SUMMARY
Modern transportation networks have facilitated the migration and mingling of previously isolated populations of plants, animals, and insects. Human activities can also influence the global distribution of microorganisms. The best understood example is yeasts associated with winemaking. Humans began making wine in the Middle East over 9,000 years ago [1, 2]. Selecting favorable fermentation products created specialized strains of Saccharomyces cerevisiae [3, 4] that were transported along with the grapevines. Today, S. cerevisiae strains residing in vineyards around the world are genetically similar, and their population structure suggests a common origin that followed the path of human migration [3–7]. Like wine, coffee and cacao depend on microbial fermentation [8, 9] and have been globally dispersed by humans. Theobroma cacao originated in the Amazon and Orinoco Basins of Colombia and Venezuela [10], was cultivated in Central America by the Mesoamerican peoples, and introduced to Europeans by Cortés in 1530 [11]. Coffea, native to Ethiopia, was disseminated by Arab traders throughout the Middle East and North Africa in the 6th century and was introduced to European consumers in the 17th century [12]. Here, we test whether the yeasts associated with coffee and cacao are genetically similar, crop-specific populations or genetically diverse, geography-specific populations. Our results uncovered populations that, while defined by niche and geography, also bear signatures of admixture between major populations in events independent of the transport of the plants. Thus, human-associated fermentations and migration may have affected the distribution of yeast involved in the production of coffee and chocolate.
eTOC Blurb
Human activity has driven the migration and mingling previously isolated populations of plants, animals, and insects. Is the same true for microorganisms? Ludlow, et al. find that yeasts associated with coffee and cacao form distinct populations with independent origins through admixtures of previously known populations, including the wine yeasts.
RESULTS
S. cerevisiae is readily isolated from unroasted cacao and coffee beans
To test the extent to which human activity may have influenced the microorganisms associated with coffee and cacao fermentation, we focused on one microbe, S. cerevisiae. The importance of yeast in cacao fermentation is clear [13]. Cacao undergoes 5–7 days of fermentation, seeded from the local flora during manipulations of the cacao pods. Successions of yeasts, lactic acid bacteria, and acetic acid bacteria digest the pectinaceous pulp surrounding the beans and trigger biochemical changes that impart flavor and color to the beans [13–15]. In contrast, the microbiota of coffee fermentation is poorly understood, and only a few studies have detected yeasts in coffee fermentations [9, 15–18]. Coffee growers use different types of fermentation to digest the cherry pulp surrounding the beans. The most common are the “wet” 24–48 hour fermentation in water [16, 18] and the “dry” 10–25 day fermentation with rounds of spreading and heaping on a platform [18]. Without direct access to the fermentations, we attempted to culture live yeast from unroasted coffee and cacao beans grown and processed in a variety of geographic locations and ecological niches. From beans grown in Central America, South America, Africa, Indonesia, or the Middle East, we obtained 78 cacao strains from 13 countries and 67 coffee strains from 14 countries (Tables 1 and S1). For brevity, we will refer to strains isolated from countries in Central and South America as “South America” to reflect similarities in climate and geographic proximity. Both cacao and coffee bean cultures also contained a variety of bacteria, yeasts, and filamentous fungi, suggesting that this approach could be readily adapted for the isolation of other microorganisms.
Table 1.
The origin and number of S. cerevisiae strains from coffee and cacao analyzed in this study. See also Table S1.
