Abstract
Tumor-induced myeloid-derived suppressor cells (MDSC) contribute to immune suppression in tumor-bearing individuals and are a major obstacle to effective immunotherapy. Reactive oxygen species (ROS) are one of the mechanisms used by MDSC to suppress T cell activation. Although ROS are toxic to most cells, MDSC survive despite their elevated content and release of ROS. Nuclear factor erythroid derived 2-like 2 (Nrf2) is a transcription factor that regulates a battery of genes which attenuates oxidative stress. Therefore, we hypothesized that MDSC resistance to ROS may be regulated by Nrf2. To test this hypothesis, we utilized Nrf2+/+ and Nrf2−/− BALB/c and C57BL/6 mice bearing 4T1 mammary carcinoma and MC38 colon carcinoma, respectively. Nrf2 enhanced MDSC suppressive activity by increasing MDSC production of H2O2, and increased the quantity of tumor-infiltrating MDSC by reducing their oxidative stress and rate of apoptosis. Nrf2 did not affect circulating levels of MDSC in tumor-bearing mice since the decreased apoptotic rate of tumor-infiltrating MDSC was balanced by a decreased rate of differentiation from bone marrow progenitor cells. These results demonstrate that Nrf2 regulates the generation, survival and suppressive potency of MDSC, and that a feedback homeostatic mechanism maintains a steady-state level of circulating MDSC in tumor-bearing individuals.
Keywords: Tumor Immunity, Tolerance/Suppression/Anergy, T cells
Introduction
The microenvironment of solid tumors (tumor microenvironment; TME) is frequently inflamed and oxidatively stressed due to hypoxia, the presence of reactive oxygen species (ROS), and multiple pro-inflammatory mediators (1). Therefore, cells in this environment must mitigate oxidative radicals in order to survive. Tumor cells survive this environment by having elevated levels of stabilized nuclear factor (erythroid-2)-related factor 2 (Nrf2), which enhances tumor cell proliferation and resistance to chemotherapy, and promotes tumor growth (2–4). Nrf2 is a basic-leucine-zipper (bZIP) transcription factor that is ubiquitously expressed in many tissues and cells (2, 5–7). Under normal redox conditions, Nrf2 is restricted to the cytoplasm by Kelch-like ECH-associated protein 1 (Keap1), which promotes the polyubiquitination of Nrf2, leading to its destruction by the 26s proteasome (8–12). Oxidative stress stabilizes Nrf2 by oxidizing key thiol residues on Keap1, which causes conformational changes of Keap1 that prevent Nrf2 polyubiquitination and subsequent degradation (13–15). Nrf2 can also be stabilized through direct phosphorylation by kinases involved in inflammatory signaling cascades (e.g. KRAS, MYC, PKC, ERK, MAPK, and p38) (16–21). Once stabilized, Nrf2 translocates to the nucleus where it heterodimerizes with other bZIP transcription factors including Jun (c-Jun, Jun-D, and Jun-B) and small Maf (MafG, MafK, and MafF) (22–27) and up-regulates genes containing an antioxidant response element (ARE) in their promoter (28–30). Activation of these antioxidant genes quenches oxidative stress and promotes detoxification, thereby protecting cells from oxidative toxicity.
Cells of the immune system are also present in the TME and must protect themselves against oxidative radicals. Myeloid-derived suppressor cells (MDSC) are immature myeloid cells that suppress T cell activation and proliferation (31), perturb naïve T cell trafficking to lymph nodes (32), impair NK cell cytotoxicity (33), induce T regulatory cells (34), and skew macrophages to a tumor promoting (type II) phenotype (35). MDSC are present in most solid tumors where they contribute to oxidative stress by their production of superoxide (36). Superoxide produced by MDSC rapidly reacts with a large number of molecules to form ROS such as hydrogen peroxide (H2O2), hydroxyl radical, hypochlorous acid, and peroxynitrate (ONOO−; e.g. PNT), which damage proteins, lipids, and nucleic acids, enhance inflammation, and promote apoptosis. H2O2 reduces T cell expression of CD3ζ chain thereby limiting their ability to become activated and mediate anti-tumor immunity (37, 38). Nitration/nitrosylation of T cell receptors (39) and MHC class I molecules (40) by PNT disrupts T cell-tumor antigen interactions and renders tumor cells resistant to CTL-mediated lysis. Despite their high intracellular content of ROS and their secretion of ROS, which constantly exposes them to oxidative stress, MDSC accumulate and function in tumor-bearing patients and animals. Given that tumor cells are protected from oxidative stress by Nrf2, we hypothesized that Nrf2 may also protect MDSC from oxidative stress.
To elucidate the role of Nrf2 in MDSC, we examined the survival time of tumor-bearing BALB/c and C57BL/6 Nrf2+/+ and Nrf2−/− mice, and the generation, survival, suppressive potency, and tumor-infiltration of MDSC in these mice. Wild type tumor-bearing mice have a decreased survival time and have more tumor-infiltrating and more suppressive MDSC compared to Nrf2-deficient mice. The increase in tumor-infiltrating MDSC is the result of a reduced rate of MDSC apoptosis. However, Nrf2 does not affect the level of MDSC in the periphery because a homeostatic regulatory mechanism increases MDSC generation from bone marrow progenitor cells.
Materials and Methods
Mice
BALB/c (Nrf2+/+), BALB/c Nrf2−/−, C57BL/6 (Nrf2+/+), and C57BL/6 Nrf2−/− mice were bred in the UMBC animal facility from stock obtained from The Jackson Laboratory (C57BL/6 and BALB/c) or provided by Dr. Masayuki Yamamoto (RIKEN, Japan; BALB/c Nrf2+/−) and Dr. Shyam Biswal (Johns Hopkins School of Public Health; C57BL/6 Nrf2−/−). BALB/c Nrf2−/− mice were generated by mating BALB/c Nrf2+/− × BALB/c Nrf2+/− mice. BALB/c Nrf2−/− and Nrf2+/+ offspring were identified by PCR typing (Supplemental Fig. 1A, 1B). DNA was isolated from pups (Qiagen, QIAamp DNA blood mini kit, per the manufacturer’s protocol), and amplified using primers specific to Nrf2 and lac Z under the following conditions: 94°C melting for 30 seconds, 56°C annealing for 30 seconds, 72°C extension for one minute, for 30 cycles. Homozygous knockouts have a 400kb band; heterozygotes 400 and 734kb bands; and homozygous wild type mice a 734kb band. BALB/c Nrf2+/+ littermates from these matings served as controls. C57BL/6 Nrf2−/− mice were generated by crossing C57BL/6 Nrf2−/− × C57BL/6 Nrf2−/− mice. Nrf2-deficiency was further verified by qPCR analysis glutamate-cysteine ligase, modifier subunit (GCLM), heme oxygenase 1 (HO-1), catalase, and NAD(P)H dehydrogenase, quinone 1(NQO1), genes that are regulated by Nrf2 (Supplemental Fig. 1B, 1C). 107 MDSC from 4T1-bearing Nrf2+/+ or Nrf2−/− mice were suspended in IMDM supplemented with 10% Fetal Clone I (Thermo Scientific, Waltham, MA), 1% penicillin-streptomycin, 1% glutamax, and 0.1% gentamycin, and plated in 35mm petri dishes in the presence of tert-butylhydroquinone (tbHQ, 50 μM; Sigma Aldrich, St. Louis, MO) or vehicle control (DMSO). Cultures were incubated for 6 hours (37°C, 5% CO2) and harvested. RNA was isolated with TRIzol reagent (ThermoFisher, Grand Island, NY) and chloroform extraction. cDNA was synthesized using a Maxima First Strand cDNA Synthesis Kit (ThermoFisher) as per the manufacture’s protocol. Quantitative PCR was performed using KiCqStart SYBR Green qPCR ReadyMix (Sigma Aldrich) with 100 nM of forward and reverse primers (Supplemental Fig. 1B) on a CFX96 Real-Time System (Bio-Rad, Hercules, CA) under the following conditions: 95°C melting for 10 seconds, 57°C annealing/extension for 30 seconds, for 40 cycles. Data were analyzed using CFX Manager (Bio-Rad), and Nrf2-regulated genes were normalized to the housekeeping gene L32 by the ΔCT method.
Tumor cells and tumor growth
BALB/c-derived 4T1 mammary carcinoma and C57BL/6-derived MC38 colon carcinoma were maintained as described (41), and have been in the authors’ lab for more than 15 and 8 years, respectively. 4T1 cells were originally obtained from Dr. Fred Miller (Karmanos Cancer Center) and MC38 cells from Dr. Dmitry Gabrilovich (Wistar Institute). Cell lines were routinely checked for mycoplasma and early freeze-downs were preferentially used. Mice were inoculated in the abdominal mammary gland with 100μL DMEM containing 7×103 or 105 4T1 cells, or in the flank with 5×105 MC38 cells. Primary tumors were measured as described (42). Survival time was recorded when mice became moribund and were euthanized. All animal procedures were approved by the UMBC Institutional Animal Care and Use Committee.
