Abstract
The adsorption of single-component bovine serum albumin (BSA), bovine fibrinogen (Fgn) and bovine immunoglobulin G (IgG) films, as well as mult--component bovine plasma films onto bare and sodium styrene sulfonate (NaSS)-grafted gold substrates was characterized. The adsorption isotherms, measured via X-ray photoelectron spectroscopy showed that at low solution concentrations all three single-component proteins adsorb with higher affinity onto gold surfaces compared to NaSS surfaces. However, at higher concentrations, NaSS surfaces adsorb the same or more total protein than gold surfaces. This may be because proteins that adsorb onto NaSS undergo structural rearrangements, resulting in a larger fraction of irreversibly adsorbed species over time. Still, with the possible exception of BSA adsorbed onto gold, neither surface appeared to have saturated at the highest protein solution concentration studied. Principal component analysis of amino acid mass fragments from time-of-flight secondary ion mass spectra distinguished between the same protein adsorbed onto NaSS and gold surfaces, suggesting that proteins adsorb differently on NaSS and gold surfaces. Explored further using peak ratios for buried/surface amino acids for each protein, we found that proteins denature more on NaSS surfaces than on gold surfaces. Also, using peak ratios for asymmetrically distributed amino acids, potential structural differences were postulated for BSA and IgG adsorbed onto NaSS and gold surfaces. Principal component (PC) modeling, used to track changes in plasma adsorption with time, suggests that plasma films on NaSS and Au surfaces become more Fgn-like with increasing adsorption time. However, the PC models included only three proteins, where plasma is composed of hundreds of proteins. Therefore, while both gold and NaSS appear to adsorb more Fgn with time, further study is required to confirm that this is representative of the final state of the plasma films.
Graphical Abstract
I. Introduction
The performance of biomedical implants is largely determined by how proteins interact with the implant surface.1–3 This is because proteins are the first biomolecules adsorbed onto the surface, with all subsequent interactions between the body and the implant mediated by the adsorbed protein layer.4–6 As such, understanding how proteins interact with different surface chemistries is important in aiding development of next-generation biomaterials.
Polymer grafting is an effective way of making controlled changes to surface chemistry.7–8 Furthermore, advancements in polymer chemistry, such as the development of ARGET ATRP (Activators are continuously ReGenerated by Electron Transfer Atom Transfer Radical Polymerization), have simplified the grafting process and greatly expanded the available combination of polymers and functional groups with which to modify surfaces.9–10
Surface functionalization with sodium styrene sulfonate (NaSS) has recently been shown to increase proliferation and adhesion of fibroblasts,11 MG63 osteoblast-like cells,12–13 and human mandibular osteoblasts14 in vitro, with promising increases in osseointegration shown in vivo.14–16 It has been hypothesized that an observed increase in osseointegration is due the ability of NaSS to preferentially adsorb specific plasma proteins in an orientation and conformation that promotes formation of new bone.17 However, detailed studies using a multi-technique approach using state-of-the-art methodology to directly examine the structure of protein films on NaSS surfaces are lacking. We have recently developed a procedure for grafting NaSS from a number of oxide and metal surfaces to test this hypothesis.18–19 Beyond the potential for biomedical applications, NaSS is an interesting system as it allows us to study protein interaction with a very specific surface chemistry: namely, sulfonated styrene with a sodium counter ion. This can be used as a platform for future work to study effects such as changing the counter ion from sodium to potassium or the head group from a negatively charged sulfonate to a positively charged quaternary amine. Thus, controlled changes in surface chemistry can be related to the properties of adsorbed protein films in a continued effort to investigate the important factors controlling protein-surface interactions20.
In the current work, we investigate the adsorption of single-component bovine serum albumin (BSA), bovine fibrinogen (Fgn) bovine immunoglobulin G (IgG) films, as well as multi-component bovine plasma films onto bare and NaSS-grafted gold surfaces. Protein adsorption isotherms were measured using X-ray photoelectron spectroscopy (XPS) to follow changes in surface nitrogen composition with increasing protein solution concentration (5, 25, and 100 μg/ml). Since gold and NaSS do not contain nitrogen, surface nitrogen composition is directly related to the amount of protein adsorbed. A combination of time-of-flight secondary ion mass spectrometry (ToF-SIMS), principal component analysis (PCA), and visual molecular dynamics (VMD) was used to analyze amino acid distributions in the adsorbed proteins and provide new insights into their adsorbed structure on both surfaces. Principal component (PC) modeling was used to qualitatively describe changes in bovine plasma films with time.
II. Experimental
A. Materials
Silicon wafers (Silicon Valley Microelectronics Inc., San Jose, CA) were diced into 1×1 cm2 substrates using a diamond saw. Gold substrates were fabricated by depositing a 5 nm titanium adhesion layer followed by a 100 nm gold layer onto the diced and cleaned silicon substrates via electron-beam deposition at room temperature and pressures < 1 x10−6 torr. Methanol, acetone, dichloromethane, phosphate buffered saline (PBS; 0.01 M phosphate, 0.138 M sodium chloride, 0.0027 M potassium chloride, pH 7.4), BSA (≥ 99%), bovine Fgn (65–85%), bovine IgG (≥ 95%), bovine plasma, Cu(II) bromide (>99.0%), 2,2′-bipyridine (bpy) (≥99%), vitamin C (≥98%), and NaSS (≥90%) were all purchased from Sigma. Ethanol (200 proof) was purchased from Decon Laboratories. Bromoisobutyrate undecyl disulfide (99%) was purchased from Asemblon. All chemicals were used as received.
B. Substrate cleaning and ATRP-initiator functionalization
Prior to the electron-beam deposition, substrates were cleaned by sonicating twice for 5 minutes in each of dichloromethane, acetone, and methanol. The electron-beam deposited gold substrates were rinsed with ethanol and immersed in a 1 mM solution of bromoisobutyrate undecyl disulfide in ethanol for 18 – 24 hrs. The substrates were then rinsed with ethanol, sonicated for 1 – 3 minutes to remove any unbound bromoisobutyrate undecyl disulfide, and rinsed once more with ethanol. Gold functionalization reactions were performed at room temperature in N2-backfilled glass test tubes stoppered with rubber septa—one sample per test tube.
