Abstract
Objectives:
To assess messenger RNA (mRNA) expression of POU2AF1 and Spi-B and their potential regulatory microRNAs (miRNAs) in natalizumab-treated patients with multiple sclerosis and in therapy-associated progressive multifocal leukoencephalopathy (PML).
Methods:
Expression of POU2AF1/Spi-B was analyzed by using real-time reverse transcription PCR assays on isolated B/CD8+ T lymphocytes and peripheral blood mononuclear cells (PBMCs) from cohorts of untreated and natalizumab-treated patients with and without PML. Longitudinal expression analysis was performed on CD4+, CD8+ T and B cells from 14 patients who interrupted natalizumab therapy for 8 weeks. The miRNA profiling was conducted in PBMCs from 5 untreated and 5 natalizumab-treated patients using low-density arrays followed by validation with single miRNAs assays in untreated and natalizumab-treated patients.
Results:
POU2AF1 and Spi-B mRNAs were upregulated in B and CD8+ T cells from natalizumab-treated patients, which was validated in PBMCs from different cohorts of natalizumab-treated patients with and without PML, with a noteworthy higher expression of Spi-B in patients with PML. In contrast, downregulation of POU2AF1/Spi-B expression was measured in B and CD8+ T cells after natalizumab discontinuation. Seventeen differentially expressed miRNAs including miR-10b, a regulator of POU2AF1 mRNA, were identified in long-term natalizumab-treated patients compared with untreated ones.
Conclusions:
Upregulation of POU2AF1 and Spi-B, known transactivators of the JC virus, the causative agent for PML, and its association with occurrence of PML in natalizumab-treated patients, corroborates POU2AF1/Spi-B as potential biomarkers for PML risk, which merits further evaluation.
Multiple sclerosis (MS) is a chronic, disabling autoimmune disorder of the CNS characterized by an inflammation-mediated demyelination leading to axonal loss and neuronal damage.1 Among the diverse disease-modifying therapies currently available for the treatment of relapsing-remitting MS (RRMS), natalizumab is regarded as one of the most effective drugs that reduces annualized relapse rates and disease activity.2,3
A relevant side effect of natalizumab treatment is the development of progressive multifocal leukoencephalopathy (PML), a severe opportunistic infection of the CNS caused by reactivation of the latent JC virus (JCV).4 JCV seropositivity, increased treatment duration, and a history of immunosuppressive therapies are defined risk factors that are commonly used for guiding therapeutic strategies.5–7 Additional predictive markers for individual PML risk assessment including JCV-AI8 and immunologic biomarkers such as CD62L9 or circulating JCV-specific activated effector memory T cells10 have been proposed.11 Also, specific microRNA (miRNA) expression profiles have been suggested as possible biomarkers for PML risk.12
The miRNAs are short noncoding RNA molecules that regulate gene expression at the posttranscriptional level.13 In a previous study performed on CD4+ T cells,14 we uncovered an effect of natalizumab on the expression of miR-126 and its potential target POU2AF1,15 a critical regulator of Spi-B,16 which binds unique sequences of JCV and drives virus activity.17–19 Here, we extend our investigations on expression of POU2AF1/Spi-B and potential regulating miRNAs in various lymphocyte subpopulations during natalizumab treatment and in therapy-associated PML.
METHODS
Participants.
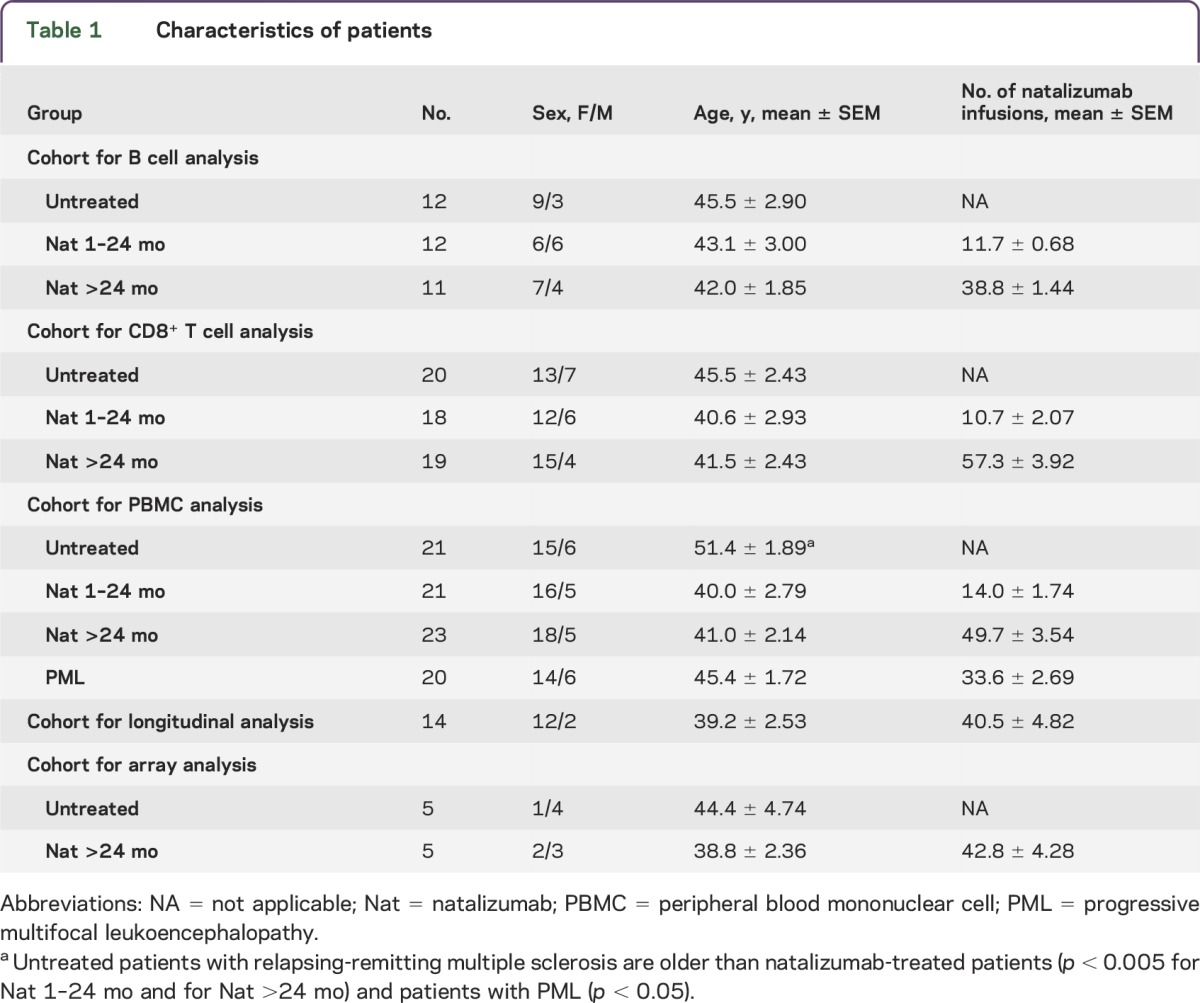
