Abstract
p97 (also known as valosin-containing protein (VCP) in mammals or Cdc48p in S. cerevisiae) is an evolutionarily conserved ATPase present in all eukaryotes and archaebacteria. In conjunction with a collection of cofactors and adaptors, p97/Cdc48p performs an array of biological functions mostly through modulating the stability of ‘client’ proteins. Using energy from ATP hydrolysis, p97/Cdc48p segregates these molecules from immobile cellular structures such as protein assemblies, membrane organelles, and chromatin. Consequently, the released polypeptides can be efficiently degraded by the ubiquitin proteasome system or recycled. This review summarizes our current understanding of the structure and function of this essential cellular chaperoning system.
A genetic screen conducted three decades ago in S. cerevisiae identified several alleles of Cdc48 that affects cell growth at non-permissive temperatures due to a cell cycle arrest at the G2-M transition stage (Moir et al., 1982). The mammalian homolog of Cdc48p was later reported as a 97 kDa protein precursor for the small peptide valosin, and therefore named as valosin-containing protein (VCP) or p97 (Koller and Brownstein, 1987). Although it turned out that valosin is a purification artifact unrelated to p97 (Gill et al., 1989), the VCP nomenclature is still being used in the literature. In some species, the name transitional endoplasmic reticulum ATPase (TER ATPase) is used given the localization and function of a fraction of this enzyme at the endoplasmic reticulum (ER) (see below). In this review, we use p97 and Cdc48p for the mammalian and yeast homologs, respectively.
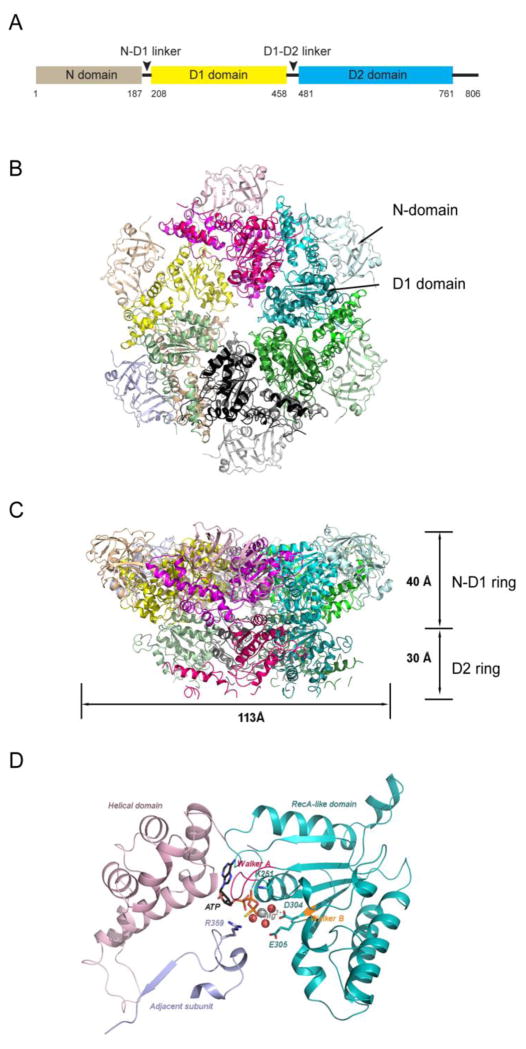
p97/Cdc48p belongs to a large ATPase family termed AAA+ (extended family of ATPases associated with various cellular activities) ATPase. Enzymes of this family function in all species from bacteria to humans, often as essential chaperones that promote protein folding or unfolding. p97/Cdc48p is a type II AAA+ ATPase because it has two AAA ATPase domains in tandem (named D1 and D2, respectively) (Figure 1A). A short polypeptide linker (D1–D2 linker) connects the two ATPase domains and another linker (N-D1 linker) joins the D1 domain to a large amino-terminal domain (N-domain). The carboxyl-terminus of the D2 domain is appended with a short tail containing ~40 residues. Interaction of p97/Cdc48p with its partners is mostly mediated by the N-domain, although a few proteins bind p97/Cdc48p via the C-terminal tail (Buchberger et al., 2015; Ogura and Wilkinson, 2001). The D1 and D2 domains are homologous both in sequence and in structure. However, they have distinct functions. For example, the hexameric assembly of p97 only requires the D1 but not the D2 domain (Wang et al., 2003).
Figure 1. Structure of p97/Cdc48p.
(A) The schematic domain organization of p97/Cdc48. (B) The structure of hexameric p97 (PDB: 3CF2 in the ADP-bound form) is viewed down the 6-fold symmetry axis showing the N-D1 ring. The six subunits are shown as cartoon diagrams in different colors. Domains of each subunit are also shaded differently. The D1 domain and the N-domain are indicated with arrows and labeled for one of the six subunits. (C) The side view of p97 is presented with indicated width and height. (D) The structure of the D1 AAA domain of a p97 subunit with bound ATPγS is presented in the ribbon format (PDB:4KO8). An AAA domain consists of a RecA-like domain (cyan) and a characteristic helical domain (purple). An ATPγS, bound at the interface between the two domains, is shown as stick model. The Mg2+ ion and three conserved water molecules are shown as silver and red balls, respectively. The Walker A motif or P-loop is highlighted in red and the conserved lysine residue K251 is shown as stick model and labeled. The Walker B motif is shown in orange and the two conserved acidic residues D304 and E305 are represented by stick models. The nucleotide-binding site communicates with a neighboring subunit through the SRH (second region of homology, in light blue) motif, where a conserved Arg-finger residue R359 is in contact with the bound nucleotide.
In mammalian cells, p97 is localized mainly to the cytoplasm with a fraction associated with the membranes of subcellular organelles such as the ER, Golgi, mitochondria, and endosomes (Acharya et al., 1995; Latterich et al., 1995; Rabouille et al., 1995; Ramanathan and Ye, 2012; Xu et al., 2011). The membrane localizations are probably mediated by membrane-associated receptors, whose identity is largely unknown in most cases. A fraction of p97/Cdc48p is also present in the nucleus and serves essential roles in chromatin-associated events and nuclear protein quality control (PQC) (see below) (Madeo et al., 1998).
As one of the most abundant proteins in eukaryotic cells, p97 is ubiquitously expressed in multicellular organisms. In humans, the mRNA expression of p97 was moderately increased in certain types of cancer, and the expression level to some extent correlates with the sensitivity of cancer cells to a potent p97 inhibitor that is currently evaluated as a potential anti-cancer drug (Anderson et al., 2015).
Most known substrates of p97/Cdc48p are conjugated with ubiquitin chains and degraded by the 26S proteasome. Accordingly, many p97/Cdc48p cofactors/adaptors are capable of recognizing ubiquitin conjugates (Ye, 2006). It has been thought that the interplay between ubiquitin and the p97 system is critical for p97 functions, although the precise role awaits further elucidation. Some p97 cofactors are ubiquitin ligase or deubiquitinase that can process ubiquitin chains, but the vast majority serves as adaptors that link p97/Cdc48p to specific subcellular compartment or substrate.
More recently, genetic studies have linked mutations in p97 to several human diseases including IBMPFD (Inclusion Body Myopathy associated with Paget’s disease of the bone and Frontotemporal Dementia) and amyotrophic lateral sclerosis (ALS). These findings have stimulated a flurry of investigations on the structure and function of p97/Cdc48p.
Structural organization of p97
Structural Features of p97
Structural information on p97 was initially obtained with negatively stained samples examined by electron microscopy (EM). The results provided the basic shape of a ring-like hexameric molecule (Peters et al., 1990; Zhang et al., 1994). Higher resolution structures were later obtained by cryo-EM (Rouiller et al., 2000) and by crystallization of a N-D1 fragment (Zhang et al., 2000). These studies confirmed the hexameric assembly of p97, but also showed that unlike many bacterial AAA+ proteins, the assembly of p97 does not depend on the presence of nucleotide.
Subsequent high resolution structural studies were primarily carried out with X-ray crystallography (Table 1) focusing on full-length wild-type p97 (Davies et al., 2008; DeLaBarre and Brunger, 2003; DeLaBarre and Brunger, 2005; Huyton et al., 2003) as well as several disease-associated mutants (Tang et al., 2010; Tang and Xia, 2012; Tang and Xia, 2013). More recently, three additional reports on the structure of full-length p97 appeared. One featured crystal structures of the full-length p97 bearing mutations in the D2 domain with nominal improvements in resolution (Hanzelmann and Schindelin, 2016b). The other two studies used the latest development in electron microscopic technology to obtain higher resolution structures of p97 (Banerjee et al., 2016; Schuller et al., 2016). The interactions of p97 with adaptors were also extensively studies with X-ray crystallography (Dreveny et al., 2004; Hanzelmann et al., 2011; Hanzelmann and Schindelin, 2011; Hanzelmann and Schindelin, 2016a; Kim et al., 2011; Kim and Kim, 2014; Lee et al., 2013; Qiu et al., 2010; Schaeffer et al., 2014; Zhao et al., 2007). These studies show that p97 forms two concentric rings (Figure 1B, C); the N-D1 ring has a larger radius than the D2 ring owing to the laterally attached N-domain. Like other AAA+ ATPases, the AAA module of p97 features a highly conserved RecA-like domain and a characteristic helical domain (Figure 1D). Each RecA-like domain in a protomer bears an active site, which is situated at the interface between two adjacent promoters in the hexameric assembly. The active site is formed by the classical Walker A (P-loop; G(x)4GKT) and Walker B motifs (hhhhDE, h for hydrophobic amino acid), responsible for nucleotide binding and hydrolysis, respectively. The configuration of the active site allows an arginine-finger residue (R359 for the D1 ring and R635 for the D2 ring) from an adjacent subunit to stimulate ATP hydrolysis.