| Cacao Origin | Isolates | Coffee Origin | Isolates |
|---|---|---|---|
| Colombia | 1 | Colombia | 6 |
| Costa Rica | 11 | Costa Rica | 3 |
| Dominican Republic | 7 | Ethiopia | 3 |
| Ecuador | 12 | Guatemala | 2 |
| Ghana1 | 5 | Honduras | 14 |
| Haiti | 7 | Indonesia | 12 |
| Ivory Coast | 6 | Kenya | 2 |
| Madagascar | 1 | Mexico | 2 |
| Nicaragua | 2 | Nicaragua | 3 |
| Nigeria | 7 | Peru | 5 |
| Papua New Guinea | 2 | Rwanda | 5 |
| Peru | 9 | Uganda | 4 |
| Venezuela | 8 | United States | 1 |
| Yemen | 5 |
Coffee and cacao yeasts form discrete, genetically diverse populations
To measure the genetic diversity of the yeast strains associated with coffee and cacao beans, we used RAD-seq [19] to sequence the same 3% of each strain’s genome. We compared this sequence to polymorphism information across the same regions of the genome for 35 wine strains from a previously published RAD-seq dataset [20]. Our results (Figure 1A and 1B) show a sharp contrast between the wine, coffee, and cacao strains. Similar to previous studies [4–6], we observed limited genetic diversity between wine strains, with a median pairwise p-distance of 1.28e-3 (Experimental Procedures). The yeast strains isolated from both coffee and cacao beans exhibited significantly greater diversity than that of the wine strains, with median pairwise p-distances of 3.39e-3 and 2.95e-3, respectively (p=9.5e-162 and p=4.6e-156, Mood’s Median Test).
Figure 1.
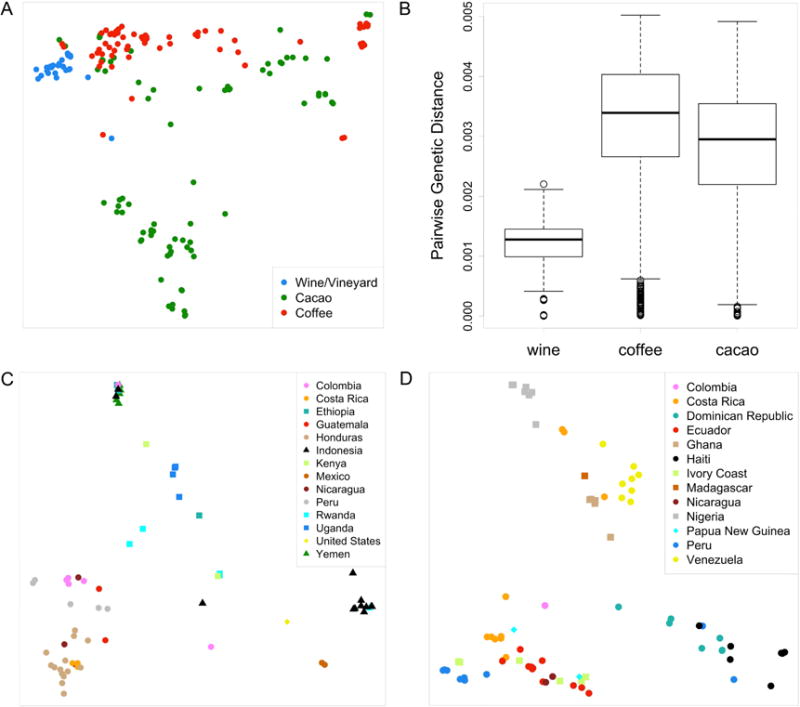
Genetic diversity of yeast strains isolated from different niches. A. MDS (multidimensional scaling) plot of genetic distance (Euclidian), between all coffee, cacao and vineyard strains using first and second principal coordinates (x- and y- axes), which explain 18% and 7% of variation among strains, respectively. B. Distributions of the pairwise P-distances between strains. C. MDS plot of genetic distance for coffee strains where the first and second principal coordinates (x- and y- axes) explain 22% and 8% of variation among strains, respectively. D. MDS plot of genetic distance between cacao strains where the first and second principal coordinates (x- and y- axes) explain 18% and 11% of variation among strains, respectively. See also Table S4.
The greater genetic diversity of the coffee and cacao strains relative to the wine strains is less consistent with a crop-specific origin of these yeasts, but is by no means conclusive evidence against it. To explore this question in more detail, we examined the genetic similarity of strains isolated from geographically proximal locations. Both coffee and cacao strains show strong country-level clustering, even though bean samples were obtained from multiple suppliers in different places at different times (Figure 1C and 1D). In fact, virtual predictions of sample origin (where the provenance of a strain is predicted based on that of the most genetically similar strain) were accurate 86% of the time for cacao strains and 79% of the time for coffee (Supplemental Information). These results support the hypothesis that the origin of yeasts isolated from imported bean samples is the original local fermentation rather than the result of cross contamination during distribution and suggests the existence of coffee and cacao-specific yeasts that differ based on the location where the beans were grown and fermented.