Five different cohorts were used for the study (table 1). The blood samples were collected during routine visits of the study patients, years 2010–2014 and years 2008–2012 (PML cases). For B cell analysis, 12 untreated and 23 natalizumab-treated (n = 12 treated up to 2 years, and n = 11 treated longer than 2 years) patients with RRMS were included. For CD8+ T cell analysis, 20 untreated and 37 natalizumab-treated (n = 18 treated up to 2 years, and n = 19 treated longer than 2 years) patients with RRMS were included. For peripheral blood mononuclear cell (PBMC) analysis, 21 untreated and a total of 44 natalizumab-treated (n = 21 treated up to 24 months and n = 23 treated longer than 24 months) patients with RRMS were included. A group of 20 natalizumab-treated patients who developed PML was also included in this cohort. The JCV serostatus was available from almost all (62/64) natalizumab-treated patients of the PBMC cohort. Patients with PML were all JCV seropositive (20/20); 10 short-term–treated patients without PML (1–24 months, 10/21) and 10 long-term–treated patients without PML (>24 months, 10/23) were JCV seropositive. In 14 patients who discontinued natalizumab therapy, PBMCs were available from their last day of natalizumab infusion (baseline) and after an 8-week washout period. An additional cohort of 5 untreated and 5 long-term natalizumab-treated patients with RRMS was used for miRNA profiling. No untreated patients had immunomodulatory or other MS-specific treatments in the 6 months before or during the study. Patient characteristics are presented in table 1.
Table 1.
Characteristics of patients
Standard protocol approvals, registrations, and patient consents.
Written informed consent was obtained from all patients. The study was approved by the Cantonal Institutional Review Board of Basel City and Basel Country.
Cell separation.
For PBMC isolation and CD4+ T/CD8+ T/B cell subset separations, we used the same methodologies as the ones employed in our previous reports.14,20–22 Briefly, PBMCs were isolated by density gradient centrifugation (Lymphoprep; Axon Lab, Baden-Dättwil, Switzerland). CD4+ T/CD8+ T and B cell subpopulations were separated from PBMCs using MACS technology (CD4 and CD8 MicroBeads, human, B cell negative enrichment kit II; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany) according to manufacturer's protocol. Purity of isolated CD4+ T, CD8+ T, and B cells was analyzed with Attune Focusing Flow Cytometer (Applied Biosystems, Darmstadt, Germany).
RNA isolation.
PBMCs and isolated cell subpopulations were lysed in QIAzol (QIAGEN AG, Hombrechtikon, Switzerland). Total RNA (including miRNA) was extracted using miRNeasy Mini Kit (QIAGEN) according to manufacturer's instructions.
miRNA profiling.
The TaqMan Human MicroRNA Panel A (Life Technologies, Zug, Switzerland), which contains 377 assays and 4 controls, was used for miRNA profiling in PBMCs from untreated and long-term–treated patients with RRMS. Expression suite (Life Technologies), Partek Genomic Suite (Partek, St. Louis, MO), and Ingenuity Pathway Analysis (QIAGEN) software were used for expression and pathway analysis.
miRNA/messenger RNA expression analysis.
Analyses of miRNA/messenger RNA (mRNA) expression were assessed as previously described.14,21 Briefly, for miRNA studies, single miRNA assays for real-time reverse transcription–PCR (RT-PCR) (Applied Biosystems, Zug, Switzerland) were used. Megaplex Primer Pool A v2.1 (Applied Biosystems) was used for RT-PCR according to manufacturer's recommendations. Expressions of POU2AF1 and Spi-B were analyzed with quantitative real-time RT-PCR by using Assay-On-Demand reagents (Applied Biosystems). As previously validated,20 RNU44 miRNA and PUM1 mRNA were used as references for normalization and relative expression analyses. The comparative cycle threshold method (Applied Biosystems) was used for calculations of relative quantitation of targets. Quantitative PCR probe sequences for miRNAs and mRNAs are presented in table e-1 at Neurology.org/nn.
Target prediction.