Table 1.
Available structures of p97 deposited in the PDB.
| PDB | Construct | Mutant | N-Conform | Bound Adaptor/Inhibitor | Bound Ligand | Res (Å) | Date Deposit | Note | References |
|---|---|---|---|---|---|---|---|---|---|
| 3CF3 | FL | WT | Down | - | ADP(D1,D2) | 4.25 | 2008 | 1YQIa | Davies et al., 2008; DeLaBarre & Brunger, 2005 |
| 1R7R | FL | WT | Down | - | ADP(D1) | 3.6 | 2003 | D2 PlyAb | Huyton et al., 2003 |
| 3CF1 | FL | WT | Down | - | ADP(D1), ADP•AlF (D2) | 4.4 | 2008 | 1OZ4a 1YQ0a |
Davies et al., 2008; DeLaBarre & Brunger, 2003 & 2005 |
| 3CF2 | FL | WT | Down | - | ADP (D1) AMP-PNP (D2) |
3.5 | 2008 | 1YPWa | Davies et al., 2008; DeLaBarre & Brunger, 2003 & 2005 |
| 3C19 | FL | α9-D4c | Down | - | None(D1, D2) | 4.2 | 2016 | Hanzelmann et al., 2016a | |
| 3C1A | FL | α9-D4c | Down | - | ATPγS (D1,D2) | 3.8 | 2016 | Hanzelmann et al., 2016a | |
| 3C18 | FL | Δ(709–728)d | Down | - | ATPγS (D1,D2) | 3.30 | 2016 | Hanzelmann et al., 2016a | |
| 5C1B | FL | Δ(709–728)d | Down | SHP | ATPγS (D1,D2) | 3.08 | 2016 | Hanzelmann et al., 2016b | |
| 5FTJ | FL | WT | Down | - | ADP(D1,D2) | 2.4 | 2016 | Banerjee et al., 2016 | |
| 5FTK | FL | WT | Down | UPCDC30245 | ADP(D1,D2) | 2.3 | 2016 | Banerjee et al., 2016 | |
| 5FTLe | FL | WT | Down | - | ADP(D1,D2) | 3.3 | 2016 | Banerjee et al., 2016 | |
| 5FTMe | FL | WT | Down | - | ADP(D1), ATPγS(D2) | 3.2 | 2016 | Banerjee et al., 2016 | |
| 5FTNe | FL | WT | Up | - | ATPγS (D1,D2) | 3.3 | Banerjee et al., 2016 | ||
| 3QQ7 | N | WT | - | - | 2.65 | 2011 | Hanzelmann et al., 2011a | ||
| 3QQ8 | N | WT | - | FAF1-UXB | - | 2.0 | 2011 | Hanzelmann et al., 2011a | |
| 4KDI | N | WT | - | OTU1-UBX | - | 1.86 | 2013 | Kim et al., 2014 | |
| 4KDL | N | WT | - | OTU1-UBX | - | 1.81 | 2013 | Kim et al., 2014 | |
| 3QC8 | N | WT | - | FAF1-UBX | - | 2.2 | 2011 | Kim et al., 2011 | |
| 3QWZ | N | WT | - | FAF1-UBX | - | 2.0 | 2011 | Lee et al., 2013 | |
| 3TIW | N | WT | - | Gp78-VIM | - | 1.8 | 2011 | Hanzelmann et al., 2011b | |
| 1E32 | N-D1 | WT | Down | - | ADP (D1) | 2.9 | 2000 | Zhang et al., 2000 | |
| 4KO8 | N-D1 | R155H | Up | - | ATPγS (D1) | 1.98 | 2014 | Tang et al., 2013 | |
| 4KLN | N-D1 | A232E | Up | - | ATPγS (D1) | 2.62 | 2014 | Tang et al., 2013 | |
| 4KOD | N-D1 | R155H | Down | - | ADP (D1) | 2.96 | 2014 | Tang et al., 2013 | |
| 3HU3 | N-D1 | R155H | Up | - | ATPγS (D1) | 2.2 | 2009 | Tang et al., 2010 | |
| 3HU1 | N-D1 | R95G | Up | - | ATPγS (D1) | 2.81 | 2009 | Tang et al., 2010 | |
| 3HU2 | N-D1 | R86A | Up | - | ATPγS (D1) | 2.85 | 2009 | Tang et al., 2010 | |
| 5DYG | N-D1 | L198W | Down | - | ADP (D1) | 2.20 | 2015 | Tang et al., 2016 | |
| 5DYi | N-D1 | WT | Down | - | ADP (D1) | 3.71 | 2015 | Tang et al., 2016 | |
| 1S3S | N-D1 | WT | Down | P47-UBX | ADP (D1) | 2.9 | 2004 | Dreveny et al., 2004 | |
| 3CF0 | D2 | WT | - | - | ADP (D2) | 3.0 | 2008 | 7merf | Davies et al., 2008 |
| 3EBB | C-10mer | WT | - | PLAA PUL | - | 1.9 | 2008 | Qiu et al., 2010 | |
| 2HPL | C-10mer | WT | - | PNGase | - | 1.8 | 2006 | Zhao et al., 2007 | |
| 4POA | C-10mer | WT | - | HOIP PUB | 2.3 | 2014 | Schaeffer et al., 2014 | ||
| 2PJH | N | WT | - | Npl4-UBD | 2011 | NMR | Isaacson et al., 2007 |
superseded. b – D2 domain is a polyA model. c – isolated D2 domain forms heptamer in solution.
D2 domain is a polyA model.
full-length p97 with four mutations N750D/R753D/M757D/Q760D.
full-length p97 with the deletion in a loop in the D2 domain from residues 709–729.
in the presence of 5 mM ATPγS during experiments.
isolated D2 domain forms a heptamer in crystal.
Nucleotide-Driven Conformational Changes in p97
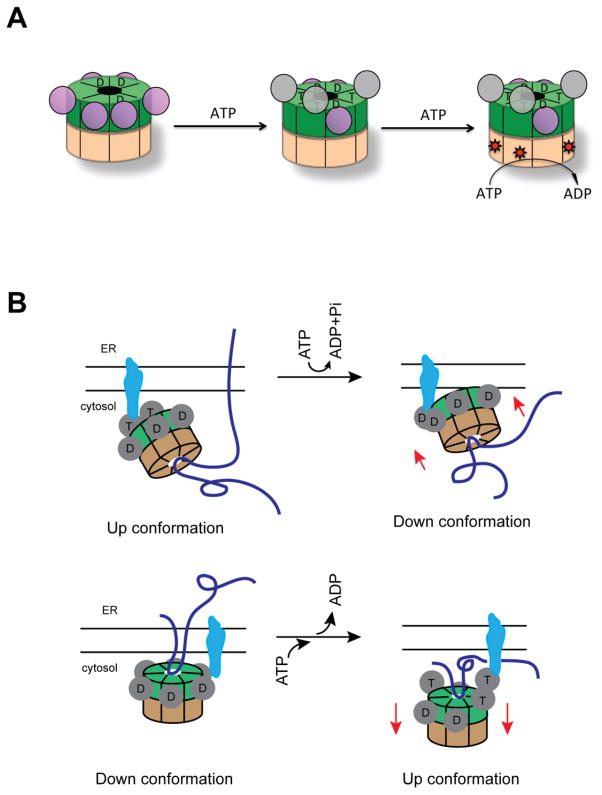
It is generally believed that p97 undergoes dramatic conformational changes during the nucleotide hydrolysis cycle (Beuron et al., 2006; Beuron et al., 2003; DeLaBarre and Brunger, 2005; Rouiller et al., 2002; Tang et al., 2010). Mechanical force generated by these conformational changes would be applied to substrate molecules to influence their stability and function. As a type II AAA+ ATPase, each p97 hexamer contains 12 ATPase domains and 6 N-domains. If each nucleotide-binding site were capable of producing 3 distinct conformations for apo-, ADP- and ATP-state, there would be a total of 312 different conformations assuming each subunit operates independently. Reported structural conformations and interactions among different subunits are far less than the theoretical possibilities due to inter-subunit communications and coordination. Nevertheless, the conformational dynamics of p97 is difficult to study because the six ATPase domains within each ring are not synchronized in ATP hydrolysis (see below).
The conformational changes driven by ATP hydrolysis have been sought by various biochemical and biophysical methods (Davies et al., 2005). Initially, low-resolution cryo-EM structures revealed that upon ATP hydrolysis moderate rotational movement occurs between the two ATPase rings, associated with either closure or opening of the D1 and D2 central pores (Rouiller et al., 2002), but subsequent studies suggested other modes of domain movement (Beuron et al., 2003). Due to resolution limitation, domain assignment in EM-reconstruction has been unreliable, and therefore, it is not feasible to consolidate these structural data into a consistent model that explains the action of p97. Moreover, because insufficient resolution prevents accurate determination of the p97 nucleotide-binding state, the interpretation of the EM results relied on the assumption that all 12 ATP-binding sites are occupied by the added nucleotide in a homogenous manner. Consequently, the EM studies failed to consider the heterogeneity in nucleotide association, not to mention the nucleotide pre-bound to the D1 ring.
