Abstract
While the notion that RNAs can function as regulators dates back to early molecular studies of gene regulation of the lac operon, it is only over the last decade that the ubiquity and diversity of regulatory RNAs are being realized. Advancements in high throughput sequencing and the adoption of these approaches to rapidly sequence genomes and transcriptomes and to examine gene expression and RNA binding protein specificity have revealed an ever-expanding RNA world. In this review, we focus on recent studies revealing that RNA fragments cleaved from larger coding or noncoding RNAs can have regulatory functions. Additionally, we discuss examples of riboswitches that function in trans as mRNA or protein-binding sRNAs, upending the traditional thinking that these are exclusively cis-acting elements.
Graphical Abstract
Introduction
sRNAs in bacteria can act by modulating the activity of a particular protein(s) or regulating mRNA stability and/or translation by base-pairing with near complementary sequence in the targeted message [reviewed in [1]]. Among the most well studied base-pairing sRNAs are those in gram-negative bacteria that require the RNA chaperone Hfq for stability and function. Most Hfq-dependent sRNAs identified by empirical and bioinformatic methods are primary transcripts, but some are produced from larger transcripts by endoribonucleolytic cleavages [2-7]. However, the vast majority of these sRNA fragments have not been well characterized. Similarly, regulatory RNA elements that can bind small molecules and/or metabolites called riboswitches have mainly been studied when found in mRNA leader sequences. However, some riboswitches have been identified antisense to the regulated gene and others are completely isolated from the regulated gene, i.e. “marooned” [8].
Several recent studies have revealed that RNA fragments cleaved from larger coding or noncoding RNAs indeed have regulatory functions and that RNA regulatory elements such as marooned riboswitches can function as independent sRNAs that regulate gene expression in trans by base-pairing with mRNAs or titrating RNA binding proteins. In this review, we will focus on recent studies examining these two major types of newly unmasked sRNAs - RNA fragments and trans-acting riboswitches.
RNA derived fragments
In the early 1970's, it was discovered that stable RNAs (rRNAs and tRNAs) were transcribed as larger molecules that were processed to generate the mature RNAs [9-12]. The fact that RNA fragments were removed during maturation raised several questions. As stated by Altman and Robertson in 1973 [13], “A major question about these molecules is that of the function of the “extra” segments”. Another question was how are these precursors processed to generate the mature tRNA and rRNA species. Studies over the subsequent four decades have primarily focused on the latter question. Over the last several years, transcriptomic studies in bacteria [3,14,15], archaea [16], and eukaryotes [17] have detected a plethora of fragments derived from tRNA, rRNAs, mRNAs, and riboswitches that have revived the important functional question.
tRNA derived fragments
Hints to a role for tRNA derived fragments (tRFs) in regulating RNA stability through base-pairing with target RNAs came from multiple studies that identified tRNAs among the RNAs that bind to the RNA chaperone Hfq in E. coli [3,6]. Hfq is a key player in the posttranscriptional regulation of gene expression in many gram-negative and some gram-positive bacteria [reviewed in [18]]. Hfq binds both base-pairing sRNAs and their target mRNAs in random order [19,20] and facilitates their annealing, which in turn alters mRNA translation and/or stability. Despite the known role for Hfq in the regulation of gene expression by facilitating sRNA/mRNA interactions, the binding of Hfq to tRNAs was initially interpreted as Hfq playing a role in tRNA maturation [3,21,22].
In a recent paper, Lalaouna et al. discovered a regulatory function for these Hfq associated tRFs in E. coli [23]. These authors sought to identify mRNA targets for the Hfq-binding RyhB and RybB sRNAs by specifically immunoprecipitating the MS2-tagged sRNA and identifying co-purifying RNAs by high throughput sequencing. Surprisingly, they recovered in addition to known and potential mRNA targets, RNA fragments that were excised during tRNA maturation. Interestingly, tRFs derived from the leuZ primary transcripts base-paired with RyhB leading to a reduction in its levels, and in turn, to up-regulation of its mRNA targets (Figure 1A). Furthermore, additional tRFs were found to bind other sRNAs. The ability of tRFs to act as base-pairing sRNAs is not unique to bacteria. Other studies have found that specific tRFs in human cells have a regulatory function, i.e., promoting cell proliferation [17] and regulating mRNAs by base-pairing [24]. While tRFs have also been discovered in archaea, those characterized so far act as stress-induced noncoding RNAs that inhibit protein synthesis by binding to the ribosome [25].
