Nussenzweig et al. use a novel mutant mouse lacking MHC class II expression on conventional dendritic cells (cDCs) to demonstrate the importance of cDCs in the maintenance of intestinal homeostasis.
Abstract
Conventional dendritic cells (cDCs) play an essential role in host immunity by initiating adaptive T cell responses and by serving as innate immune sensors. Although both innate and adaptive functions of cDCs are well documented, their relative importance in maintaining immune homeostasis is poorly understood. To examine the significance of cDC-initiated adaptive immunity in maintaining homeostasis, independent of their innate activities, we generated a cDC-specific Cre mouse and crossed it to a floxed MHC class II (MHCII) mouse. Absence of MHCII on cDCs resulted in chronic intestinal inflammation that was alleviated by antibiotic treatment and entirely averted under germ-free conditions. Uncoupling innate and adaptive functions of cDCs revealed that innate immune functions of cDCs are insufficient to maintain homeostasis and antigen presentation by cDCs is essential for a mutualistic relationship between the host and intestinal bacteria.
Conventional DCs (cDCs) are specialized immune cells that link the innate and adaptive immune system. Innate features of cDCs allow them to recognize and respond to pathogens by producing essential cytokines such as IL-6, IL-12, IL-23, and TNF. These cytokines contribute to the activation of other immune cells, including T and B cells and cells of the innate immune system. For example, in the intestine, cDCs sense bacteria and produce IL-23, which induces type III innate lymphoid cells (ILC3s) to produce IL-22, which in turn stimulates production of antimicrobial peptides (AMPs; Sonnenberg et al., 2011; Kinnebrew et al., 2012; Satpathy et al., 2013; Bernink et al., 2015). In addition to their innate functions, cDCs initiate adaptive immune responses by ingesting, processing, and presenting antigens to T cells (Nussenzweig et al., 1980; Steinman et al., 2003). In the intestine, cDCs are responsible for transport of antigen to the draining mesenteric LNs (mLNs). Under physiological conditions, the capacity of cDCs to migrate from tissue to draining LNs distinguishes them from more sessile macrophages (Schreiber et al., 2013). The importance of cDCs in adaptive immune function is exemplified by the fact that cDC depletion during viral and bacterial infection results in impaired T cell immunity and increased susceptibility to infection (Jung et al., 2002; Kassim et al., 2006; Hildner et al., 2008; Satpathy et al., 2013; Schreiber et al., 2013).
In mice, expression of Itgax (CD11c) is a hallmark of the DC lineage, and its expression has been used to label (CD11cYFP), deplete (CD11cDTR), and conditionally target (CD11cCre) cDCs (Jung et al., 2002; Lindquist et al., 2004; Caton et al., 2007; Stranges et al., 2007). However, CD11c is also expressed by plasmacytoid DCs (pDCs), activated monocytes, macrophages, and some NK cells, and therefore CD11c-based labeling and targeting strategies are not entirely cDC specific (Serbina et al., 2003; Hohl et al., 2009; Meredith et al., 2012; Schreiber et al., 2013). Higher levels of specificity can be achieved by deletion of genes that regulate the development of specific subsets of cDCs, such as Batf3, Irf4, and Notch2; however, these methods have yet to be applied to dissect the relative contributions of the innate and adaptive functions of cDCs to immune homeostasis (Caton et al., 2007; Hildner et al., 2008; Persson et al., 2013; Schlitzer et al., 2013).
To investigate the adaptive functions of cDCs independent of innate activities, we produced a cDC-restricted Cre mouse wherein Cre expression is directed by the Zbtb46 gene (zDCCre) and used it to delete MHCII in cDCs in vivo. These mice exhibited profound intestinal inflammation that was directly related to the presence of intestinal bacteria, as antibiotic-treated or germ-free mice lacking MHCII on cDCs showed no signs of intestinal inflammation. Colonization of germ-free mice allowed us to monitor adaptive immune responses against intestinal bacteria and revealed that mice lacking MHCII on cDCs have a defect in inducing proper adaptive immune responses against commensals. Collectively, our studies reveal the importance of the adaptive function of cDCs in maintaining intestinal homeostasis.
RESULTS
Generation of zDCCre mouse
Zbtb46 (zDC) is expressed in pre-DCs and their progeny, but not in monocytes, macrophages, and other myeloid cells (Fig. 1 A; Meredith et al., 2012; Satpathy et al., 2012). To express Cre recombinase in cells that actively transcribe zDC, we inserted Cre into the 3′ UTR of zDC (Fig. 1 B; Meredith et al., 2012). Cell type–specific expression of Cre was confirmed by crossing zDCCre to Rosa26lSlYFP mice (zDCCreRosalSlYFP), wherein Cre-mediated excision of a transcriptional Stop element results in YFP expression. The majority of cDCs (Lin−CD11chighMHCII+) in the spleen of zDCCreRosalSlYFP mice were YFP+. In contrast, B and T lymphocytes, pDCs (CD11clowBST2+), red pulp macrophages (RPMs; Lin−CD11blowF4/80+), and monocytes (Lin−CD11b+Ly6C+CD115+) expressed little or no YFP (Fig. 1, C and D). In contrast, in CD11cCreRosalSlYFP mice, cDCs, as well as the majority of macrophages, and pDCs were labeled in the spleen (Fig. 1 D). Similar results were also obtained after administration of polyinosinic:polycytidylic acid (pIC; Fig. 1 E) and with zDCCre+IRF4wt/fl mice that express GFP upon Cre-mediated IRF4 excision (Klein et al., 2006; Fig. S1).
Figure 1.
zDCCre mouse. (A) Diagram shows development of mononuclear phagocytes, with zDC-expressing cells in blue. (B) Schematic diagram of the Zbtb46 locus. 5′ and 3′ UTRs are shown in black, and coding regions are shown in white. (C) Flow cytometric analysis of splenic cDCs (Lin−CD11chighMHCII+), pDCs (CD11clowBST2+), RPMs (Lin−CD11b−F4/80+), and monocytes (Lin−CD11b+Ly6C+CD115+). Histograms show percentage of cells expressing YFP in zDCCreRosalSlYFP mice (open histograms) or RosalSlYFP control mice (filled gray histograms). (D) Graph depicts percentage of various immune cell populations in the spleen expressing YFP in zDCCreRosalSlYFP mice (blue symbols) and CD11cCreRosalSlYFP mice (black symbols). Each data point represents one individual mouse (n = 4–13). (E) Splenocytes were isolated 16 h after i.p. administration of 10 µg pIC. Graph depicts percentage of various immune cell populations expressing YFP in zDCCreRosalSlYFP mice (black symbols) and zDCCreRosalSlYFP mice that received pIC (red symbols). Each data point represents one individual mouse (n = 3–6). (F) Flow cytometric analysis of small intestine lamina propria mononuclear phagocytes (Lin−CD11c+MHCII+). Histogram shows YFP expression of DC subsets in zDCCreRosalSlYFP mice. (G and H) Histograms show expression of YFP in zDCCreRosalSlYFP mice (G, open histograms) and CD11cCreRosalSlYFP mice (H, open histograms) or RosalSlYFP control mice (filled gray histograms). Graphs depict percentage of CD64− and CD64+ DCs expressing YFP in zDCCreRosalSlYFP (G) and CD11cCreRosalSlYFP (H) mice, respectively. Each data point represents one individual mouse (n = 5–9).
In the intestine, the phenotype of monocytes and cDCs is altered (Caballero and Pamer, 2015). For example, small intestine lamina propria CD11c+MHCII+ cells represent a mixture of cDCs and monocytes that can be further subdivided based on expression of CD103, CD11b, and CD64 (Fig. 1 F; Bogunovic et al., 2009; Tamoutounour et al., 2012; Schreiber et al., 2013). Whereas all of these cell types are labeled in CD11cCreRosalSlYFP mice, only pre-DC–derived CD64− cells were YFP+ in zDCCreRosalSlYFP indicator mice (Fig. 1, G and H). We did not observe a difference in YFP expression in CD103+/CD11b− and CD103+/CD11b+ CD64− cells in zDCCreRosalSlYFP mice (Fig. 1 F). We conclude that zDCCre mice are suitable for cDC-specific conditional gene deletion.