In vivo generation of MDSC
MDSC were harvested from the peripheral blood of tumor-bearing mice as described (41). Briefly, 4T1 tumor-bearing Nrf2+/+ and Nrf2−/− mice with tumors >10mm in diameter were bled by submandibular venipuncture into 500μL of a 0.008% heparin solution. Red blood cells were removed by lysis, and the remaining cells were analyzed by flow cytometry for MDSC. Cell populations containing >90% Gr1+CD11b+ cells were utilized in all functional assays.
Flow cytometry reagents and antibodies
Monoclonal antibodies rat anti-mouse CCR2-PE, CD3-FITC, CD4-APC-Cy7, CD8-APC, CD11b-APC, CD11b-APC-Cy7, CD11c-FITC, CD45-PB, CD45R-PE, CD62L-PE, CXCR4-PE, Ly6C-PE, Gr1-APC-Cy7, Ly6G-Alexa 647, isotype rat IgG2b-PE, dichlorofluorescein diacetate (DCFDA), propidium iodide (PI), Annexin V, and 7AAD were from BD Pharmingen (San Diego, CA) or BioLegend (San Diego, CA). Cells were stained with antibodies, with Annexin V and PI, or with DCFDA as described (43, 44). Samples were analyzed on a Beckman/Coulter Cyan ADP flow cytometer using Summit software.
Tumor infiltrating cells
Tumors were dissociated using a modified protocol from the Tissue Dissociation Kit (protocol 2.2.1; Miltenyi Biotech, Bergisch Gladbach, Germany) with a GentleMACS Dissociator. Tumors 8 to 12 mm in diameter were resected from 4T1-bearing mice, cut in half, and each half placed into a GentleMACS C tube containing 5mL of dissociation medium (DMEM with 300U/mL collagenase IV, 0.1% hyaluronidase, and 2kU/mL DNase I). Tumors were then minced with scissors into 2–4mm pieces, processed on the GentleMACS Dissociator with the program m_impTumor_02, and then rotated (10 rpm; Glas-Col Rotator) at 37°C for 40 minutes. Samples were then processed twice on the GentleMACS Dissociator using the program m_impTumor_03. The resulting material was filtered through a 70μM mesh filter and the cells that passed through the filter were washed twice with 10 mL DMEM (Beckman Allegra 6R centrifuge, 500g for 3 minutes), resuspended in 4mL DMEM, and subjected to ficoll-plaque density gradient centrifugation (Beckman Allegra 6R centrifuge, 1400g for 20 min at 20°C). Live cells were isolated from the ficoll-aqueous interface, washed twice with DMEM, stained with 7AAD and for F4/80, Gr1 or Ly6G and Ly6C, CD3, CD4, CD8, CD11b, CD11c, CD45, and CD45R (B220), and assessed by flow cytometry. Cell percentages were calculated as a percentage of 7AAD−CD45+ cells.
MDSC differentiation from bone marrow
MDSC were generated from bone marrow progenitors as described (45). Briefly, bone marrow was flushed aseptically from the femurs of naïve mice. RBCs were lysed with Gey’s solution and the resulting cells were assayed for the percentage of Gr1+CD11b+ cells, and cultured for four days (37°C, 5% CO2) at 4.2 × 105 cells/2 mL RPMI medium supplemented with 10% FCS, 80 ng/mL IL-6, and 80 ng/mL GM-CSF/2 ml/well in 6 well plates. At the end of culture, the total number of cells was determined, and the percentage of granulocytic (Ly6GhiLy6CloCD11b+; PMN-MDSC) and monocytic (Ly6GloLy6ChiCD11b+; M-MDSC) MDSC was determined by flow cytometry. Number of MDSC = [(total number of cells) × (% M-MDSC and/or PMN-MDSC)]. Ratio of MDSC from Nrf2−/− vs Nrf2+/+ mice = (Total MDSC from Nrf2−/− mice)/(Total MDSC from Nrf2+/+ mice). Ratio PMN-MDSC to M-MDSC = (Total PMN-MDSC)/(Total M-MDSC).
Apoptosis Assay
Live MDSC were identified as 7AAD− or PI− and Gr1+CD11b+ or CD11b+Ly6G+, or CD11b+Ly6C+ cells. The percent decrease in apoptosis = 100 × [1-(% Annexin V+ MDSC from Nrf2+/+ mice/% Annexin V+ MDSC from Nrf2−/− mice)]. For some experiments, MDSC were harvested from the blood of 4T1-tumor bearing mice, re-suspended in HL-1 media (supplemented with 1% penicillin-streptomycin, 1% glutamax, and 0.1% gentamycin), and plated in 6cm petri dishes. Gr1+CD11b+ cells were assessed for viability by PI staining.
ROS detection
Reactive oxygen species (ROS) were measured by H2O2 detection using an Amplex Red Hydrogen Peroxide Assay Kit (Invitrogen) as described (46). Briefly, MDSC were suspended in Dulbecco’s PBS at 2.5 × 106/mL and 5 × 104 cells/50 μl were plated per well in 96 well black, flat-bottom plates (Greiner Bio-One, Monroe, NC). Thirty ng/ml PMA and 50 μl Amplex Red Reagent were added to each well. Plates were incubated at 37°C and fluorescence (excitation at 530 nm, emission at 590 nm) was measured for one hour at 5-minute intervals using a Biotek Synergy 2 microplate plate reader (Winooski, VT, USA). A standard curve was generated by serial dilutions of 20 μM H2O2.
T cell activation
T cell activation was measured as described (44). Briefly, 105 splenocytes from DO11.10 (ovalbumin323–339-specific, I-Ad-restricted), TS1 (hemagglutinin110–119-specific, I-Ed-restricted), Clone4 (hemagglutinin518–526-specific, H-2Kd-restricted), or OT1 (ovalbumin257–264-specific, H-2Kb-restricted) transgenic mice were cultured with their respective cognate peptides and varying concentrations of irradiated (20Gy) MDSC from the blood of 4T1 tumor-bearing mice. Catalase (1000 or 500 U/ml; Sigma Aldrich), sodium pyruvate (5 or 2.5 μM; Sigma Aldrich), nor-NOHA (500 μM; Calbiochem, CA), or L-NMMA (500 μM; Calbiochem) were included in some assays. Reversal of suppression = −100% × [1 – (CPMNo Inhibitor/CPMInhibitor)]. For some experiments, Nrf2+/+ MDSC were either initially cultured with splenocytes, or added after the addition of cognate peptide. Fresh MDSC were used for experiments in which MDSC were added to overnight splenocyte cultures. Cultures were pulsed with 3H-thymidine (1nCu/250μL) on day 4 and harvested on day 5. Peptides were synthesized at the University of Maryland Baltimore (UMB) Biopolymer Core Facility.
MDSC-macrophage cross-talk
Peritoneal macrophages were prepared from tumor-free mice as described (35) and were >95% CD11b+ F4/80+ cells as assessed by flow cytometry. MDSC and macrophage cross-talk experiments were performed as described (47). Supernatants were analyzed for IL-10 using ELISA kits (R&D Systems and eBioscience, San Diego, CA) per the manufacture’s protocol. NO was assayed by Griess assay as described (47).
Statistical Analyses
Student’s t-test and Tukey’s Honestly Significant Difference (HSD) test were performed using Microsoft Excel 2013. Values denoted with different letters (e.g. a, b, c, etc.) are significantly different from each other; values with the same letter are not significantly different. Tumor growth and ROS data were analyzed using the Mann-Whitney test on the www.VassarStats.net website. Survival data were analyzed using the log-rank test from the Walter and Eliza Hall Institute of Medical Research Bioinformatics webpage (http://bioinf.wehi.edu.au/software/russell/logrank/). Values of p<0.05 were considered statistically significant. Values are ± SD.