Crystallographic studies of wild-type p97, using either full-length or N-D1 fragment, showed that D1 domains of all six subunits always have ADP bound and all N-domains are coplanar with the D1-ring, forming the so called Down conformation (Davies et al., 2008; DeLaBarre and Brunger, 2003; DeLaBarre and Brunger, 2005; Zhang et al., 2000). On the other hand, nucleotide bound to the D2 domains could be either ADP, AMP-PNP or ADP*AlFx. The D2 domain can also exist with no nucleotide bound (Apo state) (Huyton et al., 2003). Thus, structural studies of wild-type p97 by crystallography could only reveal conformational changes associated with the D2 nucleotide cycle. To date, one of the most significant structural changes associated with the ATP cycle of the D2 domain is the opening of the D2 pore, but whether pore opening is triggered by ATP binding or hydrolysis in the D2 ring has been controversial (Banerjee et al., 2016; Davies et al., 2008; Davies et al., 2005; Hanzelmann and Schindelin, 2016b; Pye et al., 2006; Rouiller et al., 2002; Schuller et al., 2016). Part of the D2 domain also seems to undergo an order-to-disorder transition, propelling small movements of the other domains in the molecule. In this regard, it is worth noting that even under the same nucleotide-binding conditions, conformational asymmetry exists among different subunits in D2-domain, and the magnitude of such difference can be as large as that experienced by the same subunit undergoing nucleotide exchange (Davies et al., 2008). Although structural studies fail to generate a conclusive model on how ATP hydrolysis in D2 triggers motion, it has become clear that at any given time, the six D2 domains in a p97 hexamer can exist in different nucleotide-binding states, suggesting that these ATPase domains do not act in a concerted manner. Indeed, biochemical studies of wild-type p97 ATPase activity demonstrated positive cooperation between subunits of the D2 domain (Nishikori et al., 2011).
Despite the repeated observations that p97 D2 mutants with an intact D1 domain have a low but discernable ATPase activity, there had been a tendency to ignore the D1 ATPase activity. Accordingly, the D1 domain has been assumed to function only in structural assembly of p97/Cdc48p without the need to hydrolyze ATP. However, recent studies using D2 specific p97 ATPase inhibitors showed convincingly that in the presence of an energy regenerating system, the D1 domain contributes significantly to the overall ATPase activity of p97 (~30%) (Anderson et al., 2015; Chou et al., 2014). The discrepancy may be caused by the accumulation of ADP when the assay was carried out without simultaneously converting it back to ATP because ADP seems to inhibit the D1 ATPase activity (Anderson et al., 2015). Thus, the new findings have a far reaching impact because it implicates a D1-dependent ATP hydrolysis cycle in p97 function, which is consistent with genetic evidence that Cdc48p D1 mutants with a functional D2 domain is unable to complement the growth defect of Cdc48 temperature sensitive mutant cells (Ye et al., 2003). Nevertheless, conformational changes associated with D1 ATP hydrolysis has not been observed by X-ray crystallographic studies for wild-type p97, mainly because purified recombinant p97 has ADP exclusively bound to the D1 domain (Davies et al., 2005; Tang and Xia, 2013), which cannot be removed completely to obtain a homogeneous sample with the D1 subunit being all occupied by supplemented nucleotides.
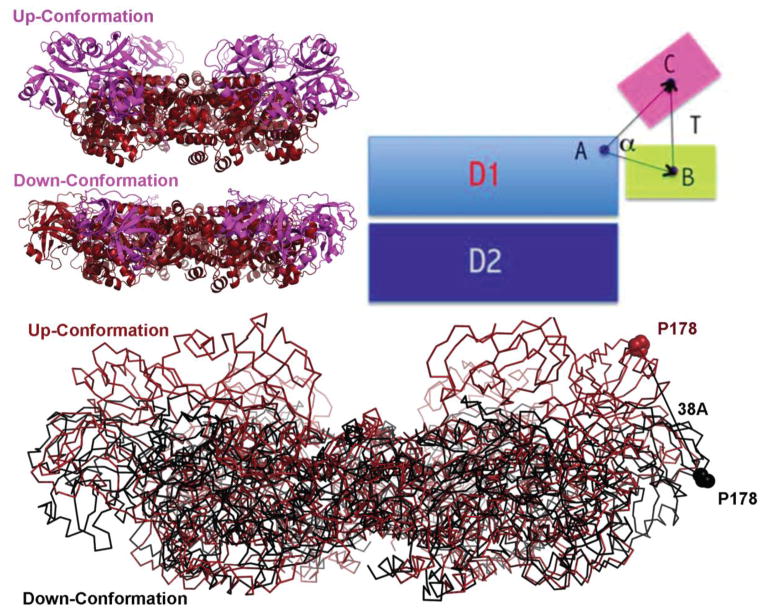
Recently, by genetic manipulation of certain regions in the D2 or N domain, the nucleotide-binding sites of the D1 ring can be altered to either empty or ATPγS-occupied state (Hanzelmann and Schindelin, 2016b; Tang et al., 2010). Strikingly, by simultaneous mutating 4 residues (N750D/R753D/M757D/Q760D) or deletion of a disordered loop in the D2 domain, Hanzelmann and colleagues were able to determine the structures of p97 with ATP binding sites all in either Apo or ATPγS-bound state. A comparison of the conformations of these two states suggests that ATP binding can open the D2 pore and also generate a rotational movement between the two ATPase rings (Hanzelmann and Schindelin, 2016b). However, the physiological relevance of these conformational changes remain to be determined because to what extent the mutations might affect p97 function is unclear (The deletion mutant has a lower ATPase activity compared to wild-type p97). Moreover, because no biochemical evidence suggests that the two ATPase rings in p97 hydrolyze ATP in a synchronized manner, intermediate conformations with the two ATPase rings in different nucleotide binding states are missing. In this regard, structural studies using another approach to remove D1 nucleotide seems to provide more biologically relevant insights into how the ATP cycle in the D1 domain might influence p97 function. The approach involved determining the structures of mutant p97 proteins carrying a single missense mutation that is associated with the IBMPFD syndrome. Because patients bearing a copy of the mutation do not suffer any developmental defects, the mutations seem to only cause suboptimal performance in the ATPase cycle that leads to gradual impairment of certain p97-dependent biological function and late on-set of the diseases in adulthood (Kimonis et al., 2000). Intriguingly, many IBMPFD mutations occurring at the interface between N- and D1-domain can weaken the affinity of ADP to the D1-domain (Tang et al., 2010). Consequently, these mutants can be purified with D1 bound by exogenously added nucleotides, permitting crystallographic studies of conformational changes associated with the D1 ATP hydrolysis cycle. Strikingly, compared to the structures in which D1 is in the ADP-bound state, when the ATP analog ATPγS occupies the D1 domain, the center of gravity of the N-domain undergoes a hinged translational movement of 13 Å with the angle (α angle in Figure 2) of 11°, followed by a further 92° rotation. As a result, the far edge of the N-domain represented by the residue Pro178 moves up by a distance of 38 Å, forming the Up-conformation (Figure 2) (Tang et al., 2010). This conformational change was also detected in wild-type p97 in solution by small-angle X-ray scattering (SAXS) (Tang et al., 2010). The difference between wild-type and mutant p97 lies in that in mutant p97, all six N-domains undergo a uniform conformational change, allowing study of p97 by X-ray crystallography, whereas in the wild-type p97 only a few subunits have their N-domains in the Up-conformation in the presence of ATP due to occluded ADP in some D1-domains. Thus, unsynchronized ATP hydrolysis appears to be a common feature for both the D1 and D2 domains. This feature may be functionally linked to the observed asymmetric binding of certain adaptor proteins to the p97 N-domain (Buchberger et al., 2015).
Figure 2. Nucleotide-dependent N-domain conformational change.
A large N-domain conformational change has been observed, being driven by the nucleotide cycle of the D1 domain. When ATP is bound, the N-domain, illustrated in ribbon diagram in magenta, goes to the Up-conformation, whereas it moves to the Down-conformation when ADP is bound to the D1 domain. During the transition between the UP- and Down-conformation, the center of gravity of the N-domain (B for the Down-conformation and C for the Up-conformation) translated by 13 Å and the α angle is 11° as defined by the triangle ABC (A is the position of G208 of the D1 domain). Additionally, to adopt the Up-conformation configuration, the N-domain needs a further 92° rotation. As a result, the residue P178 moves up by 38 Å.
The abovementioned nucleotide-dependent N-domain conformational change is further confirmed recently by structural studies of full-length wild-type p97 by cryo-EM. One of these studies showed that in the presence of ATPγS, p97 can exist in three different nucleotide-binding states in solution: one has all 12 sites occupied with ADP and the N-domains in the Down conformation, one has the six sites of the D1-ring bound with ADP and the six sites of the D2-ring taken by ATPγS with the N-domains still in the Down conformation, while in the third conformation, all 12 sites are occupied by ATPγS and the N-domains are in the Up-conformation (Banerjee et al., 2016). In another report, p97 in a solution containing AMP-PNP displays both the Up- and Down-conformations from different protomers within a hexamer as well as significant asymmetric domain movement (Schuller et al., 2016). However, it is noteworthy that the structures obtained with full-length p97 bearing D2 mutations do not reveal a similar conformational change in the N-domain between the apo and ATPγS-bound state (Hanzelmann and Schindelin, 2016b). One possible explanation is that the introduced D2 mutations may have inhibited this conformational change.
p97-interacting proteins
A large collection of p97/Cdc48p-interacting proteins has been identified through proteomic studies. These proteins either function as adaptors that link p97/Cdc48p to a specific subcellular compartment or substrate, or serve as cofactors that help to process substrates. Cofactors usually have enzymatic activities that can process protein modifiers such as N-glycan or ubiquitin conjugates that are appended to p97 substrate (e.g. N-glycanase, ubiquitin ligase, and deubiquitinase).