zDCΔMHCII mice exhibit preserved innate functions but impaired adaptive functions
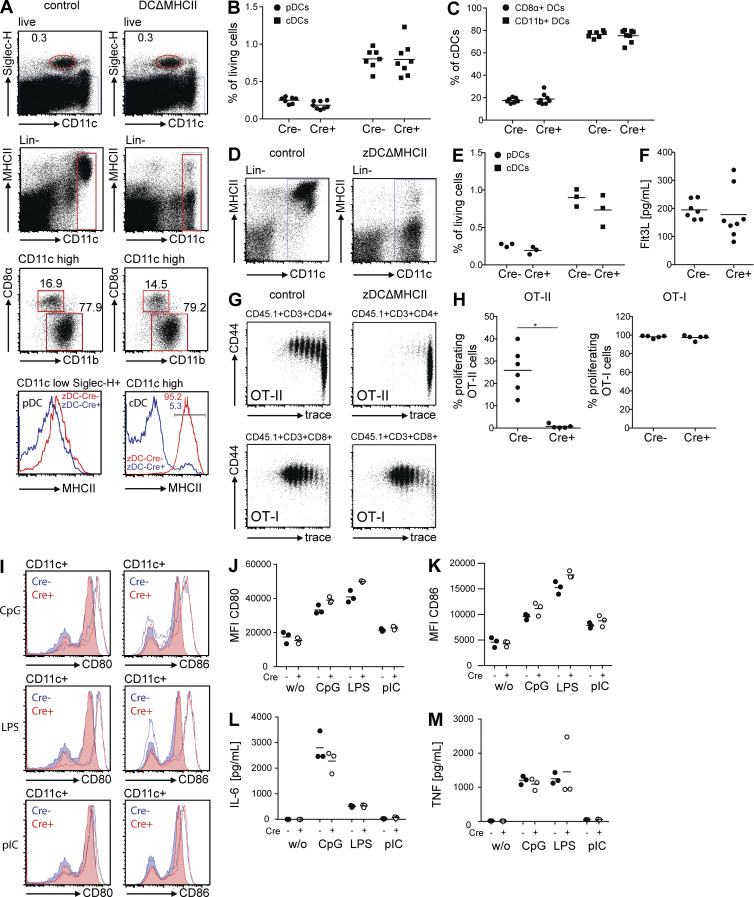
In addition to antigen processing and presentation, cDCs are essential innate sensors of pathogens and other stimuli. To distinguish between innate and adaptive functions of cDCs, we produced a cDC-specific MHCII knockout by crossing zDCCre with MHCIIfl/fl mice (zDCΔMHCII; Hashimoto et al., 2002). As expected, zDCΔMHCII mice expressed MHCII on the surface of pDCs, but not on cDCs (Fig. 2 A). Absence of MHCII expression by cDCs did not change the frequency of cDCs among splenocytes (Fig. 2 B), nor did it alter the ratio of CD8α+ and CD11b+ cDCs (Fig. 2 C). The same was true for the mLNs (Fig. 2, D and E). Moreover, the levels of circulating Flt3L in the serum, which are increased upon cDC depletion (Birnberg et al., 2008; Meredith et al., 2012), were normal in zDCΔMHCII mice (Fig. 2 F).
Figure 2.
zDCΔMHCII mice. (A) Flow cytometric analysis of splenocytes from control and zDCΔMHCII mice. Histograms depict expression of MHCII on the cell surface of cDCs (Lin−CD11chigh) and pDCs (CD11clowSiglec-H+). (B) Graph depicts frequency of pDCs and cDCs among living splenocytes. (C) Graph depicts ratio of CD8α+ and CD11b+ cDCs. Each data point represents one individual mouse (n = 7–8). (D) Flow cytometric analysis of mLN cells from control and zDCΔMHCII mice. Dot plots depict expression of MHCII on the cell surface of cDCs (Lin−CD11chigh). (E) Graph depicts frequency of pDCs and cDCs among living mLN cells. Each data point represents one individual mouse (n = 3). (F) Serum levels of Flt3L. Each data point represents one individual mouse (n = 7–8). (G and H) OT-II (top row) or OT-I (bottom row) T cells were transferred into control and zDCΔMHCII mice, respectively. Mice were immunized (i.p.) with 100 µg OVA + 10 µg pIC. Proliferation was analyzed 3 d later. (G) Dot plots show proliferation and CD44 expression of OT-II (top row) and OT-I (bottom row) T cells in LNs (pooled mesenteric and inguinal LN). (H) Graph depicts percentage of OT-II (left) and OT-I (right) cells that proliferated in response to OVA + pIC. Each data point represents one individual mouse (n = 5–6). (I–M) cDCs were isolated from spleens of control and zDCΔMHCII mice. 250,000 cells were incubated with 1 µM CpG 1668, 1 µg/ml LPS, and 1 µg/ml pIC. 16 h later, expression of CD80 and CD86 was determined by flow cytometry. Cytokines produced by cDCs after stimulation were measured in the supernatant. *, P < 0.05. (I) Histograms show expression of CD80 and CD86 on cDCs. Filled histograms show expression of CD80 and CD86 on unstimulated cells. Open histograms show levels of CD80 and CD86 after stimulation. (J and K) Graphs depict levels of expression (MFI) of CD80 and CD86 on cDCs. (L and M) Graphs depict production of IL-6 and TNF by cDCs. n = 3.
To determine whether adaptive responses are altered by cDC-specific MHCII deletion, we transferred OVA-specific CD4+ T cells (OT-II cells) to control and zDCΔMHCII mice and compared their responses to immunization with OVA. OT-II cells responded by proliferating and up-regulating CD44 expression in control but not in zDCΔMHCII mice (Fig. 2, G and H). In contrast, proliferation and activation of CD8+ OT-I T cells in response to immunization with OVA was not altered in zDCΔMHCII mice (Fig. 2, G and H). Thus, deletion of MHCII on the surface of cDCs is sufficient to impair priming of naive CD4+ T cells.
To confirm that the innate responses of cDCs are intact in zDCΔMHCII mice, we purified cDCs from the spleen, stimulated them with CpG 1668, LPS, or pIC, and measured expression of co-stimulatory molecules (CD80 and CD86) and production of cytokines (IL-6 and TNF). We found no differences between control and zDCΔMHCII cDCs, indicating that their responses to innate stimuli are intact (Fig. 2 I-M). Thus, zDCΔMHCII cDCs show intact responses to innate stimuli, but are unable to initiate MHCII-restricted adaptive immune responses.
Intestinal inflammation develops in mice lacking MHCII on cDCs
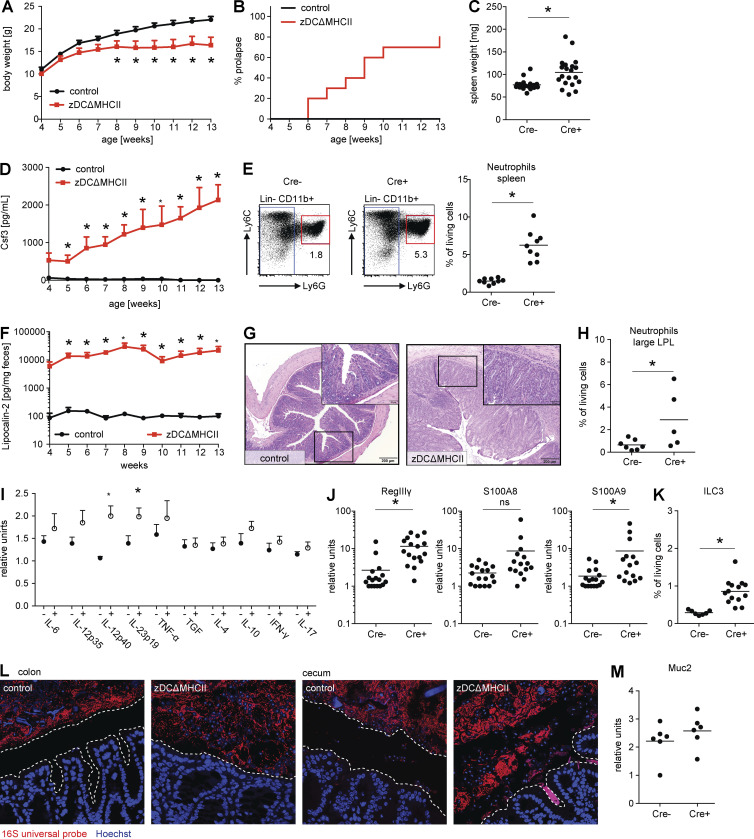
Although zDCΔMHCII mice were born at the expected Mendelian ratio, they showed significantly reduced weight gain (Fig. 3 A) and developed rectal prolapse starting at 6 wk of age (Fig. 3 B). In addition, the mutant mice concomitantly developed splenomegaly and showed increased circulating levels of the acute phase–reactant Csf3 and neutrophilia, which are indicative of a systemic process (Fig. 3, C–E). The levels of Csf1 and Csf2 remained within normal limits (unpublished data).