Results
Host expression of Nrf2 enhances tumor progression
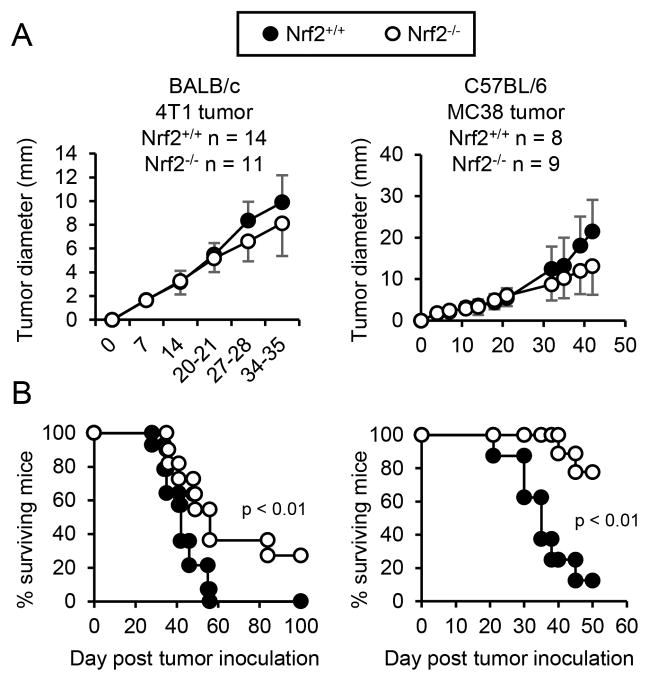
The role of host-derived Nrf2 in tumor progression has been controversial. Some studies indicate that Nrf2 supports tumor growth (48–53), while other studies suggest it deters carcinogenesis (54–58). To determine whether host Nrf2 contributes to or deters tumor growth in BALB/c and C57BL/6 mice, Nrf2+/+ and Nrf2−/− BALB/c and C57BL/6 mice were injected with syngeneic 4T1 mammary carcinoma or MC38 colon carcinoma, respectively, and followed for primary tumor growth (Fig. 1A) and survival (Fig. 1B). Nrf2 did not impact the growth rate of either primary tumor. However, tumor-bearing Nrf2+/+ mice had decreased mean survival times compared to Nrf2−/− mice (BALB/c: 42.2 vs 50.8 days; 0% and 27.27% of mice survived >100 days, respectively; C57BL/6: 35.8 vs. 43 days; 12.5% and 77.78% of mice survived >50 days, respectively), indicating that Nrf2 supports tumor progression in these mouse strains.
Figure 1. Nrf2 decreases survival time of tumor-bearing mice.
Nrf2+/+ and Nrf2−/− mice on the BALB/c or C57BL/6 backgrounds were injected with 4T1 mammary carcinoma or MC38 colon carcinoma, respectively. Mice were followed weekly for primary tumor growth (A) and survival time (B). Tumor diameter was calculated as the average measurement of tumor length and width. Data were pooled from two independent experiments. Tumor growth and survival time were tested for statistical significance by Mann-Whitney and log-rank test, respectively.
Host expression of Nrf2 enhances MDSC suppressive potency and the quantity of tumor-infiltrating MDSC
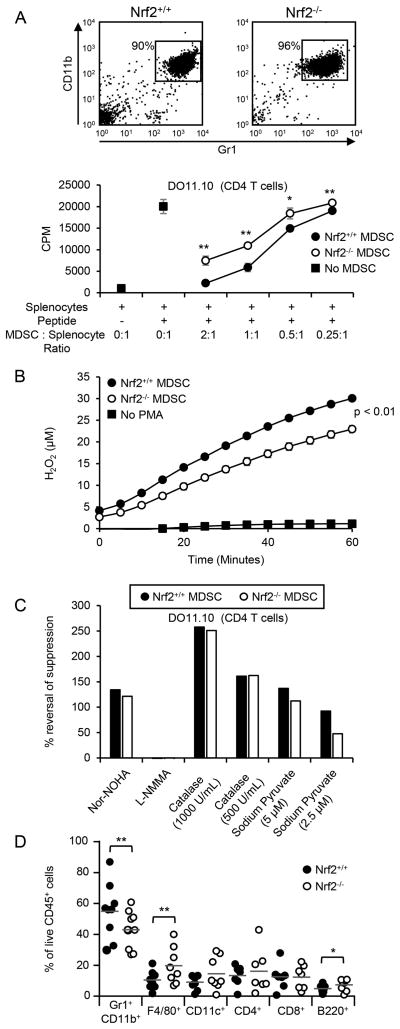
If Nrf2 mediates its effects by increasing MDSC suppressive potency, then MDSC from tumor-bearing Nrf2+/+ mice will be more suppressive than MDSC from Nrf2−/− mice. To test this possibility, titered quantities of MDSC from tumor-bearing BALB/c Nrf2+/+ and Nrf2−/− mice were co-cultured with T cells from TcR transgenic mice plus cognate peptide, and the cultures assayed for T cell activation (Fig. 2A; Supp. Fig. 2A). Both CD4+ and CD8+ T cells were suppressed more efficiently by MDSC from Nrf2+/+ mice than by MDSC from Nrf2−/− mice, suggesting that Nrf2 drives the suppressive potency of MDSC. Since differences in MDSC viability may impact suppressive potency, we determined the kinetics of MDSC-mediated suppression (Supplemental Fig. 2B) and then assessed the viability of MDSC at the time they would be active (Supplemental Fig. 2C). Addition of MDSC to splenocyte plus cognate peptide cultures at or after 19 hrs did not result in suppression, indicating that MDSC viability was only relevant at ≤16 hrs, although viability did not differ up to 24 hrs in culture. These data demonstrate that MDSC from Nrf2+/+ mice are more suppressive than MDSC from Nrf2−/− mice and that the difference is not due to differences in MDSC viability.
Figure 2. Nrf2 enhances MDSC suppressive activity and the quantity of tumor-infiltrating MDSC.
(A) Nrf2 enhances MDSC-mediated CD4+ T cell suppression. MDSC from the peripheral blood of 4T1-bearing BALB/c Nrf2+/+ and Nrf2−/− mice were assayed for their ability to suppress the antigen-activation of transgenic CD4+ (DO11.10) T cells. (B) Nrf2 enhances MDSC production of H2O2. MDSC from BALB/c Nrf2+/+ and Nrf2−/− mice with 4T1 tumors were incubated with Amplex Red reagent, stimulated with PMA, and assayed for H2O2 production over time. (C) Nrf2+/+ and Nrf2−/− MDSC suppress CD4+ T cell activation by producing arginase and H2O2. MDSC from the peripheral blood of 4T1-bearing BALB/c Nrf2+/+ and Nrf2−/− mice were assayed for their ability to suppress the antigen-activation of transgenic CD4+ (DO11.10) T cells in the presence of nor-NOHA, L-NMMA, catalase, and sodium pyruvate. (D) Nrf2 enhances the quantity of tumor-infiltrating MDSC. Each circle represents an individual mouse. Figures A, B, and D were analyzed by Students t test, Mann-Whitney test, and Wilcoxon-rank sign test, respectively. Figures A, B, and C represent one of two experiments, each with one Nrf2+/+ and one Nrf2−/− mouse per experiment. Data from figure D were pooled from 5 independent experiments; **p<.01, *p<.05.
MDSC use several mechanisms to inhibit T cells, such as the secretion of ROS, including H2O2, which decreases T cell expression of IL-2, IFNγ, and CD3ζ (36, 59), and the production of arginase which deprives T cells of the amino acid arginine, leading to CD3ζ synthesis arrest (60). MDSC also sequester cysteine which is an essential amino acid for T cell activation (61). MDSC from Nrf2+/+ mice secrete more H2O2 than MDSC from Nrf2−/− mice (Figure 2B). H2O2 contributes to the suppressive potency of MDSC since inclusion of the H2O2 scavengers catalase or sodium pyruvate in cultures of MDSC plus transgenic T cells plus cognate peptide, significantly increased T cell activation (Fig. 2C, Supplemental Fig. 2D). Neither MDSC from Nrf2+/+ nor Nrf2−/− mice produce nitric oxide (NO; Supplemental Fig. 3A), and the NOS2 inhibitor L-NMMA did not rescue T cell activation in the presence of MDSC (Fig. 2C, Supplemental Fig. 2D). MDSC from both Nrf2+/+ and Nrf2−/− mice use arginase to suppress T cell activation since inclusion of the arginase inhibitor nor-NOHA restores T cell activation (Fig. 2C; Supplemental Fig. 2D), and there is no difference in their content of arginase (Supplemental Fig. 3B). MDSC from both wild type and knockout mice express similar levels of xCT, the chain of the dimeric xc- transporter that regulates the uptake of cystine (Supplemental Fig. 3C). These results indicate that MDSC from Nrf2+/+ and Nrf2−/− mice utilize similar mechanisms to suppress T cell activation and proliferation, but MDSC from Nrf2+/+ mice are more suppressive because they produce more H2O2.
MDSC also promote tumor progression by down-regulating L-selectin on naïve T cells thereby preventing naïve T cell trafficking into lymph nodes (32), and they produce IL-10 which polarizes macrophages towards a tumor-promoting phenotype (35). MDSC from Nrf2+/+ and Nrf2−/− tumor-bearing BALB/c mice equally down-regulated T cell L-selectin (Supplemental Fig. 3D) demonstrating that Nrf2 does not affect naïve T cell entry into lymph nodes. Using MDSC and macrophages from BALB/c IL-10+/+ and IL-10−/− mice, we previously demonstrated that in co-cultures of MDSC and macrophages, MDSC are the sole producers of IL-10, and that MDSC production of IL-10 is enhanced by macrophages (35, 47, 62). Surprisingly, Nrf2 decreased MDSC production of IL-10, thus reducing the ability of MDSC to polarize macrophages towards a type 2 phenotype (Supplemental Fig. 3E). These results indicate that Nrf2 does not impact the ability of MDSC to decrease T cell homing to lymph nodes, but does reduce the ability of MDSC to polarize macrophages towards a tumor-promoting phenotype.