The microRNA.org (http://www.microrna.org/) and TargetScan (http://www.targetscan.org/) sources were used to search putative targets of various miRNAs. In addition, the microRNA target filtering tool of Ingenuity Pathway Analysis software was applied.
Data and statistical analysis.
Statistical analyses were performed on GraphPad Prism software (La Jolla, CA). Normality of the datasets was tested with the D'Agostino-Pearson omnibus normality test. For the various group comparisons, nonparametric Kruskal–Wallis test with the Dunn multiple comparison test was applied. A p value <0.05 was considered significant.
RESULTS
Natalizumab-induced upregulation of POU2AF1 and Spi-B expression in B and CD8+ T cells.
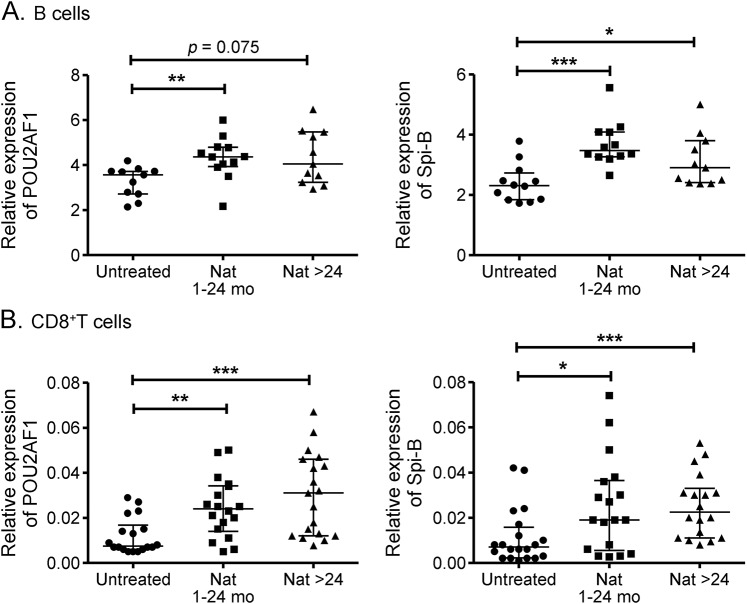
Our previous analysis of gene expression in CD4+ T cells revealed upregulation of POU2AF1 and Spi-B in natalizumab-treated patients.14 In the current study using different cohorts of patients, we examined POU2AF1 and Spi-B expression in other lymphocyte subsets, namely, B and CD8+ T cells. POU2AF1 and Spi-B relative expressions were upregulated in B cells upon natalizumab treatment (figure 1A) (***p < 0.001, **p < 0.01, *p < 0.05). A slight, but nonsignificant, decrease in the expression of Spi-B over time in B cells in natalizumab-treated patients was found. Similar upregulation of both targets was found in CD8+ T cells from both groups (Nat 1–24 and Nat >24) of natalizumab-treated compared to untreated patients (figure 1B).
Figure 1. Differential expression of POU2AF1 and Spi-B messenger RNA in B and CD8+ T cells upon natalizumab treatment.
Transcriptional expression of POU2AF1 (A and B, left panels) and Spi-B (A and B, right panels) were analyzed with real-time reverse transcription PCR in B cells (A) and CD8+ T cells (B) from untreated patients with relapsing-remitting multiple sclerosis and those who received natalizumab treatment for either less than 24 months or more than 24 months. Relative expression levels (median with interquartile range) are depicted. ***p < 0.001, **p < 0.01, *p < 0.05. Nat = natalizumab.
Differential expression of POU2AF1 and Spi-B in PBMCs from untreated and natalizumab-treated patients with and without PML.
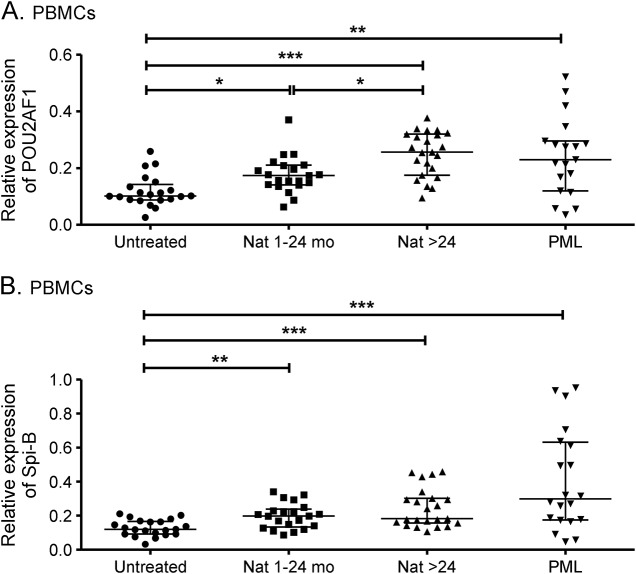
We further expanded our investigation on POU2AF1 and Spi-B expression to samples from patients with natalizumab-associated PML. The analysis was performed on PBMCs because of the limitation of sample volume. A total of 20 patients with PML whose natalizumab treatment duration varied from 17 to 62 months were included in the study. We divided our cohort of non-PML, natalizumab-treated patients into 2 groups, namely, treatment duration of 1–24 months and longer than 24 months. In accordance with the results obtained on the different lymphocyte subsets (current study and reference 14), POU2AF1 and Spi-B were upregulated upon natalizumab treatment in PBMCs from both groups of patients without PML (***p < 0.001, **p < 0.01, *p < 0.05) (figure 2). Of note, higher expression of POU2AF1 was detected in patients treated longer than 24 months with natalizumab (*p < 0.05) (figure 2). Similar expression of POU2AF1 was found in patients with and without PML treated with long-term natalizumab. It is important to note that high levels of Spi-B expression were found in patients with PML (figure 2B). In addition, no difference was found between patients with PML treated short term (1–24 months) and long term (>25 months).
