aid-anie1548>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 8.Moon JH, Mendez E, Kim Y, Kaur A. Chemical Communications. 2011;47:8370–8372. doi: 10.1039/c1cc10991j. [DOI] [PubMed] [Google Scholar]
- 9.Feng X, Lv F, Liu L, Yang Q, Wang S, Bazan GC. Advanced Materials. 2012;24:5428–5432. doi: 10.1002/adma.201202145. [DOI] [PubMed] [Google Scholar]
- 10.Vokata T, Twomey M, Mendez E, Moon JH. Journal of Polymer Science Part a-Polymer Chemistry. 2015;53:1403–1412. [Google Scholar]
- 11.Mendez E, Moon JH. Chemical Communications. 2013;49:6048–6050. doi: 10.1039/c3cc43015d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Szewczyk A, Wojtczak L. Pharmacological Reviews. 2002;54:101–127. doi: 10.1124/pr.54.1.101. [DOI] [PubMed] [Google Scholar]
- 13.Hassan SSM, Rechnitz GA. Analytical Chemistry. 1982;54:1972–1976. [Google Scholar]
- 14.Smith RAJ, Porteous CM, Gane AM, Murphy MP. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5407–5412. doi: 10.1073/pnas.0931245100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoye AT, Davoren JE, Wipf P, Fink MP, Kagan VE. Accounts of Chemical Research. 2008;41:87–97. doi: 10.1021/ar700135m. [DOI] [PubMed] [Google Scholar]
- 16.Davis W, Ronai Z, Tew KD. Journal of Pharmacology and Experimental Therapeutics. 2001;296:1–6. [PubMed] [Google Scholar]
- 17.Zinchuk V, Zinchuk O, Okada T. Acta Histochemica Et Cytochemica. 2007;40:101–111. doi: 10.1267/ahc.07002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vokata T, Moon JH. Macromolecules. 2013;46:1253–1259. doi: 10.1021/ma3019975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vercauteren D, Vandenbroucke RE, Jones AT, Rejman J, Demeester J, De Smedt SC, Sanders NN, Braeckmans K. Molecular Therapy. 2010;18:561–569. doi: 10.1038/mt.2009.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.