Since MDSC are present in most solid tumors where they can exert pro-tumor activity (63), we assessed the proportion of MDSC in 4T1 primary tumors resected from BALB/c Nrf2+/+ and Nrf2−/− mice (Fig. 2D). Tumors derived from Nrf2−/− mice had significantly fewer MDSC compared to tumors from wild type littermates. However, the ratio of tumor-infiltrating PMN-MDSC to M-MDSC was the same in Nrf2+/+ and Nrf2−/− mice (Supplemental Fig. 4A). The proportion of tumor-infiltrating CD11c+, CD4+, and CD8+ cells did not differ between the Nrf2+/+ and Nrf2−/− mice, but tumors from Nrf2−/− mice contained significantly more F4/80+ and B220+ cells (Fig. 2D).
To determine if the differences in tumor-infiltrating MDSC were due to differences in MDSC trafficking, MDSC were isolated from the peripheral blood, bone marrow, and tumor of 4T1-bearing Nrf2+/+ and Nrf2−/− mice and assayed for CCR2 and CXCR4, chemokine receptors that regulate MDSC migration (64, 65) (Supplemental Fig. 4B). CCR2 and CXCR4 expression did not differ between MDSC from Nrf2+/+ and Nrf2−/− mice. Likewise, MDSC from Nrf2+/+ and Nrf2−/− mice migrated at the same rate in a transwell chemotaxis assay (Supplemental Fig. 4C), indicating that Nrf2 does not influence MDSC trafficking.
Collectively, these data suggest that Nrf2 decreases the survival time of tumor-bearing mice by enhancing MDSC suppressive activity and by increasing the accessibility of MDSC to the TME.
Nrf2 decreases MDSC oxidative stress and apoptosis
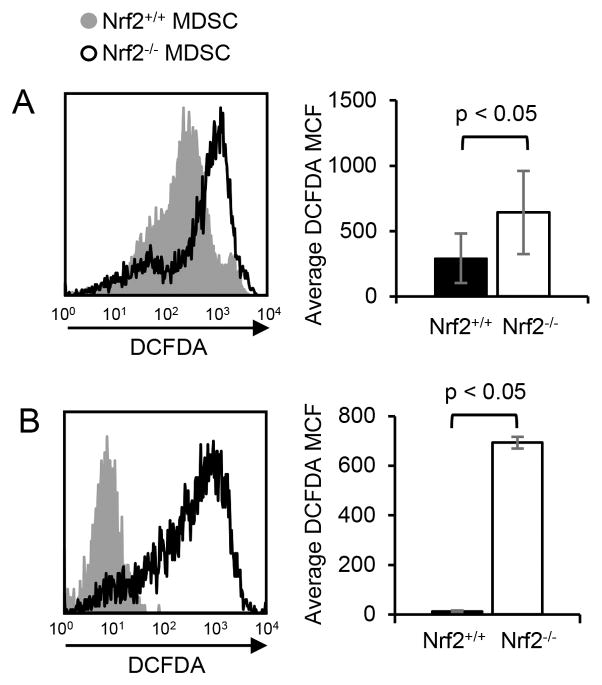
Since the increased number of tumor-infiltrating MDSC in Nrf2+/+ mice was not due to enhanced MDSC trafficking, we speculated that MDSC in Nrf2+/+ mice had lower levels of intracellular ROS and therefore were less oxidatively stressed. Therefore, we assessed intracellular ROS levels in MDSC from 4T1-bearing Nrf2+/+ and Nrf2−/− mice and in MDSC differentiated in vitro from bone marrow progenitor cells. MDSC in the blood of Nrf2+/+ 4T1-bearing mice had significantly lower levels of intracellular ROS compared to MDSC in Nrf2−/− mice as measured by DCFDA fluorescence (Fig. 3A), as did MDSC differentiated from bone marrow progenitor cells of Nrf2+/+ mice (Fig. 3B). Therefore, Nrf2 reduces intracellular ROS, consistent with the concept that MDSC in Nrf2+/+ mice survive longer due to reduced oxidative stress.
Figure 3. Nrf2 decreases intracellular MDSC oxidative stress.
(A) Circulating MDSC from tumor-bearing Nrf2-sufficient mice contain less intracellular ROS than MDSC from tumor-bearing Nrf2-deficient mice. Circulating MDSC were harvested from 4T1-bearing BALB/c Nrf2+/+ and Nrf2−/− mice, stained with DCFDA, and for Gr1 and CD11b. Gr1+CD11b+ cells were gated and analyzed by flow cytometry for DCFDA fluorescence. Left histogram: MDSC from representative individual Nrf2+/+ and Nrf2−/− mice; right graph: average MCF of DCFDA staining for MDSC from six Nrf2+/+ and five Nrf2−/− mice. (B) Nrf2 decreases intracellular ROS in MDSC differentiated in vitro from bone marrow progenitor cells. Bone marrow cells from tumor-free BALB/c Nrf2+/+ or Nrf2−/− mice were cultured under conditions favoring MDSC differentiation, and the resulting cells were stained with 7AAD and DCFDA, and for Gr1 and CD11b. 7AAD− Gr1+CD11b+ cells were gated and analyzed by flow cytometry for DCFDA fluorescence. Left histogram: MDSC from representative individual Nrf2+/+ and Nrf2−/− mice; right graph: average MCF of DCFDA staining of MDSC from three Nrf2+/+ and three Nrf2−/− mice. Data were tested for statistical significance by Student’s t test.
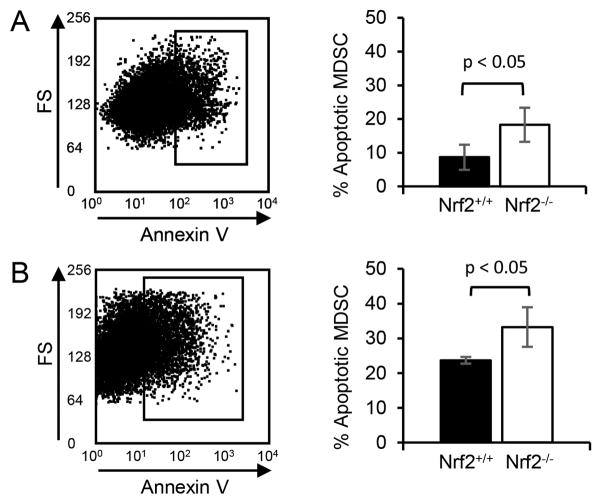
To determine if reduced levels of oxidative stress in MDSC from Nrf2+/+ mice correlate with reduced apoptosis, in vivo tumor-induced and in vitro differentiated MDSC were examined for apoptosis. Circulating MDSC from 4T1 tumor-bearing Nrf2+/+ and Nrf2−/− mice were stained for Gr1 and CD11b, and with Annexin V (Fig. 4A). MDSC from Nrf2+/+ mice were 53% less apoptotic than MDSC from Nrf2−/− mice. MDSC differentiated in vitro in bone marrow cultures were similarly analyzed except dead cells were excluded by 7AAD staining. In vitro differentiated MDSC from Nrf2+/+mice were 29% less apoptotic compared to MDSC from Nrf2−/− mice (Fig. 4B), confirming the concept that reducing oxidative stress decreases apoptosis.
Figure 4. Nrf2 protects MDSC from apoptosis.
(A) Nrf2 decreases apoptosis in circulating MDSC of tumor-bearing mice. Circulating MDSC were harvested from 4T1-bearing BALB/c Nrf2+/+ and Nrf2−/− mice, and stained for Gr1, CD11b, and with Annexin V and propidium iodide (PI) or 7AAD, and analyzed by flow cytometry. Live Gr1+CD11b+ MDSC (PI− or 7AAD−) were gated and assessed for Annexin V. Left panel: MDSC from representative individual mouse; right graph: average % annexin V+Gr1+CD11b+ MDSC from six Nrf2+/+ and five Nrf2−/− mice. (B) Nrf2 decreases apoptosis in MDSC differentiated in vitro from bone marrow progenitor cells. MDSC derived from bone marrow cell cultures were harvested, stained, and analyzed as in panel A. Averaged data are from one of three independent experiments with one Nrf2+/+ and one Nrf2−/− mouse per experiment. Data were tested for statistical significance using Student’s t test.
A decrease in apoptotic rate could result in an increase in circulating MDSC. This possibility was tested by comparing the levels of MDSC in the blood of tumor-bearing Nrf2+/+ vs. Nrf2−/− mice. Mice with the same tumor diameters were compared to eliminate MDSC differences due to different tumor burdens (Supplemental Fig. 4D). There was no difference in the level of circulating MDSC in tumor-bearing Nrf2+/+ and Nrf2−/− mice, indicating that the differential apoptotic rates did not impact MDSC accumulation in blood.