Figure 2. Differential expression of POU2AF1 and Spi-B mRNA in PBMCs upon natalizumab treatment and PML.
Transcriptional expression of POU2AF1 (A) and Spi-B (B) were analyzed with real-time reverse transcription PCR in PBMCs from untreated patients, natalizumab-treated patients with relapsing-remitting multiple sclerosis (1–24 mo, >24 mo), and patients with natalizumab-associated PML. Relative expression levels (median with interquartile range) are depicted. ***p < 0.001, **p < 0.01, *p < 0.05. Nat = natalizumab; PBMC = peripheral blood mononuclear cell; PML = progressive multifocal leukoencephalopathy.
Downregulated POU2AF1 and Spi-B expression upon natalizumab discontinuation.
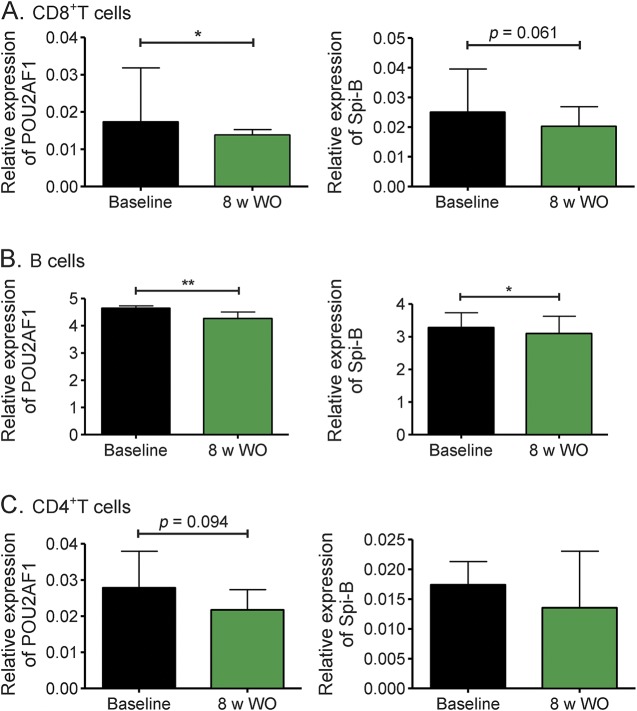
To further validate the effect of natalizumab on POU2AF1 and Spi-B expression, we analyzed relative expression levels of both genes in longitudinal samples from patients who stopped natalizumab treatment, at their last infusion day (baseline) and after 8 weeks' discontinuation (figure 3). POU2AF1 and Spi-B expression decreased in CD8+ T (figure 3A) and B (figure 3B) cells, whereas no change was measured in CD4+ T cells (figure 3C).
Figure 3. Effect of natalizumab discontinuation on expression of POU2AF1 and Spi-B messenger RNA.
Transcriptional expression of POU2AF1 and Spi-B were longitudinally analyzed with real-time reverse transcription PCR in CD8+ T cells (A), B cells (B), and CD4+ T cells (C) from 14 patients at the time of last infusion and 8 weeks after discontinuation of natalizumab therapy. Relative expression levels are depicted. **p < 0.01, *p < 0.05. 8 w WO = 8 weeks without natalizumab.
Differential miR-126 expression in PBMCs upon natalizumab treatment and PML.
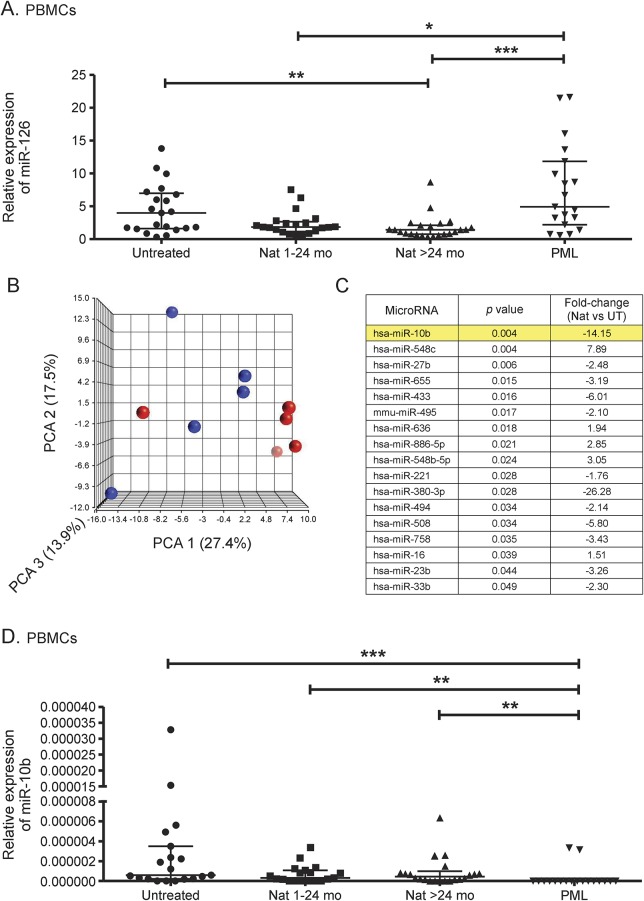
A miR-126 expression analysis of peripheral blood lymphocytes in natalizumab-treated vs untreated patients with RRMS revealed downregulation of miRNA-126 expression in CD4+ T cells.14 In the current study, we confirm downregulation of miR-126 expression in PBMCs from natalizumab-treated compared to untreated patients (**p < 0.01). Of note, miR-126 expression was reversely upregulated in patients under PML disease (***p < 0.001, *p < 0.05) (figure 4A).