To test this hypothesis more rigorously, we analyzed the population structure of the coffee and cacao yeasts using the Monte Carlo Markov chain algorithm InStruct [21]. This analysis included RAD-seq data from the coffee and cacao strains isolated in this study, a previously published set of 262 strains of S. cerevisiae from a variety of geographic origins and ecological niches [20], and 57 additional strains, including a set of 13 China tree strains (group 1), used to root and extend the phylogenetic tree (Table S1). The deviance information criterion (Experimental Procedures) suggested twelve as the most likely number of populations (Figure S1). The population structure generated (Figures 2 and S2) largely agreed with previous analyses of data generated by microarray hybridization [6], RAD-seq [20] and whole genome sequencing [5, 22] with five populations (Table S2) that, except for the addition of novel strains, are largely unchanged from our previous analysis [20]. These include the North America oak (group 3), New Zealand soil (group 11), Israel soil (group 12), Asia Mixed (group 7), and Pan Mixed 2 (group 6). The large European wine population expanded slightly to include strains previously assigned to other populations and the new Pan Mixed 1 group was formed by strains that had been placed in other groups in our previous analysis. With the exception of Pan Mixed 2, a human-associated population with extensive aneuploidy, polyploidy and heterozygosity (Supplemental Information), these populations harbor relatively few strains isolated from coffee or cacao beans (Table S3 and Figure S2).
Figure 2.
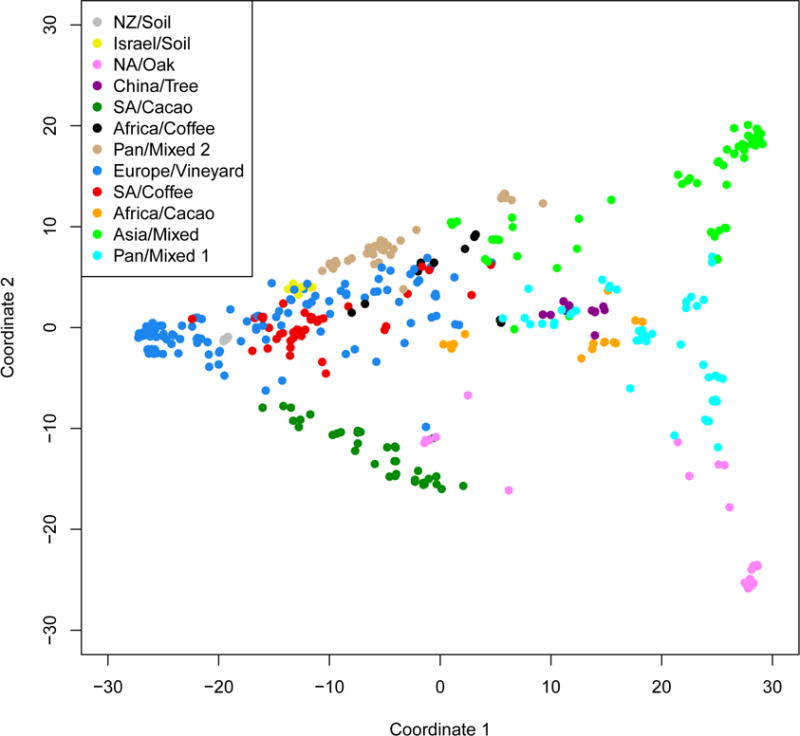
Genetic distance between yeast strains. Each strain is plotted along first and second principal coordinates obtained from multidimensional scaling of genetic distance (Euclidian). Populations are labeled by the most common source and/or geographic location from which they were originally isolated. Populations containing strains isolated from Central and South America are labeled South America. Each strain is color coded by populations inferred from InStruct with acronyms NA (North America), SA (South America), NZ (New Zealand). The first and second principal coordinates explain 22% and 7% of variation among strains, respectively. See also Figures S1–S3 and Tables S2, S3 and S5.