Nrf2-deficiency increases the rate of MDSC generation in the bone marrow
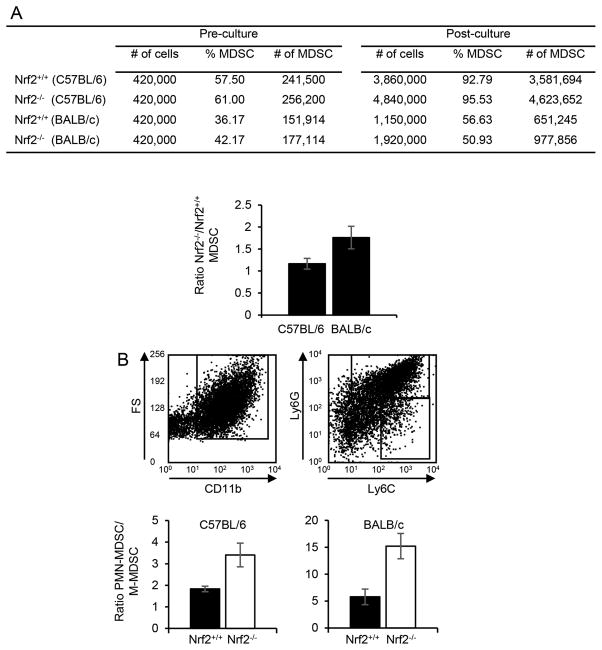
Since MDSC from Nrf2−/− mice are more apoptotic than MDSC from Nrf2+/+ mice, yet tumor-bearing Nrf2+/+ and Nrf2−/− mice have similar levels of circulating MDSC, we hypothesized that MDSC differentiate more rapidly in Nrf2−/− mice. To test this hypothesis, bone marrow cells from tumor-free Nrf2+/+ and Nrf2−/− mice were cultured under conditions to promote MDSC differentiation, and the number of resulting MDSC was quantified (Fig. 5A). C57BL/6 and BALB/c Nrf2−/− bone marrow produced 16% and 76% more MDSC than the corresponding Nrf2+/+ bone marrow, respectively. The increases were due to the expansion of PMN-MDSC (Fig. 5B). Therefore, Nrf2 reduces the rate of MDSC generation from bone marrow progenitor cells. Taken together with the apoptotic studies of Fig. 4, these data indicate that circulating MDSC levels are maintained by a balance between the generation of MDSC in the bone marrow and their turn-over in the periphery.
Figure 5. Nrf2 deficiency enhances MDSC proliferation.
MDSC were differentiated in vitro from the bone marrow of tumor-free Nrf2+/+ and Nrf2−/− BALB/c and C57BL/6 mice. The resulting cells were harvested, counted, and stained for Ly6G, Ly6C, and CD11b, and analyzed by flow cytometry. PMN-MDSC and M-MDSC were identified as LyG+Ly6C−/lowCD11b+ and Ly6G−/lowLy6C+CD11b+ cells, respectively. (A) Top: Quantity of total cells, percent of cells that are MDSC, and absolute number of MDSC pre-culture and after in vitro differentiation (post-culture). Data are representative of one of three independent experiments with one Nrf2+/+ and one Nrf2−/− mouse per experiment. Bottom: Ratio of Nrf2−/− to Nrf2+/+ MDSC from the three independent experiments. A value >1 indicates that there is more proliferation in the absence of Nrf2. (B) Nrf2 deficiency preferentially enhances differentiation of PMN-MDSC from bone marrow progenitor cells. MDSC of panel A were gated and analyzed for PMN-MDSC and M-MDSC. Top: Representative staining of M-MDSC and PMN-MDSC from individual Nrf2+/+ and Nrf2−/− mice. Bottom: Average ratio of PMN-MDSC to M-MDSC from the three independent experiments.
Discussion
For cells to survive the hostile TME, they must protect themselves against oxidative stress. Since Nrf2 regulates many genes that enable cells to survive oxidative stress, we examined the role of Nrf2 in the maintenance of MDSC suppressive activity, survival, and presence in solid tumors. Nrf2 enhanced MDSC suppressive activity by increasing MDSC production of H2O2, and increased the quantity of tumor-infiltrating MDSC by reducing their oxidative stress and apoptotic rate. Nrf2 did not affect circulating levels of MDSC in tumor-bearing mice since the decreased apoptotic rate of tumor-infiltrating MDSC was balanced by a decreased rate of differentiation from bone marrow progenitor cells. Collectively, these results provide a new avenue by which Nrf2 regulates tumor progression and add Nrf2 to the list of genes that govern MDSC accumulation, survival, and function.
Since our studies used mice globally knocked-out for Nrf2, we cannot determine if Nrf2 is directly impacting MDSC or is indirectly affecting MDSC by acting on other cells which subsequently influence MDSC development. Regardless of whether the effects are direct or indirect, Nrf2 regulates MDSC survival, function, and homeostasis.
The TME is an inflamed milieu that includes multiple cell types (e.g. tumor cells, MDSC, macrophages, dendritic cells, lymphocytes, mast cells, neutrophils cancer-associated fibroblasts, etc.). These cells participate in a complex crosstalk network that regulates the production of inflammatory mediators (47). Nrf2 increases the number of tumor-infiltrating MDSC and therefore enhances the opportunity for crosstalk between MDSC and other tumor-resident cells. Our previous studies demonstrated that macrophages enhance MDSC production of IL-10 which in turn polarizes macrophages towards a tumor-promoting phenotype (35, 47). Since Nrf2-deficiency increases both macrophage-dependent and macrophage-independent IL-10 production by MDSC, strategies aimed at limiting Nrf2 may facilitate the development of pro-tumor macrophages.
Pharmacologic down-regulation of Nrf2 may decrease the quantity of tumor-infiltrating MDSC and their suppressive potency. However, it will not reduce the level of circulating MDSC due to the homeostatic compensation by increased generation of MDSC from bone marrow progenitor cells. If MDSC predominantly mediate their suppressive effects on T cells within the tumor, then Nrf2 down-regulation may reduce MDSC-mediated suppression. However, MDSC may also mediate their effects on T cells in the periphery by suppressing circulating tumor-reactive T cells. In addition, MDSC are known to prevent the entry of naïve T cells into lymph nodes where they could become activated (32, 45). Therefore, down-regulation of Nrf2 in MDSC may only marginally reduce immune suppression and improve anti-tumor immunity because levels of circulating MDSC will remain constant due to increased generation of MDSC from bone marrow progenitor cells.
It is unlikely that the homeostatic balance of MDSC in tumor-bearing individuals is achieved by Nrf2 directly regulating genes that drive MDSC generation. Antibody-mediated depletion of MDSC results in the rebound of MDSC to levels that are higher than pre-depletion levels (66), demonstrating that some type of feed-back mechanism regulates MDSC homeostasis. Since Nrf2, like antibody-mediated depletion, alters extramedullary levels of MDSC, Nrf2 most likely also regulates MDSC homeostasis via a feedback loop rather than by a direct effect on genes within bone marrow progenitor cells.
Given that homeostasis maintains a constant level of circulating MDSC in individuals with tumor, monotherapies aimed at reducing MDSC levels by targeting circulating MDSC are unlikely to be effective. In contrast, strategies that target the induction of MDSC from progenitor cells have the potential to interrupt homeostatic regulation and thereby reduce MDSC levels. Many inducers of MDSC have been identified. These are predominantly pro-inflammatory molecules (67, 68). Since these molecules are redundant and compensate for each other in their ability to drive MDSC generation, it will be necessary to develop inhibitors that cover the full range of inducers.
Whether the depletion of MDSC by their differentiation into macrophages or other myeloid cells also results in the replacement of immune suppressive MDSC by homeostasis is unknown. However, if this process does not increase the differentiation of MDSC from bone marrow progenitors, then promoting MDSC differentiation may be therapeutic. Indeed, drugs such as CpG motifs (69), all-trans retinoic acid (70), tetrabromocinnamic acid (71), and Vitamin D3 (72) that drive the differentiation of MDSC to more mature cells have shown therapeutic effects. Interestingly, all-trans retinoic acid is a known Nrf2 inhibitor (73), suggesting that there is interplay between MDSC differentiation and Nrf2 activity.
Since Nrf2 impacts other cells in the TME in addition to MDSC, global inhibition of Nrf2 may impact overall tumor progression. For example, inhibition of Nrf2 in dendritic cells enhances MHC II and CD86 expression (74), which could result in improved antigen-presentation and therefore better activation of tumor-reactive T cells. Additionally, Nrf2 activation in tumor cells increases tumor cell proliferation and resistance to chemotherapy and radiotherapy, thereby promoting tumor progression (reviewed in (75)). Since MDSC accumulation is positively correlated with tumor burden, treatment strategies that combine Nrf2 inhibitors with conventional chemotherapy and radiotherapy could decrease tumor burden and thereby indirectly reduce MDSC levels and increase anti-tumor immunity.
