Patzke et al. create human embryonic stem cell–derived neurons that enable the generation of conditional loss-of-function mutations of L1CAM. Deletion of L1CAM impairs axonal elongation, dendritic arborization, and action potential generation.
Abstract
Hundreds of L1CAM gene mutations have been shown to be associated with congenital hydrocephalus, severe intellectual disability, aphasia, and motor symptoms. How such mutations impair neuronal function, however, remains unclear. Here, we generated human embryonic stem (ES) cells carrying a conditional L1CAM loss-of-function mutation and produced precisely matching control and L1CAM-deficient neurons from these ES cells. In analyzing two independent conditionally mutant ES cell clones, we found that deletion of L1CAM dramatically impaired axonal elongation and, to a lesser extent, dendritic arborization. Unexpectedly, we also detected an ∼20–50% and ∼20–30% decrease, respectively, in the levels of ankyrinG and ankyrinB protein, and observed that the size and intensity of ankyrinG staining in the axon initial segment was significantly reduced. Overexpression of wild-type L1CAM, but not of the L1CAM point mutants R1166X and S1224L, rescued the decrease in ankyrin levels. Importantly, we found that the L1CAM mutation selectively decreased activity-dependent Na+-currents, altered neuronal excitability, and caused impairments in action potential (AP) generation. Thus, our results suggest that the clinical presentations of L1CAM mutations in human patients could be accounted for, at least in part, by cell-autonomous changes in the functional development of neurons, such that neurons are unable to develop normal axons and dendrites and to generate normal APs.
X-linked neurodevelopmental disorders that produce intellectual disability are relatively common diseases resulting from mutations in X-chromosomal genes, with ∼1/600–1/1,000 males affected (Gécz et al., 2009). One particular gene associated with X-linked intellectual disability is L1CAM, with an estimated incidence of 1/25,000–1/60,000 males (Halliday et al., 1986; Weller and Gärtner, 2001). L1CAM is a type 1 transmembrane protein of the immunoglobulin superfamily that is conserved in vertebrates and invertebrates (Neuroglian/Sax-7) and that was initially described as a neuronal cell adhesion molecule involved in axonal growth (Rathjen and Schachner, 1984; Chang et al., 1987). L1CAM contains six extracellular N-terminal Ig domains, followed by five fibronectin type 3 (FN3) domains, a single transmembrane segment, and a short C-terminal intracellular sequence. L1CAM is the founding member of the L1CAM family that includes neurofascin, NrCAM, and CHL1, which exhibit similar domain structures and are composed of homologous sequences. L1CAM is highly expressed in the developing nervous system and has also been implicated in cancer progression (Schäfer and Altevogt, 2010; Kiefel et al., 2012; Schäfer and Frotscher, 2012).
Approximately 350 pathogenic mutations in the L1CAM gene have been described in patients with a broad spectrum of neurological abnormalities and mental retardation, summarized by the term L1 syndrome. This spectrum includes the MASA syndrome (mental retardation, aphasia, shuffling gait, and adducted thumbs), hydrocephalus due to stenosis of the aqueduct of Sylvius, agenesis of the corpus callosum, and SPG1 (X-linked hereditary spastic paraplegia type 1), which are referred to collectively as CRASH syndrome (Rosenthal et al., 1992; Stumpel and Vos, 1993; Jouet et al., 1994, 1995; Fransen et al., 1997; Weller and Gärtner, 2001; Vos et al., 2010). Besides a reported whole gene deletion (Chidsey et al., 2014), these mutations include frameshift, nonsense, and missense mutations, resulting in the production of truncated proteins or proteins with mutations in structurally defined key residues (Stumpel and Vos, 1993). Missense mutations most likely lead to alterations of intracellular trafficking and impaired function and mobility caused by additional cysteines on the surface of the molecule or aberrant ligand binding (De Angelis et al., 1999, 2002; Kenwrick et al., 2000; Schäfer et al., 2010). Pathological mutations are known to affect binding of L1CAM to itself, Neuropilin-1, Tax-1/Axonin-1, ankyrins, and integrins, or to impair triggering of epidermal growth factor receptor and Erk1/2 signaling (De Angelis et al., 1999; Schäfer and Altevogt, 2010). Overall, most of the disease-causing mutations in L1CAM appear to be loss-of-function mutations.
Interestingly, an ethanol-binding site disrupting the interface between Ig-domains 1 and 4 of L1CAM has been identified. This site might explain the inhibitory effects of ethanol on L1CAM-mediated cell adhesion and neurite outgrowth, and could contribute to neuropathological abnormalities observed in fetal alcohol spectrum disorders, which exhibit features that are similar to those observed in L1 syndrome patients (Ramanathan et al., 1996; Bearer et al., 1999; Arevalo et al., 2008). Electron microscopy studies on L1CAM and data from a crystal structure of the N-terminal Ig domains 1–4 of the L1CAM family member neurofascin, as well as a cryo-electron tomography report on liposomes supplemented with L1CAM ectodomains, revealed a horseshoe-like structure of the Ig domains 1–4 (Schürmann et al., 2001; He et al., 2009; Liu et al., 2011). Based on the structure of the Ig domains 1–4 of the L1CAM homologue Axonin-1, it has been suggested that two horseshoes on opposing cells interact in a zipper-like manner, mediating homophilic cell adhesion (Freigang et al., 2000). Ethanol, and disease-causing missense mutations in the ethanol-binding pocket (e.g., Leu-120-Val and Gly-121-Ser), likely disrupt the horseshoe-shaped structure and inhibit homophilic and heterophilic interactions of L1CAM (Bateman et al., 1996; De Angelis et al., 1999, 2002; Arevalo et al., 2008). However, in contrast to the notion that Ig domains 1–4 are essential for homophilic binding, neurons from a reported L1CAM mutant mouse line lacking only Ig domain 6, which contains the integrin-binding motif RGD, failed to attach to L1CAM in vitro (Itoh et al., 2004), suggesting a more complicated scenario for the homophilic activity of L1CAM on neurons.
Studies using constitutive L1CAM-deficient mice as a model system reported defects in axon guidance in the corticospinal tract, impaired growth of pyramidal layer V neuron apical dendrites, reduced size of the corpus callosum, malformations of the ventricular system and the cerebellar vermis, decreased association of axons with nonmyelinating Schwann cells, and reduced inhibitory synaptic transmission (Dahme et al., 1997; Cohen et al., 1998; Fransen et al., 1998; Demyanenko et al., 1999; Saghatelyan et al., 2004). Puzzlingly, mutant mice expressing L1CAM with a truncated intracellular domain that lacks the ankyrin-binding region displayed no abnormal brain development, but exhibited a dramatic decrease in L1CAM expression and defects in motor functions in adult mice (Nakamura et al., 2010). However, conditional KO (cKO) mice where the L1CAM gene is inactivated in adult brain by crossing them to a calcium/calmodulin-dependent, kinase II promoter–driven Cre-line did not display these overt morphological abnormalities, but instead exhibited an increase in basal excitatory synaptic transmission (Law et al., 2003). Thus, the precise functions of L1CAM and its mechanisms of action are not yet completely understood.
The clinical presentation of human L1CAM mutations, together with the phenotypes of the various mouse models, suggests that L1CAM contributes to the migration of neurons, neurite outgrowth, axon guidance, and axon–glia interaction. Although the mouse mutants provided important information, the overall phenotypes of the mutant mice were surprisingly mild and the effect of L1CAM mutations on human neurons remains unknown. Here, we used cre/lox technology in human embryonic stem (ES) cell–derived neurons to study the effects of L1CAM mutations on human neuronal function at the cellular level in a controlled genetic background. Importantly, this manipulation enables the direct comparison of matching cell populations in all experiments. By creating a conditional hemizygous KO (i.e., a KO of an X-chromosomal gene in a male cell), we could dissect the fundamental role of L1CAM in axonal and dendritic arborization in human neurons, and examine its function in action potential (AP) generation. Our data reveal a dramatic impairment in axon development that results in a decrease in axon length, a change in the protein composition of the axon initial segment (AIS), and a severe impairment in AP generation. In addition, we observed a less severe impairment in dendritic arborization. These results suggest that L1CAM mutations cell-autonomously impair neuronal function by impeding normal axon function.
RESULTS
Conditional hemizygous mutant human neurons as a model system for human L1CAM mutations
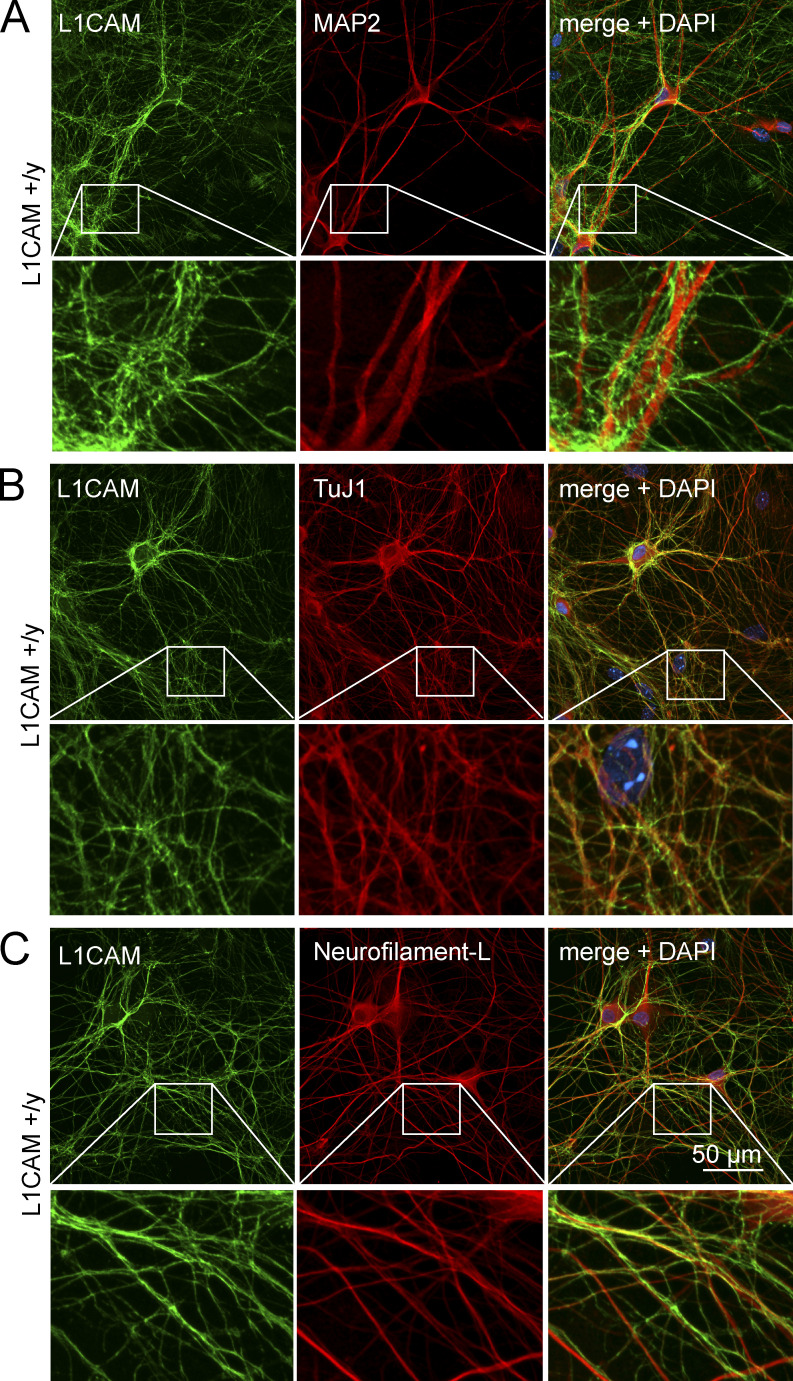
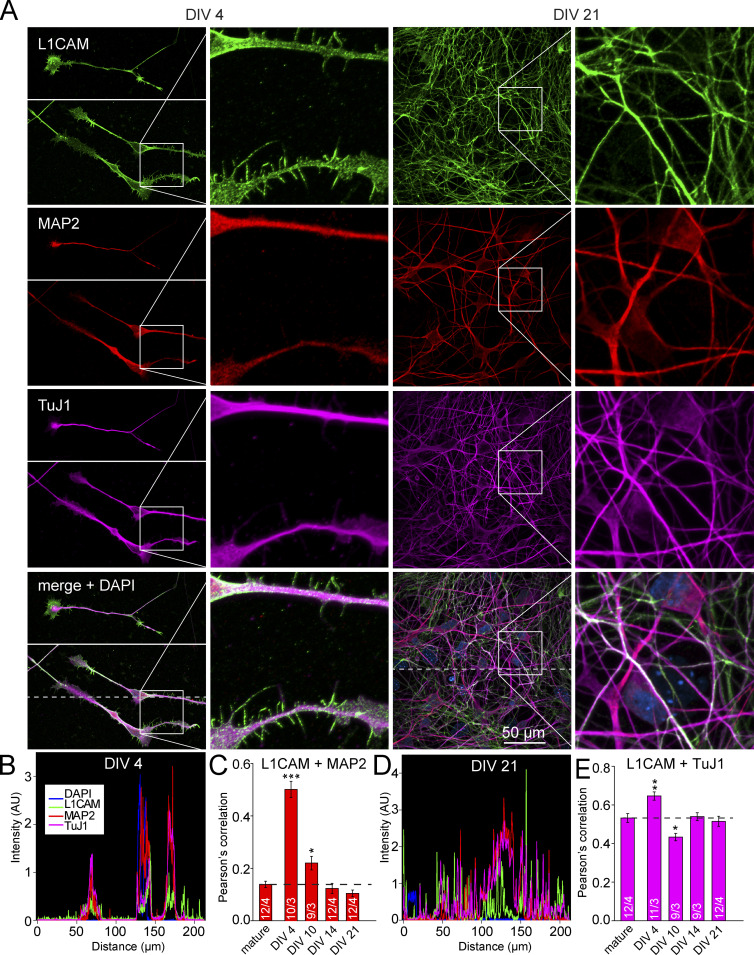
We generated human neurons by forced expression of Ngn2 in human ES cells using our previously described induced neuron (iN) cell protocol (Zhang et al., 2013). We then examined L1CAM expression in these WT neurons by immunocytochemistry (Fig. 1). We observed abundant expression of L1CAM in 5-wk-old mature neurons. L1CAM was specifically localized to axons, but was absent from MAP2-positive dendrites, suggesting that iN cells could serve as a powerful tool to study L1 syndrome–related phenotypes at the cellular level in human neurons (Fig. 1, A–C). When we assessed the distribution of L1CAM in developing iN cells, however, we detected L1CAM on all neurites at early stages (DIV 4), including MAP2-positive neurites, followed by selective L1CAM accumulation in MAP2-negative processes only at later stages (DIV 14–21 and mature neurons; Fig. 2). Thus, during neuronal development, L1CAM initially localizes to all neurites (which then differentiate into axons and dendrites), and only later becomes restricted to axons.