Figure 3.
zDCΔMHCII mice develop severe intestinal inflammation. (A) Graph depicts absolute body weight of control (black line) and zDCΔMHCII mice (red line) in grams, starting at 4 wk of age. Shown is the mean + SEM (n = 10–12). (B) Graph depicts the frequency of mice that developed rectal prolapse. Control (black line) and zDCΔMHCII mice (red line; n = 10–12). (C) Graph depicts total weight of spleen in milligrams. Each data point represents one individual mouse (n = 19–20). (D) Graph depicts concentration of Csf3 in the serum of control (black line) and zDCΔMHCII mice (red line) as the mean + SEM (n = 10–12). (E) Flow cytometric analysis of splenocytes from control and zDCΔMHCII mice. Numbers in dot plots show frequency of neutrophils (Lin−CD11b+Ly6ClowLy6G+) among living cells. Graph depicts frequency of neutrophils among living cells. Each data point represents one individual mouse (n = 9). (F) Graph depicts concentration of Lipocalin-2 in the feces of control (black line) and zDCΔMHCII mice (red line) as the mean + SEM (n = 10–12). (G) Histopathological analysis of the large intestine of control (left) and zDCΔMHCII mice (right) that were maintained under SPF conditions. The large inset represents enlarged view of small inset and shows cellular infiltrates. (H) Graph depicts frequency of neutrophils among living cells in the lamina propria of the large intestine. Each data point represents one individual mouse (n = 5–7). (I) Expression of cytokines in the large intestine (whole tissue) was determined by qPCR. Graph depicts expression as relative units. Each data point represents one individual mouse (n = 5). (J) Expression of RegIIIγ, S100A8, and S100A9 in epithelial cells isolated from the small intestine was determined by qPCR. Graphs depict expression as relative units. Each data point represents one individual mouse (n = 15–17). (K) Graph depicts frequency of ILC3s (CD45+/CD19−/B220−/NK1.1−/Gr1−/CD3−/Thy1.2+/RORγt+) among living cells in the lamina propria of the small intestine. (L) Pictures show localization of bacteria (red) and epithelial cells (blue) visualized by 16S FISH and Hoechst staining in the colon (left) and the cecum (right). Dotted line outlines the space between epithelial cells and luminal content. (M) Expression of Muc2 in epithelial cells isolated from the large intestine was determined by qPCR. Graph depicts expression as relative units. Each data point represents one individual mouse (n = 6). If not stated otherwise, mice analyzed were 8–12 wk old. *, P < 0.05
To quantitate intestinal inflammation, we measured Lipocalin-2, a sensitive biomarker for intestinal inflammation, in the feces of control and zDCΔMHCII mice (Chassaing et al., 2012). zDCΔMHCII mice showed a 100-fold increase in Lipocalin-2 compared with controls (Fig. 3 F). Consistent with this observation, histopathological analysis revealed inflammation that was primarily restricted to the large intestine, with inflammatory infiltrates, crypt hyperplasia, and villus destruction (Fig. 3 G). Neutrophil infiltration in the large intestine was also detected by flow cytometry (Fig. 3 H). In addition, we found elevated levels of the inflammatory cytokine IL-23 in the large intestine of zDCΔMHCII, but not in control mice (Fig. 3 I). To confirm that intestinal innate immune responses were intact, we measured the expression of AMPs in the intestine of zDCΔMHCII and control mice. Consistent with the inflammation found in the intestines of these mice, RegIIIγ and S100A9 were found at higher levels in zDCΔMHCII mice compared with controls (Fig. 3 J). ILC3s, whose IL-22 production is crucial for AMP expression (Sonnenberg et al., 2011; Kinnebrew et al., 2012; Satpathy et al., 2013; Bernink et al., 2015), were not affected and actually occurred at higher frequencies in zDCΔMHCII mice compared with controls (Fig. 3 K). We therefore conclude that intestinal inflammation in zDCΔMHCII mice occurs even in the presence of active cDC-dependent innate immune responses.
Normally, intestinal bacteria are separated from epithelial cells by a mucous layer. Innate immune responses and AMPs are essential for this spatial segregation (Vaishnava et al., 2011; Gallo and Hooper, 2012). To visualize bacteria in the large intestine (colon and cecum) of zDCΔMHCII mice and determine their location and proximity to the epithelial barrier, we performed 16S ribosomal RNA (rRNA) fluorescence in situ hybridization (16S FISH). We found that, in zDCΔMHCII mice, intestinal microbes and gut epithelial cells were in direct contact (Fig. 3 L). However, Muc2 expression in the large intestine was unaltered, and therefore zDCΔMHCII mice do not have a defect in mucus production (Fig. 3 M). Finally, neither orally administrated FITC-Dextran nor LPS were detectable at higher levels in the serum of zDCΔMHCII compared with control mice, indicating no gross defect in the epithelial barrier (unpublished data).
A similar phenotype was also observed in CD11cΔMHCII mice (Fig. 4 A) and in zDCΔMHCII, but not control BM chimeras (Fig. 4, B and C). Finally, BM chimeras reconstituted with a 1:1 mixture of control and zDCΔMHCII BM did not develop disease (Fig. 4 C). Therefore, intestinal inflammation in zDCΔMHCII mice is BM cell–dependent and can be rescued by WT BM cells.
Figure 4.
CD11cΔMHCII mice, zDCΔMHCII BM chimeras, and microbiome in zDCΔMHCII mice. (A) Graphs depict serum levels of Csf3, frequency of neutrophils among living cells in the spleen, and concentration of Lipocalin-2 in the feces of control and CD11cΔMHCII mice. Each data point represents one individual mouse (n = 6). Mice were analyzed at the age of 6–8 wk. (B and C) WT B6 mice were irradiated and reconstituted with BM from control or zDCΔMHCII mice. A third group received a 1:1 mix of BM from control and zDCΔMHCII mice. (B) Graph depicts percentage of cDCs expressing MHCII. (C) Graphs depict serum levels of Csf3, frequency of neutrophils among living cells in the spleen and concentration of Lipocalin-2 in the feces of mice reconstituted with BM from control, zDCΔMHCII mice, or a 1:1 mix of BM from control and zDCΔMHCII mice, respectively. Each data point represents one individual mouse (n = 10). Mice were analyzed 10 wk after irradiation. *, P < 0.05. (D) Feces was collected from control and zDCΔMHCII mice (10–12 wk old) and microbial composition was analyzed by 16S rRNA sequencing. UniFrac analysis is shown. Cage numbers indicate which mice were cohoused in the same cage.
Despite the changes in the anatomical distribution of the microbiome in the intestine, we did not observe dramatic differences in the composition of the microbiome (determined by 16S sequencing of fecal bacteria) between control and zDCΔMHCII mice that were cohoused. UniFrac analysis showed that the microbiome in individual mice clustered by the cage in which they were housed and not by genotype, suggesting that the composition of the microbiome is not dramatically altered in zDCΔMHCII mice compared with controls (Fig. 4 D). These data indicate that in addition to AMPs, MHCII expression by cDCs is required to maintain the spatial segregation between microbes and gut epithelial cells.
CD4+ T cell activation is impaired in zDCΔMHCII mice
DCs and thymic epithelial cells are both involved in the selection of mature T cells in the thymus; therefore, we examined thymic CD4+ T cells in zDCΔMHCII mice. Although there was a small increase in mature single-positive CD4+ thymocytes in zDCΔMHCII mice compared with control mice, their Vβ chain usage was comparable (Fig. 5, A and B).
Figure 5.
Thymic CD4+ T cells. (A) Flow cytometric analysis of thymocytes from control and zDCΔMHCII mice. Numbers in dot plots show frequency of single-positive CD4 and CD8 thymocytes, respectively. Graph depicts frequency of single-positive CD4 and CD8 thymocytes of living cells. Each data point represents one individual mouse (n = 6). (B) Graph depicts TCR repertoire of CD4+ T cells in the spleen and Vβ chain usage in control and zDCΔMHCII mice as means + SEM (n = 7). *, P < 0.05.
To check for the presence of autoreactive CD4+ T cells in the periphery, we analyzed T cells in the spleen, mLN, and lamina propria of the large intestine for the presence of activated and cytokine-producing CD4+ T cells. zDCΔMHCII mice showed no evidence of increased levels of such cells (Fig. 6). To the contrary, we found reduced frequencies of total CD4+ T cells, as well as increased frequencies of naive CD4+ T cells and reduced frequencies of activated CD4+ T cells, in zDCΔMHCII mice when compared with controls (Fig. 6). In addition, zDCΔMHCII mice showed lower frequencies of peripherally induced regulatory T cells (iT reg cells; defined as Nrp1−) in the mLN and lamina propria of the large intestine compared with controls. This is in accordance with a previously described cDC-mediated feedback mechanism controlling T reg cell numbers (Darrasse-Jèze et al., 2009). Despite reduced iT reg cells, the relative frequency of CD4+ T cells producing IL-2, IL-17, or IFN-γ was comparable or reduced in zDCΔMHCII mice compared with controls (Fig. 6, D–F). This was also confirmed by quantitative PCR (qPCR) analysis of the large intestine, where we found similar expression levels of IL-17 and IFN-γ (Fig. 3 I). Thus, in the absence of MHCII expression by cDCs, the defect in peripheral T reg cell induction did not result in aberrant levels of CD4+ T cell activation.
Figure 6.
CD4+ T cells in zDCΔMHCII mice are not autoreactive. Frequency and phenotype of CD4+ and CD8+ T cells in control and zDCΔMHCII mice, respectively. Dot plots show ratio of naive (CD62L+) and activated (CD44+) CD4+ and CD8+ T cells and percentage of CD4+ T reg cells (Foxp3+) in spleen (A), mLN (B), and large intestine (C) lamina propria. Graphs depict frequency of CD4+ and CD8+ T cells among living cells, frequency of CD62L+, CD44+, and Foxp3+ cells among CD4+ T cells, frequency of natural (Nrp1+) and induced (Nrp1−) T reg cells among Foxp3+ cells, frequency of cytokine producing CD4+ T cells, and frequency of CD62L−CD44+ cells among CD8+ T cells in spleen (D), mLN (E), and large intestine (F) lamina propria. Each data point represents one individual mice (n = 6). Mice analyzed were 14 wk old. *, P < 0.05.