Supplementary Material
Acknowledgments
We thank Drs. Masayuki Yamamoto (Tohoku University Graduate School of Medicine) and Shyam Biswal (Johns Hopkins) for kindly providing breeding pairs of the BALB/c Nrf2+/− and C57BL/6 Nrf2−/− mice, respectively, Dr. Tiha Long for qRT-PCR primer design, and Dr. Anil Jaiswal (University of Maryland, Baltimore) for helpful discussions.
Abbreviations used in this article
- ARE
antioxidant response element
- bZIP
basic-leucine-zipper
- DCFDA
Dichlorofluorescein diacetate
- KEAP1
Kelch-like ECH-associated protein 1
- GCLM
Glutamate-cysteine ligase, modifier subunit
- HO-1
Heme Oxygenase 1
- L32
Ribosomal protein L32
- L-NMMA
L-NG-monomethyl arginine
- PMN-MDSC
Granulocytic MDSC
- MDSC
Myeloid-derived suppressor cells
- M-MDSC
Monocytic MDSC
- nor-NOHA
Nω-hydroxy-nor-Arginine
- NOS2
Inducible nitric oxide synthase
- NQO1
NAD(P)H dehydrogenase, quinone 1
- Nrf2
Nuclear factor (erythroid-2)-related factor 2
- PI
Propidium iodide
- PNT
Peroxynitrate
- ROS
Reactive oxygen species
- tBHQ
Tertbutylhydroquinone
- TCCM
Tumor cell conditioned medium
- TME
Tumor microenvironment
Footnotes
Studies were supported by NIH RO1CA115880, RO1CA84232, and RO1GM021248 (SOR). DWB was partially supported by a pre-doctoral fellowship from the Congressionally Directed Medical Research Breast Cancer Program (W81XWH-11-1-0115).
References
- 1.Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010;44:479–496. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kawasaki Y, Ishigami S, Arigami T, Uenosono Y, Yanagita S, Uchikado Y, Kita Y, Nishizono Y, Okumura H, Nakajo A, Kijima Y, Maemura K, Natsugoe S. Clinicopathological significance of nuclear factor (erythroid-2)-related factor 2 (Nrf2) expression in gastric cancer. BMC Cancer. 2015;15:5. doi: 10.1186/s12885-015-1008-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Satoh H, Moriguchi T, Takai J, Ebina M, Yamamoto M. Nrf2 prevents initiation but accelerates progression through the Kras signaling pathway during lung carcinogenesis. Cancer Res. 2013;73:4158–4168. doi: 10.1158/0008-5472.CAN-12-4499. [DOI] [PubMed] [Google Scholar]
- 4.Homma S, Ishii Y, Morishima Y, Yamadori T, Matsuno Y, Haraguchi N, Kikuchi N, Satoh H, Sakamoto T, Hizawa N, Itoh K, Yamamoto M. Nrf2 enhances cell proliferation and resistance to anticancer drugs in human lung cancer. Clin Cancer Res. 2009;15:3423–3432. doi: 10.1158/1078-0432.CCR-08-2822. [DOI] [PubMed] [Google Scholar]
- 5.Mohler J, Mahaffey JW, Deutsch E, Vani K. Control of Drosophila head segment identity by the bZIP homeotic gene cnc. Development. 1995;121:237–247. doi: 10.1242/dev.121.1.237. [DOI] [PubMed] [Google Scholar]
- 6.Chan JY, Han XL, Kan YW. Isolation of cDNA encoding the human NF-E2 protein. Proc Natl Acad Sci U S A. 1993;90:11366–11370. doi: 10.1073/pnas.90.23.11366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moi P, Chan K, Asunis I, Cao A, Kan YW. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc Natl Acad Sci U S A. 1994;91:9926–9930. doi: 10.1073/pnas.91.21.9926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kang MI, Kobayashi A, Wakabayashi N, Kim SG, Yamamoto M. Scaffolding of Keap1 to the actin cytoskeleton controls the function of Nrf2 as key regulator of cytoprotective phase 2 genes. Proc Natl Acad Sci U S A. 2004;101:2046–2051. doi: 10.1073/pnas.0308347100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nguyen T, Sherratt PJ, Huang HC, Yang CS, Pickett CB. Increased protein stability as a mechanism that enhances Nrf2-mediated transcriptional activation of the antioxidant response element. Degradation of Nrf2 by the 26 S proteasome. J Biol Chem. 2003;278:4536–4541. doi: 10.1074/jbc.M207293200. [DOI] [PubMed] [Google Scholar]
- 10.Cullinan SB, Gordan JD, Jin J, Harper JW, Diehl JA. The Keap1-BTB protein is an adaptor that bridges Nrf2 to a Cul3-based E3 ligase: oxidative stress sensing by a Cul3-Keap1 ligase. Mol Cell Biol. 2004;24:8477–8486. doi: 10.1128/MCB.24.19.8477-8486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kobayashi A, Kang MI, Okawa H, Ohtsuji M, Zenke Y, Chiba T, Igarashi K, Yamamoto M. Oxidative stress sensor Keap1 functions as an adaptor for Cul3-based E3 ligase to regulate proteasomal degradation of Nrf2. Mol Cell Biol. 2004;24:7130–7139. doi: 10.1128/MCB.24.16.7130-7139.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang DD, Lo SC, Cross JV, Templeton DJ, Hannink M. Keap1 is a redox-regulated substrate adaptor protein for a Cul3-dependent ubiquitin ligase complex. Mol Cell Biol. 2004;24:10941–10953. doi: 10.1128/MCB.24.24.10941-10953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dinkova-Kostova AT, Holtzclaw WD, Cole RN, Itoh K, Wakabayashi N, Katoh Y, Yamamoto M, Talalay P. Direct evidence that sulfhydryl groups of Keap1 are the sensors regulating induction of phase 2 enzymes that protect against carcinogens and oxidants. Proc Natl Acad Sci U S A. 2002;99:11908–11913. doi: 10.1073/pnas.172398899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang DD, Hannink M. Distinct cysteine residues in Keap1 are required for Keap1-dependent ubiquitination of Nrf2 and for stabilization of Nrf2 by chemopreventive agents and oxidative stress. Mol Cell Biol. 2003;23:8137–8151. doi: 10.1128/MCB.23.22.8137-8151.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McMahon M, Lamont DJ, Beattie KA, Hayes JD. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc Natl Acad Sci U S A. 2010;107:18838–18843. doi: 10.1073/pnas.1007387107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloom DA, Jaiswal AK. Phosphorylation of Nrf2 at Ser40 by protein kinase C in response to antioxidants leads to the release of Nrf2 from INrf2, but is not required for Nrf2 stabilization/accumulation in the nucleus and transcriptional activation of antioxidant response element-mediated NAD(P)H:quinone oxidoreductase-1 gene expression. J Biol Chem. 2003;278:44675–44682. doi: 10.1074/jbc.M307633200. [DOI] [PubMed] [Google Scholar]
- 17.Buckley BJ, Marshall ZM, Whorton AR. Nitric oxide stimulates Nrf2 nuclear translocation in vascular endothelium. Biochem Biophys Res Commun. 2003;307:973–979. doi: 10.1016/s0006-291x(03)01308-1. [DOI] [PubMed] [Google Scholar]
- 18.Cullinan SB, Zhang D, Hannink M, Arvisais E, Kaufman RJ, Diehl JA. Nrf2 is a direct PERK substrate and effector of PERK-dependent cell survival. Mol Cell Biol. 2003;23:7198–7209. doi: 10.1128/MCB.23.20.7198-7209.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang HC, Nguyen T, Pickett CB. Phosphorylation of Nrf2 at Ser-40 by protein kinase C regulates antioxidant response element-mediated transcription. J Biol Chem. 2002;277:42769–42774. doi: 10.1074/jbc.M206911200. [DOI] [PubMed] [Google Scholar]
- 20.Yu R, Lei W, Mandlekar S, Weber MJ, Der CJ, Wu J, Kong AN. Role of a mitogen-activated protein kinase pathway in the induction of phase II detoxifying enzymes by chemicals. J Biol Chem. 1999;274:27545–27552. doi: 10.1074/jbc.274.39.27545. [DOI] [PubMed] [Google Scholar]
- 21.Zipper LM, Mulcahy RT. Inhibition of ERK and p38 MAP kinases inhibits binding of Nrf2 and induction of GCS genes. Biochem Biophys Res Commun. 2000;278:484–492. doi: 10.1006/bbrc.2000.3830. [DOI] [PubMed] [Google Scholar]
- 22.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, Oyake T, Hayashi N, Satoh K, Hatayama I, Yamamoto M, Nabeshima Y. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 23.Kwak MK, Cho JM, Huang B, Shin S, Kensler TW. Role of increased expression of the proteasome in the protective effects of sulforaphane against hydrogen peroxide-mediated cytotoxicity in murine neuroblastoma cells. Free Radic Biol Med. 2007;43:809–817. doi: 10.1016/j.freeradbiomed.2007.05.029. [DOI] [PubMed] [Google Scholar]