C. NaSS Grafting
A fresh batch of NaSS-grafted samples was prepared for each protein adsorption experiment. A batch typically consisted of 16 1×1 cm2 functionalized gold substrates and the grafting reactions were performed in 20-ml glass scintillation vials, with two substrates per vial. To each reaction vial was added 500 mg of NaSS monomer and 1.2 ml 18-Ω DI H2O. We previously established that NaSS film quality is sensitive to catalyst amount.19 Therefore, to minimize weighing error, the catalyst solution was prepared in bulk for the entire batch. For 16 samples, this entailed dissolving 8 mg CuBr2 and 28 mg bpy (1:2 CuBr2:bpy) in 19.2 ml methanol and 9.6 ml 18-Ω DI H2O. The catalyst solution (3.6 ml) was then added to each reaction vial so that the final concentration of CuBr2 and bpy was 0.5 and 1.4 mg per nominal cm2 functionalized surface area. At this point, the reaction vials were often briefly sonicated to fully dissolve the NaSS monomer. Two functionalized substrates were then added to each vial and the grafting reaction begun upon addition of 79 mg vitamin C dissolved in 1.2 ml 18-Ω DI H2O (39.5 mg vitamin C per nominal cm2 functionalized surface area). The vitamin C solution was also prepared in bulk—632 mg in 9.6 ml 18-Ω DI H2O. The total solvent volume was 6 ml (60/40 H2O/methanol). Care was taken that the substrates stayed face up and did not overlap each other. The reaction vials were capped and sealed with parafilm to minimize additional oxygen leaking into the system. After 18–24 hrs, the substrates were rinsed with 18-Ω DI H2O, soaked in PBS for a few hours, and then soaked in 18-Ω DI H2O overnight to remove any residual catalyst and monomer. Finally, the grafted substrates were rinsed again with 18-Ω DI H2O, gently dried with a stream of N2, and analyzed immediately via XPS to ensure that the reaction had succeeded and the film quality was sufficient for further use. Detailed characterization of these films has been presented elsewhere.18–19
D. Protein Adsorption
Substrates were hydrated for 30 min. in degassed PBS (pH = 7.4) prior to adsorption. The single-component protein adsorption experiments were performed for 2 hours at solution concentrations of 0, 5, 25, and 100 μg/ml to establish adsorption isotherms. Plasma adsorption experiments were performed at a solution concentration of 100 μg/ml for times varying between 5 and 120 min. All adsorption experiments were performed in 24-well plates in degassed PBS at 37°C. Two duplicate samples were prepared at each treatment, and each experiment was replicated twice, yielding four samples for each adsorption concentration (for the single-component experiments) or time point (for the plasma adsorption experiments). Following the adsorption step, the protein solution in each well is diluted eight times with fresh PBS before removing the samples to avoid pulling them through a layer of denatured proteins at the air-liquid interface. Following removal from the well-plate, the samples are rinsed twice in fresh PBS to remove any loosely bound protein, and then three times in 18-Ω deionized water to remove excess buffer salts. Samples are dried with a stream of nitrogen and then stored under nitrogen until analyzed, and also in between analyses. Samples were analyzed via XPS and then ToF-SIMS on the two subsequent days after protein adsorption.
E. X-ray photoelectron spectroscopy (XPS)
XPS compositions were determined from the average of three spots on four samples for each adsorption condition, totaling 12 analysis spots per treatment. The data were acquired on an SSI S-Probe instrument (Surface Science Instruments, Mountain View, California) with a 0° photoelectron takeoff angle (TOA), defined as the angle between the surface normal and the axis of the analyzer lens. All spectra were acquired using a monochromatic Al Kα1,2 X-ray source (hν = 1486.6 eV). The survey (0–1100 eV) and detailed scans (20 eV window) collected with analyzer pass energy of 150 eV and a 100 ms dwell time were used to determine surface elemental compositions. The survey step size was 1 eV, while 0.4 eV was used for detail scans. The high-resolution scans (20 eV window) were collected with analyzer pass energy of 50 eV and a step size of 0.065 eV. All samples were isolated from the instrument using double-sided tape and analyzed as insulators. Charge neutralization was achieved using a low-energy electron flood gun. The XPS binding energy scale was calibrated using Cu2p3/2 and Au4f7/2 peaks from clean, reference metal foils. Sample charging was corrected by setting the CHx peak in the C1s region to 284.6 eV. A linear background was subtracted for all peak area quantifications. The peak areas were normalized by the manufacturer supplied sensitivity factors, and atomic compositions calculated using the Hawk Data Analysis 7 software (Service Physics, Inc., Bend, Oregon).
F. Time-of-flight secondary ion mass spectrometry (ToF-SIMS)
Positive and negative secondary ion spectra and images were acquired on a TOF.SIMS 5–100 instrument (ION-TOF, Münster, Germany) using a pulsed 25 keV primary ion beam under static conditions (primary ion dose < 1012 ions/cm2). Five positive and three negative spectra were collected from 100 × 100 μm2 regions for four samples per adsorption condition, totaling 20 positive spectra and 12 negative spectra spots per treatment. Spectra were acquired in high-current bunch (high mass resolution) mode, over a range of 0–800 m/z at a mass resolution (m/Δm) between 4000 and 8000. Positive spectra were mass calibrated using , and peaks, and negative spectra were calibrating using CH−, OH−, C2H−, , C4H−, and peaks. Mass calibration errors were below 20 ppm.
G. Principal component analysis (PCA)
PCA of the ToF-SIMS data, previously described in detail,21 is a multivariate analysis technique that is used to identify principal sources of variation between sample spectra. Three peak lists were compiled and imported into a series of scripts written by NESAC/BIO for MATLAB22 (MathWorks, Inc., Natick, MA): all positive peaks, all negative peaks, and a peak list containing amino acid-derived mass fragments.21 Data sets were also normalized by the sum of selected peaks, mean centered, and square-root transformed to ensure that variance within the data set was due to differences in sample variances rather than in sample means. Due to detector saturation during acquisition, the sodium peak was removed from all positive peak lists prior to any data pretreatment. Obvious contaminant (e.g., PDMS, Cu, I) and isotope peaks were also typically removed prior to analysis.
III. Results and Discussion
A. Concerns Regarding NaSS Sodium Levels and Resulting Matrix Effects
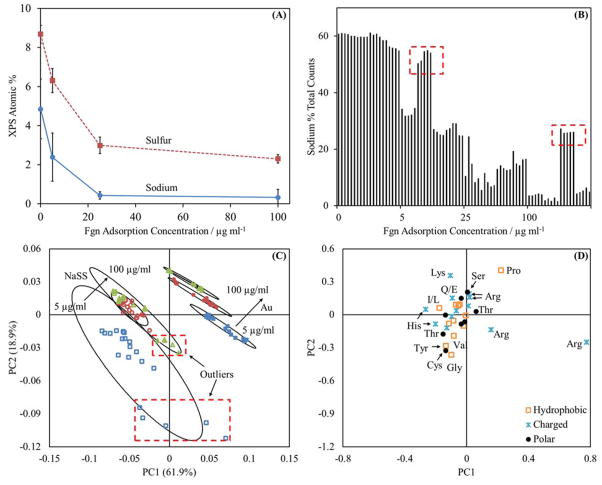
Typically, ToF-SIMS data with sodium (m/z 22.99) secondary ion counts ≥ 1% of the total counts are discounted from analysis. High sodium levels, most often due to excess buffer salts left by improper sample rinsing, are linked with matrix effects that fundamentally alter the formation of amino acid mass fragments.21 In our case, this rule is appropriate for the samples prepared on gold, where the sodium percent total counts never exceeds 0.7% (figure S1 of the supplemental information). However, sodium is part of the NaSS structure and the percentage of the total counts made up by the sodium peak ranges from 2% – 60% (figure S2 in the supplemental information). Furthermore, the matrix effects caused by sodium are part of the matrix effects of the surface. To remove the sodium would be, in effect, to study a different system.