Most coffee and cacao strains resided in four new populations of which they are the majority and in some cases exclusive members: South America cacao (group 4), Africa cacao (group 8), South America coffee (group 9), and Africa coffee (group 5). The population structure of cacao and coffee strains is significant for several reasons. First, in contrast to the wine strains, the coffee and cacao yeasts form multiple, discreet populations. The fact that not all coffee or cacao strains are related suggests independent origins for distinct populations of yeast associated with the same human-associated activities. Second, while these populations reflect ecological niche, i.e. coffee versus cacao (Table S3 and Figure S2), they also reflect geographical origin (Figure 2). The coffee strains provide the clearest example, with South American and African coffee strains each forming a single population (Table S3) that further clusters by country (Figure 1C). While the cacao strains also showed strong country level clustering (Figure 1D), each of the two major populations include strains from samples whose declared continent of origin is different from other population members (Table S3). This may reflect a more complex pattern of migration events for cacao strains than for coffee strains, although mislabeling of sample origin is also possible.
Admixture and migration events
Because their population structure suggested that the yeasts associated with coffee and cacao beans are members of new groups, we sought to understand the origin of these populations. Previous analyses had identified three major populations of S. cerevisiae (European vineyard, Asian, and North American oak, which is related to a Japanese oak population [23]) and strains with substantial admixture between them [20]. We anticipated finding novel populations of yeast by sampling new locations and ecological niches, particularly since (with the exception of vineyards) the southern hemisphere had been largely uncharacterized. Interestingly, the new coffee and cacao populations were not composed of strains with novel alleles (Figure S2), but were instead admixtures of the three known yeast populations. Furthermore, these admixtures roughly corresponded to the geographic proximity of the sample’s origin and patterns of human migration. For example, the two South American populations (SA coffee and SA cacao) share alleles with the North American oak (NA oak) population. In contrast, the allelic profiles of both African groups (coffee and cacao) show mixtures of European and Asian alleles.
To quantitatively infer historical relationships (including migration events) among the 12 populations, we used the TreeMix algorithm [24], which builds population trees and tests for the presence of gene flow between diverged populations. Using 4,966 sites with minor allele frequencies above 1%, we estimated a maximum likelihood tree (Figure 3) rooted using the China population, the likely the ancestral population of S. cerevisiae [25]. By sequentially adding migration events, we found significant improvement in fit for up to 9 events, although only the first five had admixture fractions above 5%. Without migration events, the tree structure explained 89.8% of the variance in relatedness among the populations and inclusion of the first five migration events increased the variance explained to 99.3%. Following the authors’ recommendations [24], we evaluated the inferred admixture events using a three-population test (f3) of admixture [26]. This analysis confirmed three of the five migration events, with 30.3% migration from NA Oak to SA Cacao, 18.8% from Asia to Pan Mixed 2, and 18.8% from either Asia Mixed or NA Oak to SA Coffee. With Z scores of −4.0, −2.6, −2.1, −2.8, respectively, these tests provide statistical support for models of population divergence by admixture instead of drift.
Figure 3.
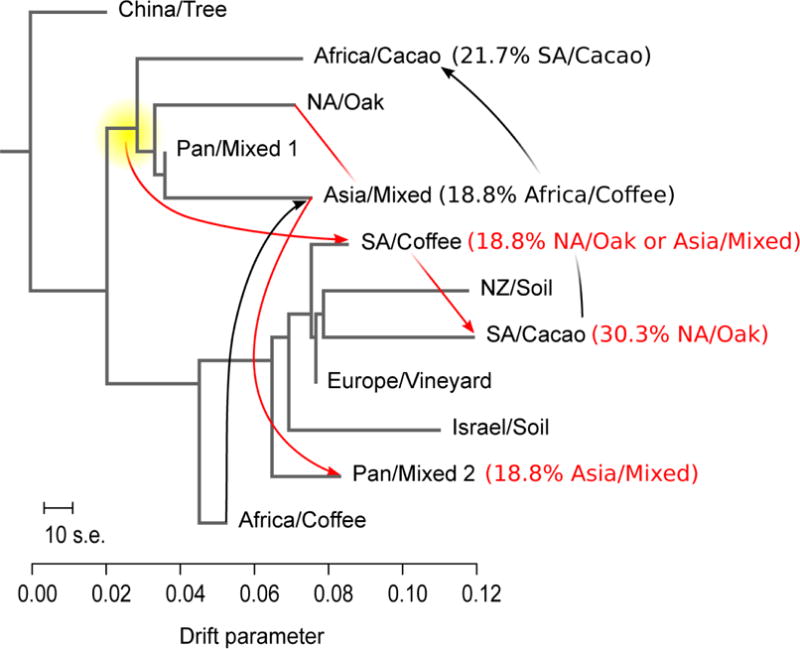
Maximum likelihood tree of genetic relationships among populations and admixture events based on the TreeMix model. Branch lengths are proportional to drift in allele frequencies between populations, shown by the scale along with the standard error (S.E.) of the sample covariance matrix. Arrows show migration events resulting in admixture. Red arrows depict migration events that passed the significance threshold of the three-population test (f3). The yellow node indicates a Treemix migration event from the cluster that contains both the NA oak and Asian groups to SA Coffee. Migration events from both of these population to SA Coffee were significant by the f3 test. Black arrows show predicted migration events that were not statistically significant in the f3 test.