Figure 1.
Axonal localization of L1CAM in human WT neurons. (A–C) Representative confocal micrographs (from three independent experiments) of L1CAM on human neurons (iN cells) obtained by forced expression of Ngn2 in ES cells and analyzed 5 wk after Ngn2 induction. Neurons were double labeled by immunofluorescence for L1CAM and MAP2 (A), TuJ1 (B), or Neurofilament-L (C). Each panel shows lower (top) and higher magnifications (bottom; boxed area in lower magnification images).
Figure 2.
L1CAM partially colocalizes with MAP2 in the dendrites of human neurons at early developmental stages. (A) Representative confocal micrographs of L1CAM on human neurons taken 4 and 21 d after Ngn2 induction. Neurons were triple-labeled by immunofluorescence for L1CAM, MAP2, and TuJ1. (B and D) Intensity profile of immunolabeling for the indicated proteins derived from line scans along the dashed line in the bottom left and second from the right images in A. (C and E) Summary graphs of Pearson’s correlation coefficients for co-localization of L1CAM versus MAP2 (C) and L1CAM versus TuJ1 (E) determined in multiple experiments at different times after Ngn2 induction as indicated (mature = 35 d after Ngn2 induction). Data are means ± SEM; statistical comparisons were performed by Student’s t test comparing various times after Ngn2 induction to mature neurons (*, P < 0.05; **, P < 0.01; ***, P < 0.001; numbers of images/independent experiments analyzed are shown in the bars).
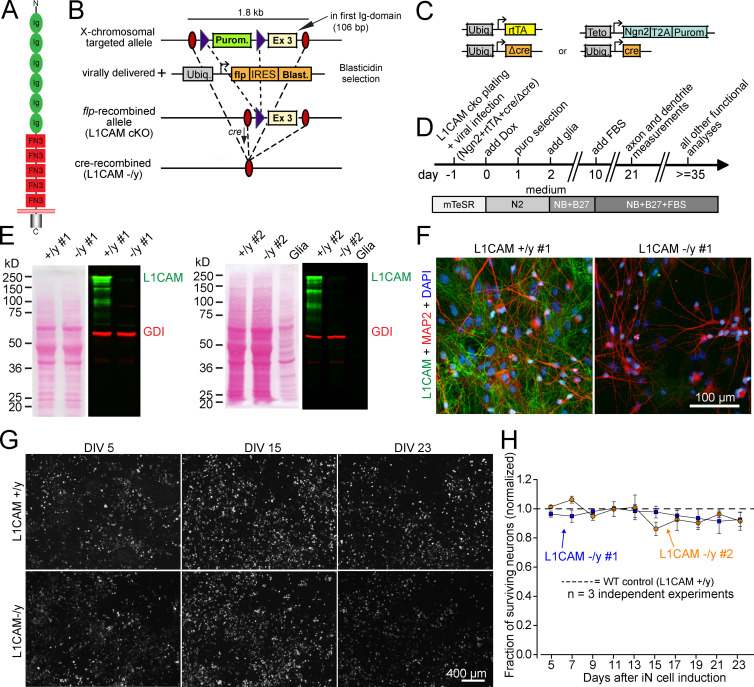
To examine the effect of L1CAM loss-of-function mutations on the properties of human neurons, we introduced a cKO mutation into the endogenous L1CAM gene of H1 ES cells (which are male, rendering a hemizygous mutation equivalent to a complete KO) by homologous recombination. For this purpose, we flanked exon 3 (which encodes part of the first Ig domain) of the L1CAM gene with loxP sites (Fig. 3, A and B; and Materials and methods). Deletion of exon 3 creates an out-of-frame junction of exons 2 to 4, thus producing a KO. Two independently targeted ES cell clones were generated to exclude possible effects by unwanted clonal variation or incidental genetic changes, and were converted into cKO cells by removal of the puromycin resistance cassette using transduced flp recombinase (Fig. 3 B).
Figure 3.
Generation of human ES cells with a conditional L1CAM gene loss-of-function mutation andeffect of L1CAM mutation on neuronal survival. (A) L1CAM contains six Ig-domains and five FN-type 3 domains followed by a single transmembrane sequence, and a highly conserved short intracellular cytoplasmic domain. (B) Targeting strategy. The L1CAM gene was mutated by homologous recombination in male H1 ES cells using a recombinant AAV, and correctly recombined mutant ES cells were verified by PCR. The introduced mutations flank anout-of-frame exon (exon 3) with loxP sites, and include a puromycin (Purom.) resistance cassette for selection; this cassette was removed using Frt-recombinase transduction to generate L1CAM cKO cells. For generation of precisely matched neurons containing or lacking L1CAM, mutant Cre-recombinase (ΔCre) or active Cre-recombinase (Cre) were expressed in iN cells during induction of neuronal differentiation as indicated. (C) Design of the lentiviruses used for conversion of ES cells into iN cells and for expression of inactive (ΔCre) or active Cre-recombinase (Cre). (D) Flow diagram of all experiments. cKO cells were infected at day –1 with lentiviruses shown in B, and differentiation was started on day 0 by activating Ngn2 expression with doxycycline. Analyses were conducted at least 21 d after induction of Ngn2. (E) Ponceau-stained blotting membranes (left) and immunoblots (right) of iN cells derived from two independent L1CAM cKO ES cell clones (#1 and #2). Immunoblots were stained with antibodies to human L1CAM (green) and GDI (red; loading control); signals were visualized with fluorescently labeled secondary antibodies (right). (F) Representative immunofluorescently labeled control and mutant L1CAM-deficient neurons. L1CAM, MAP2, and DAPI staining is shown. (G and H) Plot of the fraction of surviving neurons as a function of time in culture (G; dashed line indicates control neurons). As in all experiments, neurons from two independent L1CAM cKO ES cell clones (blue and orange) were analyzed. Data are means ± SEM (n = 3 independent biological experiments).
We produced iN cells from both L1CAM cKO ES cell clones by expression of Ngn2 delivered by recombinant lentiviruses. We also expressed WT active cre-recombinase (Cre) or mutant inactive cre-recombinase (ΔCre) in the iN cells during induction to generate precisely matching L1CAM-deficient mutant neurons and control neurons, respectively (Fig. 3, C and D; mutant neurons from the two clones are referred to as −/y #1 and −/y #2, and WT neurons as +/y #1 and +/y #2). As analyzed by immunoblotting, we found that, consistent with previous studies (Wolff et al., 1988; Schäfer and Altevogt, 2010), control neurons produced multiple L1CAM protein variants with apparent molecular weights between 220 and 65 kD (Fig. 3 E). Upon cre recombination, neurons derived from the two cKO clones showed a complete lack of all L1CAM variants, and exhibited no L1CAM expression as judged by immunocytochemistry (Fig. 3, E and F), demonstrating that the two cKO ES cell clones can be efficiently converted into control and L1CAM mutant human neurons with precisely matching backgrounds that only differ in the expression of L1CAM.
No change in survival, but decreased MAP2a/b and ankyrin expression in L1CAM mutant human neurons
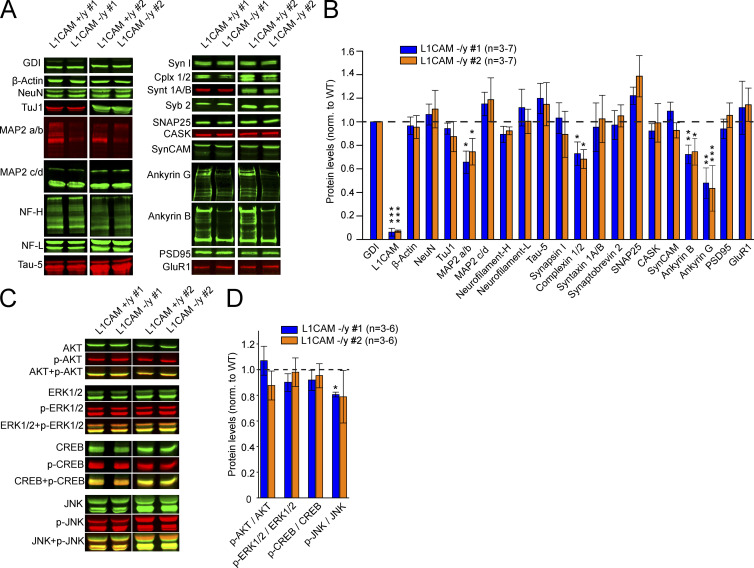
L1CAM-deficient human neurons exhibited similar survival as control neurons, with no evidence of cell death or degeneration as a result of the L1CAM deletion (Fig. 3, G and H). Furthermore, we observed no increase in phosphorylation of JNK/SAPK, which is activated in stressed cells (Fig. 4, C and D). Quantitative immunoblotting analyses revealed that the L1CAM deletion did not alter the majority of tested neuronal marker proteins; in particular, no changes in the levels of the majority of synaptic proteins were detected (Fig. 4, A and B). In contrast, we observed a decrease in the levels of the adaptor proteins ankyrinG and ankyrinB, which are axonal proteins that bind to the intracellular sequences of L1CAM (Davis and Bennett, 1994). We found a reduction of ∼50% and ∼20% of ankyrinG and ankyrinB, respectively (Fig. 4, A and B). In addition, we detected a ∼30% decrease in the dendritic marker MAP2a/b and complexins. Because it was reported that L1CAM-activated signaling stimulates the mitogen-activated protein kinase (MAPK) pathway (Schaefer et al., 1999; Schmid et al., 2000; Maness and Schachner, 2007), we examined iN cells derived from both cKO clones for changes in MAPK activation, but did not detect significant changes in phosphorylation levels of Erk 1/2 or of related components in the mutant neurons (Fig. 4, C and D). Together, our data suggest that the L1CAM deletion in human neurons does not significantly impair neuronal survival or activate MAPK signaling, but decreases the levels of the axonal proteins ankyrinB and ankyrinG, the dendritic protein MAP2a/b, and the presynaptic protein complexin.