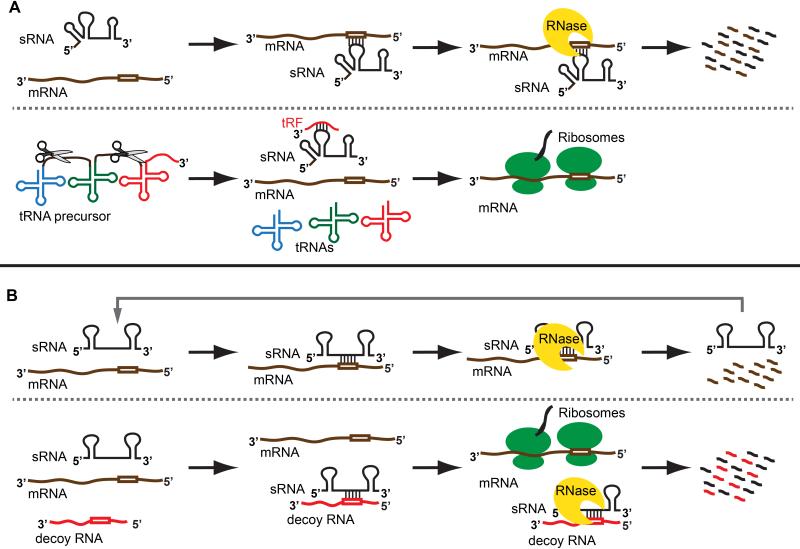
Figure 1. Mechanisms of post-transcriptional regulation of gene expression by tRNA and mRNA derived fragments.
(A) Upper panel: an sRNA base-pairs with near complementary sequence in the target mRNA leading to coupled degradation of the sRNA and mRNA. Lower panel: the primary tRNA transcript is processed by ribonucleases to generate the mature tRNAs. Fragments excised by endoribonucleases (tRFs) from the tRNA base-pair with an sRNA, preventing it from binding and regulating its target mRNA. In the absence of sRNA-mediated regulation, the mRNA is efficiently translated. (B) Upper panel: an sRNA base-pairs with a target mRNA resulting in decay of the mRNA, and the sRNA then proceeds to regulate additional targets. Lower panel: an mRNA or mRNA derived fragment (decoy RNA) base-pairs with the sRNA blocking it from binding its target mRNA, which is then efficiently translated.
mRNA derived fragments
The first global studies examining sRNA content in E. coli by sequencing cloned cDNAs from size selected RNAs or detecting Hfq bound RNAs with microarrays identified mRNA derived fragments in addition to freestanding sRNAs [2,5,6]. More recent, high throughput sequencing of Hfq-bound RNA has revealed a myriad of mRNA-derived fragments bound to Hfq [3,4]. A hint at the regulatory function for some of these mRNA-derived sRNAs came from work on ChiX, an Hfq-dependent sRNA conserved between E. coli and Salmonella enterica. Figuroa-Bossi et al. demonstrated that ChiX downregulates the mRNA encoding the chitin porin ChiP, but induction of the decoy chb mRNA leads to degradation of ChiX as a consequence of chb-ChiX base-pairing [26], resulting in up-regulation of the chitin porin encoding mRNA. Therefore, the chb mRNA acts as sponge that soaks up ChiX leading to the accumulation of its negatively regulated mRNA targets (Figure 1B). A subsequent study found that the SroC sRNA is a gltIJKL mRNA cleavage product and serves as a sponge for the GcvB sRNA, leading to reduced regulation of its target mRNAs [27]. Thus, both mRNAs and mRNA-derived sRNAs can act as molecular sponges. Not all of these mRNA-derived sRNAs act by this mechanism. Other sRNAs that are transcribed as free standing sRNAs or as part of mRNAs regulate in the traditional manner of targeting mRNAs by base-pairing [4].