Taken together, our results provide evidence for historical migration and admixture that could help explain the origin of the Pan Mixed 2 and South American populations. Two of these migration events show an interesting connection to what is known about the migration of the plants themselves. Cacao originated in South America (Columbia and Venezuela) and was transported as far north as Mexico and the Southwestern United States before being widely dispersed by Europeans. The South American cacao yeast population includes close derivatives of the European vineyard population with substantial admixture from the geographically proximal North American oak population (Figure 3). Coffee originated in Ethiopia, was transported to Yemen, and was then regionally dispersed by Arab traders before being more widely cultivated by Europeans. Several of our East African and all of our Yemeni coffee strains were present in a single population (Pan Mixed 2), which is highly related to the European vineyard population with a statistically significant migration from an Asia Mixed population (Figure 3). The ancient and continuing global traffic in yeasts associated with wine fermentation may have set the stage for subsequent mingling and admixture events that gave rise to these new populations.
DISCUSSION
Although the production of wine, coffee, and chocolate all rely on the human associated activities of cultivation and fermentation, wine production differs from coffee and cacao in a few crucial respects. First, the vessels used in wine fermentation, e.g. oak barrels, are often exported from established winemaking regions to areas of new cultivation and can serve as reservoirs of yeasts native to their country of origin [7]. Moreover, unlike wine fermentations, the use of starter cultures is not common in cacao and coffee fermentations. The more natural fermentation styles of cacao and coffee suggested that the populations associated with them might be different from yeasts found in the region where the plants originated. Indeed, our results show that, unlike wine, coffee and cacao fermentations are not typically carried out by clonal populations of yeasts common to all areas where the crops are cultivated, but rather by populations specific to geographical regions and niches, which appear to have arisen independently. Coffee and cacao yeasts appear to be the result of admixture events that generated combinations of alleles from Europe, Asia, and North America. Human activities may have fostered the establishment of these hybrid groups. In several cases, the combinations of alleles present in these groups coincide with known paths of transportation, organized cultivation and fermentation of the crops. Once established, new populations appear to have become abundant in regional coffee or cacao production. In fact, the DNA sequence of yeasts isolated from unroasted beans recovered from these fermentations can often accurately pinpoint the geographic origin of the beans themselves.
The genetic variation found in these new populations may provide a rich source of phenotypic diversity that could be exploited to enhance the products they ferment. It has long been known that different wine strains can produce vastly different fermentation results. For example, wines made from the same grape cultivar in different regions or even from the same lot of sterile grape juice in the laboratory [27] possess remarkable differences that distinguish them from one another. Bokulich et al. [28] demonstrated that the microbes found on grapes differ according to cultivar, region, and climate and posited the existence of a “microbial terroir”. In more recent work, Knight et al. [29] showed that grape fermentations using S. cerevisiae strains isolated from different locations have chemical profiles that correlate with the region of origin. Given that the organisms involved in the fermentation of cacao are known to influence flavor profiles of chocolate, and are more genetically diverse than wine strains, the idea that these local yeast populations may impart flavors that yield clues to the terroir of chocolate and, possibly coffee, is intriguing.