In any case, the sodium is likely bound to the NaSS sulfonate groups and unlikely to interfere with secondary ion fragmentation in the same way as unrinsed, free buffer salts. Evidence for this is the observed decreasing sodium signal with increasing amounts of adsorbed protein (figure 1A). Also, as amount of adsorbed protein increases (figure 2) the XPS sodium signal decreases to a greater extent than the XPS sulfur signal. As the protein solution concentration increases from 0 to 100 μg/ml the sodium concentration decreases 93% versus only 73% for the sulfur concentration. This suggests not only is the sodium signal attenuated by the adsorbed proteins, but also potentially some sodium displacement occurred during the protein adsorption process. Counter-ion displacement during protein adsorption has been observed in other charged polymer brush systems.7
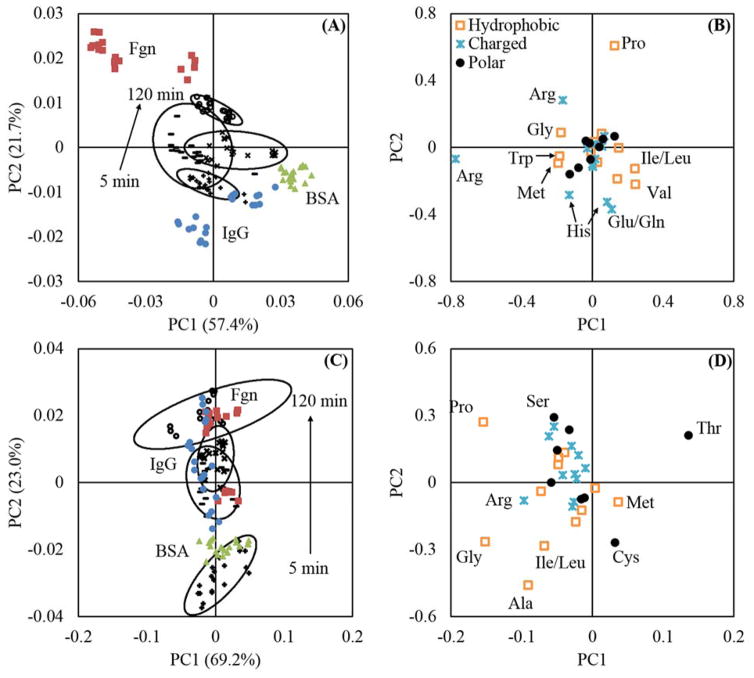
Figure 1.
(A) Sulfur and sodium XPS compositions for Fgn adsorbed onto NaSS surfaces. (B) Sodium (m/z 22.99) percent of total counts for Fgn adsorption onto NaSS surfaces. (C) and (D) amino acid peak list PCA results for Fgn adsorbed onto NaSS surfaces. For panel (C):
 represents adsorption from 5 μg/ml solutions,
represents adsorption from 5 μg/ml solutions,
 represents adsorption from 25 μg/ml solutions, and
represents adsorption from 25 μg/ml solutions, and
 represents adsorption from 100 μg/ml solutions. Open symbols are used for NaSS substrates and closed symbols are used for gold substrates. The error bars in (A) represent standard deviations and the ellipses in (C) represent 95% confidence intervals. The boxed outliers in (B) indicate samples with outlying sodium concentrations relative to their respective sample groups. The corresponding samples are boxed in panel (C). Panel (A) supports the claim that the sodium is substrate bound and has little impact on amino acid fragmentation patterns. Comparison of panels (C) and (D) with panels (C) and (D) in figure 3 shows that the NaSS sodium levels do not drastically effect the conclusions drawn from the PCA results.
represents adsorption from 100 μg/ml solutions. Open symbols are used for NaSS substrates and closed symbols are used for gold substrates. The error bars in (A) represent standard deviations and the ellipses in (C) represent 95% confidence intervals. The boxed outliers in (B) indicate samples with outlying sodium concentrations relative to their respective sample groups. The corresponding samples are boxed in panel (C). Panel (A) supports the claim that the sodium is substrate bound and has little impact on amino acid fragmentation patterns. Comparison of panels (C) and (D) with panels (C) and (D) in figure 3 shows that the NaSS sodium levels do not drastically effect the conclusions drawn from the PCA results.
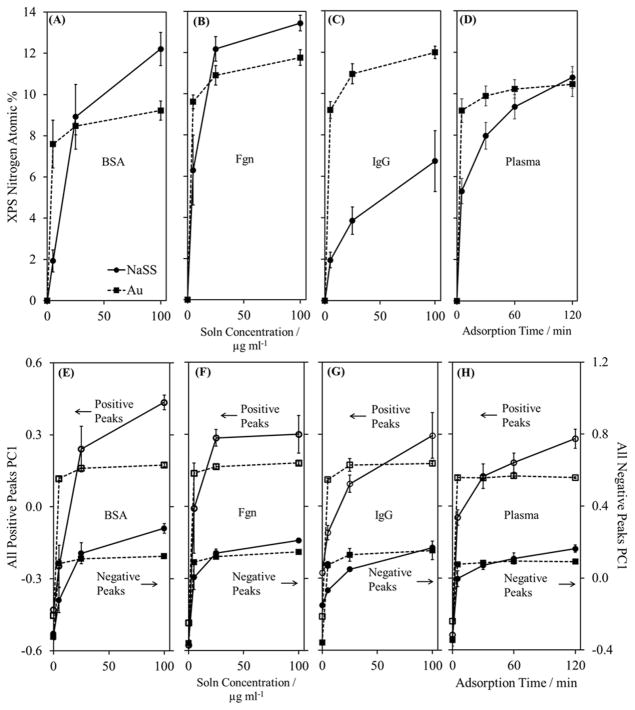
Figure 2.
XPS isotherms for (A) BSA, (B) Fgn, (C) IgG, and (D) plasma adsorption onto gold and NaSS surfaces. Panels (E) through (H) are isotherms of the PC1 scores obtained from PCA using peak lists containing all peaks that are at least 3x the background intensity. In all panels, squares and dashed lines represent gold substrates, while circles and solid lines represent NaSS substrates. In panels (E) through (H), open symbols represent scores from PCA of the positive spectra, and closed symbols represent scores from PCA of the negative spectra. Error bars represent standard deviations.
The PCA results also support the claim that the NaSS sodium levels do not interfere with secondary ion fragmentation. If sodium has a substantial impact on amino acid fragmentation, then fluctuations in sodium levels within treatment groups should yield fluctuations—or outliers—in the amino acid PCA results. For BSA adsorbed from 25 μg/ml solutions onto NaSS surfaces the average sodium atomic percent measured by XPS is 32.4% ± 3.7% (average ± standard deviation) for three of the four samples and 18.2% ± 1.1% for the remaining sample (figure S2 in the supplemental information). Similarly, for BSA adsorbed from 100 μg/ml solutions the average sodium atomic percent measured by XPS is 10.1% ± 2.4% for three of the four samples and 16.4% ± 1.0% for the remaining sample. If the sodium concentration in this system affects the amino acid fragmentation process, the sodium count fluctuations should register as two sets of five outliers in the PCA results—one set of five each in the 25 μg/ml and 100 μg/ml adsorption groups. The adsorbed BSA on NaSS results display a good deal of scatter, but there are no obvious outliers (figure 3A). However, groups of outliers, corresponding with samples with higher than average sodium counts, are observed for Fgn on NaSS (figure 1B). Separation between treatment groups does improve upon removal of these outliers (compare figures 1C and 3C); though, while the loadings change in magnitude, there are no obvious changes in loading direction. Even when the sodium levels affect the scores, removal of the outliers does not significantly change interpretation of the PCA results. Therefore, sodium appears to be primarily bound to the grafted NaSS chains and does not seem to have a significant effect on the amino acid fragmentation patterns.