Figure 4.
Protein levels in hemizygous mutant human neurons. (A and B) Representative immunoblots (A) and summary graph of protein levels (B) in iN cells produced from independent L1CAM cKO clones #1 and #2. Values in B represent mean protein levels observed in L1CAM KO neurons normalized to those of matching isogenic WT controls (dashed line), and corrected for blotting and loading variations using GDI as an internal standard. (C and D) Representative pictures and summary graph of phosphorylation levels of AKT, ERK1/2, CREB, and JNK in iN cells from clones #1 and #2 and matched controls (dashed line). Data are means ± SEM; statistical significance was assessed using Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Axonal and dendritic arborizations are reduced in human L1CAM mutant neurons
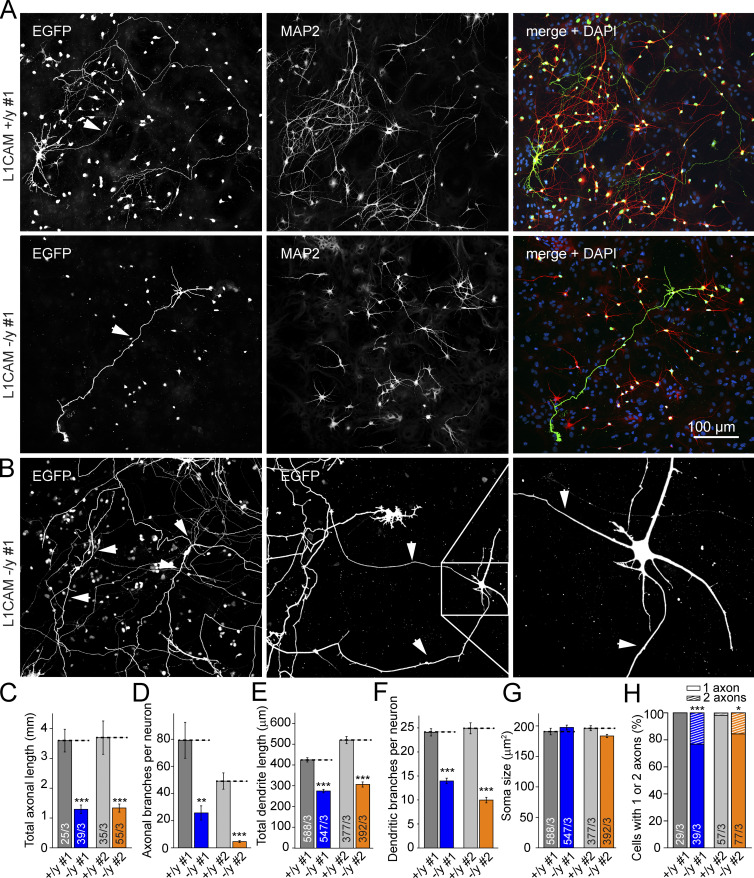
To test for changes in neuronal morphology, we next sparsely transfected (by the CaPO4 method) conditionally hemizygous mutant iN cells with a plasmid expressing TetO-EGFP and stained the neurons for MAP2a/b. Assuming that cytoplasmic EGFP visualizes the complete neuronal morphology and that MAP2a/b is selectively expressed in dendrites, this experiment allowed us to systematically distinguish between dendrites and axons (Fig. 5 A). By comparing mutant with WT cells, we observed in neurons derived from both independent cKO clones that deletion of L1CAM caused a massive decrease in the total length and number of branching points of axons, but produced no changes in cell soma size (Fig. 5, A, C, D, and G). Moreover, deletion of L1CAM decreased the total length and branch numbers of dendrites, although the effect on dendrites was smaller than that on the length and branches of axons (Fig. 5, E and F), suggesting that the partial codistribution of L1CAM and MAP2 in very young neurons is important for dendritic tree development (Fig. 2). Interestingly, ∼15–25% of L1CAM mutant neurons developed two axons, compared with only 0–3% of WT neurons (Fig. 5, B and H). To test whether this morphological phenotype persists at later developmental stages, we analyzed iN cells at 180 d after induction. Because of the immense axonal growth constraining single-cell axon analyses (Fig. 6, A and B), we measured the dendritic arborization of these “old” human neurons and detected a defect in the mutant neurons (Fig. 6, C–F). Together with the quantitative immunoblotting results and the localization of L1CAM on developing and mature human neurons, these observations indicate that loss-of-function mutations of L1CAM impair dendritic and axonal arborization of human neurons.
Figure 5.
Deletion of L1CAM in human neurons dramatically impairs axonal growth and modestly decreases dendritic arborization. (A) Representative confocal images of control (top) and L1CAM-deficient iN cells (middle and bottom) that were sparsely transfected with EGFP at day 19 after Ngn2 induction to visualize the complete axonal and dendritic arbors of neurons, and that were additionally stained for MAP2 and DAPI as indicated at day 21 after Ngn2 induction. (B) Atypical iN cells with two independent axons. The multi-axon phenotype is significantly more frequently observed in L1CAM KO than in control neurons (see H). (C–G) Summary graphs of axonal and dendritic lengths and number of branches, as well as soma size of mutant and wild-type neurons from clones #1 and #2. Statistical significance was assessed using Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Three independent biological experiments were performed for each mutant clone and matching WT control. (H) Summary graph of the percentage of cells with one or two axons. Numbers in bars indicate number of cells/independent experiments analyzed.
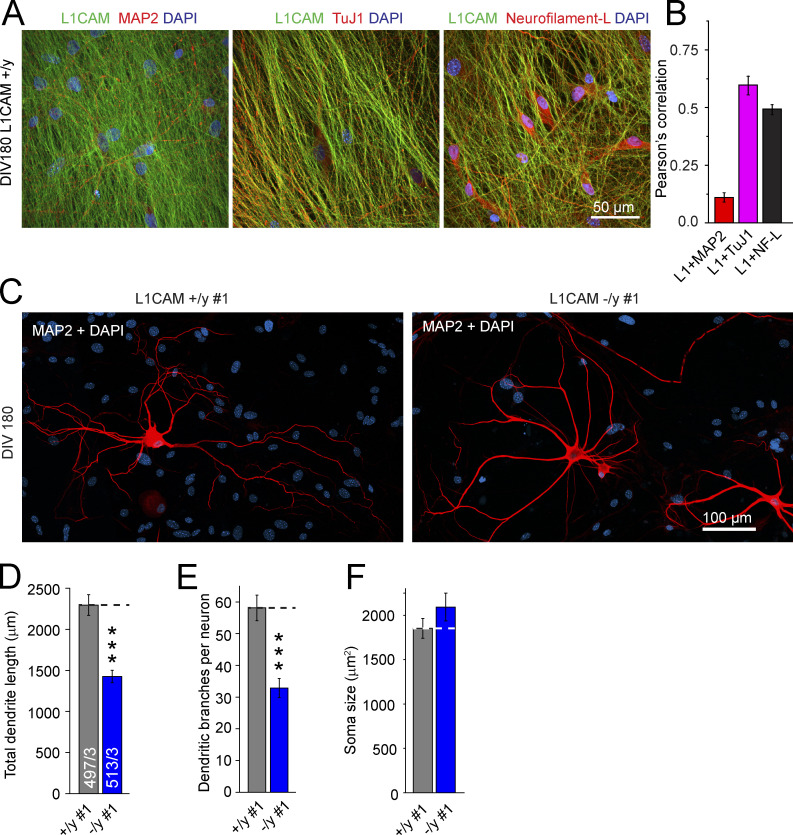
Figure 6.
Reduced dendritic arborization of L1CAM mutant human neurons after 180 d of culture. (A) Representative confocal micrographs showing WT cells 180 d after iN induction stained for L1CAM and counter-stained for MAP2a/b, TuJ1, or Neurofilament-L. (B) Summary graph of Pearson’s correlation coefficients for co-localization of the indicated proteins analyzed by double immunofluorescence labeling. Values are means of three different pictures ± SEM from a single experiment. (C) Confocal micrograph of dendritic tree visualized by MAP2a/b in mutant and matching control neurons 180 d after iN induction. (D–F) Summary graphs of dendritic lengths and number of branches, as well as soma size of mutant and control neurons. Statistical significance was assessed using Student’s t test (***, P < 0.001) of cells from three independent experiments.
L1CAM deletion selectively decreases ankyrinG levels in the AIS
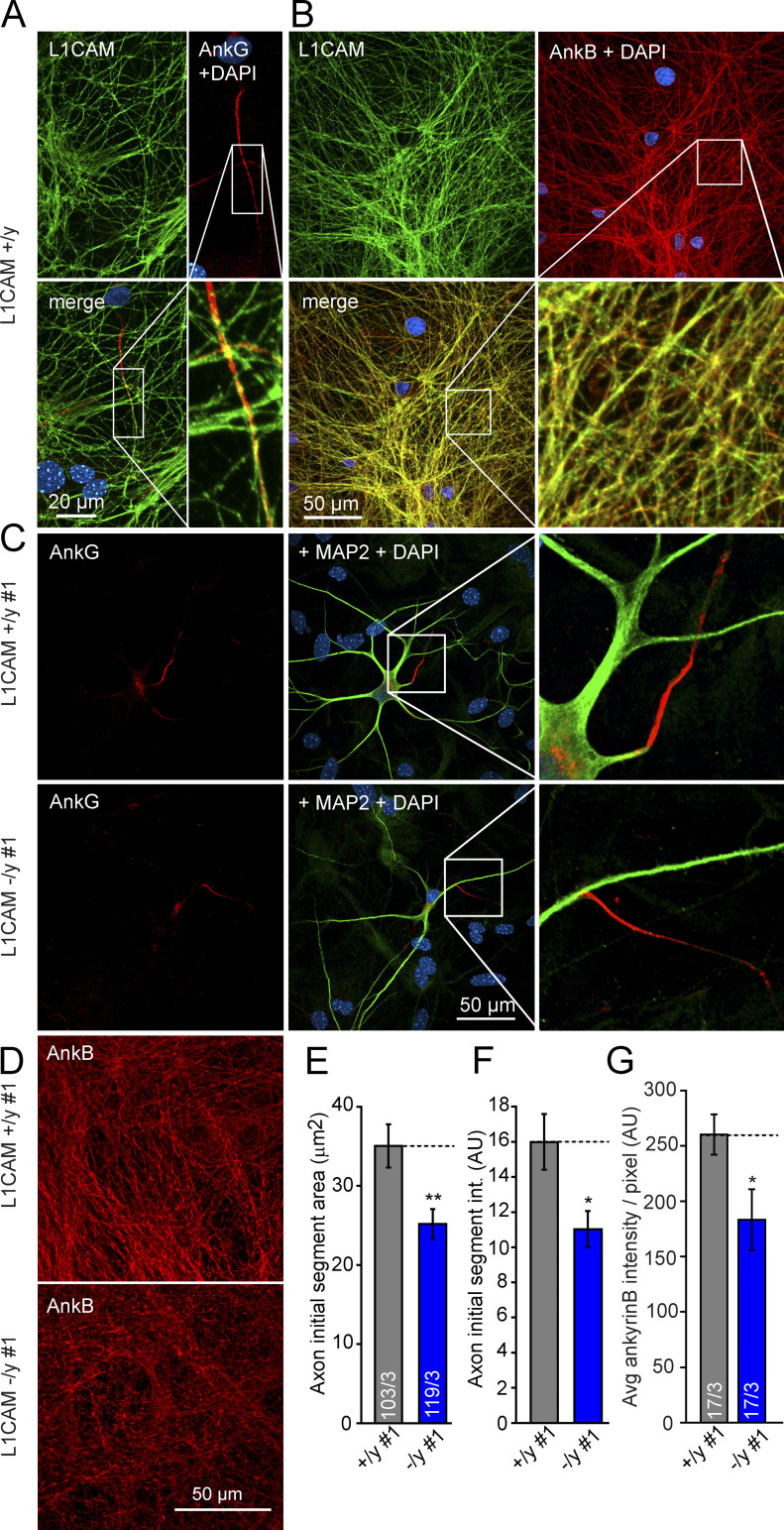
The marked decrease of ankyrinG levels in hemizygous mutant neurons (Fig. 4, A and B) suggests a potential defect in the AIS because ankyrinG is a key adaptor molecule that clusters Na+ and K+ channels at the AIS and partially co-localizes with L1CAM (Fig. 7 A; Zhou et al., 1998; Garrido et al., 2003; Kole and Stuart, 2012). To assess this aspect, we measured the area and intensity of ankyrinG staining in the AIS and found that both parameters decreased in L1CAM mutant human neurons (Fig. 7, C, E, and F). Investigating ankyrinB, which also co-localizes with L1CAM in axons (Fig. 7 B), we additionally detected a ∼30% decrease in ankyrinB staining intensity in mutant axons (Fig. 7, D and G). Together with the changes in axonal arborization (Fig. 5), these results suggest a fundamental role of L1CAM in the morphological development of axons in human neurons.