EXPERIMENTAL PROCEDURES
S. cerevisiae isolation
With the exception of 5 previously described Ghana cacao strains [14, 20], all coffee and cacao strains (Table S1) were isolated from cultures containing 8–10 cacao beans or 30–50 coffee beans using a previously described isolation method [20]. Unroasted cocoa beans were obtained from Theo Chocolate (Seattle, WA) and Chocolate Alchemy (Eugene, OR). Green coffee bean samples were obtained from Victrola Coffee Roasters (Seattle, WA), Sweet Maria’s (Oakland, CA) and Burman Coffee Traders (Madison, WI).
RAD sequencing and analysis
RAD-seq was performed on 140 coffee and cacao strains and 57 additional strains from the U.S. Department of Agriculture ARS Culture Collection or from remote locations in China [25] (Table S1) that were used to root and extend the phylogenic tree. Strains having a p-distance of less than 5×104 were removed from the analysis unless they had been isolated from independent samples. Genomic DNA was isolated and RAD-seq libraries prepared as described [20]. Pooled libraries were sequenced on a HiSeq 2000 (Illumina) with 50 base pair paired-end reads (Northwest Genomics Center, University of Washington, Seattle WA. USA). The read sequences generated for this study are available at the European Nucleotide Archive (ENA) under accession number PRJEB12530. Reads were aligned to the S. cerevisiae reference genome (chromosome accessions: NC_001133.8, NC_001134.7, NC_001135.4, NC_001136.8, NC_001137.2, NC_001138.4, NC_001139.8, NC_001140.5, NC_001141.1, NC_001142.7, NC_001143.7, NC_001144.4, NC_001145.2, NC_001146.6, NC_001147.5, NC_001148.3) using BWA (version 0.5.8c) [30] with up to 6 mismatches allowed, and the resulting read alignments were merged with those from a previous study [20]. From these alignments, single nucleotide polymorphisms (SNPs) were called using GATK’s unified genotyper (version 2.7-4) with a prior of 0.005 for heterozygous sites and with ploidy of two [31]. A total of 223311 total and 15,426 variable sites across the 438 strains remained after filtering to remove those sites with more than 20% missing genotype values, a GATK genotype quality score (Phred-scaled probability that the genotype assignment is incorrect) less than 60 or displaying more than two alleles (population-wide). Heterozygous sites with five or fewer reads supporting the minor allele were then set to missing, resulting in the elimination of 13,701 genotype values and a final overall rate of 3.9% missing genotype values (Table S6). Non-synonymous (n = 5,233) and synonymous (n = 6,167) SNPs were identified using the variant effect predictor script from Ensembl [32].
Population analysis
Population structure was inferred using InStruct [21] with two sets of data. The first set included 438 strains and genotype information at 843 variable sites, filtered from the total 15,426 variable sites to remove sites with minor allele frequency less than 1% and sites within 5 KB of one another. The second set used the same set of sites but only included the 318 strains that were not triploid, tetraploid, or aneuploid. InStruct was run with between 6 and 30 populations, a burn of 20K iterations followed by an additional 20K iterations. We chose the optimal number of populations by requiring the change in the deviance information criterion (DIC) to be greater than the standard error in DIC from 20 independent runs.
P-distance was calculated as the proportion of non-identical genotypes observed across all 223311 typed base positions. Heterozygous-homozygous differences were given a value of 0.5.
Multidimensional scaling and cluster analysis were used to further visualize the relationships of new and admixed populations to those previously identified [20]. Multidimensional scaling was applied to all sites using Euclidian identity-by-state distance (with state distance between non- identical homozygous/hemizygous alleles encoded as 2 and between heterozygous and homozygous sites encoded as 1) and the “cmdscale” function in R. Cluster analysis was applied to 2,615 sites with minor allele frequencies of 1% or greater and less than 1% missing data using hierarchical complete linkage clustering. For comparisons between wine/vineyard, coffee and cacao yeast strains, any vineyard or winemaking strains associated with Vitis species other than Vitis vinfera were excluded.
Supplementary Material
Highlights.