Although a few proteins such as PLAA/Ufd3, PNGase, HOIP, and Ufd2 bind p97/Cdc48p through its short C-terminal tail (Bohm et al., 2011; Murayama et al., 2015; Qiu et al., 2010; Rumpf and Jentsch, 2006; Schaeffer et al., 2014; Zhao et al., 2007), the vast majority of p97-interacting proteins bind it through its N-domain (Buchberger et al., 2015). Representative N-domain-interacting proteins include Ufd1, Npl4, p47, ataxin3, and FAF1. Sequence analyses identified several frequently occurring p97-interaction patterns: such as the UBX motif (ubiquitin regulatory X) (Schuberth and Buchberger, 2008), the VIM (VCP-interacting motif) (Stapf et al., 2011), VBM (VCP-binding motif) (Boeddrich et al., 2006) and SHP box (also known as binding site 1, bs1) (Bruderer et al., 2004).
The UBX domain is an 80-residue module structurally homologous to ubiquitin, whereas the VCP-interacting motif (VIM) is a linear sequence motif (RX5AAX2R) found in a number of p97 cofactors or adaptors including gp78 (Ballar et al., 2006), SVIP (small VCP-inhibiting protein) (Ballar et al., 2007) and VIMP (VCP-interacting membrane protein) (Ye et al., 2004). The VBM domain features a different, highly polarizing linear sequence motif (RRRRXXYY) found in ataxin-3, Ufd2 and hrd1 (Boeddrich et al., 2006). The SHP box is a short amino acid stretch enriched in hydrophobic residues, which can be found in p47 (Kondo et al., 1997), Ufd1-Npl4 (Meyer et al., 2000) and Derlin-1 (Greenblatt et al., 2011; Lilley and Ploegh, 2004; Ye et al., 2004).
Biochemical studies showed that some adaptors and cofactors bind p97/Cdc48p in a mutually exclusive manner (Meyer et al., 2000; Rumpf and Jentsch, 2006), raising the possibility that cells may possess distinct populations of p97/Cdc48p complexes, each bearing a unique partner for a specific function. However, some adaptors and cofactors can bind p97 simultaneously, and for those who bind p97 in a competitive manner, they do not necessarily have to act against each other because conceptually, a hierarchical binding system that allows ordered interaction with p97 to fulfill a specific function may be orchestrated (Hanzelmann et al., 2011; Meyer et al., 2012).
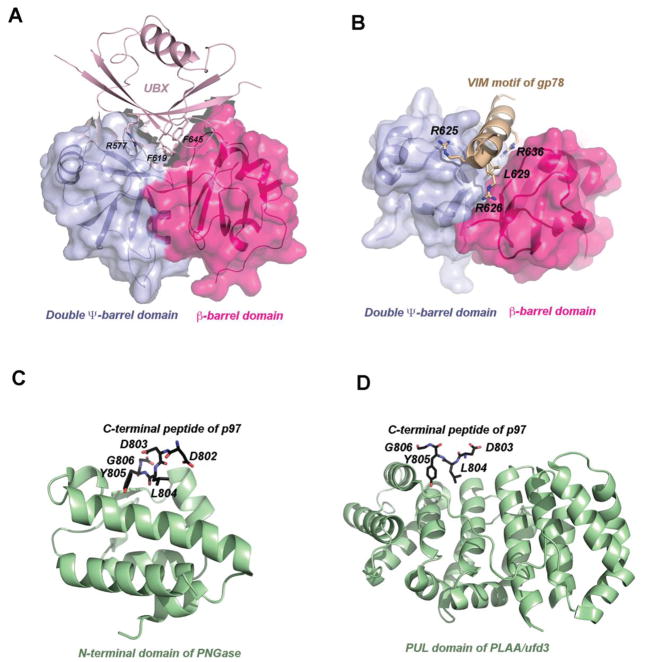
Molecular insights on adaptor or cofactor binding have been mostly obtained through crystallographic studies using isolated domains or segments in complex with either the p97 N-domain or with its C-terminal tail. One of the better characterized interactions reported to date is the p47-N-D1 complex (Dreveny et al., 2004). The structure showed that the p97 N-domain is comprised of two sub-domains of roughly equal size: an N-terminal double Ψ-barrel and a C-terminal β-barrel (Figure 3A). p47 uses a C-terminal UBX domain to bind to the N-domain of p97 at a cleft between these two sub-domains. The interaction of p47 with p97 occurs asymmetrically as only two out of the six p97 subunits are occupied by p47. Interestingly, although VIM is structurally unrelated to the UBX domain, it binds p97 at a similar location (Figure 3B) (Hanzelmann and Schindelin, 2011). More recently, Hanzelmann and colleagues described an additional surface on the N-domain of p97 for the SHP box binding, suggesting a bipartite interaction with cofactors carrying both the UBX domain and the SHP box (e.g. p47) (Hanzelmann and Schindelin, 2016a).
Figure 3. The interactions of p97 with adaptors and cofactors.
(A) Structure of the p97 N-domain in complex with the UBX domain of FAF1 (PDB:3QC8). The N-domain, depicted as a molecular surface overlaid to a ribbon representation, consists of two subdomains: N-terminal double Ψ-barrel domain (purple) and C-terminal β-barrel domain (red). The UBX domain of FAF1 is depicted as ribbon diagram in magenta. Critical residues for interaction are shown as ball-and-stick models and labeled. (B) Structure of the p97 N-domain in complex with the VIM motif of gp78 (PDB:3TIW). Here the VIM motif is shown as helix in brown and its binding to the N-domain is mostly mediated by charged residues. (C) Structure of the N-terminal domain of PNGase in complex with a C-terminal peptide of p97 (PDB:2HPL). The PNGase N-terminal domain is shown in cartoon representation in green. The bound peptide is shown as a stick model with five residues (labeled) seen in the structure. The carbon atoms are colored in black, nitrogen in blue and oxygen in red. (D) Structure of the PUL domain of FLAA/Ufd3 in complex with a C-terminal peptide of p97 (PDB:3EBB). The PLAA PUL domain is shown in cartoon representation in green. The bound peptide is shown as a stick model with four residues visible in the structure. The carbon atoms are colored in black, nitrogen in blue and oxygen in red.
Adaptors or cofactors binding to the C-terminal tail of p97 are illustrated by several crystallographic studies, exemplified by a structure of the PUB domain of the peptide-N-glycanase (PNGase) in complex with a 10-residue peptide derived from the C-terminus of p97 (Figure 3C) (Zhao et al., 2007). PNGase is responsible for the removal of N-glycosylated sugars from misfolded glycoproteins prior to their degradation by the proteasome (Blom et al., 2004). In higher eukaryotes, PNGase has acquired an additional N-terminal PUB (PNGase/UBA) domain that binds the C-terminus of p97. The latter was termed PUB-interacting motif (PIM). Crystallographic studies showed that the PUB domain forms a complex with PIM in a 1:1 stoichiometry, and the last six residues of the PIM peptide sit in a conserved surface depression of the PUB domain (Allen et al., 2006; Zhao et al., 2007). Intriguingly, the conserved residue Y805 in p97 that is essential for interaction with PNGase is subject to phosphorylation in cells, which provides a means to regulate p97-PNGase interaction (Zhao et al., 2007). Another example of adaptor recruitment by the p97 C-terminal peptide is illustrated by the structure of the complex between PLAA and the C-terminal peptide of p97 (Qiu et al., 2010). PLAA (also named Ufd3) is a phospholipase A2-activating protein implicated in maintaining cellular ubiquitin level (Johnson et al., 1995). In this structure, the critical residue Y805 is also located at the binding interface, which may allow a similar regulation by phosphorylation and de-phosphorylation (Figure 3D).
Structural studies on p97 complexes have also been attempted by cryo-EM (Bebeacua et al., 2012; Beuron et al., 2006; Pye et al., 2007; Rouiller et al., 2000), but inconsistent sample quality and insufficient resolution have led to some controversies. For example, in one study, the stoichiometry of p97 in complex with p47 was determined as six p47 to one hexameric p97 (Rouiller et al., 2000), whereas in another study (Beuron et al., 2006), p47 was shown to be a trimer in solution and the binding stoichiometry is one trimeric p47 to one hexameric p97, a ratio that is more consistent with previously published biochemical data (Kondo et al., 1997). Two EM studies on the complex of p97 and Ufd1-Npl4 (Bebeacua et al., 2012; Pye et al., 2007), a heterodimeric complex known for their involvement in the endoplasmic reticulum-associated degradation (ERAD) (see below) (Ye et al., 2001), were also reported. Collectively, these EM studies cast a general impression that the N-domain-binding adaptors contact both the N- and the D1-domain simultaneously as they sit on top of the N-D1 ring. This conclusion is further supported by an EM study on the interaction of Fas-associated factor-1 (FAF1) with p97 (Ewens et al., 2014).
A comparison between the crystal structure of wild-type N-D1 (PDB:1E32) and that with p47 bound (PDB:1S3S) shows no major domain movement on the part of p97 upon p47 binding (Dreveny et al., 2004). However, adaptor- or cofactor-induced conformational changes may have escaped detection thus far because almost all structural studies were conducted with isolated N-domains or C-terminal PIM (Hanzelmann et al., 2011; Hanzelmann and Schindelin, 2011; Isaacson et al., 2007; Kim et al., 2011; Qiu et al., 2010; Schaeffer et al., 2014; Zhao et al., 2007). On a related note, ATP-dependent conformational changes, particularly those associated with the D1 ATP hydrolysis cycle will certainly influence the position of adaptors and cofactors that bind to the N domain, which may be important for the physiological function of p97.