Figure 4. Differential miRNA expression profile during natalizumab treatment and PML.
Transcriptional expression of miR-126 was analyzed with real-time reverse transcription PCR in PBMCs from untreated patients, natalizumab-treated patients with RRMS (1–24 mo, >24 mo), and patients with natalizumab-associated PML (A). Low-density array analysis of transcriptional miRNA expression in PBMCs from untreated patients with RRMS (blue circles) compared with patients treated with long-term natalizumab (red circles) is illustrated as 3-dimensional PCA score plot of miRNA expression data (B). List of differentially expressed miRNAs in PBMCs analyzed with low-density arrays. The predicted regulator of POU2AF1, hsa-miR-10b, which was validated with other cohorts, is highlighted in yellow (C). Transcriptional expression of miR-10b was analyzed with real-time reverse transcription PCR in PBMCs from untreated patients, natalizumab-treated patients with RRMS (1–24 mo, >24 mo), and patients with natalizumab-associated PML (D). Relative expression levels (median with interquartile range) are depicted. ***p < 0.001, **p < 0.01, *p < 0.05. miRNA = microRNA; Nat = natalizumab; PBMC = peripheral blood mononuclear cell; PCA = principal component analysis; PML = progressive multifocal leukoencephalopathy; RRMS = relapsing-remitting multiple sclerosis; UT = untreated.
miRNA profiling in PBMCs from long-term natalizumab-treated vs untreated patients with RRMS.
The principal component analysis was applied to assess the quality of the samples used for miRNA profiling with low-density arrays. Uniformity of samples and datasets was confirmed (figure 4B); no outliers were identified. Transcriptional expression analysis of 377 miRNAs revealed a set of 17 differentially expressed miRNAs in long-term natalizumab-treated vs untreated patients (figure 4C). The miR-126 expression was also downregulated in patients under natalizumab treatment (1.3-fold); however, statistical significance was not reached in these datasets (data not shown). Of interest, one of the most downregulated miRNAs was miR-10b, which has a predicted targeting site in POU2AF1 sequence (microRNA.org comprehensive resource). The validation of miR-10b expression in PBMCs with different cohorts of untreated and natalizumab-treated patients revealed concordant results with low-density array data, with a trend for downregulation in treated patients (figure 4D). Expression levels of miR-10b were relatively low or even undetectable in some samples from all groups. Of note, expression of miR-10b was detectable in only 2 of 20 patients with PML, suggesting a decreased expression in PBMCs from patients with PML compared to untreated and natalizumab-treated patients without PML (figure 4D). Pathway analysis of putative miR-10b targets revealed several potential affected signaling pathways, including, e.g., “virus entry via endocytic pathways” and “NF-κB activation by viruses” (figure e-1).
DISCUSSION
Besides its appearance in patients with HIV, PML has been associated with the use of immunomodulatory therapies for the treatment of autoimmune diseases.23 In MS, incidence of PML in patients treated with natalizumab, an α4β1, 7 integrin blocking monoclonal antibody,24,25 has raised an urgent need to identify potential biomarkers for a PML risk assessment.
In a previous study conducted on CD4+ T cells,14 we showed that natalizumab induces upregulation of POU2AF1, also known as OBF-1/OCA-B/BOB.1, an originally described B cell–specific transcriptional coactivator that stimulates immunoglobulin promoter activity through its interaction with the octamer-binding transcription factors Oct-1 and Oct-2.26 Inducible POU2AF1 expression and phosphorylation has also been reported to be in T cells,27 where it was further shown to regulate T helper cell function.28 In the present study, we expanded our analysis of POU2AF1 mRNA expression to B and CD8+ T cells and also found upregulation of POU2AF1 expression in both lymphocyte subsets upon natalizumab treatment. To note, in our previous study, higher expression of POU2AF1 was found in CD4+ T cells from patients treated longer than 2 years, whereas in the present study, no treatment duration effect on the upregulation of POU2AF1 was found in B and CD8+ T cells.
In the context of PML research, these are important findings regarding the regulatory function of POU2AF1 on the transcription factor Spi-B,16 which is involved in early JCV gene expression and has a critical role in JCV activity.17,18 Spi-B is a transcription factor of the Ets family that is reported to be mainly expressed in B and T lymphocytes and is essential for B cell function and T cell–dependent humoral immune responses.29 In the present study, Spi-B mRNA expression was detected in PBMCs and isolated CD4+ T, CD8+ T, and B cells. Of note, as previously reported in CD4+ T cells14 and in line with measured natalizumab-induced POU2AF1 upregulation, Spi-B expression was found to be upregulated in both B and CD8+ T cell subsets upon treatment. These results are in accordance with a recent study19 that showed that Spi-B gene expression is upregulated in response to long-term treatment with natalizumab in CD34+ hematopoietic precursors but also in CD19+ B cells.
A specific effect of natalizumab on expression of POU2AF1 and Spi-B mRNAs was further confirmed by longitudinal expression analysis in CD8+ T and B cells from patients discontinuing treatment for 8 weeks. However, in contrast to the target downregulation observed in these cell subsets, CD4+ T cells displayed rather similar expression levels of POU2AF1 and Spi-B after therapy cessation, suggesting that a longer washout period is likely needed to remove the treatment effect. Of note, several PML cases after natalizumab withdrawal were recently reported.30
While it is commonly accepted that cells from the immune system have a critical role in the clearance of JCV infection, the mechanism by which immune cells could also participate in the pathogenesis of PML remains a matter of debate.31 So far, there is no evidence that JCV productively infects and replicates in lymphocytes. However, several lines of evidence suggest a role for B cells as a reservoir carrying JCV through the blood–brain barrier.32,33 POU2AF1-mediated increased Spi-B expression in B cells may therefore contribute to JCV infection of the CNS.