Trans-Acting Riboswitches
Over two decades ago, it was first observed that 5’ untranslated regions (5’ UTRs) of some mRNAs can bind ligands, specifically uncharged tRNAs. The cis binding elements were termed T-boxes, and their interactions with the tRNAs control whether the downstream genes encoding cognate tRNA synthases are expressed [28,29]. While in this case the interaction between the uncharged-tRNA and T-box occurs via base-pairing, RNA is also capable of forming structures that can bind other types of ligands [30,31], and 5’ UTRs that bind other ligands were subsequently discovered and termed riboswitches. A flurry of research ensued with the focus on discovering riboswitches, identifying novel ligands, and defining their regulatory mechanisms. At present, close to 20 different ligand classes of riboswitches have been discovered that bind metabolites such as lysine and glycine, coenzymes including adenosyl cobalamine (AdoCbl) and S-adenosylmethionine (SAM) and ions such as magnesium and fluoride (reviewed by [32]). Most riboswitches characterized thus far reside in the 5’ UTR and regulate in cis the transcription or translation of the downstream open reading frames (ORFs). In this classic mode of riboswitch control, ligand binding causes a change in the mRNA leader structure that either promotes or inhibits the formation of a hairpin that terminates transcription or sequesters the translational start site (reviewed by [32]). Other types of regulatory mechanisms have also been discovered for riboswitches. Here, we focus on riboswitches acting within sRNAs.
Riboswitch governed base-pairing RNAs
That riboswitches might regulate gene expression in ways other than in cis regulation of downstream ORFs was realized when riboswitches were discovered at the 3’ ends of genes [33-35]. Two of these riboswitches were molecularly characterized in detail and found to be regulating the expression of the upstream adjacent genes by controlling the generation of an antisense RNA (asRNA) [33,36]. The discovery that a riboswitch can control the generation an asRNA meant that a riboswitch did not need to be upstream of its cognate gene, but could indirectly regulate its expression via an sRNA encoded antisense or even elsewhere in the genome.
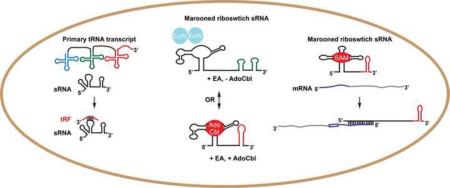
Indeed, examples of such trans-acting riboswitches were discovered in the form of SAM riboswitches, found in Listeria monocytogenes [37]. SAM riboswitches originally identified in the 5’ UTRs of operons encoding genes involved in methionine and cysteine transport and metabolism. Binding of SAM to its cognate riboswitch typically results in premature termination, preventing expression of the downstream ORFs. However, a novel discovery showed that some of these terminated RNA fragments have second lives as non-coding sRNAs influencing the expression of genes located elsewhere in the genome. Two SAM riboswitches, SreA and SreB were found to alter the levels of the prfA transcript, which encodes a major virulence regulator. Detailed molecular characterization of the SreA sRNA demonstrated that this terminated riboswitch directly binds the 5’UTR of the prfA transcript reducing its stability and translation (Figure 2A). By this mechanism, nutrient sensing by one class of RNA aptamer can simultaneously regulate both metabolism and virulence. This study showed that terminated riboswitch RNAs are not necessarily “junk” RNA, but may have secondary functions as transacting sRNAs [37] .
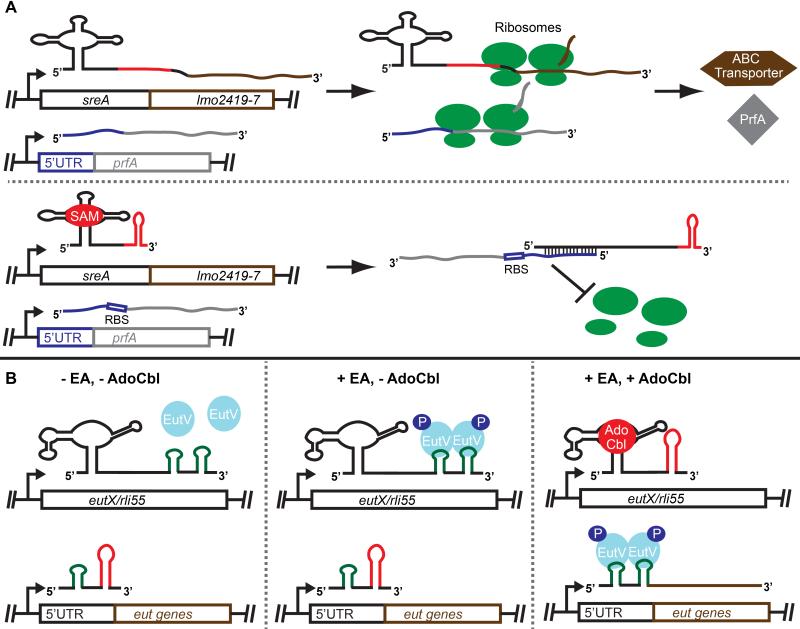
Figure 2. Mechanisms of post-transcriptional regulation of gene expression by trans-acting riboswitches.