In contrast to CD4+ T cells, we observed a higher frequency of activated CD8+ T cells (CD3+CD8+CD62L−CD44+) in the spleen and mLN of zDCΔMHCII mice (Fig. 6, D and E). Furthermore, zDCΔMHCII mice showed unaltered or slightly reduced frequencies of γδ T cells in spleen, mLN, and large intestine compared with control mice, ruling out the involvement of pathogenic γδ T cells (Park et al., 2010; not depicted). To investigate whether the phenotype in zDCΔMHCII mice might be a result of insufficient CD4+ T cell activation, we depleted CD4+ T cells in control (MHCII sufficient) mice. Within 4 wk of depletion, mice developed neutrophilia, and elevated levels of Lipocalin-2 were detectable in their feces, although mice did not develop rectal prolapse within the 4 wk of depletion (Fig. 7 A; and not depicted). To test whether the observed increase in activated CD8+ T cells contributed to the phenotype in zDCΔMHCII mice, we depleted CD8+ T cells for 4 wk. However, CD8+ T cell–depleted zDCΔMHCII mice did not differ from nondepleted zDCΔMHCII mice (Fig. 7 B). Collectively, these data suggest that the phenotype in zDCΔMHCII mice is associated with insufficient adaptive CD4+ T cell responses but not pathogenic γδ or CD8+ T cells.
Figure 7.
CD4+ T cell depletion induces neutrophilia and intestinal inflammation. CD8+ T cell depletion does not affect disease. (A) Cre− control mice were injected with PBS or 100 µg anti-CD4 antibody (clone GK1.5) every 2–3 d for 4 wk. Graphs depict frequency of neutrophils among living cells in the spleen and concentration of Lipocalin-2 in the feces. Each data point represents one individual mouse (n = 9–13). (B) Cre− control and zDCΔMHCII mice were injected with PBS or 100 µg anti-CD8 antibody (clone 53–6.72) every 2–3 d for 4 wk. Graphs depict concentration of Lipocalin-2 in the feces, serum levels of Csf3, and frequency of neutrophils among living cells in the spleen. Each data point represents one individual mouse (n = 5). *, P < 0.05.
Microbiota is required for intestinal inflammation
To test the possibility that zDCΔMHCII mice develop intestinal inflammation as a result of inefficient effector CD4+ T cell responses to intestinal bacteria, we treated the mice with broad-spectrum antibiotics. Lipocalin-2 in the feces, Csf3 in serum, and neutrophil levels in the spleen of zDCΔMHCII mice dropped to baseline levels after 4 wk of antibiotic treatment (Fig. 8, A–C). Consistent with these findings, antibiotic-treated zDCΔMHCII mice were free of intestinal inflammation and inflammatory infiltrate as determined by histology (Fig. 8 E). Moreover, maintaining zDCΔMHCII mice in a germ-free environment completely prevented the development of all signs of disease, including intestinal inflammation, high levels of Lipocalin-2, elevated Csf3 levels, neutrophilia, and increased numbers of activated CD8+ T cells (Fig. 8, A–E). These results indicate that the intestinal inflammation in zDCΔMHCII mice is dependent on the microbiota. They also suggest that rather than causing the disease, the activated CD8+ T cells in specific pathogen–free (SPF) zDCΔMHCII mice develop in response to microbiota-mediated inflammation.
Figure 8.
Treatment of zDCΔMHCII mice with antibiotics reduces intestinal inflammation. (A) Graph depicts concentration of Lipocalin-2 in the feces of control and zDCΔMHCII mice that were maintained under SPF conditions, zDCΔMHCII mice that received antibiotics with the drinking water for 4 wk, and zDCΔMHCII mice that were maintained under germ-free conditions. Each data point represents one individual mouse (n = 4–12). (B) Graph depicts serum levels of Csf3. Each data point represents one individual mouse (n = 4–12). (C) Graph depicts frequency of neutrophils among living cells in the spleen. Each data point represents one individual mouse (n = 4–12). (D) Graph depicts frequency of CD62L−CD44+ cells among CD8+ T cells in the spleen. Each data point represents one individual mouse (n = 4–8). (E) Histopathological analysis of the large intestine of control and zDCΔMHCII mice that were maintained under SPF conditions, zDCΔMHCII mice that received antibiotics in the drinking water for 4 wk, and zDCΔMHCII mice that were maintained under germ-free conditions. Large inset represents enlarged view of small inset and shows cellular infiltrates. *, P < 0.05.
zDCΔMHCII mice exhibit impaired antimicrobial adaptive immune responses
IgA, the most abundant antibody isotype at mucosal surfaces, is secreted into the intestinal lumen, where it coats bacteria that enter the host and elicit adaptive immune responses (Palm et al., 2014). mLNs, Peyer’s patches (PPs), and isolated lymphoid follicles are the primary sites of adaptive immune responses against intestinal antigens. zDCΔMHCII mice showed normal levels of B cells in the mLNs, PPs, and lamina propria of the large intestine. Whereas GC B cells were reduced in zDCΔMHCII mice, the relative frequency of IgA+ B cells was not; the only significant defect in this compartment was in the lamina propria, suggesting that zDCΔMHCII mice do not have an overall defect in class switching. Consistent with the relative paucity of GC B cells, zDCΔMHCII mice showed significantly reduced frequencies of follicular T helper (TFH) cells in both mLNs and PPs (Fig. 9, A–D). These results support the idea that zDCΔMHCII mice have aberrant antimicrobial adaptive immune responses due to insufficient CD4+ T cell help (Grajales-Reyes et al., 2015).
Figure 9.
Adaptive immune response against intestinal bacteria. (A and C) Flow cytometric analysis of TFH cells (CD3+CD4+CD62L−CD44+PD-1+CXCR5+) and GC B cells (CD19+B220+CD38−CD95+) isolated from mLNs (A) and PPs (C) of control and zDCΔMHCII mice. (B and D) Graphs depict percentage of TFH cells among CD4+ T cells, frequency of B cells among living cells, percentage of GC B cells among B cells, and frequency of IgA+ B cells in mLNs (B) and PPs (D). Each data point represents one individual mouse (n = 7–10). (E) Graphs depict frequency of B cells among living cells, percentage of GC B cells among B cells, and frequency of IgA+ B cells in the lamina propria of the large intestine (n = 6). *, P < 0.05.
To directly measure adaptive immune responses against intestinal bacteria, we colonized germ-free control and zDCΔMHCII mice with a mixture of six different commensal intestinal bacteria (Escherichia coli, Enterococcus faecalis, Bifidobacterium longum, Lactobacillus plantarum, Bacteroides vulgatus, and Ruminococcus gnavus). Colonization was similar for control and zDCΔMHCII mice as determined by PCR 8 wk after colonization (Fig. S2). Although germ-free mice have no detectable GC B or TFH cells in gut-associated lymphoid tissues, newly colonized control mice showed robust GC B and TFH cell responses in mLNs and PPs. In contrast, these responses were barely detectable in colonized zDCΔMHCII mice, even 8 wk after colonization (Fig. 10, A and B).
Figure 10.
Colonization of germ-free mice. Germ-free control and zDCΔMHCII mice were colonized with a mixture of six different intestinal bacteria. Flow cytometric analysis of TFH cells (CD3+CD4+CD62L−CD44+PD-1+CXCR5+) and GC B cells (CD19+B220+CD38−CD95+) isolated from mLNs (A) and PPs (B) of control and zDCΔMHCII mice 61 d after colonization. Graphs depict percentage of TFH cells among CD4+ T cells and percentage of GC B cells among all B cells in mLNs (A) and PPs (B). Each data point represents one individual mouse (n = 3–5). (C and D) Graphs depict amount of total IgA in feces (C) and serum (D) 61 d after colonization. Each data point represents one individual mouse (n = 3–5). (E) Bacteria were isolated from the feces of control and zDCΔMHCII mice and stained with anti–mouse-IgA. Graph depicts normalized MFI of IgA coated bacteria. Each data point represents one individual mouse (n = 3–5). *, P < 0.05.
Consistent with the near absence of GC responses, IgA levels in the feces and serum of colonized zDCΔMHCII mice were significantly lower compared with colonized control mice. Fecal IgA increased progressively in control mice starting after day 7, reaching a mean of 200–400 ng/mg feces. In contrast, fecal IgA reached a plateau of around 10–20 ng/mg feces by day 12 and remained relatively constant thereafter in zDCΔMHCII mice (Fig. 10, C and D). Consistent with the overall lower levels of serum and fecal IgA, bacteria isolated from feces of zDCΔMHCII mice were coated with far lower levels of IgA than bacteria from feces of control mice (Fig. 10 E).
Collectively, these data reveal that zDCΔMHCII mice have defective adaptive immune responses against intestinal commensals, and this defect is associated with a loss of bacterial segregation from the epithelium and severe colitis.