- 24.Alam J, Stewart D, Touchard C, Boinapally S, Choi AM, Cook JL. Nrf2, a Cap’n’Collar transcription factor, regulates induction of the heme oxygenase-1 gene. J Biol Chem. 1999;274:26071–26078. doi: 10.1074/jbc.274.37.26071. [DOI] [PubMed] [Google Scholar]
- 25.Nguyen T, Huang HC, Pickett CB. Transcriptional regulation of the antioxidant response element. Activation by Nrf2 and repression by MafK. J Biol Chem. 2000;275:15466–15473. doi: 10.1074/jbc.M000361200. [DOI] [PubMed] [Google Scholar]
- 26.Wild AC, Moinova HR, Mulcahy RT. Regulation of gamma-glutamylcysteine synthetase subunit gene expression by the transcription factor Nrf2. J Biol Chem. 1999;274:33627–33636. doi: 10.1074/jbc.274.47.33627. [DOI] [PubMed] [Google Scholar]
- 27.Venugopal R, Jaiswal AK. Nrf2 and Nrf1 in association with Jun proteins regulate antioxidant response element-mediated expression and coordinated induction of genes encoding detoxifying enzymes. Oncogene. 1998;17:3145–3156. doi: 10.1038/sj.onc.1202237. [DOI] [PubMed] [Google Scholar]
- 28.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc Natl Acad Sci U S A. 1997;94:5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prestera T, Holtzclaw WD, Zhang Y, Talalay P. Chemical and molecular regulation of enzymes that detoxify carcinogens. Proc Natl Acad Sci U S A. 1993;90:2965–2969. doi: 10.1073/pnas.90.7.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li Y, Jaiswal AK. Regulation of human NAD(P)H:quinone oxidoreductase gene. Role of AP1 binding site contained within human antioxidant response element. J Biol Chem. 1992;267:15097–15104. [PubMed] [Google Scholar]
- 31.Gabrilovich DI, Bronte V, Chen SH, Colombo MP, Ochoa A, Ostrand-Rosenberg S, Schreiber H. The terminology issue for myeloid-derived suppressor cells. Cancer Res. 2007;67:425–426. doi: 10.1158/0008-5472.CAN-06-3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hanson EM, V, Clements K, Sinha P, Ilkovitch D, Ostrand-Rosenberg S. Myeloid-derived suppressor cells down-regulate L-selectin expression on CD4+ and CD8+ T cells. J Immunol. 2009;183:937–944. doi: 10.4049/jimmunol.0804253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu C, Yu S, Kappes J, Wang J, Grizzle WE, Zinn KR, Zhang HG. Expansion of spleen myeloid suppressor cells represses NK cell cytotoxicity in tumor-bearing host. Blood. 2007;109:4336–4342. doi: 10.1182/blood-2006-09-046201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang B, Pan PY, Li Q, Sato AI, Levy DE, Bromberg J, Divino CM, Chen SH. Gr-1+CD115+ immature myeloid suppressor cells mediate the development of tumor-induced T regulatory cells and T-cell anergy in tumor-bearing host. Cancer Res. 2006;66:1123–1131. doi: 10.1158/0008-5472.CAN-05-1299. [DOI] [PubMed] [Google Scholar]
- 35.Sinha P, V, Clements K, Bunt SK, Albelda SM, Ostrand-Rosenberg S. Cross-talk between myeloid-derived suppressor cells and macrophages subverts tumor immunity toward a type 2 response. J Immunol. 2007;179:977–983. doi: 10.4049/jimmunol.179.2.977. [DOI] [PubMed] [Google Scholar]
- 36.Corzo CA, Cotter MJ, Cheng P, Cheng F, Kusmartsev S, Sotomayor E, Padhya T, McCaffrey TV, McCaffrey JC, Gabrilovich DI. Mechanism regulating reactive oxygen species in tumor-induced myeloid-derived suppressor cells. J Immunol. 2009;182:5693–5701. doi: 10.4049/jimmunol.0900092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ezernitchi AV, Vaknin I, Cohen-Daniel L, Levy O, Manaster E, Halabi A, Pikarsky E, Shapira L, Baniyash M. TCR zeta down-regulation under chronic inflammation is mediated by myeloid suppressor cells differentially distributed between various lymphatic organs. J Immunol. 2006;177:4763–4772. doi: 10.4049/jimmunol.177.7.4763. [DOI] [PubMed] [Google Scholar]
- 38.Schmielau J, Finn OJ. Activated granulocytes and granulocyte-derived hydrogen peroxide are the underlying mechanism of suppression of t-cell function in advanced cancer patients. Cancer Res. 2001;61:4756–4760. [PubMed] [Google Scholar]
- 39.Nagaraj S, Gupta K, Pisarev V, Kinarsky L, Sherman S, Kang L, Herber DL, Schneck J, Gabrilovich DI. Altered recognition of antigen is a mechanism of CD8+ T cell tolerance in cancer. Nat Med. 2007;13:828–835. doi: 10.1038/nm1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lu T, Ramakrishnan R, Altiok S, Youn JI, Cheng P, Celis E, Pisarev V, Sherman S, Sporn MB, Gabrilovich D. Tumor-infiltrating myeloid cells induce tumor cell resistance to cytotoxic T cells in mice. J Clin Invest. 2011;121:4015–4029. doi: 10.1172/JCI45862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sinha P, Parker KH, Horn L, Ostrand-Rosenberg S. Tumor-induced myeloid-derived suppressor cell function is independent of IFN-γ and IL-4Rα. Eur J Immunol. 2012;42:2052–2059. doi: 10.1002/eji.201142230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bunt SK, Sinha P, Clements VK, Leips J, Ostrand-Rosenberg S. Inflammation induces myeloid-derived suppressor cells that facilitate tumor progression. J Immunol. 2006;176:284–290. doi: 10.4049/jimmunol.176.1.284. [DOI] [PubMed] [Google Scholar]
- 43.Chornoguz O, Grmai L, Sinha P, Artemenko KA, Zubarev RA, Ostrand-Rosenberg S. Proteomic pathway analysis reveals inflammation increases myeloid-derived suppressor cell resistance to apoptosis. Mol Cell Proteomics. 2011;10:M110 002980. doi: 10.1074/mcp.M110.002980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sinha P, V, Clements K, Ostrand-Rosenberg S. Reduction of myeloid-derived suppressor cells and induction of M1 macrophages facilitate the rejection of established metastatic disease. J Immunol. 2005;174:636–645. doi: 10.4049/jimmunol.174.2.636. [DOI] [PubMed] [Google Scholar]
- 45.Parker K, Sinha P, Horn L, Clements V, Ostrand-Rosenberg S. HMGB1 enhances immune suppression by facilitating the differentiation and suppressive activity of myeloid-derived suppressor cells. Cancer Res. 2014;74:5723–5733. doi: 10.1158/0008-5472.CAN-13-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sinha P, Ostrand-Rosenberg S. Myeloid-derived suppressor cell function is reduced by Withaferin A, a potent and abundant component of Withania somnifera root extract. Cancer Immunol Immunother. 2013;62:1663–1673. doi: 10.1007/s00262-013-1470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beury DW, Parker KH, Nyandjo M, Sinha P, Carter KA, Ostrand-Rosenberg S. Cross-talk among myeloid-derived suppressor cells, macrophages, and tumor cells impacts the inflammatory milieu of solid tumors. J Leukoc Biol. 2014;96:1109–1118. doi: 10.1189/jlb.3A0414-210R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inoue D, Suzuki T, Mitsuishi Y, Miki Y, Suzuki S, Sugawara S, Watanabe M, Sakurada A, Endo C, Uruno A, Sasano H, Nakagawa T, Satoh K, Tanaka N, Kubo H, Motohashi H, Yamamoto M. Accumulation of p62/SQSTM1 is associated with poor prognosis in patients with lung adenocarcinoma. Cancer Sci. 2012;103:760–766. doi: 10.1111/j.1349-7006.2012.02216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shibata T, Kokubu A, Gotoh M, Ojima H, Ohta T, Yamamoto M, Hirohashi S. Genetic alteration of Keap1 confers constitutive Nrf2 activation and resistance to chemotherapy in gallbladder cancer. Gastroenterology. 2008;135:1358–1368. 1368 e1351–1354. doi: 10.1053/j.gastro.2008.06.082. [DOI] [PubMed] [Google Scholar]