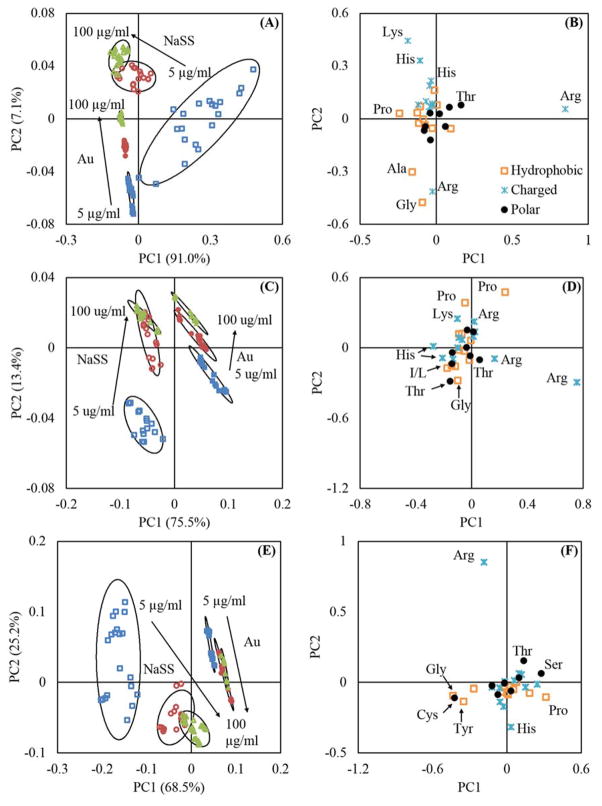
Figure 3.
Amino acid peak list PCA results. Panels (A) and (B) are the scores and loadings plots for BSA. Panels (C) and (D) are the scores and loadings plots for Fgn. Panels (E) and (F) and the scores and loadings plots for IgG. In panels (A), (C), and (E),
 represent adsorption from 5 μg/ml solutions,
represent adsorption from 5 μg/ml solutions,
 represent adsorption from 25 μg/ml solutions, and
represent adsorption from 25 μg/ml solutions, and
 represent adsorption from 100 μg/ml solutions. Open symbols are used for NaSS substrates and closed symbols are used for gold substrates. The ellipses in (A), (C), and (E) represent 95% confidence intervals.
represent adsorption from 100 μg/ml solutions. Open symbols are used for NaSS substrates and closed symbols are used for gold substrates. The ellipses in (A), (C), and (E) represent 95% confidence intervals.
B. XPS and ToF-SIMS Isotherms
Protein adsorption, at solution concentrations ranging from 5 to 100 μg/ml, was followed with XPS, plotting nitrogen atomic% as an indication of surface protein concentration (figure 2). Representative high-resolution C1s spectra for protein adsorption onto the NaSS surface are shown in figure S3 in the Supplemental Information. Caution must be taken with regard to classifying these adsorption curves as Langmuirian, or any other type, since the physics of protein adsorption are often at odds with the fundamental assumptions behind the classical isotherm models.23–24 What is clear from the shape of the adsorption isotherms, specifically the near-infinite initial slopes, is that all three proteins and plasma have a higher affinity for gold versus NaSS.23 However, except for IgG—which shows the largest disparity in the amount of protein adsorbed between these two surfaces—the NaSS surface adsorbs the same (for plasma) or more (for BSA and Fgn) total protein than the gold surface from the 100 μg/ml solutions. A possible explanation for this is that proteins in their native state have much lower affinity and form fewer initial contacts with NaSS than gold.23 Any proteins that do adsorb to NaSS undergo structural rearrangements—which occur on a time scale from 100 to potentially even 103 seconds25—increasing the number of contacts on the surface and resulting in a larger fraction of irreversibly adsorbed species.23 The differing plasma adsorption curves on NaSS and gold surfaces are further evidence in support of this argument. The plasma adsorption experiments were performed at a constant 100 μg/ml solution concentration and increasing times. At times up to 30 minutes, the gold surfaces adsorb substantially more protein than NaSS. Hence, proteins initially have higher affinity for gold than NaSS, even at high solution concentrations. At 60 minutes, however, NaSS adsorbs nearly as much protein as gold. And where gold appears to have saturated at 120 minutes, the slope of the NaSS isotherm is still fairly steep suggesting that this system has not yet reached its equilibrium surface concentration. Hence, the protein affinity for NaSS increases with time, and we postulate structural rearrangement of the adsorbed proteins is the reason for this. Still, except for possibly plasma and BSA adsorbed onto gold surfaces, none of the isotherms appear to have saturated. However, in all cases the largest and most significant changes in the protein films were observed over the solution concentrations (0 to 100 μg/ml for single-component proteins) and adsorption time period (0 to 2 hr for multi-component plasma) used in this study. That BSA adsorbed onto gold approaches saturation is confirmed by the measurements of Tidwell et al.26 who arrived at a nitrogen XPS surface composition of ~9% for adsorption from a 1 mg/ml solution of BSA. The nitrogen atomic% here is also ~9%, at a BSA solution concentration of 100 μg/ml.
Structural rearrangements may also explain the differences in amounts of adsorbed IgG. If IgG adsorbs in a higher surface area conformation onto NaSS surfaces compared to gold (due to a lower packing-density orientation and/or a greater degree of denaturation), then the total amount of adsorbed protein would be much higher on gold for the same surface coverage. This is supported by the PC1 isotherms in figure 2 (E) through (H). In the case of BSA and Fgn, the shapes of the XPS and SIMS isotherms more or less match, indicating that PC1 scores are correlated to adsorbed protein surface concentration. However, this correlation does not hold for IgG. PC1 would predict that NaSS adsorbs more IgG than gold at 100 μg/ml, even though XPS—which is the more quantitative of the two techniques—clearly shows the opposite to be the case. This is simply because the XPS sampling depth (~10 nm) is nominally five times that of SIMS (~2 nm).2 Wagner et al. reported that, relative to 125I-radiolabeling measurements, SIMS overpredicted the amount of Fgn adsorbed in a binary IgG/Fgn film. This was attributed to the differences in sampling depth between the two techniques, and the organization of the adsorbed protein film.27 While 125I-radiolabeling measured the film bulk composition, SIMS—due to its high surface sensitivity—oversampled the adsorbed Fgn, which was preferentially located at the vacuum interface. (The sampling depth of 125I-radiolabeling, for all practical purposes, can be considered infinite.)27 Similarly, in our case, the larger XPS sampling depth results in measurements that more closely represent the total amount of adsorbed protein, whereas SIMS more closely represent the fraction of covered versus exposed substrate. It is, therefore, more accurate to say that the SIMS isotherms are correlated to surface coverage, which is dependent not only on surface concentration but also on adsorbed protein structure. Thus, we propose that the SIMS isotherms indicate the IgG molecules have adsorbed in a lower packing-density arrangement, and may also have denatured and spread out to cover a higher fraction of the NaSS surface. This is compared to more intact IgG molecules covering a smaller fraction of the gold surface. Sampling depth differences also explain why the SIMS isotherms saturate more often and at lower concentrations than those measured by XPS. Similar differences in protein adsorption curves have been observed comparing XPS and atomic force microscopy results for fibronectin adsorbed onto mica surfaces.28
C. PCA of Amino Acid Mass Fragments
The analysis below was performed using a peak list containing only amino-acid derived mass fragments. PCA results for the entire peak set, containing all positive and negative peaks with intensity at least three times the background, are presented in the Supplemental Information.