DISCUSSION
cDCs link innate and adaptive immunity by sensing pathogens and initiating adaptive immune responses. Although the two physiological functions of cDCs are likely to play distinct roles in immune homeostasis, they have not previously been evaluated independently. In the gut, cDCs sense and capture gut microbes in part by extending their processes into the gut lumen (Macpherson and Uhr, 2004; Niess et al., 2005; Chieppa et al., 2006; Vallon-Eberhard et al., 2006). Microbial sensing induces cDCs to produce cytokines such as IL-23, which are required to activate innate lymphoid cells (Kinnebrew et al., 2012; Satpathy et al., 2013). In addition, the ingested microbes are carried to local lymphoid organs, such as the mLNs, processed, and presented to T cells to initiate adaptive immune responses (Macpherson and Uhr, 2004; Niess et al., 2005).
Ablation experiments using CD11cDTR mice, conditional deletion of genes (e.g., Irf4, Irf8, or Notch2) with CD11cCre mice, and Batf3-deficient mice result in loss of cDCs and cDC subsets, and therefore concomitant loss of both innate and adaptive immune functions (Hildner et al., 2008; Lewis et al., 2011; Persson et al., 2013; Schlitzer et al., 2013). For example, CD11cCre-mediated DC deletion has been described to activate a feedback loop that produces high levels of circulating Flt3L, resulting in a myeloproliferative syndrome characterized by extramedullary hematopoiesis, splenomegaly, and neutrophilia (Birnberg et al., 2008). Depletion of cDCs in this model likely has additional consequences beyond elevated Flt3L levels. Our data suggests that the phenotype may have been mediated in part by intestinal microbes. In a different study, however, CD11cCre-mediated DC depletion was associated with colitis and increased numbers of activated CD4+ T cells (Birnberg et al., 2008; Ohnmacht et al., 2009). Although cDCs play an important role for T cell selection in the thymus (Perry et al., 2014), in our model we found no evidence of activated or autoreactive CD4+ T cells. In addition to being unable to distinguish between the innate and adaptive functions of cDCs, interpretation of these earlier studies was confounded in part by deletion of CD11c expressing monocytes, macrophages, pDCs, and NK cells. In contrast, deletion of MHCII in cDCs using zDCCre selectively ablates the ability of this cell type to initiate adaptive immunity while leaving its innate activity intact. The described phenotype was specific for cDCs because zDC expression is restricted to cDCs and their precursors. zDC is first expressed in committed cDC progenitors (pre-DCs) in the BM and is maintained in all subsequent stages of cDC development, including all major subsets of cDCs (Meredith et al., 2012; Satpathy et al., 2012).
Our experiments show that cDC-induced adaptive immune responses are required to maintain homeostasis between intestinal flora and the host. Other MHCII-expressing antigen-presenting cells, such as activated monocytes, macrophages, or monocyte-derived DCs, are unable to compensate for this requirement, because they are not specialized for CD4+ T cell priming. Absence of MHCII on cDCs in zDCΔMHCII mice is associated with intestinal inflammation and elevated serum levels of Csf3, an acute phase reactant. This phenotype is dependent on bacteria, as Csf3 levels dropped to normal levels when the mice were treated with antibiotics or housed under germ-free conditions. These results highlight a previously underappreciated role for cDC-mediated adaptive immune responses to commensal bacteria. Mice lacking MHCII on cDCs may therefore be considered immunocompromised, as they are unable to efficiently prime CD4+ T cells. Consistent with our observations, mice lacking CD4+ T cells and HIV-infected humans that suffer CD4+ T cell loss show increased microbial translocation and systemic immune activation (Brenchley et al., 2006; Sandler and Douek, 2012).
cDC-initiated CD4+ T cell responses are likely to help keep the gut microbiome in check by at least several distinct mechanisms. They are required to activate and induce differentiation of T cells, such as protective TH17/TH22 cells, which prevent invasion of enterobacteria (Mangan et al., 2006; Curtis and Way, 2009; Ivanov et al., 2009; Basu et al., 2012). Consistent with this idea, T cell–deficient mice and humans show increased risk for intestinal inflammation (Tamura et al., 2005). In addition, cDCs are also essential in initiating TFH responses that are required to produce germinal centers and high-affinity IgA (Crotty, 2011; Maruya et al., 2013; Kato et al., 2014; Kubinak et al., 2015). Finally, MHCII expression by cDC is required to maintain the physiological number of induced T reg cells (Darrasse-Jèze et al., 2009). In zDCΔMHCII mice, reduced numbers of T reg cells did not result in increased numbers of activated CD4+ T cells because their activation requires MHCII expression on cDCs. Nevertheless, decreased numbers of induced T reg cells could contribute to the severity of the phenotype by altering innate immune responses.
Colonization of germ-free mice allowed us to follow the immune response against a limited number of commensal bacteria. This controlled experiment showed that zDCΔMHCII mice have diminished TFH, GC, and IgA responses to intestinal bacteria. Mice that are unable to secrete polymeric antibodies into the intestinal lumen show elevated serum IgG levels specific for commensal bacteria, suggesting more translocation of intestinal bacteria in the absence of luminal IgA (Johansen et al., 1999). In addition, translocation of bacteria was observed in germ-free B cell–deficient mice that were colonized with SPF microbiota (Macpherson and Uhr, 2004), emphasizing the importance of the adaptive response in containing the microbiota. However, the phenotype observed in zDCΔMHCII mice cannot be mediated solely by a defective IgA response against intestinal bacteria. AID−/− mice, which are unable to switch to IgA, do not develop the phenotype in our SPF facility, possibly because they have unimpaired TFH and germinal center responses and IgM can compensate in part for IgA. In zDCCre+MHCIIfl/fl mice, however, CD4+ TFH cell priming is defective, resulting in diminished GCs. This idea is supported by the observation that CD4+ T cell depletion partially phenocopies zDCCre+MHCIIfl/fl mice.
cDCs in zDCΔMHCII mice show intact innate microbial sensing and cytokine production. These features are essential for activation and normal function of innate lymphoid cells (Bernink et al., 2015). The observation that zDCΔMHCII mice develop intestinal inflammation indicates that intact innate mechanisms are insufficient to maintain the normal balance between host and intestinal commensals. Transferable risk for colitis has been described for mice lacking components of the inflammasome, which has been attributed to alterations in intestinal microbiota (Elinav et al., 2011; Palm et al., 2014). In our experiments, control mice did not develop any signs of inflammation, although they were cohoused with zDCΔMHCII mice. The fact that cohoused control mice did not get sick, along with the finding that they share a similar microbiome, indicates that commensal bacteria that can be controlled and are usually harmless in mice with an unimpaired immune system may be responsible for disease in zDCΔMHCII mice that have defective adaptive immune responses. Collectively, the data indicate that in the steady state, cDC-mediated CD4+ T cell priming and subsequent adaptive immune responses are essential to maintain homeostasis and prevent intestinal commensals from producing pathology.
MATERIALS AND METHODS
Mice
zDCCre knock-in mice were generated by homologous recombination in C57BL/6 albino embryonic stem cells at The Rockefeller University Gene Targeting Resource Center (New York, NY). The targeting strategy was the same as for the previously described zDCDTR mice (Meredith et al., 2012). zDCCre mice were maintained on a C57BL/6 background. ItgaxCre (CD11cCre; stock number 008068), Rosa26lSlYFP (Rosa26lSlYFP; stock number 006148), iabneo (MHCIIfl/fl; stock number 013181), and Irf4fl/fl (IRF4fl/fl; stock number 009380) mice were purchased from The Jackson Laboratory. Mice were housed in The Rockefeller University Comparative Bioscience Center (New York, NY) under SPF and germ-free conditions, respectively. All experiments were performed in accordance with the National Institutes of Health guidelines and approved by The Rockefeller University Animal Care and Use Committee. Germ-free mice were obtained by transfer of newborn pups (Cesarean section) into germ-free isolators. Germ-free status was confirmed by plating feces on a broad range of nutrient agars, as well as by qPCR analysis (16S rRNA). zDCΔMHCII mice were bred with MHCIIfl/fl mice. This way we could compare Cre+ (zDCΔMHCII) and Cre− (control) littermates. It is also important to note that zDCΔMHCII males were bred with MHCIIfl/fl females. This way, the fostering female is healthy and does not develop the described phenotype, and development of pups is not affected by the genotype of the fostering female. Littermates were cohoused during all experiments. Mice were generally analyzed at 8–12 wk of age.
BM chimeras
BM chimera recipients were irradiated with two doses of 525 rad 3 h apart and injected with 5–10 × 106 BM cells. BM chimeras were analyzed 10 wk after irradiation.