- 50.Shibata T, Ohta T, Tong KI, Kokubu A, Odogawa R, Tsuta K, Asamura H, Yamamoto M, Hirohashi S. Cancer related mutations in NRF2 impair its recognition by Keap1-Cul3 E3 ligase and promote malignancy. Proc Natl Acad Sci U S A. 2008;105:13568–13573. doi: 10.1073/pnas.0806268105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singh A, Boldin-Adamsky S, Thimmulappa RK, Rath SK, Ashush H, Coulter J, Blackford A, Goodman SN, Bunz F, Watson WH, Gabrielson E, Feinstein E, Biswal S. RNAi-mediated silencing of nuclear factor erythroid-2-related factor 2 gene expression in non-small cell lung cancer inhibits tumor growth and increases efficacy of chemotherapy. Cancer Res. 2008;68:7975–7984. doi: 10.1158/0008-5472.CAN-08-1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solis LM, Behrens C, Dong W, Suraokar M, Ozburn NC, Moran CA, Corvalan AH, Biswal S, Swisher SG, Bekele BN, Minna JD, Stewart DJ, Wistuba Nrf2 and Keap1 abnormalities in non-small cell lung carcinoma and association with clinicopathologic features. Clin Cancer Res. 2010;16:3743–3753. doi: 10.1158/1078-0432.CCR-09-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yamamoto T, Yoh K, Kobayashi A, Ishii Y, Kure S, Koyama A, Sakamoto T, Sekizawa K, Motohashi H, Yamamoto M. Identification of polymorphisms in the promoter region of the human NRF2 gene. Biochem Biophys Res Commun. 2004;321:72–79. doi: 10.1016/j.bbrc.2004.06.112. [DOI] [PubMed] [Google Scholar]
- 54.Fahey JW, Haristoy X, Dolan PM, Kensler TW, Scholtus I, Stephenson KK, Talalay P, Lozniewski A. Sulforaphane inhibits extracellular, intracellular, and antibiotic-resistant strains of Helicobacter pylori and prevents benzo[a]pyrene-induced stomach tumors. Proc Natl Acad Sci U S A. 2002;99:7610–7615. doi: 10.1073/pnas.112203099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Khor TO, Huang MT, Prawan A, Liu Y, Hao X, Yu S, Cheung WK, Chan JY, Reddy BS, Yang CS, Kong AN. Increased susceptibility of Nrf2 knockout mice to colitis-associated colorectal cancer. Cancer Prev Res. 2008;1:187–191. doi: 10.1158/1940-6207.CAPR-08-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Osburn WO, Karim B, Dolan PM, Liu G, Yamamoto M, Huso DL, Kensler TW. Increased colonic inflammatory injury and formation of aberrant crypt foci in Nrf2-deficient mice upon dextran sulfate treatment. Int J Cancer. 2007;121:1883–1891. doi: 10.1002/ijc.22943. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki T, Shibata T, Takaya K, Shiraishi K, Kohno T, Kunitoh H, Tsuta K, Furuta K, Goto K, Hosoda F, Sakamoto H, Motohashi H, Yamamoto M. Regulatory nexus of synthesis and degradation deciphers cellular Nrf2 expression levels. Mol Cell Biol. 2013;33:2402–2412. doi: 10.1128/MCB.00065-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xu C, Huang MT, Shen G, Yuan X, Lin W, Khor TO, Conney AH, Kong AN. Inhibition of 7,12-dimethylbenz(a)anthracene-induced skin tumorigenesis in C57BL/6 mice by sulforaphane is mediated by nuclear factor E2-related factor 2. Cancer Res. 2006;66:8293–8296. doi: 10.1158/0008-5472.CAN-06-0300. [DOI] [PubMed] [Google Scholar]
- 59.Mazzoni A, Bronte V, Visintin A, Spitzer JH, Apolloni E, Serafini P, Zanovello P, Segal DM. Myeloid suppressor lines inhibit T cell responses by an NO-dependent mechanism. J Immunol. 2002;168:689–695. doi: 10.4049/jimmunol.168.2.689. [DOI] [PubMed] [Google Scholar]
- 60.Rodriguez PC, Quiceno DG, Zabaleta J, Ortiz B, Zea AH, Piazuelo MB, Delgado A, Correa P, Brayer J, Sotomayor EM, Antonia S, Ochoa JB, Ochoa AC. Arginase I production in the tumor microenvironment by mature myeloid cells inhibits T-cell receptor expression and antigen-specific T-cell responses. Cancer Res. 2004;64:5839–5849. doi: 10.1158/0008-5472.CAN-04-0465. [DOI] [PubMed] [Google Scholar]
- 61.Srivastava MK, Sinha P, Clements VK, Rodriguez P, Ostrand-Rosenberg S. Myeloid-derived suppressor cells inhibit T-cell activation by depleting cystine and cysteine. Cancer Res. 2010;70:68–77. doi: 10.1158/0008-5472.CAN-09-2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bunt SK, V, Clements K, Hanson EM, Sinha P, Ostrand-Rosenberg S. Inflammation enhances myeloid-derived suppressor cell cross-talk by signaling through Toll-like receptor 4. J Leukoc Biol. 2009;85:996–1004. doi: 10.1189/jlb.0708446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gabrilovich DI, Ostrand-Rosenberg S, Bronte V. Coordinated regulation of myeloid cells by tumours. Nat Rev Immunol. 2012;12:253–268. doi: 10.1038/nri3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lesokhin AM, Hohl TM, Kitano S, Cortez C, Hirschhorn-Cymerman D, Avogadri F, Rizzuto GA, Lazarus JJ, Pamer EG, Houghton AN, Merghoub T, Wolchok JD. Monocytic CCR2(+) myeloid-derived suppressor cells promote immune escape by limiting activated CD8 T-cell infiltration into the tumor microenvironment. Cancer Res. 2012;72:876–886. doi: 10.1158/0008-5472.CAN-11-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tsai JJ, Dudakov JA, Takahashi K, Shieh JH, Velardi E, Holland AM, Singer NV, West ML, Smith OM, Young LF, Shono Y, Ghosh A, Hanash AM, Tran HT, Moore MA, van den Brink MR. Nrf2 regulates haematopoietic stem cell function. Nat Cell Biol. 2013;15:309–316. doi: 10.1038/ncb2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Condamine T, Kumar V, Ramachandran IR, Youn JI, Celis E, Finnberg N, El-Deiry WS, Winograd R, Vonderheide RH, English NR, Knight SC, Yagita H, McCaffrey JC, Antonia S, Hockstein N, Witt R, Masters G, Bauer T, Gabrilovich DI. ER stress regulates myeloid-derived suppressor cell fate through TRAIL-R-mediated apoptosis. J Clin Invest. 2014;124:2626–2639. doi: 10.1172/JCI74056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ostrand-Rosenberg S, Sinha P. Myeloid-derived suppressor cells: linking inflammation and cancer. J Immunol. 2009;182:4499–4506. doi: 10.4049/jimmunol.0802740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Parker KH, Beury DW, Ostrand-Rosenberg S. Myeloid-Derived Suppressor Cells: Critical Cells Driving Immune Suppression in the Tumor Microenvironment. Adv Cancer Res. 2015;128:95–139. doi: 10.1016/bs.acr.2015.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zoglmeier C, Bauer H, Norenberg D, Wedekind G, Bittner P, Sandholzer N, Rapp M, Anz D, Endres S, Bourquin C. CpG blocks immunosuppression by myeloid-derived suppressor cells in tumor-bearing mice. Clin Cancer Res. 2011;17:1765–1775. doi: 10.1158/1078-0432.CCR-10-2672. [DOI] [PubMed] [Google Scholar]
- 70.Iclozan C, Antonia S, Chiappori A, Chen DT, Gabrilovich D. Therapeutic regulation of myeloid-derived suppressor cells and immune response to cancer vaccine in patients with extensive stage small cell lung cancer. Cancer Immunol Immunother. 2013;62:909–918. doi: 10.1007/s00262-013-1396-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cheng P, Kumar V, Liu H, Youn JI, Fishman M, Sherman S, Gabrilovich D. Effects of notch signaling on regulation of myeloid cell differentiation in cancer. Cancer Res. 2014;74:141–152. doi: 10.1158/0008-5472.CAN-13-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wiers KM, Lathers DM, Wright MA, Young MR. Vitamin D3 treatment to diminish the levels of immune suppressive CD34+ cells increases the effectiveness of adoptive immunotherapy. J Immunother. 2000;23:115–124. doi: 10.1097/00002371-200001000-00014. [DOI] [PubMed] [Google Scholar]
- 73.Wang XJ, Hayes JD, Henderson CJ, Wolf CR. Identification of retinoic acid as an inhibitor of transcription factor Nrf2 through activation of retinoic acid receptor alpha. Proc Natl Acad Sci U S A. 2007;104:19589–19594. doi: 10.1073/pnas.0709483104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Al-Huseini LM, Aw Yeang HX, Sethu S, Alhumeed N, Hamdam JM, Tingle Y, Djouhri L, Kitteringham N, Park BK, Goldring CE, Sathish JG. Nuclear factor-erythroid 2 (NF-E2) p45-related factor-2 (Nrf2) modulates dendritic cell immune function through regulation of p38 MAPK-cAMP-responsive element binding protein/activating transcription factor 1 signaling. J Biol Chem. 2013;288:22281–22288. doi: 10.1074/jbc.M113.483420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jaramillo MC, Zhang DD. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013;27:2179–2191. doi: 10.1101/gad.225680.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.