PCA, using only amino-acid-fragment peaks, is able to distinguish between the same protein adsorbed onto gold or NaSS surfaces, and also between protein films adsorbed from different solution concentrations on the same surface (Panels A, C, and E of figure 3). This suggests that proteins adsorb differently onto NaSS and gold surfaces, and that the adsorbed protein structure changes with surface concentration. To probe these differences, we first broadly classified each amino acid as charged, polar, or hydrophobic (Panels B, D, and F of figure 3). We found no obvious trends using these broad categories. For example, no trend was observed that suggested coulombic attractions between negatively charged sulfonate groups and positively charged amino acids yielded a preferred protein orientation on NaSS substrates.
VMD—used to visualize the Protein Data Bank (PDB) crystal structures for BSA (PDB ID: 4F5S), Fgn (PDB ID: 3GHG), and IgG29—confirmed that each amino acid classification (charged, polar, or hydrophobic) is roughly evenly distributed across the surface of each protein (data not shown). Therefore, it is not surprising that none of these three general categories is correlated with adsorbed protein conformation or orientation. Instead, trends in specific peak-intensity ratios for strategically chosen, heterogeneously distributed amino acids were used to further explore the PCA results. Baugh et al. showed that structural information about surface immobilized proteins can be gleaned from such ratios.30
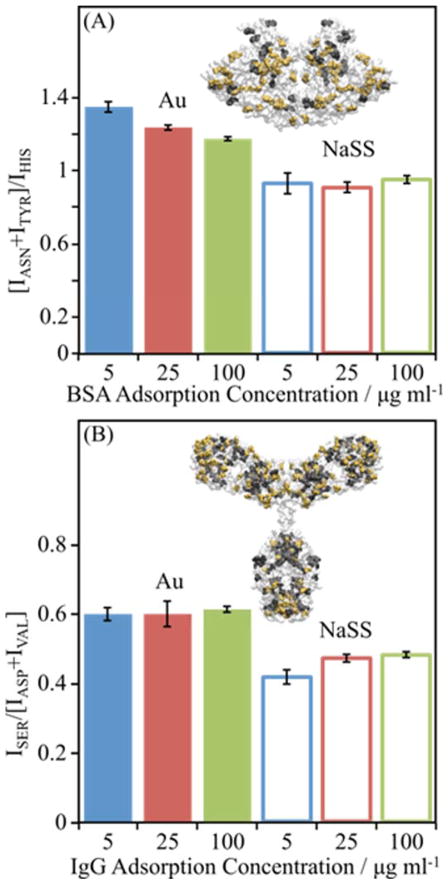
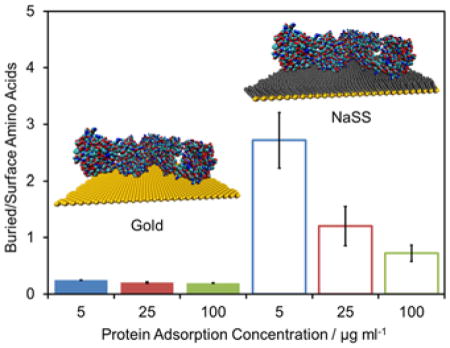
To assess the level of denaturation, the ratio of certain buried/surface amino acids was examined for each protein. An increase in this ratio indicates an increase in protein denaturation. The specific residues chosen for each protein are listed in table 1 and highlighted in the insets of figure 4, where the relevant buried and surface amino acids are highlighted in yellow and black, respectively. The peak intensity (I) ratios used are shown in Eq. 1.
Table 1.
Amino acids selected for protein structural analysis.
| Amino Acids for Denaturation Analysis | Amino Acids for Orientation Analysis | |||
|---|---|---|---|---|
|
| ||||
| Buried (m/z) | Surface (m/z) | Bottom* (m/z) | Top* (m/z) | |
|
| ||||
| BSA | Cys (76.0) | Glu (84.0) | Asn, Tyr (70.0, 107) | His (81.0, 82.0) |
| Fgn | Val (83.1) | Pro, Arg (70.1) | - | - |
| IgG | Cys (76.0) | Arg (100.1) | Asp, Val (72.0) | Ser (60.0) |
Bottom and top refer to the protein with the orientation shown in figure 5.
Figure 4.
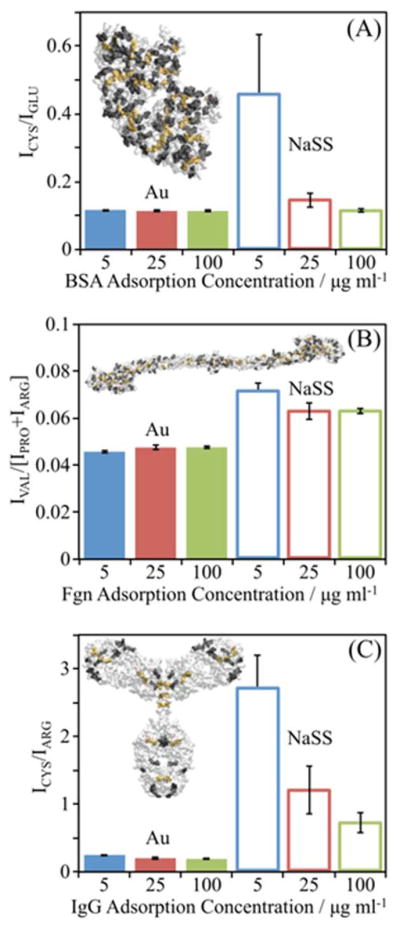
ToF-SIMS peak intensity ratios suggest levels of denaturation of (A) BSA, (B) Fgn, (C) and IgG adsorbed onto gold and NaSS surfaces. Insets show the crystal structure of each protein. Bars represent mean amino acid peak ratios for proteins adsorbed from 5 (blue), 25 (red), and 100 (green) μg/ml solutions onto Au surfaces (solid bars) or NaSS surfaces (open bars). Peak ratios are the sum of intensities of secondary ions from amino acids buried, highlighted in yellow in inset protein, divided by the sum of intensities from amino acids on the surface of the protein, highlighted in black. Error bars represent standard deviations.
| (Eq. 1) |
For all three proteins, the buried/surface amino acid ratios are higher on NaSS than gold (figure 4), particularly for samples prepared from the lowest solution concentrations—i.e., low surface coverage and packing density. This suggests that (1) proteins denature more on NaSS, and (2) that denaturation decreases with increasing packing density at higher surface concentrations.31–32 The largest difference in the buried/surface amino acid ratio between the two surfaces is for IgG (figure 4C), again, especially at the lowest surface coverages. Consistent with the XPS and ToF-SIMS isotherms discussed above, this suggests that IgG adopts a more highly denatured, higher surface-area conformation (e.g., thinner and more spread out) when adsorbed onto NaSS surfaces compared gold surfaces.
Scatter in the scores plots (figure 3) also support both these conclusions. As the scatter within a treatment group increases, so does the variability among the constituent spectra. Higher scatter is expected for a highly denatured protein film with a random sampling of amino acids at the vacuum interface. And the scatter in the scores plots is considerably higher for NaSS surfaces verses gold surfaces. This is true across all three proteins and solution concentrations. Thus, adsorbed proteins denature more on NaSS than on gold. The scatter also decreases at higher protein solution concentrations for all three proteins, indicating that denaturation decreases with increasing adsorbed-protein packing density. This trend is less obvious on gold due to the high affinity of each protein for gold surfaces—i.e., high packing densities are obtained even at the lowest solution concentration studied.