Flow cytometry and cell sorting
For FACS analysis, cells were stained using antibodies against B220 (RA3-6B2), BST2 (eBio927), CXCR5 (L138D7), F4/80 (BM8), Foxp3 (FJK-16s), Gr-1 (RB6-8C5), IFN-γ (181157), IL-2 (JES6-5H4), IL-17A (TC11-18H10), Ly6C (AL-21), Ly6G (1A8), MHCII (M5/114.15.2), NK1.1 (PK136), Nrp1 (3DS304M), PD-1 (J43), RORg(t) (B2D), Siglec-H (440c), TCRβ (H57-597), CD3 (145-2C11), CD4 (RM4-5), CD8 (53–6.7), CD11b (M1/70), CD11c (N418), CD19 (1D3), CD38 (90), CD44 (1M7), CD45 (30-F110), CD62L (MEL-14), CD64 (X54-5/7.1), CD95 (Jo2), CD103 (2E7), CD115 (AFS98), TCRγδ (GL3), CD80 (16-10A1), and CD86 (GL1). TCR Vβ chain distribution was assessed with a FITC-labeled screening panel (BD). PI (Sigma-Aldrich), DAPI (Invitrogen), or Aqua live/dead (Invitrogen) was added to exclude dead cells from analysis. For intracellular cytokine staining, cells were stimulated for 4 h using Leukocyte Activation Cocktail with GolgiPlug (BD). Surface stained cells were fixed with Cytofix/Cytoperm (BD) and washed with Perm/Wash buffer (BD). Cells were analyzed on a LSR Fortessa flow cytometer (BD). Analysis was performed using FlowJo software (Tree Star).
Spleen, LN, thymus, and PP cell isolation
For isolation of mononuclear phagocytes, spleen and LNs were mechanically disrupted and incubated with HBSS (Gibco) + 5% FBS + 0.5 mg/ml Collagenase D (Roche) at 37°C for 30 min. Cells were filtered through 100 µM cell strainers and washed. Erythrocytes were lysed with ACK Lysing buffer (Gibco). For isolation of T and B cells, spleen, LNs, thymi, and PP were mechanically disrupted, filtered through 100-µM cell strainers, and washed. Erythrocytes were lysed with ACK Lysing buffer (Gibco).
In vitro stimulation of DCs
250,000 cDCs (CD11c MACS enrichment) were stimulated with 1 µM CpG 1668, 1 µg/ml LPS, and 1 µg/ml pIC, respectively. Expression of CD80 and CD86 were determined by flow cytometry 16 h later. Cytokines were measured using the Cytometric Bead Array kit (BD).
Intestinal cell isolation
Small and large intestine were excised and freed of mesentery and fat. Feces were removed and PPs were excised from the small intestine. Both small and large intestine were opened longitudinally and washed in HBSS, followed by HBSS + 1 mM DTT (Sigma-Aldrich). Tissue was cut into 1-cm pieces and incubated with 25 ml of HBSS + 5% FBS + 5 mM EDTA for 15 min at 37°C at 230 rpm. Supernatant containing intraepithelial lymphocytes was decanted and spun down (for RNA isolation from epithelial cells, see next section). Tissue was washed with 10 ml of HBSS + 5% FBS for 15 min at 37°C at 230 rpm. The gut tissues were then finely chopped and digested in HBSS + 5% FBS + 1X Sodium Pyruvate + 25 mM Hepes + 50 µg/ml DNaseI + 0.05 mg/ml Collagenase VIII for 1 h at 37°C at 100 rpm. Cells were separated by discontinuous Percoll gradient (70%/35%) centrifugation. Cells were isolated from the interphase.
RNA isolation and qPCR
Epithelial cells were resuspended in TRIzol (Invitrogen), RNA was isolated according to the manufacturer’s protocol, and cDNA was generated using Superscript II (Invitrogen) and random primers. GAPDH forward, 5′-CTACAGCAACAGGGTGGTGG-3′, GAPDH reverse, 5′-AGATGGGGCATCTTTCTTGG-3′; S100A8 forward, 5′-TGCGATGGTGATAAAAGTGG-3′, S100A8 reverse, 5′-GGCCAGAAGCTCTGCTACTC-3′; S100A9 forward, 5′-CACCCTGAGCAAGAAGGAAT-3′, S100A9 reverse, 5′-TGTCATTTATGAGGGCTTCATTT-3′; Muc2 forward, 5′-ACAAAAACCCCAGCAACAAG-3′, Muc2 reverse, 5′-GAGCAAGGGACTCTGGTCTG-3′. All qPCR reactions were performed with Brilliant SYBR Green (Agilent Technologies) on an Mx3005P system (Agilent Technologies).
ELISA
Serum levels of Flt3L and Csf3 were determined by ELISA (R&D Systems) according to the manufacturer’s protocol. Lipocalin-2 levels were measured in feces by ELISA according to the manufacturer’s protocol (R&D Systems). IgA concentrations were determined by ELISA according to the manufacturer’s protocol (eBioscience).
Histology
Tissue was fixed in 10% Formalin. Tissue was embedded, cut, and stained by the Tri-Institutional Laboratory of Comparative Pathology (New York, NY). A pathologist evaluated the tissue sections in a blinded fashion and took pictures representing the phenotype of the respective group.
OT-I and OT-II T cell transfer
CD8+ and CD4+ T cells were isolated from spleen and LNs of CD45.1+ OT-I and OT-II mice, respectively (MACS enrichment), labeled with violet tracem, and transferred into control and zDCΔMHCII mice (CD45.2) by i.v. injection (3–5 × 106 cells/mouse). Mice were immunized (i.p.) with 100 µg OVA + 10 µg pIC. 3 d later, proliferation and expression of activation markers was analyzed by flow cytometry (pooled mesenteric and inguinal LN).
CD4+ and CD8+ T cell depletion
To deplete CD4+ T cells, mice were injected i.p. with 100 µg anti-CD4 (GK1.5; Bio X Cell) every 2–3 d for 4 wk. To deplete CD8+ T cells mice were injected i.p. with 100 µg anti-CD8 (53–6.72; Bio X Cell) every 2–3 d for 4 wk.
Abx treatment
Ampicillin (1 mg/ml), Neomycin (1 mg/ml), Vancomycin (0.5 mg/ml), and Metronidazole (1 mg/ml; Sigma-Aldrich) were administered in the drinking water. Mice were put on Abx drinking water at the age of 8–10 wk and kept on Abx for 4 wk.
Time course/aging experiments
Body weight was measured weekly, as well as occurrence of rectal prolapse. Feces and serum samples were collected and stored at –20°C until further use.
16S FISH
16S rRNA FISH was performed with a universal probe (EUB388; Cy3-GCTGCCTCCCGTAGGAGT-Cy3) on sections fixed with Methacarn solution as described previously (Salzman et al., 2002; Swidsinski et al., 2005). In brief, intestinal tissues (∼1 cm), including intestinal contents, were fixed in Methacarn solution (60% methanol, 30% chloroform, and 10% glacial acetic acid) for 6 h at 4°C. After three washes with 70% ethanol, tissues were embedded in paraffin. 5-µm sections were cut and deparaffinized with xylene, and subsequently dehydrated. Probe was diluted (5 ng/µl) in hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl, pH 7.2, and 0.1% SDS), and hybridization was performed at 50°C for 3 h in. After hybridization, tissues were washed with wash buffer (0.9 M NaCl and 20 mM Tris-HCl, pH 7.2) and nuclei were stained with Hoechst stain.
Bacteria isolation and IgA staining
Bacteria were isolated from feces as described previously (Palm et al., 2014). Coating of bacteria with endogenous IgA was determined by staining bacteria with anti–mouse-IgA-PE (clone mA-6E1). Where indicated, MFIs were normalized to the sample with the lowest MFI to be able to compare different time points.
DNA extraction, V4-V5 16S rRNA gene amplification, multiparallel sequencing, and sequence analysis
Fecal samples and intestinal contents were frozen immediately after collection in a dry ice/ethanol slurry and stored at −80°C. DNA was extracted using the phenol-chloroform and bead beating method described previously (Ubeda et al., 2010). The V4-V5 region of the 16S rRNA gene was amplified and sequenced with the Illumina Miseq platform, as described previously (Buffie et al., 2015). Sequences were analyzed using version 1.33.3 of the MOTHUR pipeline (Schloss et al., 2009) as described in Buffie et al. (2015). Sequences with distance-based similarity of at least 97% were assigned the same OTU (operational taxonomic unit) and representative OTUs were classified using a modified Greengenes reference database (DeSantis et al., 2006).
Colonization of germ-free mice
Germ-free control and zDCΔMHCII mice were colonized with a mix of six intestinal bacteria (E. coli [LF82], E. faecalis, B. longum, L. plantarum, B. vulgatus, and R. gnavus). Colonization was assessed by PCR using following primers: E. faecalis forward, 5′-CCCTTATTGTTAGTTGCCATCATT-3′ E. faecalis reverse, 5′-ACTCGTTGTACTTCCCATTGT-3′; B. longum forward, 5′-GGGTGGTAATGCCGGATG-3′ B. longum reverse, 5′-TAAGCGATGGACTTTCACACC-3′; L. plantarum forward, 5′-AGCAGTAGGGAATCTTCCA-3′ L. plantarum reverse, 5′-CACCGCTACACATGGAG-3′; B. vulgatus forward, 5′-GGTGTCGGCTTAAGTGCCAT-3′ B. vulgatus reverse, 5′-CGGAYGTAAGGGCCGTGC-3′; R. gnavus forward, 5′-GGAGCAGACCAGATCCAAAT-3′ R. gnavus reverse, 5′-CCAATATACATTCCCGGTCTTT-3′; E. coli forward, 5′-CCATTCATGCAGCAGCTCTTT-3′ E. coli reverse, 5′-ATCGGACAACATTAGCGGTGT-3′. Coating of bacteria with endogenous IgA and amount of total IgA in the serum and feces was determined at indicated time points.