Figure 7.
Double labeling of L1CAM and ankyrins at the AIS and axons of WT and KO human neurons. (A and B) Confocal micrographs of L1CAM and ankyrinB-positive axons, as well as of the ankyrinG-positive AIS of axons in WT neurons. (C) Representative confocal micrographs of an L1CAM-deficient and control iN cell illustrating the AIS. Dendrites are labeled by anti-MAP2a/b. (D) Confocal micrographs of L1CAM hemizygous control and mutant iN cell labeling ankyrinB. The Pearson’s correlation for co-localization of L1CAM and ankyrinB is 0.77 ± 0.02 (determined from three independent experiments with three to four images each). (E and F) Summary graphs of the ankyrinG-labeled AIS area (E) and the ankyrinG staining intensity of the AIS (F). Data are means ± SEM; numbers in bars indicate the numbers of cells/independent experiments performed; statistical comparisons were done using Student’s t test (*, P < 0.05; **, P < 0.01). (G) Summary graph of mean ankyrinB intensities (per ankyrinB-positive pixel), indicating a reduction of ∼30% in the axons of L1CAM-deficient human neurons. Data are means ± SEM; statistical comparisons were performed by Student’s t test (*, P < 0.05; numbers of images/independent experiments analyzed are shown in the bars).
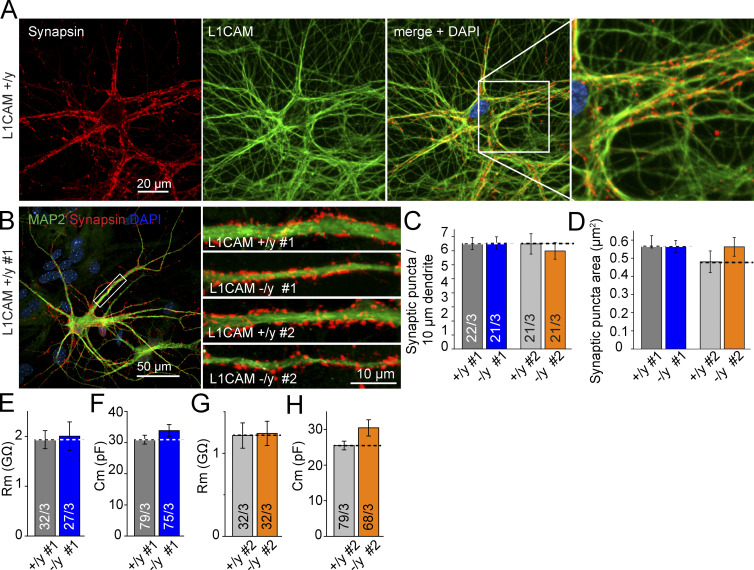
No decrease in synapse density in human mutant neurons
To test the hypothesis that L1 syndrome symptoms and its associated severe intellectual disability are caused by morphological changes at the synaptic level in human neurons, we quantified presynaptic synapsin-positive puncta (which partially overlap with L1CAM on axons from WT neurons; Fig. 8 A) on post-synaptic MAP2-positive dendrites. Both the size and the number of synapses per dendritic segment in WT and mutant neurons were compared (Fig. 8 B), and no significant differences could be detected (Fig. 8, C and D). No alterations of the intrinsic electrical properties were detected (Fig. 8, E–H), suggesting no alterations in the membrane composition. In summary, our results suggest that L1CAM is not required for synapse development as such, although we cannot exclude any functional synaptic changes at this point.
Figure 8.
Hemizygous L1CAM KO does not alter synapse density. (A) Micrographs of double-labeling by immunofluorescence showing L1CAM and Synapsin in axons of WT human neurons. (B) Representative images of dendrites from control and hemizygous L1CAM mutant iN cells stained for MAP2a/b and Synapsin to visualize presynaptic terminals. Clones #1 and #2 are shown. (left) Zoom-out of L1CAM +/y #1. (C and D) Summary graphs of synaptic density and area of Synapsin-positive puncta on dendrites from mutant and matching WT. (E–H) Input resistance and capacitance of hemizygous L1CAM mutant iN cells. Data are means ± SEM; no statistically significant difference was detected using Student’s t test.
L1CAM deletion impairs AP generation and decreases neuronal excitability in mutant human neurons
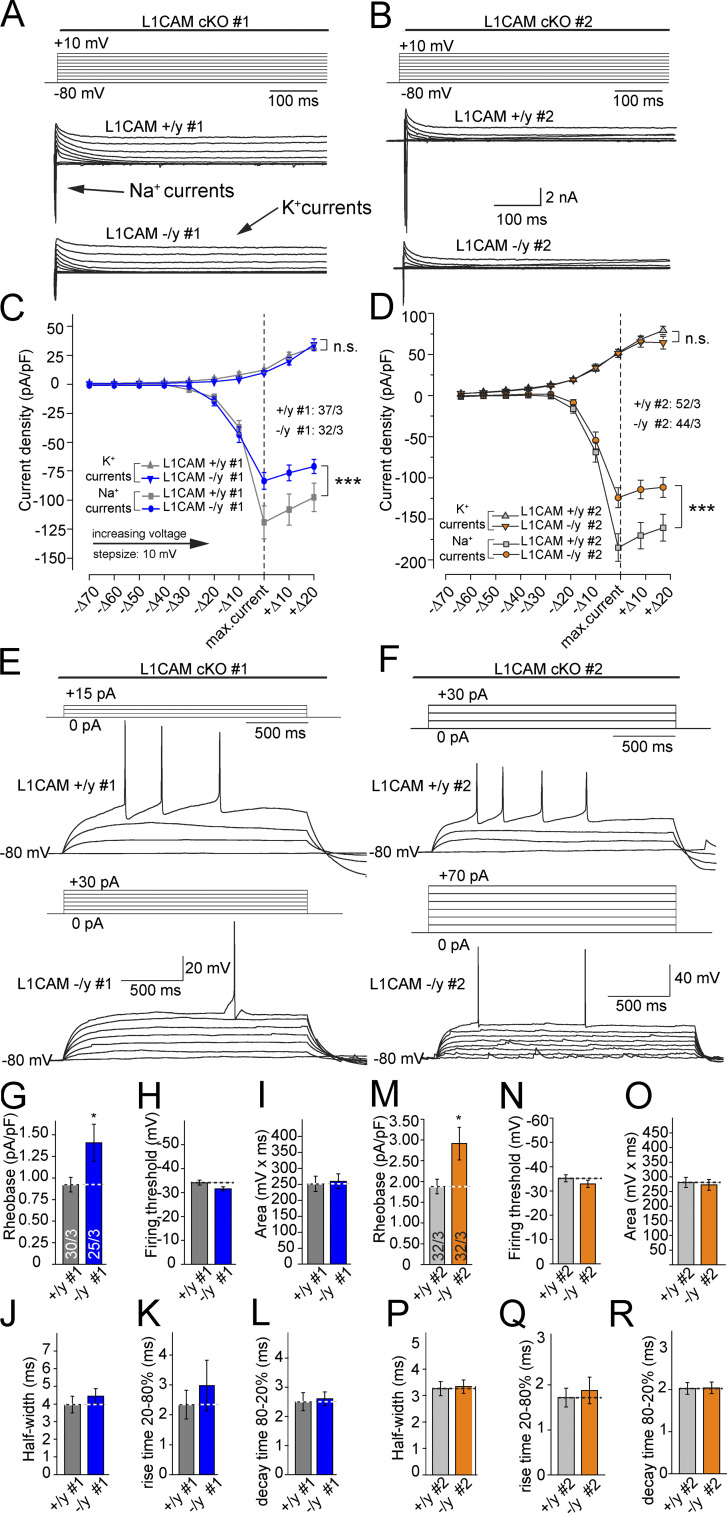
To investigate if L1CAM deficiency, and thus a decrease of ankyrinG at the AIS, leads to functional consequences in AP firing, we performed patch-clamp recordings and measured the whole-cell density of Na+ and K+ currents in 5-wk-old iN cells in voltage-clamp configuration (Fig. 9, A–D). Cells were held at −80 mV, and then the membrane potential was sequentially depolarized in 10-mV steps for 2 s (Fig. 9, A and B). We observed a significant decrease in the peak amplitude of Na+ currents (inward), whereas K+ currents (outward) measured in the same experiments remained unchanged (Fig. 9, C and D). Thus, the removal of L1CAM seems to selectively reduce the total density of functional Na+ channels in human neurons.
Figure 9.
Decreased Na+ currents and reduced excitability of mutant iN cells. (A and B) Protocol used for voltage-clamp experiments and representative traces of whole-cell voltage-clamp Na+ and K+ currents recorded in control and L1CAM mutant neurons. Cells were subjected to 10-mV step depolarizations (2 s each) from −80 mV to +10 mV. (C and D) Quantification of I/V curves of Na+ and K+ current densities. Currents were aligned for the maximal Na+ current densities. Data are means ± SEM; statistical comparisons were performed by ANOVA (***, P < 0.001; number of cells from three independent experiments is indicated). (E and F) Representative traces and protocol of analyses of the AP firing properties of control and L1CAM mutant neurons. Neurons were maintained at near −80 mV in current-clamp mode and were depolarized with increasing current pulses (2–20 pA increments) for 2 s, until an AP was triggered. (G and M) Summary graph showing cell excitability normalized by the capacitance of the iN cells. (H–L and N–R) Summary graphs analyzing the properties of the first AP. Data in G–L and M–R are means ± SEM; statistical comparisons were performed Student’s t test (*, P < 0.05; numbers of cells/independent experiments analyzed are shown in the bars of G and M). Access resistance was on average 9.5 ± 0.3 MΩ and 8 ± 0.2 MΩ for the control and 9.0 ± 0.2 MΩ and 7.4 ± 0.2 MΩ for the mutant cells of clones #1 and #2, respectively. Leak current was 25.2 ± 3.5 pA and 36 ± 4.4 pA for the control cells and 27.4 ± 4.6 pA and 36.5 ± 5.2 pA for the mutant cell of clones #1 and #2, respectively.