Biological functions of p97/Cdc48p
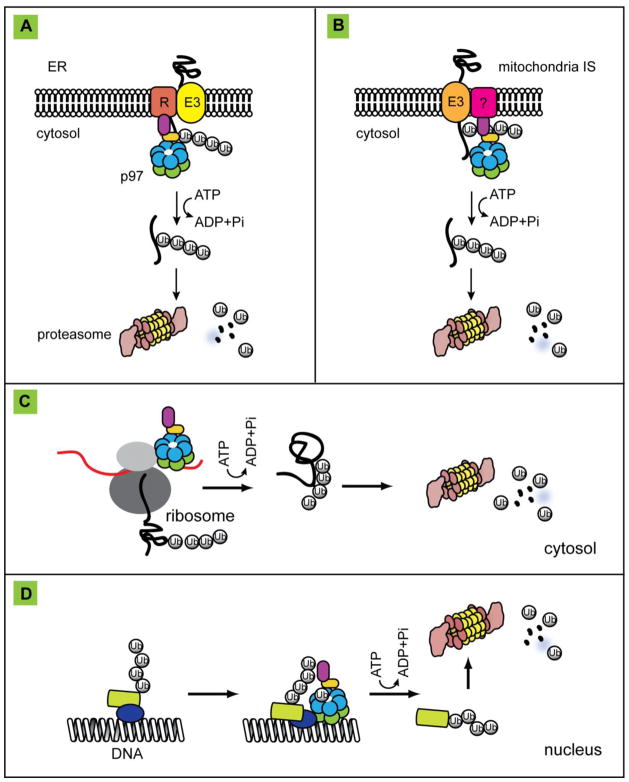
The diverse biological functions of p97 have been extensively reviewed (Dantuma and Hoppe, 2012; Franz et al., 2014; Meyer et al., 2012; Meyer and Weihl, 2014; Yamanaka et al., 2012). Therefore, we only highlight a few key established functions in this review. In general, p97 uses ATP hydrolysis to segregate polypeptides from large protein assemblies or immobile cellular structures such as membranes or chromatin, and therefore, facilitates the degradation of the released polypeptides by the 26S proteasome. To date, the major functions of p97 can be summarized into the following three categories (Figure 4).
Figure 4. The established segregase function of p97/Cdc48p.
p97 collaborates with the proteasome in degradation of misfolded ER proteins (the ERAD pathway) (A), misfolded proteins in the mitochondrial outer membrane (B), defective translocation products (C), and chromatin-associated proteins (D). In each scenario, p97 uses energy from ATP hydrolysis to release polypeptides from either the membranes, the ribosome, or DNA, and then target them to the proteasome for degradation. E3, ubiquitin ligase, R, retrotranslocation complex, mitochondrial IS, mitochondrial inter-membrane space.
Roles in protein homeostasis regulation
A key function of p97/Cdc48p is to maintain protein homeostasis through a network of protein quality control processes (Meyer et al., 2012). In this context, the best studied process is endoplasmic reticulum-associated degradation (ERAD), a pathway that degrades misfolded proteins of the endoplasmic reticulum (ER) (Christianson and Ye, 2014; Ruggiano et al., 2014; Smith et al., 2011). In ERAD, p97/Cdc48p is recruited to the ER membrane via association with membrane adaptors including Derlins and VIMP in mammalian cells or Ubxd2 in S. cerevesiae (Lilley and Ploegh, 2004; Neuber et al., 2005; Schuberth and Buchberger, 2005; Ye et al., 2004). Membrane-associated p97 captures misfolded proteins once they have emerged through a putative retrotranslocation channel (Carvalho et al., 2010). Misfolded proteins are then ubiquitinated and extracted from the membranes upon p97 ATP hydrolysis (Bays et al., 2001; Braun et al., 2002; Flierman et al., 2003; Garza et al., 2009; Jarosch et al., 2002; Rabinovich et al., 2002; Ye et al., 2001; Ye et al., 2003; Zhong et al., 2004), and subsequently targeted to the proteasome for degradation (Figure 4A) (Zhang and Ye, 2014). Besides misfolded ER proteins, p97 and Cdc48p are both capable of releasing certain membrane-bound transcription factors (Hitchcock et al., 2001; Radhakrishnan et al., 2014; Rape et al., 2001; Shcherbik and Haines, 2007). These factors apparently are not degraded by the proteasome. Instead, once released from the membrane, they translocate into the nucleus to influence gene expression in response to specific environmental insults.
Besides ERAD, ATP hydrolysis by p97 is also involved in extracting polypeptides from the mitochondrial outer membrane to facilitate mitochondria-associated degradation (Figure 4B) (Hemion et al., 2014; Heo et al., 2010; Xu et al., 2011). This process can regulate mitochondrial protein homeostasis if the client proteins to be degraded are aberrant molecules. Additionally, factors controlling the turnover of damaged mitochondria by the mitophagy process (e.g. mitofusin) can be degraded in a p97-dependent manner (Tanaka et al., 2010). Intriguingly, p97 and its co-factors Ufd1 and Npl4 are highly enriched on the surface of mitochondria that are damaged by a protonophore, and loss of either p97 or these co-factors causes a defect in mitophagy-mediated clearance of damaged mitochondria (Kimura et al., 2013). These findings suggest a critical role for p97 in mitochondrial homeostasis regulation.
p97 can also release defective translation products stalled on ribosome in a process termed ribosome-associated degradation (Figure 4C) (Brandman et al., 2012; Defenouillere et al., 2013; Verma et al., 2013). It appears that only after extraction from ribosome, can these aberrant polypeptides be degraded efficiently by the proteasome. In addition to the proposed “segregase” function, p97 might play a role in shuttling aberrant polypeptides to the proteasome for degradation. This chaperoning function seems to be particularly important for degradation of certain aggregation-prone misfolded proteins in the nucleus in budding yeast (Gallagher et al., 2014).
Several lines of evidence also implicate p97 and Cdc48p in autophagy, a stress adaptation process that turns over cellular proteins (including misfolded ones) by engulfment of cytosol into double-membrane-surrounded vesicles named autophagosome. However, the precise role of p97 in this process is unclear (Bug and Meyer, 2012; Ju et al., 2009; Ju and Weihl, 2010). In S. cerevisiae, the Cdc48p adaptor Shp1p interacts with Atg8p, an essential autophagy regulator, and the complex of Cdc48p and Shp1p is required for macroautophagy (Krick et al., 2010). Whether the homologous p97-p47 complex interacts with the Atg8 homolog LC3 to regulate autophagy in mammalian cells is unclear. Lastly, p97 was recently demonstrated to play a pivotal role in clearance of non-translating messenger ribonucleoprotein complexes accumulated in stress granules (Buchan et al., 2013).
Chromatin-associated functions
By releasing protein molecules from chromatins in a manner analogous to that in ERAD, p97 also participates in a series of degradation processes in the nucleus collectively known as chromatin-associated degradation (Figure 4D) (Dantuma et al., 2014). Established nuclear substrates of p97/Cdc48p include transcriptional repressor α2 (Wilcox and Laney, 2009), RNA polymerase (Pol) II complex (Verma et al., 2011) and CMG DNA helicase (Maric et al., 2014) in budding yeast, and the DNA replicating licensing factor CDT1 (Franz et al., 2011; Raman et al., 2011), a replisome component Mcm7 (Moreno et al., 2014), DNA repairing proteins DDB2, XPC, and Rad52 (Bergink et al., 2013; Puumalainen et al., 2014), mitosis regulator Aurora B (Ramadan et al., 2007), certain DNA polymerases (Davis et al., 2012; Mosbech et al., 2012) and the polycomb protein L3MBTL1 (Acs et al., 2011) in mammals. These substrates have linked p97 function to diverse nuclear events ranging from gene transcription to DNA replication and repair.
Membrane fusion and vesicular trafficking
Biochemical and genetic evidence also implicated p97 in fusion of vesicles that lead to the re-formation of the Golgi apparatus at the end of mitosis (Uchiyama and Kondo, 2005). This process depends on the ubiquitin binding adaptor p47 (Kondo et al., 1997; Meyer et al., 2002), a related adaptor p37 that lacks ubiquitin binding activity (Uchiyama et al., 2006), and a p97-associated deubiquitinase named VCIP135 (Uchiyama et al., 2002). However, the precise role of p97 in vesicle fusion is unclear due to lack of information on its substrate(s).
A role in endocytosis for p97 has also been revealed two recent studies that used a proteomic approach to explore p97-interacting proteins in mammalian cells (Bug and Meyer, 2012; Kirchner et al., 2013; Ramanathan and Ye, 2012). One study identified the early endosome-associated antigen 1 (EEA1) as a p97 interacting protein and showed that inhibition of p97 results in a delay in lysosomal targeting of an endocytosis cargo. p97 inhibition also generated an endosome clustering and swelling phenotype, which appeared to be caused by increased EEA1 oligomerization and thus uncontrolled endosome tethering and fusion (Ramanathan and Ye, 2012). In a second study, the plasma membrane protein caveolin was found to interact with p97 and its cofactor UbxD1. In p97-inhibited cells, enlargement of endosome was similarly observed and the trafficking of caveolin to late endosome is affected (Ritz et al., 2011). These results not only suggest a general function for p97 in regulating the clustering and fusion of membrane vesicles, but also imply that it can control the trafficking itinerary of a specific endocytic cargo, perhaps by regulating its partition between different lipid domains.