Regarding CD8+ T cells, a link between better prognosis of PML and the presence of JCV-specific cytotoxic CD8+ T cell response was reported.34,35 However, one cannot rule out the hypothesis of a contribution of CD8+ T cells to PML development. A direct transport of the JCV by CD8+ T cells is unlikely since the virus is reported to bind to primary human glial cells, tonsillar stromal cells, and B lymphocytes, but not to T cells.36 However, as suggested in our previous study on CD4+ T cells,14 transfer of genetic material, e.g., mRNAs, can occur between cells via small microvesicles also known as exosomes.37,38 Exosomal Spi-B mRNA, derived from CD8+ T cells, could therefore be transferred into recipient cells, e.g., CD4+ T or B lymphocytes, where it would promote JCV activity. Natalizumab treatment has been shown to induce a reduction in the CSF CD4+/CD8+ T cell ratio,39 suggesting a prominent incidence of CD8+ T cells over CD4+ T cells.
Compared to isolated cell subsets, whole blood or PBMCs would be an easier source for biomarker research discovery, diagnosis, and disease monitoring. We already reported upregulation of POU2AF1 and Spi-B in our previous longitudinal gene expression profile analysis conducted in peripheral blood from patients with MS treated long term with natalizumab.40 In the current study, we confirmed upregulation of both genes in PBMCs from natalizumab-treated patients. Noteworthy, higher expression of Spi-B mRNA was detected in samples from patients with PML, therefore strengthening the potential use of Spi-B as a biomarker of PML risk. Furthermore, the differential expression of Spi-B in our PBMC cohorts of patients with and without PML was concordant with the JCV serostatus, a known risk factor for PML. Extended investigations on longitudinal samples from patients before PML diagnosis and at PML are needed to evaluate Spi-B as a predictive marker for PML.
The naturally following question that arises is the mechanism by which natalizumab effects expression of POU2AF1 and Spi-B. In our previous study performed on CD4+ T cells,14 we identified miR-126 as a potential POU2AF1 regulator whose expression was downregulated under natalizumab therapy. In the present study, we verified miR-126 downregulation in PBMCs from natalizumab-treated patients. However, miR-126 expression was upregulated in PBMCs from patients with PML, probably reflecting major deregulated mechanisms occurring under the disease process. Furthermore, no change of miR-126 expression was detected in CD8+ T and B cells (data not shown), suggesting a different mechanism of target regulation in these cells. Also, analysis of miR-126 expression in NK (natural killer) cells and monocytes could be informative.
By further performing a miRNA expression profiling in PBMCs from long-term natalizumab-treated vs untreated patients with RRMS, we uncovered and then validated differential expression of miR-10b, a potential regulator of POU2AF1 expression. Of note, in contrast to miR-126 expression, miR-10b remained downregulated in samples from patients with PML, suggesting its sustained regulatory effect on expression of POU2AF1 and subsequently Spi-B in these cells. Canonical pathway analysis of all putative target mRNAs of miR-10b indicated several affected biological processes, involving viral contributions, further supporting the importance of gene deregulation by natalizumab in PML disease pathophysiology. Additional studies are needed to evaluate whether these pathways are activated in patients with PML and thus confirm the potential link with strong downregulation of miR-10b. Longitudinal pre- and onset-PML samples of the same individuals are also needed to validate the significance of the expression of miR-10b as a predictive marker for PML risk. Another study suggested the differential expression of miR-320, -320b, and -629 as predictive markers already at 12 months for a PML risk in 2 patients, who developed PML after 28 and 49 infusions.12
Taken together, we have shown that natalizumab treatment has an effect on expression of POU2AF1 and Spi-B and regulating miRNAs, miR-126, and miR-10b in various immune cells of patients with MS. In particular, our results revealed a sustained pattern of gene expression, which is treatment duration dependent and more pronounced in patients treated long term (>2 years). Furthermore, these gene expression changes are consistent in patients with PML. The pathway analysis revealed complex physiologic processes, involving also activated viral functions, suggesting that natalizumab has various effects on cellular and viral activities. Our results provide more insights into the development and pathophysiology of PML. More studies on longitudinal “pre-” and post-PML samples are needed to confirm the link between deregulated genes and activated viral pathways and to evaluate them as potential predictive markers for PML.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Mrs. Marguerite Limberg, Mrs. Hedwig Wariwoda, and Mrs. Heidi Bodmer for excellent technical assistance.
GLOSSARY
- JCV
JC virus
- miRNA
microRNA
- mRNA
messenger RNA
- MS
multiple sclerosis
- PBMC
peripheral blood mononuclear cell
- PML
progressive multifocal leukoencephalopathy
- RRMS
relapsing-remitting multiple sclerosis
- RT-PCR
reverse transcription–PCR
Footnotes
Supplemental data at Neurology.org/nn
AUTHOR CONTRIBUTIONS
Dr. M. Meira designed the study, performed experimental work, and wrote and edited the manuscript. Dr. C. Sievers and Mrs. F. Hoffmann contributed in performing experimental work. A. Haghikia, Dr. M. Rasenack, Dr. B. F. Décard, Dr. J. Kuhle, Prof. T. Derfuss, and Prof. L. Kappos critically revised the manuscript and contributed to sample collection/management. Prof. R. L. P. Lindberg designed and supervised the study, collected and managed samples, performed statistical analyses, and wrote and edited the manuscript.