To test whether the reduction in Na+ channels impacts AP generation, we performed current-clamp recordings and sequentially depolarized the cells by injecting positive current. We found that L1CAM hemizygous mutant iN cells required larger current injections to reach AP threshold than precisely matching WT iN cells (Fig. 9, E–G, and M). We also assessed the firing threshold and the shape of the APs, but found no statistically significant differences between control and mutant cells (Fig. 9, H–L and N–R). These results indicate that deletion of L1CAM expression in human neurons leads to a partial loss of ankyrinG from the AIS, a decrease in the density of Na+ channels, and a change in a neuron’s response to somatic current injections.
Ankyrin-binding site of L1CAM is required for normal expression levels of ankyrinG and ankyrinB in human neurons
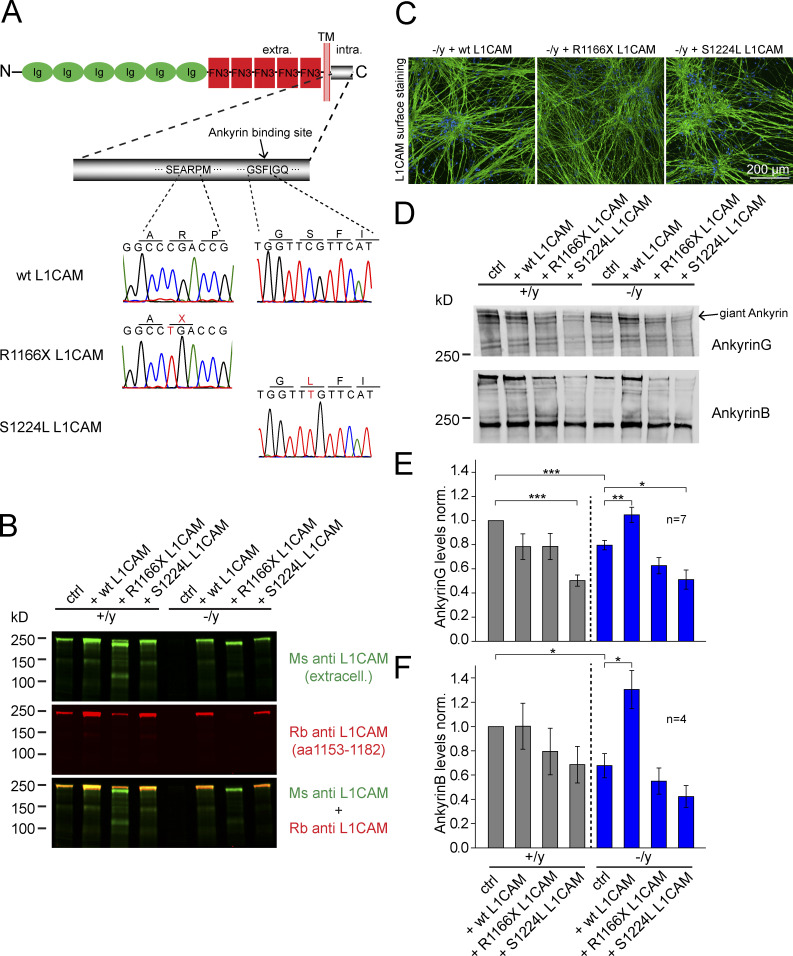
To evaluate the mechanistic role of the binding of ankyrins to L1CAM in maintaining expression levels of ankyrins, we next performed rescue experiments by lentiviral-mediated expression (infection on day −1; Fig. 3 D) in iN cells of WT L1CAM, or mutant L1CAM. The two pathological point mutations, R1166X and S1224L, either completely lack the ankyrin-binding site or contain a substitution in a key residue of the ankyrin-binding site (Fig. 10 A; Vos and Hofstra, 2010). Overexpressed WT, as well as R1166X and S1224L mutant, L1CAM could be detected by immunoblots at similar levels, and were both expressed on the surface of human neurons (Fig. 10, B and C). Interestingly, in contrast to WT L1CAM, R1166X, and S1224L mutant L1CAM did not rescue the decrease of ankyrin levels in mutant human neurons (which in this experiment were suppressed to ∼20% for ankyrinG and ∼30% for ankyrinB), suggesting a requirement of the L1CAM ankyrin protein interaction for the stabilization of ankyrins (Fig. 10, D–F).
Figure 10.
WT, but not mutant, L1CAM can rescue reduced ankyrinG and ankyrinB levels in L1CAM-deficient human neurons, and mutant L1CAM suppresses ankyrinG levels in WT neurons. (A) Schematic representation of rescue constructs. S1224L produces an amino acid substitution in the intracellular ankyrin-binding site of L1CAM, whereas R1166X introduces a stop codon N-terminal to the ankyrin-binding site. (B) Immunoblots showing expression of rescue L1CAM constructs in hemizygous mutant and matched control human neurons. Note that in contrast to the monoclonal mouse L1CAM antibody that binds to the extracellular part of L1CAM, the polyclonal rabbit L1CAM antibody that binds to intracellular sequences (aa 1153–1182) does not detect R1166X. (C) Confocal micrographs (from three independent experiments) of anti-L1CAM surface staining of hemizygous mutant neurons expressing the rescue constructs or WT L1CAM. (D) Immunoblots of iN cells as shown in B, detecting the levels of ankyrinG and ankyrinB. (E and F) Summary graphs of ankyrinG (top) and ankyrinB protein levels (bottom), corrected for blotting and loading variations using GDI as an internal standard. Data are means ± SEM; statistical significance was assessed using Student’s t test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
DISCUSSION
In this study, we used the cre/lox technology in human neurons to investigate the effect of hemizygous L1CAM loss-of-function mutations on neuronal function. This experimental design eliminates potentially confounding effects induced by genetic background changes or selection of cell clones. So far, hundreds of mutations in the L1CAM gene (which is X-chromosomal) were reported in patients with severe developmental malformations, neurological abnormalities, and intellectual disability (Rosenthal et al., 1992; Stumpel and Vos, 1993; Jouet et al., 1994, 1995; Fransen et al., 1997; Vos et al., 2010; Adle-Biassette et al., 2013; Chidsey et al., 2014). Most of the disease-causing L1CAM mutations likely represent loss-of-function mutations. We found that conditionally L1CAM-deficient human neurons manifest three types of defects: first, a large reduction in the size of axons, a smaller decrease in dendritic arborizations, and a significant reduction in the size of the AIS; second, a decrease in the levels of ankyrinG and ankyrinB; and third, impairment of AP generation. It seems likely that these three types of defects are commonly produced by the loss of the L1CAM complex with ankyrins upon deletion of L1CAM, and that these defects are responsible, at least in part, for the symptoms of patients with L1 syndrome.
In mutant mice and in in vitro systems, it has been well established that the intracellular segment of L1CAM binds to ankyrins (Davis and Bennett, 1994; Bennett and Chen, 2001). In general, it has been shown that ankyrinB binding promotes neuritogenesis by coupling L1CAM to the actin cytoskeleton (Nishimura et al., 2003). Binding L1CAM to ankyrinG, conversely, is thought to recruit L1CAM to the AIS, a highly specialized compartment in the proximal axon where APs are generated (Malhotra et al., 1998; Yoshimura and Rasband, 2014). Both ankyrins are thought to recruit and compartmentalize L1CAM and other L1CAM family members, such as neurofascin (Zhou et al., 1998; Boiko et al., 2007). For example, it has been reported that L1CAM and ankyrinB codistribute on axons in neonatal mouse brains before myelination. Mice deficient for ankyrinB exhibit reduced levels of L1CAM on these axons and, because of their hypoplasia of the corpus callosum and dilated ventricles, share features of L1 syndrome patients (Scotland et al., 1998). Furthermore, it has been shown that the cytoplasmic SFIGQY motif of L1CAM (aa 1224–1229, which is present in all L1CAM family members and is highly conserved from invertebrates to vertebrates) is essential for ankyrin binding (Garver et al., 1997; Needham et al., 2001). By introducing two pathological missense mutations, namely S1224L and Y1229H, reduced recruitment of ankyrin to the plasma membrane in vitro was observed (Needham et al., 2001). Knock-in mice expressing the missense mutation Y1229H exhibit mistargeting of mutant ganglion cell axons from the ventral retina to lateral sites in the contralateral superior colliculus (Buhusi et al., 2008). Additionally, observations of neonatal L1CAM-deficient mice suggest a decrease of ankyrinB expression in thalamocortical and corticothalamic axons, together with axonal hyperfasciculation and path-finding errors (Wiencken-Barger et al., 2004). However, mice with a truncated intracellular L1CAM sequence lacking this ankyrin-binding region exhibited morphologically normal brains (Nakamura et al., 2010), raising questions about the precise role of the intracellular L1CAM sequence.
In the present study, we show that L1CAM codistributes with ankyrinG and ankyrinB and that the deletion of L1CAM decreases ankyrins on the subcellular levels (ankyrinG at the AIS, ankyrinB on axons; Fig. 7), and on the global protein level (as revealed by quantitative immunoblot analyses; Fig. 4, A and B), suggesting that L1CAM may actually recruit and stabilize ankyrins to specific subcellular localizations. In addition to previous studies, our data show that on human axons L1CAM is not only important for the expression of ankyrinB and axonal arborizations but it is also crucial to the levels of ankyrinG at the AIS and its proper function in Na+-mediated AP generation. It is tempting to speculate that L1CAM and ankyrins are interdependent subunits of a complex stabilizing each other, and thereby regulating axonal morphological and functional development. Evidence for this notion comes from our rescue experiments, suggesting that the integrity of the ankyrin-binding site is important for the stabilization of ankyrins (Fig. 10).
AnkyrinG is a key component of the AIS and has been shown to be responsible for clustering Na+ and K+ channels at the AIS (Zhou et al., 1998; Garrido et al., 2003; Kole and Stuart, 2012). In the absence of ankyrinG in mouse neurons, Na+ currents are highly reduced and the shape of the AP is altered (Zhou et al., 1998; Hedstrom et al., 2008; Barry et al., 2014). This phenotype is similar to the Na+ channel phenotype we observed in L1CAM-deficient human neurons. AnkyrinG mutations in patients have been associated with severe cognitive deficits, autism, sleeping disorder, severe attention deficit hyperactivity disorder, and bipolar disorder (Ferreira et al., 2008; Schulze et al., 2009; Iqbal et al., 2013).
To date, a change in neuronal excitability has not been observed in L1CAM mouse mutants and could represent a feature that is more easily distinguished in human neurons (Dahme et al., 1997; Cohen et al., 1998; Fransen et al., 1998; Demyanenko et al., 1999; Law et al., 2003; Saghatelyan et al., 2004; Nakamura et al., 2010). However, a disintegration of the AIS, APs with modified waveform upon stimulation, abolition of spontaneous firing in Purkinje cells, and deficits in motor learning have been reported for mice deficient for neurofascin in adult cerebellar Purkinje cells. But in developing neurons, neurofascin was shown to be dispensable for AIS assembly, while ankyrinG staining intensities were unchanged (Zonta et al., 2011). Neurofascin is thought to be essential for the clustering of Na+ channels at the nodes of Ranvier, another excitable region of the axon, by its binding to ankyrinG (Sherman et al., 2005).