Before moving on, it is important to note the effect of the analysis conditions on protein conformation. Air drying and the ultra-high vacuum analysis environment are known to cause protein denaturation.33 However, in the case where sample drying accounts for the most of the conformational changes, a given protein would be expected to look the same on both surfaces: completely denatured. But as discussed above, significant differences in extent of denaturation are observed between proteins adsorbed onto the two surfaces. Therefore, we hypothesize that, while the adsorbed proteins may denature a bit further upon air or vacuum drying, most of the conformation changes occur during the adsorption process. Testing this hypothesis will require future work employing techniques (e.g., trehalose coating or frozen hydrated analysis conditions) to minimize the impact of sample drying on adsorbed protein structure.33–34
To further investigate the impact of the different surface chemistries on adsorbed protein structure, a similar ratio analysis was performed using amino acids on opposite sides of each protein. A relative increase in the ratio of amino acids enriched on one end of a protein to amino acids enriched on the opposite end suggests orientational differences in the adsorbed protein on the two surfaces. The key amino acids for BSA and IgG are listed in table 1, and the ratios are found in Eq. 2. Due to the symmetry and even distribution of amino acids, we were unable to find an analogous ratio for Fgn.
| (Eq. 2) |
For both proteins adsorbed from all solution concentrations, these ratios are different for the two surfaces (figure 5). This is consistent with the PCA results (figure 3), which show proteins adsorb differently onto NaSS and gold surfaces. The ratios also shed some light on relative concentrations of certain amino acids at the vacuum interface. These amino acid peak ratios also suggest concentration-dependent orientation changes, as the ratios trend with concentration for BSA on gold and IgG on NaSS. This is again consistent with the PCA results (figure 3), which show there are significant concentration-dependent structural differences for a given protein adsorbed onto a given surface. However, the concentration dependence is more obvious in PCA since changes in the intensity of all amino acid fragments in the peak list are simultaneously considered. Concentration-dependent changes in adsorbed protein orientation—e.g., side on vs. end on—have been postulated elsewhere in the literature.35–36 While the trends in the amino acid ratios suggest such orientation changes, interpretation of these ratios is complicated by the simultaneous, concentration-dependent conformation changes (i.e., extent of denaturation) discussed above. Still, this analysis may be useful in guiding future molecular dynamics and other modeling studies.
Figure 5.
ToF-SIMS peak intensity ratios reveal changes in adsorbed protein structure for (A) BSA and (B) IgG adsorbed onto gold and NaSS surfaces. Insets show the crystal structure of each protein. In (A) Asn and Tyr are highlighted in yellow while His is highlighted in black. In (B) Ser is highlighted in yellow while Asp and Val are highlighted in black. Plotted data represent average amino acid ratios for proteins absorbed from 5 (blue), 25 (red), and 100 (green) μg/ml solutions onto Au (solid bars) or NaSS (open bars) surfaces. The peak ratios are the sum of intensities of secondary ions from amino acids with asymmetric distributions, highlighted in yellow and black in the inset protein. Error bars represent standard deviations.
D. PCA Modeling of Multi-component Plasma Films
PC models are useful for characterizing protein films adsorbed from complex mixtures, such as plasma.21 Wagner et al. constructed such a model from ToF-SIMS data from single-component fibronectin, Fgn, IgG, BSA, and γ-globulins films on mica surfaces. Spectra from bovine plasma films, adsorbed onto mica for times ranging from 5 – 120 minutes, were then projected into the model. These experiments were done to investigate time-dependent changes in adsorbed plasma films. For Wagner et al., at short adsorption times, the plasma films were more Fgn-like in character. But at longer times, the plasma films became more γ-globulins and IgG-like than Fgn.21
We replicated this procedure, constructing separate models from the single-component BSA, Fgn, and IgG ToF-SIMS data on both gold and NaSS surfaces. The amino acid peak list was used, and only two PCs were retained since retention of additional PCs had no effect on the results or conclusions of the models. Spectra from bovine plasma films, adsorbed for times ranging from 5 – 120 minutes, were then projected into the PC model for the corresponding surface. On gold, the variance in the data is dominated by differences between the single-component protein spectra, obscuring any time-dependent trends in the plasma data (Figure S10A in the supplemental information). On NaSS, only a weak trend is observed suggesting that the plasma film changes from being more BSA-like to more IgG and Fgn-like with increasing adsorption time (Figure S10C in the supplemental information).
We found that clearer trends could be obtained by inverting the procedure above—that is, constructing the PC models from the plasma spectra and projecting in the single-component protein data. That the two procedures are mathematically different is evident since they produce distinct scores and loadings plots. However, both allow qualitative observations to be made about time-dependent changes in complex, adsorbed plasma film composition. In particular, both procedures (using either signal-component data or plasma time-dependent data as the models) produce the same trends for NaSS surfaces: the plasma films are more BSA-like at short adsorption times, and more IgG and Fgn-like at longer adsorption times (Figure 6C). With the plasma time-dependent model, clear trends are also now observable for the gold surfaces: the plasma films are more IgG-like and BSA-like at short to intermediate times, becoming more Fgn-like at long times (Figure 6A). Therefore, gold and NaSS both apparently adsorb increasing amounts of fibrinogen with time. This is the opposite of what was observed by Wagner et al. on mica substrates.21 However, Fgn is the heaviest of the three proteins investigated in this study, and its increase with increasing adsorption time may be related to a slower rate of diffusion from bulk solution to the surface. As Fgn arrives at the surface, it displaces the smaller BSA and IgG proteins that were the initial proteins to arrive and adsorb onto gold and NaSS. This is known as the Vroman effect.25 Also, mica is a rigid, oxide surface; gold, a rigid, metallic surface; NaSS, a soft, polymeric surface. It is not surprising that the composition of adsorbed plasma films changes with different surface chemistries and structures. However, our PC models are overly simplistic since they contain only three proteins, whereas plasma is composed of hundreds of proteins.37 Therefore, while both gold and NaSS appear to adsorb more Fgn with time, further study with a more extensive set of proteins is required to confirm that this finding is truly representative of the final state of the adsorbed plasma films.
Figure 6.
PC models for gold (A and B) and NaSS (C and D) surfaces. The models were constructed using the amino acids peaks from 100 μg/ml plasma adsorption ToF-SIMS data. Single-component BSA (
 ), Fgn (
), Fgn (
 ), and IgG (
), and IgG (
 ) spectra were projected into the models. In panels (A) and (C) all black symbols represent the plasma adsorption data used to construct the model: + represent the 5 minute adsorption group, − represent the 30 minute adsorption group, × represent the 60 minute adsorption group, and + represent the 120 minute adsorption group. The ellipses in (A) and (C) represent 95% confidence intervals. Panel (A) shows that on gold: the plasma films are more like adsorbed IgG and BSA at short to intermediate times, and more like Fgn at long times. Panel (C) shows that on NaSS: the plasma films are more like BSA at short times, then more like IgG and Fgn at long times.