Statistical analysis
Results are shown as means + SEM. Comparisons between groups was done using Student’s two-tailed t test analysis. Statistical significance was determined as p-values <0.05.
Online supplemental material
Fig. S1 shows flow cytometric analysis of zDCCre+IRF4wt/fl reporter mice. Fig. S2 show bacteria-specific PCR to confirm colonization of germ-free mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20160062/DC1.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to members of the Nussenzweig laboratory for helpful discussion, reagents, and critical reading of the manuscript.
This work was supported in part by National Institutes of Health grant number AI 13013. M.C. Nussenzweig is a Howard Hughes Medical Institute investigator.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- AMP
- antimicrobial peptides
- cDC
- conventional DC
- FISH
- fluorescence in situ hybridization
- ILC3
- type III innate lymphoid cell
- mLN
- mesenteric LN
- pDC
- plasmacytoid DC
- pIC
- polyinosinic:polycytidylic acid
- PP
- Peyer’s patches
- qPCR
- quantitative PCR
- RPM
- red pulp macrophage
- rRNA
- ribosomal RNA
- SPF
- specific pathogen–free
References
- Basu R., O’Quinn D.B., Silberger D.J., Schoeb T.R., Fouser L., Ouyang W., Hatton R.D., and Weaver C.T.. 2012. Th22 cells are an important source of IL-22 for host protection against enteropathogenic bacteria. Immunity. 37:1061–1075. 10.1016/j.immuni.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernink J.H., Krabbendam L., Germar K., de Jong E., Gronke K., Kofoed-Nielsen M., Munneke J.M., Hazenberg M.D., Villaudy J., Buskens C.J., et al. . 2015. Interleukin-12 and -23 control plasticity of CD127+ group 1 and group 3 innate lymphoid cells in the intestinal lamina propria. Immunity. 43:146–160. 10.1016/j.immuni.2015.06.019 [DOI] [PubMed] [Google Scholar]
- Birnberg T., Bar-On L., Sapoznikov A., Caton M.L., Cervantes-Barragán L., Makia D., Krauthgamer R., Brenner O., Ludewig B., Brockschnieder D., et al. . 2008. Lack of conventional dendritic cells is compatible with normal development and T cell homeostasis, but causes myeloid proliferative syndrome. Immunity. 29:986–997. 10.1016/j.immuni.2008.10.012 [DOI] [PubMed] [Google Scholar]
- Bogunovic M., Ginhoux F., Helft J., Shang L., Hashimoto D., Greter M., Liu K., Jakubzick C., Ingersoll M.A., Leboeuf M., et al. . 2009. Origin of the lamina propria dendritic cell network. Immunity. 31:513–525. 10.1016/j.immuni.2009.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley J.M., Price D.A., Schacker T.W., Asher T.E., Silvestri G., Rao S., Kazzaz Z., Bornstein E., Lambotte O., Altmann D., et al. . 2006. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat. Med. 12:1365–1371. 10.1038/nm1511 [DOI] [PubMed] [Google Scholar]
- Buffie C.G., Bucci V., Stein R.R., McKenney P.T., Ling L., Gobourne A., No D., Liu H., Kinnebrew M., Viale A., et al. . 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 517:205–208. 10.1038/nature13828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caballero S., and Pamer E.G.. 2015. Microbiota-mediated inflammation and antimicrobial defense in the intestine. Annu. Rev. Immunol. 33:227–256. 10.1146/annurev-immunol-032713-120238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton M.L., Smith-Raska M.R., and Reizis B.. 2007. Notch-RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J. Exp. Med. 204:1653–1664. 10.1084/jem.20062648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassaing B., Srinivasan G., Delgado M.A., Young A.N., Gewirtz A.T., and Vijay-Kumar M.. 2012. Fecal lipocalin 2, a sensitive and broadly dynamic non-invasive biomarker for intestinal inflammation. PLoS One. 7:e44328 10.1371/journal.pone.0044328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chieppa M., Rescigno M., Huang A.Y., and Germain R.N.. 2006. Dynamic imaging of dendritic cell extension into the small bowel lumen in response to epithelial cell TLR engagement. J. Exp. Med. 203:2841–2852. 10.1084/jem.20061884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663. 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Curtis M.M., and Way S.S.. 2009. Interleukin-17 in host defence against bacterial, mycobacterial and fungal pathogens. Immunology. 126:177–185. 10.1111/j.1365-2567.2008.03017.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darrasse-Jèze G., Deroubaix S., Mouquet H., Victora G.D., Eisenreich T., Yao K.H., Masilamani R.F., Dustin M.L., Rudensky A., Liu K., and Nussenzweig M.C.. 2009. Feedback control of regulatory T cell homeostasis by dendritic cells in vivo. J. Exp. Med. 206:1853–1862. 10.1084/jem.20090746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis T.Z., Hugenholtz P., Larsen N., Rojas M., Brodie E.L., Keller K., Huber T., Dalevi D., Hu P., and Andersen G.L.. 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elinav E., Strowig T., Kau A.L., Henao-Mejia J., Thaiss C.A., Booth C.J., Peaper D.R., Bertin J., Eisenbarth S.C., Gordon J.I., and Flavell R.A.. 2011. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 145:745–757. 10.1016/j.cell.2011.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallo R.L., and Hooper L.V.. 2012. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 12:503–516. 10.1038/nri3228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajales-Reyes G.E., Iwata A., Albring J., Wu X., Tussiwand R., Kc W., Kretzer N.M., Briseño C.G., Durai V., Bagadia P., et al. . 2015. Batf3 maintains autoactivation of Irf8 for commitment of a CD8α+ conventional DC clonogenic progenitor. Nat. Immunol. 16:708–717. 10.1038/ni.3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto K., Joshi S.K., and Koni P.A.. 2002. A conditional null allele of the major histocompatibility IA-beta chain gene. Genesis. 32:152–153. 10.1002/gene.10056 [DOI] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. . 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100. 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohl T.M., Rivera A., Lipuma L., Gallegos A., Shi C., Mack M., and Pamer E.G.. 2009. Inflammatory monocytes facilitate adaptive CD4 T cell responses during respiratory fungal infection. Cell Host Microbe. 6:470–481. 10.1016/j.chom.2009.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I.I., Atarashi K., Manel N., Brodie E.L., Shima T., Karaoz U., Wei D., Goldfarb K.C., Santee C.A., Lynch S.V., et al. . 2009. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 139:485–498. 10.1016/j.cell.2009.09.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen F.E., Pekna M., Norderhaug I.N., Haneberg B., Hietala M.A., Krajci P., Betsholtz C., and Brandtzaeg P.. 1999. Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J. Exp. Med. 190:915–922. 10.1084/jem.190.7.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung S., Unutmaz D., Wong P., Sano G., De los Santos K., Sparwasser T., Wu S., Vuthoori S., Ko K., Zavala F., et al. . 2002. In vivo depletion of CD11c+ dendritic cells abrogates priming of CD8+ T cells by exogenous cell-associated antigens. Immunity. 17:211–220. 10.1016/S1074-7613(02)00365-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kassim S.H., Rajasagi N.K., Zhao X., Chervenak R., and Jennings S.R.. 2006. In vivo ablation of CD11c-positive dendritic cells increases susceptibility to herpes simplex virus type 1 infection and diminishes NK and T-cell responses. J. Virol. 80:3985–3993. 10.1128/JVI.80.8.3985-3993.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato L.M., Kawamoto S., Maruya M., and Fagarasan S.. 2014. Gut TFH and IgA: key players for regulation of bacterial communities and immune homeostasis. Immunol. Cell Biol. 92:49–56. 10.1038/icb.2013.54 [DOI] [PubMed] [Google Scholar]
- Kinnebrew M.A., Buffie C.G., Diehl G.E., Zenewicz L.A., Leiner I., Hohl T.M., Flavell R.A., Littman D.R., and Pamer E.G.. 2012. Interleukin 23 production by intestinal CD103+CD11b+ dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 36:276–287. 10.1016/j.immuni.2011.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U., Casola S., Cattoretti G., Shen Q., Lia M., Mo T., Ludwig T., Rajewsky K., and Dalla-Favera R.. 2006. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat. Immunol. 7:773–782. 10.1038/ni1357 [DOI] [PubMed] [Google Scholar]