We observed an ∼20–50% decrease in ankyrinG levels, possibly leading to a decrease in the concentration of voltage-gated Na+ channels at the AIS. This result might explain the rather modest reduction of Na+ currents and the lack of significant changes in the shape of APs. However, this points to an important aspect of L1 syndrome because the missing L1CAM–ankyrin interactions likely contribute to the clinical presentations of L1CAM and ankyrin mutations. Future work could use this model system to test ways of restoring neuronal excitability, for example, by reducing the activity of resting K+ channels at the AIS. It has been shown that pharmacological inhibition of Cyclin-dependent kinase 2 or 5 (Cdk2 or Cdk5) by roscovitine,enriched K+ channels at the AIS (Vacher et al., 2011). One possible way to reduce K+ channel activity would be to pharmacologically activate Cdk2 or Cdk5 function.
Overall, our study using cre/lox technology on human neurons presents a first step in understanding L1 syndrome at the neuronal level. Our results confirm a morphological phenotype previously observed in animal models (Dahme et al., 1997; Cohen et al., 1998; Demyanenko et al., 1999), but also presents an additional component in the human mutant neuron’s reduced ability to generate APs.
MATERIALS AND METHODS
Viral constructs
The following lentiviral constructs were used: FUW-TetO-Ng2-T2A-puromycin expressing TetO-Ng2-T2A-puromycin cassette (TetO promoter drives expression of full-length mouse Ngn2 and of puromycin via the cleavage-peptide sequence T2A; Fig. 3 B); F-Ubiquitin-W (FUW)-rtTA containing rtTA; FUW-TetO-EGFP expressing EGFP; FSW-NLS-mCherry expressing mCherry preceded by a nuclear localization sequence (under the control of neuron-specific human synapsin promoter) to monitor cell death; FUW-Flp to express Flp-recombinase; FUW-GFP::Cre to express Cre-recombinase to delete L1CAM exon 3 and create via frameshift a null-allele or GFP::ΔCre for the wild-type control; FUW-TetO-wild-type-L1CAM; FUW-TetO-R1166X-L1CAM; and FUW-TetO-S1224L-L1CAM. One adeno-associated virus (AAV) construct was used for gene targeting of X-chromosomal L1CAM allele of the male hESCs H1 (also shown in Fig. 3 B): the construct contains sequences from the region encoding exon 3 flanked by loxP sites and frt-sites flanked puromycin resistance cassette (Frt-PGK-Puro-SV40polyA-Frt) adjacent to the 5′ loxP site. For homologous recombination, the 5′ arm of the construct includes 1.6 kb of sequences located upstream of exon 3. The 3′ arm contains 1.4 kb of sequences located downstream of exon 3.
Virus generation
Lentiviruses were produced as previously described (Zhang et al., 2013) in HEK293T cells (ATCC) by co-transfection with three helper plasmids (pRSV-REV, pMDLg/pRRE, and vesicular stomatitis virus G protein expression vector) with 12 µg of lentiviral vector DNA and 6 µg of each of the helper plasmid DNA per 75 cm2 culture area) using calcium phosphate. Lentiviruses were harvested in the medium 48 h after transfection, pelleted by centrifugation (49,000 g for 90 min), resuspended in MEM, aliquoted, and frozen at –80°C. Only virus preparations with >90% infection efficiency as assessed by EGFP expression or puromycin resistance were used for experiments. AAV-DJ was used to deliver the targeting construct for generation of cKO cells. AAV-DJ was produced in HEK293T cells by co-transfection of pHelper, pDJ, and AAV vector (8.5 µg of DNA per 75 cm2 culture area) using calcium phosphate. Cells were harvested 72 h after transfection in PBS/1 mM EDTA and after one freezing/thawing cycle. AAVs were collected from cytoplasm using Benzonase nuclease at a final concentration of 50 U/ml at 37°C for 30 min. After clearing the suspension from cell debris by slow centrifugation (3,000 g for 30 min) AAVs were isolated after fast centrifugation (400,000 g for 120 min) in iodixanol (gradient from 15–60%) from the 40% layer and further concentrated using centricon concentrating tube (100,000 MWCO; EMD Millipore) according to the manufacturer’s suggested protocol.
Cell culture
Experiments were performed as previously described (Zhang et al., 2013). H1 ES cells (WiCell Research Resources) were maintained as feeder-free cells in mTeSR1 medium (Stem Cell Technologies). Mouse glial cells were cultured from the forebrain of newborn WT CD1 mice. In brief, newborn mouse forebrain homogenates were digested with papain and EDTA for 30 min, and cells were dissociated by harsh trituration to avoid growing of neurons, and then plated onto T75 flasks in DMEM supplemented with 10% FBS. Upon reaching confluence, glial cells were trypsinized and replated at lower density a total of three times to remove potential trace amounts of mouse neurons before the glia cell cultures were used for co-culture experiment with iN cells.
Gene targeting in ES cells
H1 ES cells were transduced by AAVs to generate conditional hemizygous mutant ES cells by flanking exon 3 (ENSE00003680685, coding the aa MEPPVITEQSPRRLVVFPTDDISLKCEASGKPEV in the first Ig domain), which results in a frameshift and a premature stop codon upon cre recombination. Puromycin (1 µg/ml) was added and kept in mTeSR1 medium 2 d after transduction. The surviving ES cells were allowed to grow into colonies, and then individually picked. 7 correctly targeted colonies out of 76 colonies in total were confirmed by PCR screening using oligo sequences 5′-ACAGATGACATCAGCCTCAAGTGTGAGG-3′ and 5′-GACTGGGAGATGGCGAGGACTTG-3′, resulting in a 240-bp band for a correctly targeted clone or 185 bp for the untargeted WT. Correctly mutated alleles were confirmed after flp and cre recombination and iN differentiation.
Standard protocol for generation of iN cells from conditional mutant human ES cells and for activating the conditional mutations
iN cell generation has been previously described (Zhang et al., 2013). In brief, targeted and flp-recombined (after blasticidin selection) human ES cells were treated with Accutase (Innovative Cell Technologies) and plated as dissociated cells in 24-well plates (104 cells/well) on day −1 (Fig. 3 C). Cells were plated on Matrigel-coated (BD) coverslips in mTeSR1 containing 2 mM thiazovivin (Bio Vision). At the same time point, lentiviruses prepared as described in the paragraph "Virus generation"(0.3 µl/well of 24-well plate) were added. Two different types of lentiviruses were co-infected: the lentiviruses used for iN cell induction as described, and lentiviruses expressing either Cre-recombinase (to create a null allele) under control of the ubiquitin promoter or an inactive mutated ΔCre-recombinase for the WT control (Fig. 3 C). On day 0, the culture medium was replaced with N2/DMEM/F12/NEAA (Invitrogen) containing human brain-derived neurotrophic factor (BDNF; 10 ng/ml; PeproTech), human NT-3 (10 ng/ml; PeproTech), and mouse Laminin-1 (0.2 µg/ml; Invitrogen). Doxycycline (2 µg/ml; Takara Bio Inc.) was added on day 0 to induce TetO gene expression and retained in the medium until the end of the experiment. On day 1, a 24-h puromycin selection (1 µg/ml) period was started. On day 2, mouse glia cells were added in neurobasal medium supplemented with B27/Glutamax (Invitrogen) containing BDNF, NT3, and Laminin-1; Ara-C (2 µM; Sigma-Aldrich) was added to the medium to inhibit astrocyte proliferation. After day 2, 50% of the medium in each well was exchanged every 2 d. FBS (2.5%) was added to the culture medium on day 10 to support astrocyte viability, and iN cells were assayed after at least 21 d or as indicated.
Quantification of neuronal morphology
iN cells were sparsely transfected by calcium phosphate on day 19 with a TetO-EGFP expression vector. On day 21, cells were fixed and stained for EGFP and MAP2a/b to discriminate between dendrites and axons. Fixation at later time points would result in too abundant axonal growth. All MAP2- and EGFP-positive neurites were considered to be dendrites and all MAP2-negative but EGFP-positive neurites to be axons. Images were acquired using an A1RSi confocal microscope system (Nikon), with a 10× or 20× objective at room temperature. Images of ∼10–30 neurons per condition (n = 1 for axons; n = ∼100 in case dendrites) were reconstructed using the MetaMorph neurite application, scoring for total dendritic length, dendritic branch points, total axonal length, axonal branch points, and soma area. In the case of old human neurons, cells were fixed after DIV180 and immunolabeled.
Immunofluorescence and immunoblotting experiments
Immunofluorescence experiments were performed essentially as previously described (Zhang et al., 2013). In brief, cultured iN cells were fixed in 4% paraformaldehyde and 4% sucrose in PBS for 20 min at room temperature, washed three times with PBS, and incubated in 0.2% Triton X-100 in PBS for 10 min at room temperature. Cells were blocked in PBS containing 5% goat serum for 1 h at room temperature. Primary antibodies were applied overnight at 4°C, cells were washed in PBS three times, and fluorescently labeled secondary antibodies (Alexa Fluor; 1:1,000) were applied for 1 h at room temperature. The following antibodies were used in immunocytochemisty experiments: MAP2 a/b (Sigma-Aldrich; 1:1,000), MAP2 a/b (AB5622; EMD Millipore; 1:500), MAP2 (CPCA-MAP2; EnCor; 1:2,000), Synapsin (E028; 1:1,000), ankyrinG (106/36; Neuromab; 1:500), ankyrinB (105/13; Neuromab; 1:500), Neurofilament-L (171002; Synaptic Systems; 1:500), and GFP (Invitrogen; A11122; 1:2,000). To label L1CAM on human neurons, in some cases (Fig. 7) a post-fix staining using polyclonal Rb antibody (10140-R004-50; Sinobiological) was performed. In all other cases, a live staining was performed. The human-specific clone UJ127.11 (Sigma-Aldrich) was given into fresh culturing media for 20 min at 37°C (10 µg/ml). Next, cells were fixed and washed as described in this paragraph. Images were taken using an A1RSi confocal microscope system (Nikon), with a 10×, 20×, or 60× objective at room temperature. All quantitative immunoblotting experiments were performed with fluorescently labeled secondary antibodies (LiCor; 1:5,000). Samples were separated by SDS-PAGE under reducing conditions and transferred onto nitrocellulose membranes. Blots were blocked in Tris-buffered saline containing 0.1% Tween 20 (Sigma-Aldrich) and 5% fat-free milk for 2 h at room temperature. The blocked membrane was incubated in blocking buffer containing the primary antibody overnight at 4°C, followed by three to five washes. The washed membrane was incubated in blocking buffer containing secondary antibody for 2 h at room temperature. Blots were scanned with the Odyssey-system (LiCor), followed by quantification with ImageStudio software (LiCor). For immunodetection, the following antibodies were used: NeuN (ABN78; EMD Millipore), TuJ1 (MMS-435P; Covance), Complexin 1/2 (L668), SNAP25 (P913), Synaptobrevin-2 (P939), Synapsin (E028), Syntaxin-1 (438B), β-Actin (A1978; Sigma-Aldrich), Synaptophysin (Synaptic Systems, 7.2), GDP-dissociation inhibitor (Synaptic Systems; GDI; 81.2), calmodulin-associated serine/threonine kinase (BD; clone 7/CASK), L1CAM (UJ127.11; Sigma-Aldrich), L1CAM (ab123990; Abcam), SynCAM (T2412), MAP2 a/b (AB5622; EMD Millipore; 1:500), MAP2 a/b/c/d (Sigma-Aldrich; SAB4300658; 1:500), Neurofilament-L (171002,SySy; 1:500), Neurofilament-H (Abcam; RT97; 1:500), ankyrinG (Neuromab; 106/20, 1:500), ankyrinB (105/13; Neuromab; 1:500), Tau-5 (EMD Millipore; MAB361; 1:1,000), PSD95 (Abcam; ab76115; 1:500), GluR1 (Abcam; ab1504; 1:500), CREB (Cell Signaling Technology; 86B10; 1:1,000), phospho-CREB (Cell Signaling Technology; 87G3; 1:1,000), Erk1/2 (Cell Signaling Technology; 137F5; 1:1,000), phospho-Erk1/2 (Cell Signaling Technology; E10; 1:1,000), JNK (Cell Signaling Technology; 9252; 1:1,000), phospho-JNK (Cell Signaling Technology; G9; 1:1000), Akt (Cell Signaling Technology; 5G3; 1:1,000), phospho-Akt (Cell Signaling Technology; 9271; 1:1,000).