(A) Upper panel: In the absence of the riboswitch ligand (SAM), gene expression of the downstream genes occurs as well as expression of prfA at an independent locus. Lower panel: In the presence of the riboswitch ligand, termination occurs preventing downstream gene expression and generating an sRNA that acts in trans to prevent expression of prfA. (B) Left panel: In the absence of EA and AdoCbl, EutV is inactive, full-length EutX/Rli55 is generated and eut gene expression is off. Middle panel: In presence of only EA, EutV is active, but sequestered by full-length EutX/Rli55, and eut gene expression is off. Right panel: In the presence of both EA and AdoCbl, EutV is active, a short form of EutX/Rli55, unable to sequester EutV, is formed due to termination by the ligand-bound riboswitch, and eut gene expression is induced by EutV antitermination.
Riboswitch dependent protein binding RNAs
Very recently, a coenzyme B12 (AdoCbl) riboswitch associated with the ethanolamine (EA) utilization (eut) loci of Enterococcus faecalis and L. monocytogenes was shown to operate primarily as a non-coding, trans-acting RNA. However, rather than binding and down-regulating mRNAs, this riboswitch-RNA, EutX/Rli55, binds and sequesters a response regulator [38-40]. The response regulator, EutV, was previously characterized as RNA-binding family of AmiR and NasR Transcriptional Antiterminator Regulators (ANTARs) [41,42]. Upon sensing EA, the histidine sensor kinase, EutW, autophosphorylates and phospho-transfers to EutV, which causes dimerization and RNA-binding activity [43-45]. Within the eut locus of E. faecalis and preceding several ORFs are constitutively active promoters followed by transcriptional terminators that prevent downstream gene expression under non-inducing conditions. When EA is present, active EutV binds to specific sites overlapping the terminators thereby preventing terminator formation and promoting read-through. However, it was noted that induction of the eut genes could not be induced only with EA; a co-factor required for EA breakdown, AdoCbl, was also required. A riboswitch, that binds AdoCbl, was discovered in an intergenic region of the eut locus [44]. While several ideas were proposed for how this riboswitch ensures that gene expression only occurs when AdoCbl is present in addition to EA [44,46], the actual mechanism was only revealed when a potential EutV binding site, not associated with a terminator, was located downstream of the riboswitch aptamer. Studies carried out in both E. faecalis and in L. monocytogenes demonstrated that this binding site and the riboswitch comprise a non-coding RNA, EutX/Rli55, that binds and sequesters active EutV when EA is available, but not AdoCbl (Figure 2B). When AdoCbl is also present, it binds to the riboswitch and causes termination, resulting in a truncated RNA lacking the EutV sequestering site [38-40].
Other examples of riboswitches controlling access to protein binding sites have been identified recently. Instead of regulating a Rho-independent terminator, one Mg2+ riboswitch controls access to a Rho-dependent binding site [47,48]. Both a lysine and a Mg2+ riboswitch have been characterized as controlling access to RNase E cleavage sites, thereby promoting or protecting a transcript from degradation [49-51].
Conclusions
These recent studies highlight the notion that very little goes to waste in cells. RNA segments generated from spacers excised during tRNA processing, mRNA cleavage products, and terminated riboswitches have second lives as regulatory RNAs. Furthermore, the riboswitches observed to be marooned, i.e., not located close to the loci they regulate [8] are unlikely to be junk waiting to be mutated into nonexistence, but may control the expression of sRNAs that regulate the relevant genes in trans. In other instances, these riboswitch-containing non-coding RNAs may function to titrate RNA binding proteins that regulate the expression of these ORFs. Altogether, these findings suggest that many other RNA cleavage products, terminated transcripts, and pseudogenes may be important regulatory molecules hiding in plain sight.
Highlights.
small noncoding RNAs (sRNAs) are an important class of regulators in bacteria.
Several new classes of sRNAs were revealed by recent studies.
tRNA, mRNA, and riboswitch derived fragments have a second life as sRNA regulators.