) spectra were projected into the models. In panels (A) and (C) all black symbols represent the plasma adsorption data used to construct the model: + represent the 5 minute adsorption group, − represent the 30 minute adsorption group, × represent the 60 minute adsorption group, and + represent the 120 minute adsorption group. The ellipses in (A) and (C) represent 95% confidence intervals. Panel (A) shows that on gold: the plasma films are more like adsorbed IgG and BSA at short to intermediate times, and more like Fgn at long times. Panel (C) shows that on NaSS: the plasma films are more like BSA at short times, then more like IgG and Fgn at long times.
IV. Conclusions
In this study, we investigated the adsorption of BSA, bovine Fgn, bovine IgG, and bovine plasma onto bare and NaSS-grafted gold surfaces using a multi-technique approach to obtain new insights to the structure of protein films formed on these surfaces. We measured adsorption isotherms, using XPS to follow changes in surface nitrogen composition with increasing protein solution concentration. Using a combination of ToF-SIMS, PCA, and VMD we investigated differences in adsorbed protein structure between both surfaces. We also used PC modeling to qualitatively describe changes in bovine plasma adsorbed films with time.
Our conclusions are as follows:
All proteins have a higher initial affinity for gold than NaSS surfaces. However, except for IgG, NaSS surfaces adsorb the same (for plasma) or more (for BSA and Fgn) total protein than gold surfaces from the 100 μg/ml solution concentration. This may be because any proteins that do adsorb to NaSS undergo structural rearrangements over time, increasing the number of contacts with the surface and resulting in a larger fraction of the surface covered by irreversibly adsorbed species. Nevertheless, with the exception of BSA and plasma adsorption onto gold, neither surface appeared to have completely saturated after two hours at the 100 μg/ml protein solution concentration.
PCA using just the amino acid peaks is able to distinguish between the same protein adsorbed onto gold or NaSS surfaces, and also between protein films adsorbed from different solution concentrations on the same surface. This suggests that proteins adsorb differently on NaSS and gold surfaces, and that the structure of the adsorbed proteins changes with surface concentration.
To assess the level of denaturation, the ratio of certain buried/surface amino acids was examined for each protein. For all three proteins, the buried/surface amino acid ratios are higher on NaSS than gold surfaces, particularly at low surface coverage and packing density. This suggests that proteins denature more on NaSS surfaces, and that denaturation decreases with increasing packing density.
To further investigate the impact of the different surface chemistries on adsorbed protein structure, a similar ratio analysis was performed using amino acids on opposite sides of each protein. For both BSA and IgG, these ratios are significantly different between the two surfaces at all adsorption solution concentrations. Due to the symmetry and even distribution of amino acids, we were unable to find an analogous ratio for Fgn.
The PC models suggest the following changes in the character of proteins films adsorbed from plasma at times ranging from 5–120 minutes. For both surfaces the plasma films become more Fgn-like at long times. However, our PC models only contain three proteins, whereas plasma is composed of hundreds of proteins. Therefore, while both gold and NaSS surfaces appear to adsorb more Fgn with time, further study is required to confirm that this is truly representative of the final state of the adsorbed plasma films.
Supplementary Material
Acknowledgments
This study was supported by NIH grant EB-002027 to NESAC/BIO. E.T.H. was also supported by NIH grant EB-014516. We thank Professor Veronique Migonney for stimulating discussions about the preparation and biological applications of grafted NaSS films.
ABBREVIATIONS
- BSA
bovine serum albumin
- Fgn
fibrinogen
- IgG
immunoglobulin G
- NaSS
sodium styrene sulfonate
- ARGET ATRP
Activators are continuously ReGenerated by Electron Transfer Atom Transfer Radical Polymerization
- XPS
X-ray photoelectron spectroscopy
- ToF-SIMS
time-of-flight secondary ion mass spectrometry
- PCA
principal component analysis
- PC
principal component
- VMD
visual molecular dynamics
- PBS
phosphate buffered saline
- DI
deionized
- TOA
takeoff angle
- PDMS
polydimethyl siloxane
- PDB
protein data bank
Footnotes
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
Supporting Information. Discussion and PCA results using full peak lists composed of all ToF-SIMS peaks with intensity 3x the background. Representative high-resolution C1s spectra are also shown. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bierbaum S, Hintze V, Scharnweber D. Functionalization of biomaterial surfaces using artificial extracellular matrices. Biomatter. 2012;2(3):132–141. doi: 10.4161/biom.20921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castner DG, Ratner BD. Biomedical surface science: Foundations to frontiers. Surface Science. 2002;500(1–3):28–60. [Google Scholar]
- 3.Fang F, Szleifer I. Kinetics and Thermodynamics of Protein Adsorption: A Generalized Molecular Theoretical Approach. Biophysical Journal. 2001;80(6):2568–2589. doi: 10.1016/S0006-3495(01)76228-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brash JL. Exploiting the current paradigm of blood–material interactions for the rational design of blood-compatible materials. Journal of Biomaterials Science, Polymer Edition. 2000;11(11):1135–1146. doi: 10.1163/156856200744237. [DOI] [PubMed] [Google Scholar]
- 5.Gray JJ. The interaction of proteins with solid surfaces. Current Opinion in Structural Biology. 2004;14(1):110–115. doi: 10.1016/j.sbi.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 6.Haynes CA, Norde W. Globular proteins at solid/liquid interfaces. Colloids and Surfaces B: Biointerfaces. 1994;2(6):517–566. [Google Scholar]
- 7.Bittrich E, Burkert S, Eichhorn K-J, Stamm M, Uhlmann P. Control of Protein Adsorption and Cell Adhesion by Mixed Polymer Brushes Made by the “Grafting-To” Approach. In: Horbett TA, Brash JL, Norde W, editors. Proteins at Interfaces III: State of the Art. Vol. 1120. American Chemical Society; Washington DC: 2012. pp. 179–193. [Google Scholar]
- 8.Fang F, Satulovsky J, Szleifer I. Kinetics of Protein Adsorption and Desorption on Surfaces with Grafted Polymers. Biophysical Journal. 2005;89(3):1516–1533. doi: 10.1529/biophysj.104.055079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matyjaszewski K, Dong H, Jakubowski W, Pietrasik J, Kusumo A. Grafting from Surfaces for “Everyone”: ARGET ATRP in the Presence of Air. Langmuir. 2007;23(8):4528–4531. doi: 10.1021/la063402e. [DOI] [PubMed] [Google Scholar]
- 10.Matyjaszewski K. Atom Transfer Radical Polymerization (ATRP): Current Status and Future Perspectives. Macromolecules. 2012;45(10):4015–4039. [Google Scholar]