Quantification of synaptic density and survival
For synapsin puncta analyses, images were acquired using a A1RSi confocal microscope system (Nikon; 60× objective at room temperature), and the puncta density was determined using the software NIS-Elements (Nikon). Analyses of the survival of iN cells was directly monitored using images of iN cell nuclei expressing mCherry at the same position of the culture dish taken every other day, using a DFC400 digital camera (Leica) attached to a DMIL LED inverted microscope with a 5× objective (Leica), driven by Application Suite image-acquisition software (Leica). Number of cells was determined using ImageJ software (National Institutes of Health). For each experimental condition, the mean of the pictures from 20–30 culture wells (of a 96-well culture plate) was considered as n = 1. In total, a mean of n = 3 was calculated.
Electrophysiology
All electrophysiological recordings were performed using whole-cell patch clamp. In brief, 5 wk after iN induction, on the day of recording, a coverslip containing relatively low-density induced human neurons was placed in a recording chamber mounted onto an Axioskop 2F upright microscope (Zeiss) equipped with DIC and fluorescence capabilities. Cells were approached under DIC with ∼2 MΩ pipettes pulled from borosilicate glass (Warner Instruments, Inc.) using a vertical PC-10 puller (Narishige), and impaled until a high-resistance seal (GΩ) was formed between the recording pipette and the cell membrane. Next, whole-cell configuration was established by gentle application of negative pressure trough the recording pipette. The recording pipette contained (in mM): 125 K-gluconate, 20 KCl, 10 Hepes, 0.5 EGTA, 4 ATP-Magnesium, 0.3 GTP-Sodium, and 10 Na-Phosphocreatine; osmolarity, 312 mOsm, pH 7.2, adjusted with KOH. Electrical signals were recorded at 25 kHz with a two-channel Axoclamp 700B amplifier (Axon Instruments) and digitalized with a Digidata 1440 digitizer (Molecular Devices) that was, in turn, controlled by Clampex 10.1 (Molecular Devices). All recordings were performed at ∼24°C.
For whole-cell voltage-clamp recordings, neurons were maintained at −80 mV holding potentials. Series resistance varied between 4–10 MΩ. All iNs in which series resistance was higher than that were not included in the analysis. In all current-clamp experiments, the membrane potential was maintained approximately −80 mV by constant injection of negative current through the recording pipette. Samples in the recording chamber were continuously perfused with oxygenated (95% O2/5%CO2) bath solution containing (in mM): 125 NaCl, 2.5 KCl, 1 MgCl2, 2 CaCl2, 25 glucose, 1.25 NaH2PO4, 0.4 ascorbic acid, 3 myo-inositol, 2 Na-pyruvate, and 25 NaHCO3, pH 7.4, and 315 mOsm.
Data presentation and statistics
All data shown are means ± SEMs. Number of measured cells and independent experiments is indicated inside each bar, or mentioned in the figure legend.
All statistical analyses were performed using either two-tailed Student’s t test or two-way ANOVA, comparing the test sample to the control sample examined in the same experiments.
Study approval
The present study was approved by Stem Cell Research Oversight (SCRO) at the Stanford University Research Compliance Office (SCRO 518: Studying brain diseases affecting synaptic transmission by using human iNs). Experiments involving animals were approved by the Stanford University Institutional Animal Care and Use Committee, Administrative Panel on Laboratory Animal Care Research Compliance Office.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (MH092931 to M. Wernig and AG010770 to S. Prusiner) and from the California Institute for Regenerative Medicine (RT2 02061 to M. Wernig), and by a postdoctoral fellowship to C. Patzke (DFG PA 2110/1-1) and L.R. Giam (National Science Foundation Award 1202829 and the Helen Hay Whitney Foundation). M. Wernig is a New York Stem Cell Foundation-Robertson Investigator.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AAV
- adeno-associated virus
- AIS
- axon initial segment
- AP
- action potential
- cKO
- conditional KO
- ES
- embryonic stem
- iN
- induced neuron
References
- Adle-Biassette H., Saugier-Veber P., Fallet-Bianco C., Delezoide A.L., Razavi F., Drouot N., Bazin A., Beaufrère A.M., Bessières B., Blesson S., et al. 2013. Neuropathological review of 138 cases genetically tested for X-linked hydrocephalus: evidence for closely related clinical entities of unknown molecular bases. Acta Neuropathol. 126:427–442. 10.1007/s00401-013-1146-1 [DOI] [PubMed] [Google Scholar]
- Arevalo E., Shanmugasundararaj S., Wilkemeyer M.F., Dou X., Chen S., Charness M.E., and Miller K.W.. 2008. An alcohol binding site on the neural cell adhesion molecule L1. Proc. Natl. Acad. Sci. USA. 105:371–375. 10.1073/pnas.0707815105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barry J., Gu Y., Jukkola P., O’Neill B., Gu H., Mohler P.J., Rajamani K.T., and Gu C.. 2014. Ankyrin-G directly binds to kinesin-1 to transport voltage-gated Na+ channels into axons. Dev. Cell. 28:117–131. 10.1016/j.devcel.2013.11.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A., Jouet M., MacFarlane J., Du J.S., Kenwrick S., and Chothia C.. 1996. Outline structure of the human L1 cell adhesion molecule and the sites where mutations cause neurological disorders. EMBO J. 15:6050–6059. [PMC free article] [PubMed] [Google Scholar]
- Bearer C.F., Swick A.R., O’Riordan M.A., and Cheng G.. 1999. Ethanol inhibits L1-mediated neurite outgrowth in postnatal rat cerebellar granule cells. J. Biol. Chem. 274:13264–13270. 10.1074/jbc.274.19.13264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett V., and Chen L.. 2001. Ankyrins and cellular targeting of diverse membrane proteins to physiological sites. Curr. Opin. Cell Biol. 13:61–67. 10.1016/S0955-0674(00)00175-7 [DOI] [PubMed] [Google Scholar]
- Boiko T., Vakulenko M., Ewers H., Yap C.C., Norden C., and Winckler B.. 2007. Ankyrin-dependent and -independent mechanisms orchestrate axonal compartmentalization of L1 family members neurofascin and L1/neuron-glia cell adhesion molecule. J. Neurosci. 27:590–603. 10.1523/JNEUROSCI.4302-06.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhusi M., Schlatter M.C., Demyanenko G.P., Thresher R., and Maness P.F.. 2008. L1 interaction with ankyrin regulates mediolateral topography in the retinocollicular projection. J. Neurosci. 28:177–188. 10.1523/JNEUROSCI.3573-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S., Rathjen F.G., and Raper J.A.. 1987. Extension of neurites on axons is impaired by antibodies against specific neural cell surface glycoproteins. J. Cell Biol. 104:355–362. 10.1083/jcb.104.2.355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidsey B.A., Baldwin E.E., Toydemir R., Ahles L., Hanson H., and Stevenson D.A.. 2014. L1CAM whole gene deletion in a child with L1 syndrome. Am. J. Med. Genet. A. 164A:1555–1558. 10.1002/ajmg.a.36474 [DOI] [PubMed] [Google Scholar]
- Cohen N.R., Taylor J.S., Scott L.B., Guillery R.W., Soriano P., and Furley A.J.. 1998. Errors in corticospinal axon guidance in mice lacking the neural cell adhesion molecule L1. Curr. Biol. 8:26–33. 10.1016/S0960-9822(98)70017-X [DOI] [PubMed] [Google Scholar]
- Dahme M., Bartsch U., Martini R., Anliker B., Schachner M., and Mantei N.. 1997. Disruption of the mouse L1 gene leads to malformations of the nervous system. Nat. Genet. 17:346–349. 10.1038/ng1197-346 [DOI] [PubMed] [Google Scholar]
- Davis J.Q., and Bennett V.. 1994. Ankyrin binding activity shared by the neurofascin/L1/NrCAM family of nervous system cell adhesion molecules. J. Biol. Chem. 269:27163–27166. [PubMed] [Google Scholar]
- De Angelis E., MacFarlane J., Du J.S., Yeo G., Hicks R., Rathjen F.G., Kenwrick S., and Brümmendorf T.. 1999. Pathological missense mutations of neural cell adhesion molecule L1 affect homophilic and heterophilic binding activities. EMBO J. 18:4744–4753. 10.1093/emboj/18.17.4744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Angelis E., Watkins A., Schäfer M., Brümmendorf T., and Kenwrick S.. 2002. Disease-associated mutations in L1 CAM interfere with ligand interactions and cell-surface expression. Hum. Mol. Genet. 11:1–12. 10.1093/hmg/11.1.1 [DOI] [PubMed] [Google Scholar]
- Demyanenko G.P., Tsai A.Y., and Maness P.F.. 1999. Abnormalities in neuronal process extension, hippocampal development, and the ventricular system of L1 knockout mice. J. Neurosci. 19:4907–4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M.A., O’Donovan M.C., Meng Y.A., Jones I.R., Ruderfer D.M., Jones L., Fan J., Kirov G., Perlis R.H., Green E.K., et al. Wellcome Trust Case Control Consortium . 2008. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat. Genet. 40:1056–1058. 10.1038/ng.209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fransen E., Van Camp G., Vits L., and Willems P.J.. 1997. L1-associated diseases: clinical geneticists divide, molecular geneticists unite. Hum. Mol. Genet. 6:1625–1632. 10.1093/hmg/6.10.1625 [DOI] [PubMed] [Google Scholar]
- Fransen E., D’Hooge R., Van Camp G., Verhoye M., Sijbers J., Reyniers E., Soriano P., Kamiguchi H., Willemsen R., Koekkoek S.K., et al. 1998. L1 knockout mice show dilated ventricles, vermis hypoplasia and impaired exploration patterns. Hum. Mol. Genet. 7:999–1009. 10.1093/hmg/7.6.999 [DOI] [PubMed] [Google Scholar]
- Freigang J., Proba K., Leder L., Diederichs K., Sonderegger P., and Welte W.. 2000. The crystal structure of the ligand binding module of axonin-1/TAG-1 suggests a zipper mechanism for neural cell adhesion. Cell. 101:425–433. 10.1016/S0092-8674(00)80852-1 [DOI] [PubMed] [Google Scholar]
- Garrido J.J., Giraud P., Carlier E., Fernandes F., Moussif A., Fache M.P., Debanne D., and Dargent B.. 2003. A targeting motif involved in sodium channel clustering at the axonal initial segment. Science. 300:2091–2094. 10.1126/science.1085167 [DOI] [PubMed] [Google Scholar]
- Garver T.D., Ren Q., Tuvia S., and Bennett V.. 1997. Tyrosine phosphorylation at a site highly conserved in the L1 family of cell adhesion molecules abolishes ankyrin binding and increases lateral mobility of neurofascin. J. Cell Biol. 137:703–714. 10.1083/jcb.137.3.703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gécz J., Shoubridge C., and Corbett M.. 2009. The genetic landscape of intellectual disability arising from chromosome X. Trends Genet. 25:308–316. 10.1016/j.tig.2009.05.002 [DOI] [PubMed] [Google Scholar]