Saccharomyces cerevisiae can be isolated from unroasted coffee and cacao beans
Yeasts associated with coffee and cacao are more diverse than wine yeasts
Coffee and cacao yeasts form distinct populations specific to geographic locations
Genomic signatures of migration and admixture events provide clues to their origin
Acknowledgments
The authors thank Amir Sherman (Agricultural Research Organization) for helpful discussions. Dan Ollis and Perry Hook of Victrola Coffee in Seattle, WA for gifts of coffee beans. Andy McShea, of Theo Chocolate, Seattle, WA and John Nanci of Chocolate Alchemy, Eugene, OR for gifts of cacao beans. Feng-Yan Bai (Chinese Academy of Sciences) and James Swezey, (USDA/ARS) for providing strains. This work was funded by a strategic partnership between the University of Luxembourg and the Institute for Systems Biology and NIH grant GM080669 to J.F.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
Conceptualization, A.M.D., C.L.L. and C.G.T.; Methodology, G.A.C., J.F., E.J. and C.L.L.; Investigation, G.A.C., J.F., C.F., M.H., E.J., C.L.L., A.S. and C.G.T.; Writing – Original Draft, C.L.L.; Writing – Review & Editing, A.M.D. and C.L.L.; Funding Acquisition, A.M.D. and J.F.; Supervision, A.M.D.
References
- 1.Cavalieri D, McGovern PE, Hartl DL, Mortimer R, Polsinelli M. Evidence for S. cerevisiae fermentation in ancient wine. Journal of molecular evolution. 2003;57(Suppl 1):S226–232. doi: 10.1007/s00239-003-0031-2. [DOI] [PubMed] [Google Scholar]
- 2.McGovern PE. Ancient wine: the search for the origins of viniculture. Princeton, New Jersey: Princeton University Press; 2003. [Google Scholar]
- 3.Fay JC, Benavides JA. Evidence for domesticated and wild populations of Saccharomyces cerevisiae. PLoS genetics. 2005;1:66–71. doi: 10.1371/journal.pgen.0010005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Legras JL, Merdinoglu D, Cornuet JM, Karst F. Bread, beer and wine: Saccharomyces cerevisiae diversity reflects human history. Mol Ecol. 2007;16:2091–2102. doi: 10.1111/j.1365-294X.2007.03266.x. [DOI] [PubMed] [Google Scholar]
- 5.Liti G, Carter DM, Moses AM, Warringer J, Parts L, James SA, Davey RP, Roberts IN, Burt A, Koufopanou V, et al. Population genomics of domestic and wild yeasts. Nature. 2009;458:337–341. doi: 10.1038/nature07743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schacherer J, Shapiro JA, Ruderfer DM, Kruglyak L. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature. 2009;458:342–345. doi: 10.1038/nature07670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goddard MR, Anfang N, Tang R, Gardner RC, Jun C. A distinct population of Saccharomyces cerevisiae in New Zealand: evidence for local dispersal by insects and human-aided global dispersal in oak barrels. Environmental microbiology. 2010;12:63–73. doi: 10.1111/j.1462-2920.2009.02035.x. [DOI] [PubMed] [Google Scholar]
- 8.Papalexandratou Z, Falony G, Romanens E, Jimenez JC, Amores F, Daniel HM, De Vuyst L. Species diversity, community dynamics, and metabolite kinetics of the microbiota associated with traditional ecuadorian spontaneous cocoa bean fermentations. Applied and environmental microbiology. 2011;77:7698–7714. doi: 10.1128/AEM.05523-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Silva CF, Batista LR, Abreu LM, Dias ES, Schwan RF. Succession of bacterial and fungal communities during natural coffee (Coffea arabica) fermentation. Food microbiology. 2008;25:951–957. doi: 10.1016/j.fm.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Motamayor JC, Lachenaud P, da Silva EMJW, Loor R, Kuhn DN, Brown JS, Schnell RJ. Geographic and genetic population differentiation of the Amazonian chocolate tree (Theobroma cacao L) PloS one. 2008;3:e3311. doi: 10.1371/journal.pone.0003311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Young AM. The chocolate tree: a natural history of cacao. Washington, DC: Smithsonian Institution Press; 1994. [Google Scholar]
- 12.Pendergrast M. Uncommon grounds: the history of coffee and how it transformed our world. New York: Basic Books; 1999. [Google Scholar]