Molecular basis of force generation
Although intensely studied, the molecular mechanism underlying the “segregase” activity of p97/Cdc48p remains poorly defined. Major unresolved issues are whether or not p97/Cdc48p unfolds its client proteins, and therefore disrupting their interactions with protein assemblies, membranes, or chromatin; if so, whether or not it can act as a ‘translocase’ that moves substrates through the central pore.
The ATP hydrolysis cycle of p97/Cdc48P
Isolated full-length p97 has a moderate ATPase activity that turns over 1–5 ATP molecules per hexmer per second (Meyer et al., 1998; Song et al., 2003; Tang and Xia, 2013; Ye et al., 2003). Mutations in the D1 Walker A (K251A/T) motif led to a huge reduction in ATPase activity, indicating that nucleotide binding in the D1 domain is important for the activity of the D2 domain (Nishikori et al., 2011; Tang and Xia, 2013; Ye et al., 2003). Conversely, Walker A or Walker B mutations in the D2 domain almost completely abolish the p97 ATPase activity (Nishikori et al., 2011; Tang and Xia, 2013; Ye et al., 2003), suggesting that nucleotide binding and hydrolysis by the D2 domain are required for activating the D1 domain. Apparently, there is a communication mechanism between the D1 and D2 domains. Based on these observations, it was proposed that the two ATPase rings may alternate in ATP hydrolysis (Ye et al., 2003). The feedback from the D2 domain to the D1 domain is likely mediated by the D1–D2 linker, because a p97 mutant carrying both the N- and D1-domains shows no ATPase activity when this linker is absent (Chou et al., 2014; Tang and Xia, 2013; Tang and Xia, 2016; Ye et al., 2003).
The N-domain appears to suppress p97 ATPase activity via communications with the D1 domain. Deletion of the N-domain in VAT, a VCP-like ATPase from Thermoplasma acidophilum, increases its ATPase activity significantly (Rothballer et al., 2007), and fixing the N-domain to the Down-conformation by an engineered disulfide linker reduces p97 ATPase activity (Niwa et al., 2012). p97 mutants carrying single IBMPFD mutations (see below) have 2–3 fold increase in ATPase activity. These mutations are all mapped to the N-D1 interface, and therefore likely affect the N-D1 interactions (Halawani et al., 2009; Niwa et al., 2012; Tang and Xia, 2013; Weihl et al., 2006; Zhang et al., 2015). The slow deteriorating pathology associated with these mutations suggests that the p97 ATPase activity needs to be tightly regulated to achieve optimal function in cells.
The ATPase activity of p97 can also be influenced by many other factors in vitro. For example, it can be stimulated by elevated temperature (Song et al., 2003) or by a putative substrate protein (DeLaBarre et al., 2006). Association with different cofactors can have either positive or negative impact on p97 ATPase activity (Meyer et al., 1998; Zhang et al., 2015), but the physiological relevance of these observations is unclear. How cells regulate the p97 ATPase activity is also largely unknown.
Is p97 an unfoldase?
By definition, a protein unfoldase unfolds substrates into linear peptides. A “segregase” may act with or without unfolding its client proteins (Yamanaka et al., 2012). Despite that many hexameric ring-like AAA+ proteins are classical examples of protein unfoldases (e.g. ClpA and ClpX) that unfold polypeptides by threading them through the central pores (Singh et al., 2000), full-length wild-type p97 cannot unfold GFP-ssrA, a model aberrant protein (Rothballer et al., 2007). Nevertheless, it is interesting to note that VAT, a thermoplasma acidophilum p97 homolog, is able to unfold GFP-ssrA (Gerega et al., 2005), but it does so with high efficiency only when the N-domain is removed (Barthelme and Sauer, 2012; Gerega et al., 2005). In addition, a N-domain-deleted VAT variant can cooperate with the 20S proteasome to efficiently degrade GFP-ssrA in vitro. A comparison of the sequences between p97 and VAT identified a motif (KYYG) at the D1 pore of VAT, which is replaced with KLAG in p97. When combined with the removal of the N-domain, tyrosine substitution to either Leu or Ala allows p97 to unfold GFP-ssrA, which causes its degradation by the 20S proteasome (Barthelme and Sauer, 2013; Rothballer et al., 2007). These findings suggest that the widely observed cooperation between AAA rings and the 20S proteasome is an ancient molecular design for unfolding and degradation of aberrant polypeptides, but the divergent evolution of the N-domain and the D1 pore of p97 seems to allow p97 to operate by a new mode.
Although eukaryotic p97/Cdc48p does not appear to thread substrates through the central pore, it may still act as an unfoldase to promote protein turnover. In cells, the requirement of p97/Cdc48 for degradation of a ubiquitin fusion protein can be bypassed if a flexible peptide was appended to the C-terminus of this substrate (Beskow et al., 2009). This observation was taken to indicate that p97/Cdc48p acts to initiate protein unfolding, which exposes a loosely-folded initiation peptide for engaging the proteasome. Intriguingly, the involvement of Cdc48p in the ubiquitin fusion degradation pathway coincides tightly with the requirement for Lys48- and Lys29-linked ubiquitination in the ubiquitin fusion part (Godderz et al., 2015), suggesting that the function of p97/Cdc48p is intimately linked to substrate ubiquitination. This is consistent with the observation that many p97/Cdc48p interacting-proteins contain independent ubiquitin binding site.
Mechanism of force generation
How do conformational changes in p97 account for the proposed “segregase” activity? By far, the most consistent conformational changes observed are the close-and-open of the D2 pore, which seems to be associated with a rotational movement between the D1 and D2 rings during the D2 ATPase cycle, and more dramatically, the up-and-down swing motion of the N-domain, driven by nucleotide hydrolysis in the D1 domain (Figure 5A). If the two ATPase domains indeed alternate in ATP hydrolysis, it is conceivable that the opening of the D2 ring, if coupled to the ATP-bound state of the D1 ring and therefore to the Up-conformation (Banerjee et al., 2016; Tang et al., 2010), may allow substrates to bind to the D2 pore, as suggested by a mutagenesis study (DeLaBarre et al., 2006). The alternate ATPase cycle could then lead to a piston-like, up-and-down motion for the D1–D2 rings, which could be coupled to substrate binding and release. Taking ERAD as an example, since the N-domain is anchored to the immobile ER membranes via adaptors, the downside swing of the N-domain would naturally be translated into a pulling force that moves the D1–D2 rings toward the membrane, leading to the extraction of substrates bound to the D2 pore (Figure 5B, top panel). The model is equally applicable if p97 engages substrates with only the D1 ring (bottom panel). Although these models provide a plausible means for p97 to separate a client protein from its interaction partners, at the moment, we cannot rule out other models, as proposed previously (Pye et al., 2006).
Figure 5. Force generation coupled to ATP hydrolysis.
(A) An ATP hydrolysis model for p97. A p97 hexamer is represented as two concentric rings with D1 ring in green and D2 ring in brown. The N-domains in the Down-conformation are shown as magenta balls. D1 domains with pre-bound ADP are labeled with the letter D. ATP molecules introduced into the system will first go to the D1 domains with no pre-bound nucleotide, which leads the N-domains to the Up-conformation. Occupation of ATP to the D1 domain renders the cognate D2 domain capable of hydrolyzing ATP, which is labeled with a red *. The D1 domain probably hydrolyzes ATP once a few D2 domains have been converted to the ADP bound state. (B) A proposed model of force generation by a N-domain conformational change in ERAD. p97 is anchored to the ER membrane by association with a membrane adaptor (blue) using its N-domain. If substrate is bound to the central pore of the D2 domain when the D1 domain is in the Up-conformation (top panel), the swing of the N-domain to the Down-conformation after ATP hydrolysis will pull the D1 ring closer to the membrane, leading to the extraction of substrate out of the membranes. Alternatively, if the substrate is bound to the D1 domain (bottom panel), a switch from the Down-conformation to the Up-conformation following nucleotide exchange in D1 will move the ATPase domains away from the membrane, pulling substrate out of the membrane.
p97 inhibitors and cancer therapy
By screening and characterizing compounds that inhibit the degradation of a fluorescence-labeled ERAD substrate, the first p97 inhibitor Eeyarestatin (EerI) was reported a few years ago (Fiebiger et al., 2004; Wang et al., 2008; Wang et al., 2010). Structure-activity relationship studies suggested that EerI contains two functional modules: a nitrofuran ring that binds the D1 domain of p97 and an aromatic ring-containing module that recruits EerI to cellular membranes including the ER. Once localized to the ER membrane, the target selectivity of EerI is significantly enhanced. Consequently, it mainly targets p97 that is membrane-bound (Wang et al., 2010). Although the mechanism of p97 inhibition by EerI is unclear, it induces several key phenotypes associated with p97 inhibition such as polyubiquitinated protein accumulation, ERAD inhibition, ER stress induction, and apoptosis (Wang et al., 2009). Importantly, EerI displays significant cancer-killing activity in vitro preferentially against cancer cells isolated from patients, and it can synergize with the proteasome inhibitor Bortezomib to kill cancer cells (Wang et al., 2009). These observations raise the possibility of targeting p97 as a potential cancer therapy. Subsequent studies on several ATP competitive and allosteric inhibitors of p97 confirmed this hypothesis (Chou et al., 2011; Chou et al., 2013; Magnaghi et al., 2013). Importantly, a potent and specific p97 inhibitor CB-5083 has been developed recently, which elicits promising anti-cancer effect in mouse xenograft tumor models (Anderson et al., 2015; Zhou et al., 2015). The mode of interaction by p97 inhibitors has been studied by both structural modeling as well as by cryo-EM technology (Anderson et al., 2015; Banerjee et al., 2016; Magnaghi et al., 2013). The information may be helpful for design of more potent and specific p97 inhibitors.