STUDY FUNDING
The study was supported by the Swiss National Science Foundation (SNSF) (grant 310030_132644) and the Swiss Multiple Sclerosis Society (SMSG).
DISCLOSURE
M. Meira, C. Sievers, F. Hoffmann, A. Haghikia, and M. Rasenack report no disclosures. B. Décard's institution received advisory board and/or speaker fees from Biogen, Teva, and Novartis; he received travel support from Biogen. J. Kuhle's institution received speaker honoraria from the Swiss MS Society, Biogen, Novartis, Roche, and Genzyme, travel expenses from Merck Serono and Novartis, research support from Bayer (Schweiz) AG, Genzyme, Novartis, Swiss National Research Foundation, ECTRIMS Research Fellowship Programme, University of Basel, and Swiss MS Society. T. Derfuss served on the scientific advisory board for Biogen Idec, Novartis Pharma, Genzyme, Merck Serono, Bayer Schering, Octapharma, GeNeuro, and Roche, received travel and speaker honoraria from Bayer Schering, Biogen Idec, Merck Serono, Novartis Pharma, and Genzyme, is on the editorial board of PLoS One, his spouse is employed at Novartis Pharma, he is a member of the steering committee for Mitsubishi Pharma and GeNeuro, is on the speakers bureaus for Biogen Idec, Novartis Pharma, and Merck Serono, is a member of the executive board of ECTRIMS, received research support from Novartis Pharma, Merck Serono, Biogen Idec, Swiss National Foundation, and Swiss MS Society. L. Kappos is on the editorial board for Multiple Sclerosis Journal, Multiple Sclerosis and Related Disorders, and Journal of Neurology, received research support from Actelion, Alkermes, Almirall, Bayer, Biogen Idec, Excemed, GeNeuro SA, Genzyme, Merck, Mitsubishi, Novartis, Receptos, Pfizer, Roche, Sanofi-Aventis, Santhera, Teva, and UCB (all payments have been transferred to the research account of the university hospital), received research support from Swiss National Research Foundation, European Union, Swiss MS Society, Novartis, and Roche Research Foundations. R. L. P. Lindberg received research support from Swiss Multiple Sclerosis Society, Swiss National Science Foundation, and Roche Postdoctoral Fellowship (RPF) program. Go to Neurology.org/nn for full disclosure forms.
REFERENCES
- 1.Compston A, Coles A. Multiple sclerosis. Lancet 2008;372:1502–1517. [DOI] [PubMed] [Google Scholar]
- 2.Kappos L, O'Connor PW, Polman CH, et al. Clinical effects of natalizumab on multiple sclerosis appear early in treatment course. J Neurol 2013;260:1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinstock-Guttman B, Galetta SL, Giovannoni G, et al. Additional efficacy endpoints from pivotal natalizumab trials in relapsing-remitting MS. J Neurol 2012;259:898–905. [DOI] [PubMed] [Google Scholar]
- 4.Carruthers RL, Berger J. Progressive multifocal leukoencephalopathy and JC virus-related disease in modern neurology practice. Mult Scler Relat Disord 2014;3:419–430. [DOI] [PubMed] [Google Scholar]
- 5.Bloomgren G, Richman S, Hotermans C, et al. Risk of natalizumab-associated progressive multifocal leukoencephalopathy. N Engl J Med 2012;366:1870–1880. [DOI] [PubMed] [Google Scholar]
- 6.Tyler KL. Progressive multifocal leukoencephalopathy: can we reduce risk in patients receiving biological immunomodulatory therapies? Ann Neurol 2010;68:271–274. [DOI] [PubMed] [Google Scholar]
- 7.Fox RJ, Rudick RA. Risk stratification and patient counseling for natalizumab in multiple sclerosis. Neurology 2012;78:436–437. [DOI] [PubMed] [Google Scholar]
- 8.Plavina T, Subramanyam M, Bloomgren G, et al. Anti-JC virus antibody levels in serum or plasma further define risk of natalizumab-associated progressive multifocal leukoencephalopathy. Ann Neurol 2014;76:802–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwab N, Schneider-Hohendorf T, Posevitz V, et al. L-selectin is a possible biomarker for individual PML risk in natalizumab-treated MS patients. Neurology 2013;81:865–871. [DOI] [PubMed] [Google Scholar]
- 10.Hendel-Chavez H, de Goer de Herve MG, Giannesini C, et al. Immunological hallmarks of JC virus replication in multiple sclerosis patients on long-term natalizumab therapy. J Virol 2013;87:6055–6059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antoniol C, Stankoff B. Immunological markers for PML prediction in MS patients treated with natalizumab. Front Immunol 2014;5:668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munoz-Culla M, Irizar H, Castillo-Trivino T, et al. Blood miRNA expression pattern is a possible risk marker for natalizumab-associated progressive multifocal leukoencephalopathy in multiple sclerosis patients. Mult Scler 2014;20:1851–1859. [DOI] [PubMed] [Google Scholar]
- 13.Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem 2010;79:351–379. [DOI] [PubMed] [Google Scholar]
- 14.Meira M, Sievers C, Hoffmann F, et al. MiR-126: a novel route for natalizumab action? Mult Scler 2014;20:1363–1370. [DOI] [PubMed] [Google Scholar]