Marooned riboswitches have a life on their own as modulators of gene expression.
Acknowledgements
This work was supported by the National Institutes of Health grants R01AI076406, R01AI110432 (D.A.G.) and The University of Texas Health Science Center at Houston startup funds (N.R.D).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Argaman L, Hershberg R, Vogel J, Bejerano G, Wagner EG, Margalit H, Altuvia S. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–950. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- 3*.Bilusic I, Popitsch N, Rescheneder P, Schroeder R, Lybecker M. Revisiting the coding potential of the E. coli genome through Hfq co-immunoprecipitation. RNA Biol. 2014;11:641–654. doi: 10.4161/rna.29299. [Via deep sequencing of RNAs co-immunoprecipitated with Hfq, these authors demonstrated that in addition to classical trans-acting sRNAs binding Hfq, tRNAs, antisense sRNAs, and intragenic RNAs bind Hfq in vivo. The results of this study suggested that Hfq may also mediate the action of anti-sense RNAs and that other nontraditional sRNAs may exist in E. coli.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chao Y, Papenfort K, Reinhardt R, Sharma CM, Vogel J. An atlas of Hfq-bound transcripts reveals 3' UTRs as a genomic reservoir of regulatory small RNAs. EMBO J. 2012;31:4005–4019. doi: 10.1038/emboj.2012.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vogel J, Bartels V, Tang TH, Churakov G, Slagter-Jager JG, Huttenhofer A, Wagner EG. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 2003;31:6435–6443. doi: 10.1093/nar/gkg867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wassarman KM, Repoila F, Rosenow C, Storz G, Gottesman S. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 2001;15:1637–1651. doi: 10.1101/gad.901001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang A, Wassarman KM, Rosenow C, Tjaden BC, Storz G, Gottesman S. Global analysis of small RNA and mRNA targets of Hfq. Mol Microbiol. 2003;50:1111–1124. doi: 10.1046/j.1365-2958.2003.03734.x. [DOI] [PubMed] [Google Scholar]
- 8.Mellin JR, Cossart P. Unexpected versatility in bacterial riboswitches. Trends Genet. 2015;31:150–156. doi: 10.1016/j.tig.2015.01.005. [DOI] [PubMed] [Google Scholar]
- 9.Altman S. Isolation of tyrosine tRNA precursor molecules. Nat New Biol. 1971;229:19–21. doi: 10.1038/newbio229019a0. [DOI] [PubMed] [Google Scholar]
- 10.Altman S, Smith JD. Tyrosine tRNA precursor molecule polynucleotide sequence. Nat New Biol. 1971;233:35–39. doi: 10.1038/newbio233035a0. [DOI] [PubMed] [Google Scholar]
- 11.Dahlberg AE, Peacock AC. Studies of 16 and 23 S ribosomal RNA of Escherichia coli using composite gel electrophoresis. J Mol Biol. 1971;55:61–74. doi: 10.1016/0022-2836(71)90281-6. [DOI] [PubMed] [Google Scholar]
- 12.Hecht NB, Woese CR. Separation of bacterial ribosomal ribonucleic acid from its macromolecular precursors by polyacrylamide gel electrophoresis. J Bacteriol. 1968;95:986–990. doi: 10.1128/jb.95.3.986-990.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altman S, Robertson HD. RNA precursor molecules and ribonucleases in E. coli. Mol Cell Biochem. 1973;1:83–93. doi: 10.1007/BF01659941. [DOI] [PubMed] [Google Scholar]
- 14.Ghosal A, Upadhyaya BB, Fritz JV, Heintz-Buschart A, Desai MS, Yusuf D, Huang D, Baumuratov A, Wang K, Galas D, et al. The extracellular RNA complement of Escherichia coli. Microbiologyopen. 2015 doi: 10.1002/mbo3.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haiser HJ, Karginov FV, Hannon GJ, Elliot MA. Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res. 2008;36:732–741. doi: 10.1093/nar/gkm1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heyer R, Dorr M, Jellen-Ritter A, Spath B, Babski J, Jaschinski K, Soppa J, Marchfelder A. High throughput sequencing reveals a plethora of small RNAs including tRNA derived fragments in Haloferax volcanii. RNA Biol. 2012;9:1011–1018. doi: 10.4161/rna.20826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee YS, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23:2639–2649. doi: 10.1101/gad.1837609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sauer E. Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol. 2013;10:610–618. doi: 10.4161/rna.24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamson DN, Lim HN. Essential requirements for robust signaling in Hfq dependent small RNA networks. PLoS Comput Biol. 2011;7:e1002138. doi: 10.1371/journal.pcbi.1002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sagawa S, Shin JE, Hussein R, Lim HN. Paradoxical suppression of small RNA activity at high Hfq concentrations due to random-order binding. Nucleic Acids Res. 2015 doi: 10.1093/nar/gkv777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee T, Feig AL. The RNA binding protein Hfq interacts specifically with tRNAs. RNA. 2008;14:514–523. doi: 10.1261/rna.531408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheibe M, Bonin S, Hajnsdorf E, Betat H, Morl M. Hfq stimulates the activity of the CCA-adding enzyme. BMC Mol Biol. 2007;8:92. doi: 10.1186/1471-2199-8-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23**.Lalaouna D, Carrier MC, Semsey S, Brouard JS, Wang J, Wade JT, Masse E. A 3' external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol Cell. 2015;58:393–405. doi: 10.1016/j.molcel.2015.03.013. [In this seminal work, these authors demonstrate that fragments derived from precursor tRNAs function in regulating the activity of certain sRNAs. The implications of this work is that other RNA fragments thought of as byproducts of RNA metabolism may have important regulatory roles.] [DOI] [PubMed] [Google Scholar]
- 24.Kumar P, Anaya J, Mudunuri SB, Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gebetsberger J, Zywicki M, Kunzi A, Polacek N. tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012;2012:260909. doi: 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Figueroa-Bossi N, Valentini M, Malleret L, Fiorini F, Bossi L. Caught at its own game: regulatory small RNA inactivated by an inducible transcript mimicking its target. Genes Dev. 2009;23:2004–2015. doi: 10.1101/gad.541609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27*.Miyakoshi M, Chao Y, Vogel J. Cross talk between ABC transporter mRNAs via a target mRNA-derived sponge of the GcvB small RNA. EMBO J. 2015;34:1478–1492. doi: 10.15252/embj.201490546. [These authors show that pairing of the GcvB sRNA to the 5'UTR of the gltIJKL mRNA leads to translational repression and RNase E mediated decay that generates a second stable sRNA derived from an intragenic region between gltI and gltJ. This mRNA derived sRNA pairs with the GcvB sRNA leading to destabilization of the sRNA. Altogether this study highlights the complex interactions that occur between sRNAs and their target.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grundy FJ, Turinsky AJ, Henkin TM. Catabolite regulation of Bacillus subtilis acetate and acetoin utilization genes by CcpA. J Bacteriol. 1994;176:4527–4533. doi: 10.1128/jb.176.15.4527-4533.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grundy FJ, Henkin TM. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993;74:475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- 30.von Ahsen U, Davies J, Schroeder R. Antibiotic inhibition of group I ribozyme function. Nature. 1991;353:368–370. doi: 10.1038/353368a0. [DOI] [PubMed] [Google Scholar]
- 31.Yarus M. A specific amino acid binding site composed of RNA. Science. 1988;240:1751–1758. doi: 10.1126/science.3381099. [DOI] [PubMed] [Google Scholar]
- 32.Breaker RR. Prospects for riboswitch discovery and analysis. Mol Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mellin JR, Tiensuu T, Becavin C, Gouin E, Johansson J, Cossart P. A riboswitch-regulated antisense RNA in Listeria monocytogenes. Proc Natl Acad Sci U S A. 2013;110:13132–13137. doi: 10.1073/pnas.1304795110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the vitamin B12 metabolism and regulation in prokaryotes. J Biol Chem. 2003;278:41148–41159. doi: 10.1074/jbc.M305837200. [DOI] [PubMed] [Google Scholar]
- 35.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res. 2004;32:3340–3353. doi: 10.1093/nar/gkh659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andre G, Even S, Putzer H, Burguiere P, Croux C, Danchin A, Martin-Verstraete I, Soutourina O. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res. 2008;36:5955–5969. doi: 10.1093/nar/gkn601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loh E, Dussurget O, Gripenland J, Vaitkevicius K, Tiensuu T, Mandin P, Repoila F, Buchrieser C, Cossart P, Johansson J. A trans-acting riboswitch controls expression of the virulence regulator PrfA in Listeria monocytogenes. Cell. 2009;139:770–779. doi: 10.1016/j.cell.2009.08.046. [DOI] [PubMed] [Google Scholar]