- Halliday J., Chow C.W., Wallace D., and Danks D.M.. 1986. X linked hydrocephalus: a survey of a 20 year period in Victoria, Australia. J. Med. Genet. 23:23–31. 10.1136/jmg.23.1.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Jensen G.J., and Bjorkman P.J.. 2009. Cryo-electron tomography of homophilic adhesion mediated by the neural cell adhesion molecule L1. Structure. 17:460–471. 10.1016/j.str.2009.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedstrom K.L., Ogawa Y., and Rasband M.N.. 2008. AnkyrinG is required for maintenance of the axon initial segment and neuronal polarity. J. Cell Biol. 183:635–640. 10.1083/jcb.200806112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal Z., Vandeweyer G., van der Voet M., Waryah A.M., Zahoor M.Y., Besseling J.A., Roca L.T., Vulto-van Silfhout A.T., Nijhof B., Kramer J.M., et al. 2013. Homozygous and heterozygous disruptions of ANK3: at the crossroads of neurodevelopmental and psychiatric disorders. Hum. Mol. Genet. 22:1960–1970. 10.1093/hmg/ddt043 [DOI] [PubMed] [Google Scholar]
- Itoh K., Cheng L., Kamei Y., Fushiki S., Kamiguchi H., Gutwein P., Stoeck A., Arnold B., Altevogt P., and Lemmon V.. 2004. Brain development in mice lacking L1-L1 homophilic adhesion. J. Cell Biol. 165:145–154. 10.1083/jcb.200312107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouet M., Rosenthal A., Armstrong G., MacFarlane J., Stevenson R., Paterson J., Metzenberg A., Ionasescu V., Temple K., and Kenwrick S.. 1994. X-linked spastic paraplegia (SPG1), MASA syndrome and X-linked hydrocephalus result from mutations in the L1 gene. Nat. Genet. 7:402–407. 10.1038/ng0794-402 [DOI] [PubMed] [Google Scholar]
- Jouet M., Moncla A., Paterson J., McKeown C., Fryer A., Carpenter N., Holmberg E., Wadelius C., and Kenwrick S.. 1995. New domains of neural cell-adhesion molecule L1 implicated in X-linked hydrocephalus and MASA syndrome. Am. J. Hum. Genet. 56:1304–1314. [PMC free article] [PubMed] [Google Scholar]
- Kenwrick S., Watkins A., and De Angelis E.. 2000. Neural cell recognition molecule L1: relating biological complexity to human disease mutations. Hum. Mol. Genet. 9:879–886. 10.1093/hmg/9.6.879 [DOI] [PubMed] [Google Scholar]
- Kiefel H., Bondong S., Hazin J., Ridinger J., Schirmer U., Riedle S., and Altevogt P.. 2012. L1CAM: a major driver for tumor cell invasion and motility. Cell Adhes. Migr. 6:374–384. 10.4161/cam.20832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kole M.H., and Stuart G.J.. 2012. Signal processing in the axon initial segment. Neuron. 73:235–247. 10.1016/j.neuron.2012.01.007 [DOI] [PubMed] [Google Scholar]
- Law J.W., Lee A.Y., Sun M., Nikonenko A.G., Chung S.K., Dityatev A., Schachner M., and Morellini F.. 2003. Decreased anxiety, altered place learning, and increased CA1 basal excitatory synaptic transmission in mice with conditional ablation of the neural cell adhesion molecule L1. J. Neurosci. 23:10419–10432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Focia P.J., and He X.. 2011. Homophilic adhesion mechanism of neurofascin, a member of the L1 family of neural cell adhesion molecules. J. Biol. Chem. 286:797–805. 10.1074/jbc.M110.180281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra J.D., Tsiotra P., Karagogeos D., and Hortsch M.. 1998. Cis-activation of L1-mediated ankyrin recruitment by TAG-1 homophilic cell adhesion. J. Biol. Chem. 273:33354–33359. 10.1074/jbc.273.50.33354 [DOI] [PubMed] [Google Scholar]
- Maness P.F., and Schachner M.. 2007. Neural recognition molecules of the immunoglobulin superfamily: signaling transducers of axon guidance and neuronal migration. Nat. Neurosci. 10:19–26. 10.1038/nn1827 [DOI] [PubMed] [Google Scholar]
- Nakamura Y., Lee S., Haddox C.L., Weaver E.J., and Lemmon V.P.. 2010. Role of the cytoplasmic domain of the L1 cell adhesion molecule in brain development. J. Comp. Neurol. 518:1113–1132. 10.1002/cne.22267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needham L.K., Thelen K., and Maness P.F.. 2001. Cytoplasmic domain mutations of the L1 cell adhesion molecule reduce L1-ankyrin interactions. J. Neurosci. 21:1490–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K., Yoshihara F., Tojima T., Ooashi N., Yoon W., Mikoshiba K., Bennett V., and Kamiguchi H.. 2003. L1-dependent neuritogenesis involves ankyrinB that mediates L1-CAM coupling with retrograde actin flow. J. Cell Biol. 163:1077–1088. 10.1083/jcb.200303060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan R., Wilkemeyer M.F., Mittal B., Perides G., and Charness M.E.. 1996. Alcohol inhibits cell-cell adhesion mediated by human L1. J. Cell Biol. 133:381–390. 10.1083/jcb.133.2.381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathjen F.G., and Schachner M.. 1984. Immunocytological and biochemical characterization of a new neuronal cell surface component (L1 antigen) which is involved in cell adhesion. EMBO J. 3:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenthal A., Jouet M., and Kenwrick S.. 1992. Aberrant splicing of neural cell adhesion molecule L1 mRNA in a family with X-linked hydrocephalus. Nat. Genet. 2:107–112. 10.1038/ng1092-107 [DOI] [PubMed] [Google Scholar]
- Saghatelyan A.K., Nikonenko A.G., Sun M., Rolf B., Putthoff P., Kutsche M., Bartsch U., Dityatev A., and Schachner M.. 2004. Reduced GABAergic transmission and number of hippocampal perisomatic inhibitory synapses in juvenile mice deficient in the neural cell adhesion molecule L1. Mol. Cell. Neurosci. 26:191–203. 10.1016/j.mcn.2004.01.008 [DOI] [PubMed] [Google Scholar]
- Schaefer A.W., Kamiguchi H., Wong E.V., Beach C.M., Landreth G., and Lemmon V.. 1999. Activation of the MAPK signal cascade by the neural cell adhesion molecule L1 requires L1 internalization. J. Biol. Chem. 274:37965–37973. 10.1074/jbc.274.53.37965 [DOI] [PubMed] [Google Scholar]
- Schäfer M.K., and Altevogt P.. 2010. L1CAM malfunction in the nervous system and human carcinomas. Cell. Mol. Life Sci. 67:2425–2437. 10.1007/s00018-010-0339-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer M.K., and Frotscher M.. 2012. Role of L1CAM for axon sprouting and branching. Cell Tissue Res. 349:39–48. 10.1007/s00441-012-1345-4 [DOI] [PubMed] [Google Scholar]
- Schäfer M.K., Nam Y.C., Moumen A., Keglowich L., Bouché E., Küffner M., Bock H.H., Rathjen F.G., Raoul C., and Frotscher M.. 2010. L1 syndrome mutations impair neuronal L1 function at different levels by divergent mechanisms. Neurobiol. Dis. 40:222–237. 10.1016/j.nbd.2010.05.029 [DOI] [PubMed] [Google Scholar]
- Schmid R.S., Pruitt W.M., and Maness P.F.. 2000. A MAP kinase-signaling pathway mediates neurite outgrowth on L1 and requires Src-dependent endocytosis. J. Neurosci. 20:4177–4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze T.G., Detera-Wadleigh S.D., Akula N., Gupta A., Kassem L., Steele J., Pearl J., Strohmaier J., Breuer R., Schwarz M., et al. NIMH Genetics Initiative Bipolar Disorder Consortium . 2009. Two variants in Ankyrin 3 (ANK3) are independent genetic risk factors for bipolar disorder. Mol. Psychiatry. 14:487–491. 10.1038/mp.2008.134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann G., Haspel J., Grumet M., and Erickson H.P.. 2001. Cell adhesion molecule L1 in folded (horseshoe) and extended conformations. Mol. Biol. Cell. 12:1765–1773. 10.1091/mbc.12.6.1765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotland P., Zhou D., Benveniste H., and Bennett V.. 1998. Nervous system defects of AnkyrinB (−/−) mice suggest functional overlap between the cell adhesion molecule L1 and 440-kD AnkyrinB in premyelinated axons. J. Cell Biol. 143:1305–1315. 10.1083/jcb.143.5.1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman D.L., Tait S., Melrose S., Johnson R., Zonta B., Court F.A., Macklin W.B., Meek S., Smith A.J., Cottrell D.F., and Brophy P.J.. 2005. Neurofascins are required to establish axonal domains for saltatory conduction. Neuron. 48:737–742. 10.1016/j.neuron.2005.10.019 [DOI] [PubMed] [Google Scholar]
- Stumpel C., and Vos Y.J.. 1993. L1 Syndrome. In GeneReviews(R). Pagon R.A., Adam M.P., Ardinger H.H., Wallace S.E., Amemiya A., Bean L.J.H., Bird T.D., Dolan C.R., Fong C.T., Smith R.J.H., and Stephens K., editors. University of Washington, Seattle, Seattle WA. [PubMed] [Google Scholar]
- Vacher H., Yang J.W., Cerda O., Autillo-Touati A., Dargent B., and Trimmer J.S.. 2011. Cdk-mediated phosphorylation of the Kvβ2 auxiliary subunit regulates Kv1 channel axonal targeting. J. Cell Biol. 192:813–824. 10.1083/jcb.201007113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos Y.J., and Hofstra R.M.. 2010. An updated and upgraded L1CAM mutation database. Hum. Mutat. 31:E1102–E1109. 10.1002/humu.21172 [DOI] [PubMed] [Google Scholar]
- Vos Y.J., de Walle H.E., Bos K.K., Stegeman J.A., Ten Berge A.M., Bruining M., van Maarle M.C., Elting M.W., den Hollander N.S., Hamel B., et al. 2010. Genotype-phenotype correlations in L1 syndrome: a guide for genetic counselling and mutation analysis. J. Med. Genet. 47:169–175. 10.1136/jmg.2009.071688 [DOI] [PubMed] [Google Scholar]
- Weller S., and Gärtner J.. 2001. Genetic and clinical aspects of X-linked hydrocephalus (L1 disease): Mutations in the L1CAM gene. Hum. Mutat. 18:1–12. 10.1002/humu.1144 [DOI] [PubMed] [Google Scholar]
- Wiencken-Barger A.E., Mavity-Hudson J., Bartsch U., Schachner M., and Casagrande V.A.. 2004. The role of L1 in axon pathfinding and fasciculation. Cereb. Cortex. 14:121–131. 10.1093/cercor/bhg110 [DOI] [PubMed] [Google Scholar]
- Wolff J.M., Frank R., Mujoo K., Spiro R.C., Reisfeld R.A., and Rathjen F.G.. 1988. A human brain glycoprotein related to the mouse cell adhesion molecule L1. J. Biol. Chem. 263:11943–11947. [PubMed] [Google Scholar]
- Yoshimura T., and Rasband M.N.. 2014. Axon initial segments: diverse and dynamic neuronal compartments. Curr. Opin. Neurobiol. 27:96–102. 10.1016/j.conb.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Pak C., Han Y., Ahlenius H., Zhang Z., Chanda S., Marro S., Patzke C., Acuna C., Covy J., et al. 2013. Rapid single-step induction of functional neurons from human pluripotent stem cells. Neuron. 78:785–798. 10.1016/j.neuron.2013.05.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Lambert S., Malen P.L., Carpenter S., Boland L.M., and Bennett V.. 1998. AnkyrinG is required for clustering of voltage-gated Na channels at axon initial segments and for normal action potential firing. J. Cell Biol. 143:1295–1304. 10.1083/jcb.143.5.1295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonta B., Desmazieres A., Rinaldi A., Tait S., Sherman D.L., Nolan M.F., and Brophy P.J.. 2011. A critical role for Neurofascin in regulating action potential initiation through maintenance of the axon initial segment. Neuron. 69:945–956. 10.1016/j.neuron.2011.02.021 [DOI] [PMC free article] [PubMed] [Google Scholar]