Relevance to human diseases
p97 has received much attention recently also because genetic studies have linked mutations in p97 to pathogenesis of several human diseases including Inclusion Body Myopathy associated with Paget’s disease of the bone and Frontotemporal Dementia (IBMPFD) and amyotrophic lateral sclerosis (ALS) (Johnson et al., 2010; Watts et al., 2004).
IBMPFD is an autosomal dominant, progressive, and ultimately fatal disorder with initial symptoms typically appearing in adulthood (Kimonis et al., 2000). The disease mainly affects one or more of the following tissue types: the muscles (myopathy), the bones (Paget’s disease of the bone), and the brain (frontotemporal dementia). Among them, myopathy is the most common clinical menifestation found in approximately 85% of the patients, while the bone pathology and frontotemporal dementia are observed in approximately 50% and 30% of the patients, respectively. However, there is no clear genotype-phenotype corrletion as patients from the same family with the same mutation can show different symptoms.
Phenotypic characterizations of muscle samples from IBMPFD patient revealed rimmed vacuoles that were stained positively for both ubiquitin and p97 (Watts et al., 2004). In brain tissues from patients, nuclear inclusions were detected in neurons and they also contain ubiquitin and p97 (Kimonis and Watts, 2005). More recent studies found inclusions positive for TAR DNA-binding Protein-43 (TDP-43) in patient samples (Weihl et al., 2008). These findings suggest that defects in p97-mediated PQC network may contribute to the etiology of this disease.
To date, more than 30 missense mutations in p97 have been reported in IBMPFD patients, involving amino acids at 17 different positions (Mehta et al., 2013; Nalbandian et al., 2011). Intriguingly, these mutations are all mapped to the interface between the N- and D1-domain of p97, suggesting that communications between the N-domain and the D1 domain may be crucial for p97 function.
Autosomal dominantly inherited amyotrophic lateral sclerosis (ALS) is another disorder genetically linked to mutations in p97. ALS is clinically characterized by progressive dysfunction in motor neurons, resulting in death from respiratory failure. Unlike IBMPFD, ALS-associated p97 mutations account for ~1%–2% of all familial ALS cases (Johnson et al., 2010). The pathological hallmark of the disease, loss of motor neurons, is often associated with the presence of ubiquitin-positive inclusions or deposition of TDP-43 aggregates in motor neurons (Johnson et al., 2010), once again, linking the ALS pathology to defects in quality control of misfolded proteins. However, how mutations in p97 cause ALS has remained unclear.
Conclusions and Perspective
Through years of studies, we have significantly deepened our understanding on the structure and function of p97/Cdc48p. Most noticeable is the rapid expansion in our knowledge on cofactors and adaptors and the corresponding new functions of this essential chaperone system. Nonetheless, several fundamental questions regarding the mechanistic action of p97/Cdc48p remain unanswered. The most important one is whether the conformational changes observed in the reported studies are truly physiologically relevant, and if so, how these conformational changes generate force to carry out the “segregase” function. To better address this question, we would need a robust in vitro assay that recapitulates an in vivo function of p97. This appears to be an extremely challenging task as it has apparently been tried by many laboratories without much success. The recently identified p97 substrates may offer new hope along this direction. Another key issue is to understand the hierarchical organization of adaptors/cofactors binding in the context of a given p97/Cdc48p dependent pathway. Animal studies are also in urgent need to better understand the mechanistic links between p97 and human diseases. Finally, given the demonstrated favorable anti-cancer activity in mouse model for a p97 inhibitor, it is anticipated that more p97 inhibitors may be sought, and studies along this line may one day lead to a new class of anti-cancer agents.
Highlights.
p97/Cdc48p participates in various cellular pathways by functioning as a “segregase” to separate polypeptide substrates from large immobile cellular structures.
Mutations in p97 have been associated with various human diseases.
Significant progress has been made in obtaining atomic resolution structures of p97 in different conformations.
Controversy remains as to the mechanism underlying force generation by p97.
Acknowledgments
This review and the corresponding Gene Wiki article are written as part of the Gene Wiki Review series—a series resulting from a collaboration between the journal GENE and the Gene Wiki Initiative. The Gene Wiki Initiative is supported by National Institutes of Health (GM089820). The research in the laboratories of D. Xia and Y. Ye is supported by the Intramural Research Program of the National Cancer Institute and of the National Institute of Diabetes, Digestive & Kidney Diseases at the National Institutes of Health.
Abbreviations
- VCP
valosin-containing protein
- TER ATPase
transitional endoplasmic reticulum ATPase
- AAA+
extended family of ATPases associated with various cellular activities
- ER
endoplasmic reticulum
- PQC
protein quality control
- IBMPEF
Inclusion Body Myopathy associated with Paget’s disease of the bone and Frontotemporal Dementia
- ALS
amyotrophic lateral sclerosis
- EM
electron microscopy
- ATP
Adenosine triphosphate
- ADP
Adenosine diphosphate
- AMP-PNP
Adenylyl-imidodiphosphate
- ERAD
ER-associated protein degradation
- VAT
VCP-like ATPase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acharya U, Jacobs R, Peters JM, Watson N, Farquhar MG, Malhotra V. The formation of Golgi stacks from vesiculated Golgi membranes requires two distinct fusion events. Cell. 1995;82:895–904. doi: 10.1016/0092-8674(95)90269-4. [DOI] [PubMed] [Google Scholar]
- Acs K, Luijsterburg MS, Ackermann L, Salomons FA, Hoppe T, Dantuma NP. The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nature structural & molecular biology. 2011;18:1345–1350. doi: 10.1038/nsmb.2188. [DOI] [PubMed] [Google Scholar]
- Allen MD, Buchberger A, Bycroft M. The PUB domain functions as a p97 binding module in human peptide N-glycanase. The Journal of biological chemistry. 2006;281:25502–25508. doi: 10.1074/jbc.M601173200. [DOI] [PubMed] [Google Scholar]
- Anderson DJ, Le Moigne R, Djakovic S, Kumar B, Rice J, Wong S, Wang J, Yao B, Valle E, Kiss von Soly S, Madriaga A, Soriano F, Menon MK, Wu ZY, Kampmann M, Chen Y, Weissman JS, Aftab BT, Yakes FM, Shawver L, Zhou HJ, Wustrow D, Rolfe M. Targeting the AAA ATPase p97 as an Approach to Treat Cancer through Disruption of Protein Homeostasis. Cancer cell. 2015;28:653–665. doi: 10.1016/j.ccell.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballar P, Shen Y, Yang H, Fang S. The role of a novel p97/valosin-containing protein-interacting motif of gp78 in endoplasmic reticulum-associated degradation. The Journal of biological chemistry. 2006;281:35359–35368. doi: 10.1074/jbc.M603355200. [DOI] [PubMed] [Google Scholar]
- Ballar P, Zhong Y, Nagahama M, Tagaya M, Shen Y, Fang S. Identification of SVIP as an Endogenous Inhibitor of Endoplasmic Reticulum-associated Degradation. The Journal of biological chemistry. 2007;282:33908–33914. doi: 10.1074/jbc.M704446200. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Bartesaghi A, Merk A, Rao P, Bulfer SL, Yan Y, Green N, Mroczkowski B, Neitz RJ, Wipf P, Falconieri V, Deshaies RJ, Milne JL, Huryn D, Arkin M, Subramaniam S. 2.3 A resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016 doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Sauer RT. Identification of the Cdc48*20S proteasome as an ancient AAA+ proteolytic machine. Science. 2012;337:843–846. doi: 10.1126/science.1224352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelme D, Sauer RT. Bipartite determinants mediate an evolutionarily conserved interaction between Cdc48 and the 20S peptidase. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3327–3332. doi: 10.1073/pnas.1300408110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays NW, Wilhovsky SK, Goradia A, Hodgkiss-Harlow K, Hampton RY. HRD4/NPL4 is required for the proteasomal processing of ubiquitinated ER proteins. Molecular biology of the cell. 2001;12:4114–4128. doi: 10.1091/mbc.12.12.4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebeacua C, Forster A, McKeown C, Meyer HH, Zhang X, Freemont PS. Distinct conformations of the protein complex p97-Ufd1-Npl4 revealed by electron cryomicroscopy. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:1098–1103. doi: 10.1073/pnas.1114341109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergink S, Ammon T, Kern M, Schermelleh L, Leonhardt H, Jentsch S. Role of Cdc48/p97 as a SUMO-targeted segregase curbing Rad51-Rad52 interaction. Nature cell biology. 2013;15:526–532. doi: 10.1038/ncb2729. [DOI] [PubMed] [Google Scholar]
- Beskow A, Grimberg KB, Bott LC, Salomons FA, Dantuma NP, Young P. A conserved unfoldase activity for the p97 AAA-ATPase in proteasomal degradation. Journal of molecular biology. 2009;394:732–746. doi: 10.1016/j.jmb.2009.09.050. [DOI] [PubMed] [Google Scholar]