- Kubinak J.L., Petersen C., Stephens W.Z., Soto R., Bake E., O’Connell R.M., and Round J.L.. 2015. MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe. 17:153–163. 10.1016/j.chom.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis K.L., Caton M.L., Bogunovic M., Greter M., Grajkowska L.T., Ng D., Klinakis A., Charo I.F., Jung S., Gommerman J.L., et al. . 2011. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 35:780–791. 10.1016/j.immuni.2011.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindquist R.L., Shakhar G., Dudziak D., Wardemann H., Eisenreich T., Dustin M.L., and Nussenzweig M.C.. 2004. Visualizing dendritic cell networks in vivo. Nat. Immunol. 5:1243–1250. 10.1038/ni1139 [DOI] [PubMed] [Google Scholar]
- Macpherson A.J., and Uhr T.. 2004. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 303:1662–1665. 10.1126/science.1091334 [DOI] [PubMed] [Google Scholar]
- Mangan P.R., Harrington L.E., O’Quinn D.B., Helms W.S., Bullard D.C., Elson C.O., Hatton R.D., Wahl S.M., Schoeb T.R., and Weaver C.T.. 2006. Transforming growth factor-β induces development of the TH17 lineage. Nature. 441:231–234. 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]
- Maruya M., Kawamoto S., Kato L.M., and Fagarasan S.. 2013. Impaired selection of IgA and intestinal dysbiosis associated with PD-1-deficiency. Gut Microbes. 4:165–171. 10.4161/gmic.23595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meredith M.M., Liu K., Darrasse-Jeze G., Kamphorst A.O., Schreiber H.A., Guermonprez P., Idoyaga J., Cheong C., Yao K.H., Niec R.E., and Nussenzweig M.C.. 2012. Expression of the zinc finger transcription factor zDC (Zbtb46, Btbd4) defines the classical dendritic cell lineage. J. Exp. Med. 209:1153–1165. 10.1084/jem.20112675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niess J.H., Brand S., Gu X., Landsman L., Jung S., McCormick B.A., Vyas J.M., Boes M., Ploegh H.L., Fox J.G., et al. . 2005. CX3CR1-mediated dendritic cell access to the intestinal lumen and bacterial clearance. Science. 307:254–258. 10.1126/science.1102901 [DOI] [PubMed] [Google Scholar]
- Nussenzweig M.C., Steinman R.M., Gutchinov B., and Cohn Z.A.. 1980. Dendritic cells are accessory cells for the development of anti-trinitrophenyl cytotoxic T lymphocytes. J. Exp. Med. 152:1070–1084. 10.1084/jem.152.4.1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnmacht C., Pullner A., King S.B., Drexler I., Meier S., Brocker T., and Voehringer D.. 2009. Constitutive ablation of dendritic cells breaks self-tolerance of CD4 T cells and results in spontaneous fatal autoimmunity. J. Exp. Med. 206:549–559. 10.1084/jem.20082394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm N.W., de Zoete M.R., Cullen T.W., Barry N.A., Stefanowski J., Hao L., Degnan P.H., Hu J., Peter I., Zhang W., et al. . 2014. Immunoglobulin A coating identifies colitogenic bacteria in inflammatory bowel disease. Cell. 158:1000–1010. 10.1016/j.cell.2014.08.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.G., Mathur R., Long M., Hosh N., Hao L., Hayden M.S., and Ghosh S.. 2010. T regulatory cells maintain intestinal homeostasis by suppressing γδ T cells. Immunity. 33:791–803. 10.1016/j.immuni.2010.10.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry J.S., Lio C.W., Kau A.L., Nutsch K., Yang Z., Gordon J.I., Murphy K.M., and Hsieh C.S.. 2014. Distinct contributions of Aire and antigen-presenting-cell subsets to the generation of self-tolerance in the thymus. Immunity. 41:414–426. 10.1016/j.immuni.2014.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson E.K., Uronen-Hansson H., Semmrich M., Rivollier A., Hägerbrand K., Marsal J., Gudjonsson S., Håkansson U., Reizis B., Kotarsky K., and Agace W.W.. 2013. IRF4 transcription-factor-dependent CD103+CD11b+ dendritic cells drive mucosal T helper 17 cell differentiation. Immunity. 38:958–969. 10.1016/j.immuni.2013.03.009 [DOI] [PubMed] [Google Scholar]
- Salzman N.H., de Jong H., Paterson Y., Harmsen H.J., Welling G.W., and Bos N.A.. 2002. Analysis of 16S libraries of mouse gastrointestinal microflora reveals a large new group of mouse intestinal bacteria. Microbiology. 148:3651–3660. 10.1099/00221287-148-11-3651 [DOI] [PubMed] [Google Scholar]
- Sandler N.G., and Douek D.C.. 2012. Microbial translocation in HIV infection: causes, consequences and treatment opportunities. Nat. Rev. Microbiol. 10:655–666. 10.1038/nrmicro2848 [DOI] [PubMed] [Google Scholar]
- Satpathy A.T., Kc W., Albring J.C., Edelson B.T., Kretzer N.M., Bhattacharya D., Murphy T.L., and Murphy K.M.. 2012. Zbtb46 expression distinguishes classical dendritic cells and their committed progenitors from other immune lineages. J. Exp. Med. 209:1135–1152. 10.1084/jem.20120030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy A.T., Briseño C.G., Lee J.S., Ng D., Manieri N.A., Kc W., Wu X., Thomas S.R., Lee W.L., Turkoz M., et al. . 2013. Notch2-dependent classical dendritic cells orchestrate intestinal immunity to attaching-and-effacing bacterial pathogens. Nat. Immunol. 14:937–948. 10.1038/ni.2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A., McGovern N., Teo P., Zelante T., Atarashi K., Low D., Ho A.W., See P., Shin A., Wasan P.S., et al. . 2013. IRF4 transcription factor-dependent CD11b+ dendritic cells in human and mouse control mucosal IL-17 cytokine responses. Immunity. 38:970–983. 10.1016/j.immuni.2013.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss P.D., Westcott S.L., Ryabin T., Hall J.R., Hartmann M., Hollister E.B., Lesniewski R.A., Oakley B.B., Parks D.H., Robinson C.J., et al. . 2009. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75:7537–7541. 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber H.A., Loschko J., Karssemeijer R.A., Escolano A., Meredith M.M., Mucida D., Guermonprez P., and Nussenzweig M.C.. 2013. Intestinal monocytes and macrophages are required for T cell polarization in response to Citrobacter rodentium. J. Exp. Med. 210:2025–2039. 10.1084/jem.20130903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serbina N.V., Salazar-Mather T.P., Biron C.A., Kuziel W.A., and Pamer E.G.. 2003. TNF/iNOS-producing dendritic cells mediate innate immune defense against bacterial infection. Immunity. 19:59–70. 10.1016/S1074-7613(03)00171-7 [DOI] [PubMed] [Google Scholar]
- Sonnenberg G.F., Monticelli L.A., Elloso M.M., Fouser L.A., and Artis D.. 2011. CD4+ lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 34:122–134. 10.1016/j.immuni.2010.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinman R.M., Hawiger D., and Nussenzweig M.C.. 2003. Tolerogenic dendritic cells. Annu. Rev. Immunol. 21:685–711. 10.1146/annurev.immunol.21.120601.141040 [DOI] [PubMed] [Google Scholar]
- Stranges P.B., Watson J., Cooper C.J., Choisy-Rossi C.M., Stonebraker A.C., Beighton R.A., Hartig H., Sundberg J.P., Servick S., Kaufmann G., et al. . 2007. Elimination of antigen-presenting cells and autoreactive T cells by Fas contributes to prevention of autoimmunity. Immunity. 26:629–641. 10.1016/j.immuni.2007.03.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swidsinski A., Weber J., Loening-Baucke V., Hale L.P., and Lochs H.. 2005. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 43:3380–3389. 10.1128/JCM.43.7.3380-3389.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoutounour S., Henri S., Lelouard H., de Bovis B., de Haar C., van der Woude C.J., Woltman A.M., Reyal Y., Bonnet D., Sichien D., et al. . 2012. CD64 distinguishes macrophages from dendritic cells in the gut and reveals the Th1-inducing role of mesenteric lymph node macrophages during colitis. Eur. J. Immunol. 42:3150–3166. 10.1002/eji.201242847 [DOI] [PubMed] [Google Scholar]
- Tamura T., Tailor P., Yamaoka K., Kong H.J., Tsujimura H., O’Shea J.J., Singh H., and Ozato K.. 2005. IFN regulatory factor-4 and -8 govern dendritic cell subset development and their functional diversity. J. Immunol. 174:2573–2581. 10.4049/jimmunol.174.5.2573 [DOI] [PubMed] [Google Scholar]
- Ubeda C., Taur Y., Jenq R.R., Equinda M.J., Son T., Samstein M., Viale A., Socci N.D., van den Brink M.R., Kamboj M., and Pamer E.G.. 2010. Vancomycin-resistant Enterococcus domination of intestinal microbiota is enabled by antibiotic treatment in mice and precedes bloodstream invasion in humans. J. Clin. Invest. 120:4332–4341. 10.1172/JCI43918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaishnava S., Yamamoto M., Severson K.M., Ruhn K.A., Yu X., Koren O., Ley R., Wakeland E.K., and Hooper L.V.. 2011. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science. 334:255–258. 10.1126/science.1209791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vallon-Eberhard A., Landsman L., Yogev N., Verrier B., and Jung S.. 2006. Transepithelial pathogen uptake into the small intestinal lamina propria. J. Immunol. 176:2465–2469. 10.4049/jimmunol.176.4.2465 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.