- 11.Pavon-Djavid G, Gamble LJ, Ciobanu M, Gueguen V, Castner DG, Migonney V. Bioactive Poly(ethylene terephthalate) Fibers and Fabrics: Grafting, Chemical Characterization, and Biological Assessment. Biomacromolecules. 2007;8(11):3317–3325. doi: 10.1021/bm070344i. [DOI] [PubMed] [Google Scholar]
- 12.Helary G, Noirclere F, Mayingi J, Migonney V. A new approach to graft bioactive polymer on titanium implants: Improvement of MG 63 cell differentiation onto this coating. Acta Biomaterialia. 2009;5(1):124–133. doi: 10.1016/j.actbio.2008.07.037. [DOI] [PubMed] [Google Scholar]
- 13.Helary G, Noirclere F, Mayingi J, Bacroix B, Migonney V. A bioactive polymer grafted on titanium oxide layer obtained by electrochemical oxidation. Improvement of cell response. Journal of Materials Science: Materials in Medicine. 2010;21(2):655–663. doi: 10.1007/s10856-009-3892-0. [DOI] [PubMed] [Google Scholar]
- 14.Vaquette C, Viateau V, Guérard S, Anagnostou F, Manassero M, Castner DG, Migonney V. The effect of polystyrene sodium sulfonate grafting on polyethylene terephthalate artificial ligaments on in vitro mineralisation and in vivo bone tissue integration. Biomaterials. 2013;34(29):7048–7063. doi: 10.1016/j.biomaterials.2013.05.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou J, Manassero M, Migonney V, Viateau V. Evaluation clinique et biologique d’un ligament synthetique bioactif chez la brebis. IRBM. 2009;30(4):153–155. [Google Scholar]
- 16.Kerner S, Migonney V, Pavon-Djavid G, Helary G, Sedel L, Anagnostou F. Bone tissue response to titanium implant surfaces modified with carboxylate and sulfonate groups. Journal of Materials Science: Materials in Medicine. 2010;21(2):707–715. doi: 10.1007/s10856-009-3928-5. [DOI] [PubMed] [Google Scholar]
- 17.Felgueiras HP, Sommerfeld SD, Murthy NS, Kohn J, Migonney V. Poly(NaSS) Functionalization Modulates the Conformation of Fibronectin and Collagen Type I To Enhance Osteoblastic Cell Attachment onto Ti6Al4V. Langmuir. 2014;30(31):9477–9483. doi: 10.1021/la501862f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Foster RN, Keefe AJ, Jiang S, Castner DG. Surface initiated atom transfer radical polymerization grafting of sodium styrene sulfonate from titanium and silicon substrates. Journal of Vacuum Science & Technology A: Vacuum, Surfaces, and Films. 2013;31(6):06F103–9. doi: 10.1116/1.4819833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Foster RN, Johansson PK, Tom NR, Koelsch P, Castner DG. Experimental design and analysis of activators regenerated by electron transfer-atom transfer radical polymerization experimental conditions for grafting sodium styrene sulfonate from titanium substrates. Journal of Vacuum Science & Technology A: Vacuum, Surfaces, and Films. 2015;33(5):05E131. doi: 10.1116/1.4929506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry M, Dupont-Gillain C, Bertrand P. Conformation change of albumin adsorbed on polycarbonate membranes as revealed by ToF-SIMS. Langmuir. 2003;19(15):6271–6276. [Google Scholar]
- 21.Wagner MS, Castner DG. Characterization of Adsorbed Protein Films by Time-of-Flight Secondary Ion Mass Spectrometry with Principal Component Analysis. Langmuir. 2001;17(15):4649–4660. doi: 10.1002/1097-4636(20011205)57:3<432::aid-jbm1186>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 22.Graham DJ, Castner DG. Multivariate Analysis of ToF-SIMS Data from Multicomponent Systems: The Why, When, and How. Biointerphases. 2012;7(1–4):1–12. doi: 10.1007/s13758-012-0049-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haynes CA, Sliwinsky E, Norde W. Structural and Electrostatic Properties of Globular Proteins at a Polystyrene-Water Interface. Journal of Colloid and Interface Science. 1994;164(2):394–409. [Google Scholar]
- 24.Latour RA. The langmuir isotherm: A commonly applied but misleading approach for the analysis of protein adsorption behavior. Journal of Biomedical Materials Research Part A. 2015;103(3):949–958. doi: 10.1002/jbm.a.35235. [DOI] [PubMed] [Google Scholar]
- 25.Norde W, Thomas AH, John LB. Proteins at Interfaces III State of the Art. Vol. 1120. American Chemical Society; 2012. Proteins at Interfaces III: Introductory Overview; pp. 1–34. [Google Scholar]
- 26.Tidwell CD, Castner DG, Golledge SL, Ratner BD, Meyer K, Hagenhoff B, Benninghoven A. Static time-of-flight secondary ion mass spectrometry and x-ray photoelectron spectroscopy characterization of adsorbed albumin and fibronectin films. Surface and Interface Analysis. 2001;31(8):724–733. [Google Scholar]
- 27.Wagner MS, Horbett TA, Castner DG. Characterization of the Structure of Binary and Ternary Adsorbed Protein Films Using Electron Spectroscopy for Chemical Analysis, Time-of-Flight Secondary Ion Mass Spectrometry, and Radiolabeling. Langmuir. 2003;19(5):1708–1715. doi: 10.1016/s0142-9612(02)00612-9. [DOI] [PubMed] [Google Scholar]
- 28.Hull JR, Tamura GS, Castner DG. Structure and Reactivity of Adsorbed Fibronectin Films on Mica. Biophysical Journal. 2007;93:2852–2860. doi: 10.1529/biophysj.107.109819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. [accessed 2015–08–01];Atomic Coordinate Files for Whole IgG1. https://http://www.umass.edu/microbio/rasmol/padlan.htm.
- 30.Baugh L, Weidner T, Baio JE, Nguyen PCT, Gamble LJ, Stayton PS, Castner DG. Probing the Orientation of Surface-Immobilized Protein G B1 using ToF-SIMS, Sum Frequency Generation, and NEXAFS Spectroscopy. Langmuir. 2010;26:16434–16441. doi: 10.1021/la1007389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kochwa S, Litwak RS, Rosenfield RE, Leonard EF. BLOOD ELEMENTS AT FOREIGN SURFACES: A BIOCHEMICAL APPROACH TO THE STUDY OF THE ADSORPTION OF PLASMA PROTEINS*. Annals of the New York Academy of Sciences. 1977;283(1):37–49. [Google Scholar]
- 32.Norde W, Favier JP. Structure of adsorbed and desorbed proteins. Colloids and Surfaces. 1992;64(1):87–93. [Google Scholar]
- 33.Xia N, May CJ, McArthur SL, Castner DG. Time-of-Flight Secondary Ion Mass Spectrometry Analysis of Conformational Changes in Adsorbed Protein Films. Langmuir. 2002;18(10):4090–4097. [Google Scholar]
- 34.Kim YP, Hong MY, Kim J, Oh E, Shon HK, Moon DW, Kim HS, Lee TG. Quantitative analysis of surface-immobilized protein by TOF-SIMS: Effects of protein orientation and trehalose additive. Analytical Chemistry. 2007;79(4):1377–1385. doi: 10.1021/ac0616005. [DOI] [PubMed] [Google Scholar]
- 35.Lhoest JB, Detrait E, van den Bosch de Aguilar P, Bertrand P. Fibronectin adsorption, conformation, and orientation on polystyrene substrates studied by radiolabeling, XPS, and ToF SIMS. Journal of Biomedical Materials Research. 1998;41(1):95–103. doi: 10.1002/(sici)1097-4636(199807)41:1<95::aid-jbm12>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 36.Koutsoukos PG, Mumme-Young CA, Norde W, Lyklema J. Effect of the nature of the substrate on the adsorption of human plasma albumin. Colloids and Surfaces. 1982;5(2):93–104. [Google Scholar]
- 37.Andrade JD, Hlady V. Plasma Protein Adsorption: The Big Twelve. Annals of the New York Academy of Sciences. 1987;516(1):158–172. doi: 10.1111/j.1749-6632.1987.tb33038.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.