- 38.Chen J, Gottesman S. RNA. Riboswitch regulates RNA. Science. 2014;345:876–877. doi: 10.1126/science.1258494. [DOI] [PubMed] [Google Scholar]
- 39**.DebRoy S, Gebbie M, Ramesh A, Goodson JR, Cruz MR, van Hoof A, Winkler WC, Garsin DA. Riboswitches. A riboswitch-containing sRNA controls gene expression by sequestration of a response regulator. Science. 2014;345:937–940. doi: 10.1126/science.1255091. [Describes the discovery of the second only known trans-acting riboswitch and its mechanism of action. Rather than base-pairing with mRNA targets, this sRNA regulates by sequestering a response regulator with antiterminator activity. The work was done in Enterococcus faecalis.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40**.Mellin JR, Koutero M, Dar D, Nahori MA, Sorek R, Cossart P. Riboswitches. Sequestration of a two-component response regulator by a riboswitch-regulated noncoding RNA. Science. 2014;345:940–943. doi: 10.1126/science.1255083. [This work was published back-to-back with reference 39 and describes the same discovery. Interestingly, it was made independently by investigators working in Listeria monocytogenes.] [DOI] [PubMed] [Google Scholar]
- 41.Galperin MY. Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol. 2006;188:4169–4182. doi: 10.1128/JB.01887-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shu CJ, Zhulin IB. ANTAR: an RNA-binding domain in transcription antitermination regulatory proteins. Trends Biochem Sci. 2002;27:3–5. doi: 10.1016/s0968-0004(01)02036-9. [DOI] [PubMed] [Google Scholar]
- 43.Del Papa MF, Perego M. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J Bacteriol. 2008;190:7147–7156. doi: 10.1128/JB.00952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fox KA, Ramesh A, Stearns JE, Bourgogne A, Reyes-Jara A, Winkler WC, Garsin DA. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. PMCID: 2647976. Proc Natl Acad Sci U S A. 2009;106:4435–4440. doi: 10.1073/pnas.0812194106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramesh A, DebRoy S, Goodson JR, Fox KA, Faz H, Garsin DA, Winkler WC. The mechanism for RNA recognition by ANTAR regulators of gene expression. PLoS Genet. 2012;8:e1002666. doi: 10.1371/journal.pgen.1002666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker KA, Perego M. Transcription Antitermination by a Phosphorylated Response Regulator and Cobalamin-Dependent Termination at a B12 Riboswitch Contribute to Ethanolamine Utilization in Enterococcus faecalis. J Bacteriol. 2011;193:2575–2586. doi: 10.1128/JB.00217-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hollands K, Proshkin S, Sklyarova S, Epshtein V, Mironov A, Nudler E, Groisman EA. Riboswitch control of Rho-dependent transcription termination. Proc Natl Acad Sci U S A. 2012;109:5376–5381. doi: 10.1073/pnas.1112211109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hollands K, Sevostiyanova A, Groisman EA. Unusually long-lived pause required for regulation of a Rho-dependent transcription terminator. Proc Natl Acad Sci U S A. 2014;111:E1999–2007. doi: 10.1073/pnas.1319193111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bastet L, Dube A, Masse E, Lafontaine DA. New insights into riboswitch regulation mechanisms. Mol Microbiol. 2011;80:1148–1154. doi: 10.1111/j.1365-2958.2011.07654.x. [DOI] [PubMed] [Google Scholar]
- 50.Caron MP, Bastet L, Lussier A, Simoneau-Roy M, Masse E, Lafontaine DA. Dual-acting riboswitch control of translation initiation and mRNA decay. Proc Natl Acad Sci U S A. 2012;109:E3444–3453. doi: 10.1073/pnas.1214024109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spinelli SV, Pontel LB, Garcia Vescovi E, Soncini FC. Regulation of magnesium homeostasis in Salmonella: Mg(2+) targets the mgtA transcript for degradation by RNase E. FEMS Microbiol Lett. 2008;280:226–234. doi: 10.1111/j.1574-6968.2008.01065.x. [DOI] [PubMed] [Google Scholar]