- 15.Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proc Natl Acad Sci USA 2009;106:18704–18709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartholdy B, Du Roure C, Bordon A, Emslie D, Corcoran LM, Matthias P. The Ets factor Spi-B is a direct critical target of the coactivator OBF-1. Proc Natl Acad Sci USA 2006;103:11665–11670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marshall LJ, Dunham L, Major EO. Transcription factor Spi-B binds unique sequences present in the tandem repeat promoter/enhancer of JC virus and supports viral activity. J Gen Virol 2010;91:3042–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall LJ, Moore LD, Mirsky MM, Major EO. JC virus promoter/enhancers contain TATA box-associated Spi-B-binding sites that support early viral gene expression in primary astrocytes. J Gen Virol 2012;93:651–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marshall LJ, Ferenczy MW, Daley EL, Jensen PN, Ryschkewitsch CF, Major EO. Lymphocyte gene expression and JC virus noncoding control region sequences are linked with the risk of progressive multifocal leukoencephalopathy. J Virol 2014;88:5177–5183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindberg RL, Hoffmann F, Mehling M, Kuhle J, Kappos L. Altered expression of miR-17-5p in CD4+ lymphocytes of relapsing-remitting multiple sclerosis patients. Eur J Immunol 2010;40:888–898. [DOI] [PubMed] [Google Scholar]
- 21.Meira M, Sievers C, Hoffmann F, et al. Unraveling natalizumab effects on deregulated miR-17 expression in CD4+ T cells of patients with relapsing-remitting multiple sclerosis. J Immunol Res 2014;2014:897249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sievers C, Meira M, Hoffmann F, Fontoura P, Kappos L, Lindberg RL. Altered microRNA expression in B lymphocytes in multiple sclerosis: towards a better understanding of treatment effects. Clin Immunol 2012;144:70–79. [DOI] [PubMed] [Google Scholar]
- 23.Major EO. Progressive multifocal leukoencephalopathy in patients on immunomodulatory therapies. Annu Rev Med 2010;61:35–47. [DOI] [PubMed] [Google Scholar]
- 24.Polman CH, O'Connor PW, Havrdova E, et al. A randomized, placebo-controlled trial of natalizumab for relapsing multiple sclerosis. N Engl J Med 2006;354:899–910. [DOI] [PubMed] [Google Scholar]
- 25.Rudick R, Polman C, Clifford D, Miller D, Steinman L. Natalizumab: bench to bedside and beyond. JAMA Neurol 2013;70:172–182. [DOI] [PubMed] [Google Scholar]
- 26.Strubin M, Newell JW, Matthias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell 1995;80:497–506. [DOI] [PubMed] [Google Scholar]
- 27.Zwilling S, Dieckmann A, Pfisterer P, Angel P, Wirth T. Inducible expression and phosphorylation of coactivator BOB.1/OBF.1 in T cells. Science 1997;277:221–225. [DOI] [PubMed] [Google Scholar]
- 28.Yosef N, Shalek AK, Gaublomme JT, et al. Dynamic regulatory network controlling TH17 cell differentiation. Nature 2013;496:461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su GH, Chen HM, Muthusamy N, et al. Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J 1997;16:7118–7129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fine AJ, Sorbello A, Kortepeter C, Scarazzini L. Progressive multifocal leukoencephalopathy after natalizumab discontinuation. Ann Neurol 2014;75:108–115. [DOI] [PubMed] [Google Scholar]
- 31.Monaco MC, Major EO. Immune system involvement in the pathogenesis of JC virus induced PML: what is learned from studies of patients with underlying diseases and therapies as risk factors. Front Immunol 2015;6:159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chapagain ML, Nerurkar VR. Human polyomavirus JC (JCV) infection of human B lymphocytes: a possible mechanism for JCV transmigration across the blood-brain barrier. J Infect Dis 2010;202:184–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Frohman EM, Monaco MC, Remington G, et al. JC virus in CD34+ and CD19+ cells in patients with multiple sclerosis treated with natalizumab. JAMA Neurol 2014;71:596–602. [DOI] [PubMed] [Google Scholar]
- 34.Du Pasquier RA, Kuroda MJ, Zheng Y, Jean-Jacques J, Letvin NL, Koralnik IJ. A prospective study demonstrates an association between JC virus-specific cytotoxic T lymphocytes and the early control of progressive multifocal leukoencephalopathy. Brain 2004;127:1970–1978. [DOI] [PubMed] [Google Scholar]
- 35.Gheuens S, Bord E, Kesari S, et al. Role of CD4+ and CD8+ T-cell responses against JC virus in the outcome of patients with progressive multifocal leukoencephalopathy (PML) and PML with immune reconstitution inflammatory syndrome. J Virol 2011;85:7256–7263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wei G, Liu CK, Atwood WJ. JC virus binds to primary human glial cells, tonsillar stromal cells, and B-lymphocytes, but not to T lymphocytes. J Neurovirol 2000;6:127–136. [DOI] [PubMed] [Google Scholar]
- 37.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 2007;9:654–659. [DOI] [PubMed] [Google Scholar]
- 38.Wahlgren J, Karlson Tde L, Glader P, Telemo E, Valadi H. Activated human T cells secrete exosomes that participate in IL-2 mediated immune response signaling. PLoS One 2012;7:e49723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stuve O, Marra CM, Bar-Or A, et al. Altered CD4+/CD8+ T-cell ratios in cerebrospinal fluid of natalizumab-treated patients with multiple sclerosis. Arch Neurol 2006;63:1383–1387. [DOI] [PubMed] [Google Scholar]
- 40.Lindberg RL, Achtnichts L, Hoffmann F, Kuhle J, Kappos L. Natalizumab alters transcriptional expression profiles of blood cell subpopulations of multiple sclerosis patients. J Neuroimmunol 2008;194:153–164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.