- 13.Schwan RF, Wheals AE. The microbiology of cocoa fermentation and its role in chocolate quality. Crit Rev Food Sci Nutr. 2004;44:205–221. doi: 10.1080/10408690490464104. [DOI] [PubMed] [Google Scholar]
- 14.Jespersen L, Nielsen DS, Honholt S, Jakobsen M. Occurrence and diversity of yeasts involved in fermentation of West African cocoa beans. FEMS yeast research. 2005;5:441–453. doi: 10.1016/j.femsyr.2004.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Schwan RF, Wheals AE. Mixed microbial fermentations of chocolate and coffee. In: Boekhout T, Robert V, editors. Yeasts in Food. Boca Raton, FL: CRC Press; 2003. [Google Scholar]
- 16.Agate AD, Bhat JV. Role of pectinolytic yeasts in the degradation of mucilage layer of Coffea robusta cherries. Applied microbiology. 1966;14:256–260. doi: 10.1128/am.14.2.256-260.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Masoud W, Cesar LB, Jespersen L, Jakobsen M. Yeast involved in fermentation of Coffea arabica in East Africa determined by genotyping and by direct denaturating gradient gel electrophoresis. Yeast. 2004;21:549–556. doi: 10.1002/yea.1124. [DOI] [PubMed] [Google Scholar]
- 18.Silva CF, Schwan RF, Sousa Dias ES, Wheals AE. Microbial diversity during maturation and natural processing of coffee cherries of Coffea arabica in Brazil. Int J Food Microbiol. 2000;60:251–260. doi: 10.1016/s0168-1605(00)00315-9. [DOI] [PubMed] [Google Scholar]
- 19.Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PloS one. 2008;3:e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cromie GA, Hyma KE, Ludlow CL, Garmendia-Torres C, Gilbert TL, May P, Huang AA, Dudley AM, Fay JC. Genomic sequence diversity and population structure of Saccharomyces cerevisiae assessed by RAD-seq. G3. 2013;3:2163–2171. doi: 10.1534/g3.113.007492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao H, Williamson S, Bustamante CD. A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics. 2007;176:1635–1651. doi: 10.1534/genetics.107.072371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strope PK, Skelly DA, Kozmin SG, Mahadevan G, Stone EA, Magwene PM, Dietrich FS, McCusker JH. The 100-genomes strains, an S. cerevisiae resource that illuminates its natural phenotypic and genotypic variation and emergence as an opportunistic pathogen. Genome research. 2015;25:762–774. doi: 10.1101/gr.185538.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Almeida P, Barbosa R, Zalar P, Imanishi Y, Shimizu K, Turchetti B, Legras JL, Serra M, Dequin S, Couloux A, et al. A population genomics insight into the Mediterranean origins of wine yeast domestication. Mol Ecol. 2015;24:5412–5427. doi: 10.1111/mec.13341. [DOI] [PubMed] [Google Scholar]
- 24.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS genetics. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang QM, Liu WQ, Liti G, Wang SA, Bai FY. Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol Ecol. 2012;21:5404–5417. doi: 10.1111/j.1365-294X.2012.05732.x. [DOI] [PubMed] [Google Scholar]
- 26.Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hyma KE, Saerens SM, Verstrepen KJ, Fay JC. Divergence in wine characteristics produced by wild and domesticated strains of Saccharomyces cerevisiae. FEMS yeast research. 2011;11:540–551. doi: 10.1111/j.1567-1364.2011.00746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bokulich NA, Thorngate JH, Richardson PM, Mills DA. Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E139–148. doi: 10.1073/pnas.1317377110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Knight S, Klaere S, Fedrizzi B, Goddard MR. Regional microbial signatures positively correlate with differential wine phenotypes: evidence for a microbial aspect to terroir. Scientific reports. 2015;5:14233. doi: 10.1038/srep14233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McLaren W, Pritchard B, Rios D, Chen Y, Flicek P, Cunningham F. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 2010;26:2069–2070. doi: 10.1093/bioinformatics/btq330. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