- Beuron F, Dreveny I, Yuan X, Pye VE, McKeown C, Briggs LC, Cliff MJ, Kaneko Y, Wallis R, Isaacson RL, Ladbury JE, Matthews SJ, Kondo H, Zhang X, Freemont PS. Conformational changes in the AAA ATPase p97-p47 adaptor complex. The EMBO journal. 2006;25:1967–1976. doi: 10.1038/sj.emboj.7601055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuron F, Flynn TC, Ma J, Kondo H, Zhang X, Freemont PS. Motions and negative cooperativity between p97 domains revealed by cryo-electron microscopy and quantised elastic deformational model. Journal of molecular biology. 2003;327:619–629. doi: 10.1016/s0022-2836(03)00178-5. [DOI] [PubMed] [Google Scholar]
- Blom D, Hirsch C, Stern P, Tortorella D, Ploegh HL. A glycosylated type I membrane protein becomes cytosolic when peptide: N-glycanase is compromised. The EMBO journal. 2004;23:650–658. doi: 10.1038/sj.emboj.7600090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeddrich A, Gaumer S, Haacke A, Tzvetkov N, Albrecht M, Evert BO, Muller EC, Lurz R, Breuer P, Schugardt N, Plassmann S, Xu K, Warrick JM, Suopanki J, Wullner U, Frank R, Hartl UF, Bonini NM, Wanker EE. An arginine/lysine-rich motif is crucial for VCP/p97-mediated modulation of ataxin-3 fibrillogenesis. The EMBO journal. 2006;25:1547–1558. doi: 10.1038/sj.emboj.7601043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohm S, Lamberti G, Fernandez-Saiz V, Stapf C, Buchberger A. Cellular functions of Ufd2 and Ufd3 in proteasomal protein degradation depend on Cdc48 binding. Molecular and cellular biology. 2011;31:1528–1539. doi: 10.1128/MCB.00962-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandman O, Stewart-Ornstein J, Wong D, Larson A, Williams CC, Li GW, Zhou S, King D, Shen PS, Weibezahn J, Dunn JG, Rouskin S, Inada T, Frost A, Weissman JS. A ribosome-bound quality control complex triggers degradation of nascent peptides and signals translation stress. Cell. 2012;151:1042–1054. doi: 10.1016/j.cell.2012.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun S, Matuschewski K, Rape M, Thoms S, Jentsch S. Role of the ubiquitin-selective CDC48(UFD1/NPL4)chaperone (segregase) in ERAD of OLE1 and other substrates. The EMBO journal. 2002;21:615–621. doi: 10.1093/emboj/21.4.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruderer RM, Brasseur C, Meyer HH. The AAA ATPase p97/VCP interacts with its alternative co-factors, Ufd1-Npl4 and p47, through a common bipartite binding mechanism. The Journal of biological chemistry. 2004;279:49609–49616. doi: 10.1074/jbc.M408695200. [DOI] [PubMed] [Google Scholar]
- Buchan JR, Kolaitis RM, Taylor JP, Parker R. Eukaryotic stress granules are cleared by autophagy and Cdc48/VCP function. Cell. 2013;153:1461–1474. doi: 10.1016/j.cell.2013.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Schindelin H, Hanzelmann P. Control of p97 function by cofactor binding. FEBS letters. 2015;589:2578–2589. doi: 10.1016/j.febslet.2015.08.028. [DOI] [PubMed] [Google Scholar]
- Bug M, Meyer H. Expanding into new markets--VCP/p97 in endocytosis and autophagy. Journal of structural biology. 2012;179:78–82. doi: 10.1016/j.jsb.2012.03.003. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Stanley AM, Rapoport TA. Retrotranslocation of a misfolded luminal ER protein by the ubiquitin-ligase Hrd1p. Cell. 2010;143:579–591. doi: 10.1016/j.cell.2010.10.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TF, Brown SJ, Minond D, Nordin BE, Li K, Jones AC, Chase P, Porubsky PR, Stoltz BM, Schoenen FJ, Patricelli MP, Hodder P, Rosen H, Deshaies RJ. Reversible inhibitor of p97, DBeQ, impairs both ubiquitin-dependent and autophagic protein clearance pathways. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:4834–4839. doi: 10.1073/pnas.1015312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TF, Bulfer SL, Weihl CC, Li K, Lis LG, Walters MA, Schoenen FJ, Lin HJ, Deshaies RJ, Arkin MR. Specific inhibition of p97/VCP ATPase and kinetic analysis demonstrate interaction between D1 and D2 ATPase domains. Journal of molecular biology. 2014;426:2886–2899. doi: 10.1016/j.jmb.2014.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou TF, Li K, Frankowski KJ, Schoenen FJ, Deshaies RJ. Structure-activity relationship study reveals ML240 and ML241 as potent and selective inhibitors of p97 ATPase. ChemMedChem. 2013;8:297–312. doi: 10.1002/cmdc.201200520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Ye Y. Cleaning up in the endoplasmic reticulum: ubiquitin in charge. Nature structural & molecular biology. 2014;21:325–335. doi: 10.1038/nsmb.2793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma NP, Acs K, Luijsterburg MS. Should I stay or should I go: VCP/p97-mediated chromatin extraction in the DNA damage response. Experimental cell research. 2014;329:9–17. doi: 10.1016/j.yexcr.2014.08.025. [DOI] [PubMed] [Google Scholar]
- Dantuma NP, Hoppe T. Growing sphere of influence: Cdc48/p97 orchestrates ubiquitin-dependent extraction from chromatin. Trends Cell Biol. 2012;22:483–491. doi: 10.1016/j.tcb.2012.06.003. [DOI] [PubMed] [Google Scholar]
- Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- Davies JM, Tsuruta H, May AP, Weis WI. Conformational changes of p97 during nucleotide hydrolysis determined by small-angle X-Ray scattering. Structure. 2005;13:183–195. doi: 10.1016/j.str.2004.11.014. [DOI] [PubMed] [Google Scholar]
- Davis EJ, Lachaud C, Appleton P, Macartney TJ, Nathke I, Rouse J. DVC1 (C1orf124) recruits the p97 protein segregase to sites of DNA damage. Nature structural & molecular biology. 2012;19:1093–1100. doi: 10.1038/nsmb.2394. [DOI] [PubMed] [Google Scholar]
- Defenouillere Q, Yao Y, Mouaikel J, Namane A, Galopier A, Decourty L, Doyen A, Malabat C, Saveanu C, Jacquier A, Fromont-Racine M. Cdc48-associated complex bound to 60S particles is required for the clearance of aberrant translation products. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:5046–5051. doi: 10.1073/pnas.1221724110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLaBarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nature structural biology. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Brunger AT. Nucleotide dependent motion and mechanism of action of p97/VCP. Journal of molecular biology. 2005;347:437–452. doi: 10.1016/j.jmb.2005.01.060. [DOI] [PubMed] [Google Scholar]
- DeLaBarre B, Christianson JC, Kopito RR, Brunger AT. Central pore residues mediate the p97/VCP activity required for ERAD. Molecular cell. 2006;22:451–462. doi: 10.1016/j.molcel.2006.03.036. [DOI] [PubMed] [Google Scholar]
- Dreveny I, Kondo H, Uchiyama K, Shaw A, Zhang X, Freemont PS. Structural basis of the interaction between the AAA ATPase p97/VCP and its adaptor protein p47. The EMBO journal. 2004;23:1030–1039. doi: 10.1038/sj.emboj.7600139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens CA, Panico S, Kloppsteck P, McKeown C, Ebong IO, Robinson C, Zhang X, Freemont PS. The p97-FAF1 protein complex reveals a common mode of p97 adaptor binding. The Journal of biological chemistry. 2014;289:12077–12084. doi: 10.1074/jbc.M114.559591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiebiger E, Hirsch C, Vyas JM, Gordon E, Ploegh HL, Tortorella D. Dissection of the dislocation pathway for type I membrane proteins with a new small molecule inhibitor, eeyarestatin. Molecular biology of the cell. 2004;15:1635–1646. doi: 10.1091/mbc.E03-07-0506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flierman D, Ye Y, Dai M, Chau V, Rapoport TA. Polyubiquitin serves as a recognition signal, rather than a ratcheting molecule, during retrotranslocation of proteins across the endoplasmic reticulum membrane. The Journal of biological chemistry. 2003;278:34774–34782. doi: 10.1074/jbc.M303360200. [DOI] [PubMed] [Google Scholar]
- Franz A, Ackermann L, Hoppe T. Create and preserve: proteostasis in development and aging is governed by Cdc48/p97/VCP. Biochimica et biophysica acta. 2014;1843:205–215. doi: 10.1016/j.bbamcr.2013.03.031. [DOI] [PubMed] [Google Scholar]
- Franz A, Orth M, Pirson PA, Sonneville R, Julian Blow J, Gartner A, Stemmann O, Hoppe T. CDC-48/p97 coordinates CDT-1 degradation with GINS chromatin dissociation to ensure faithful DNA replication. Molecular cell. 2011 doi: 10.1016/j.molcel.2011.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher PS, Clowes Candadai SV, Gardner RG. The requirement for Cdc48/p97 in nuclear protein quality control degradation depends on the substrate and correlates with substrate insolubility. Journal of cell science. 2014;127:1980–1991. doi: 10.1242/jcs.141838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garza RM, Sato BK, Hampton RY. In vitro analysis of Hrd1p-mediated retrotranslocation of its multispanning membrane substrate 3-hydroxy-3-methylglutaryl (HMG)-CoA reductase. The Journal of biological chemistry. 2009;284:14710–14722. doi: 10.1074/jbc.M809607200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerega A, Rockel B, Peters J, Tamura T, Baumeister W, Zwickl P. VAT, the thermoplasma homolog of mammalian p97/VCP, is an N domain-regulated protein unfoldase. The Journal of biological chemistry. 2005;280:42856–42862. doi: 10.1074/jbc.M510592200. [DOI] [PubMed] [Google Scholar]
- Gill JS, Ghatei MA, Domin J, Bloom SR. The generation of valosin-like peptides from a precursor protein in vitro as an extraction artifact. Life sciences. 1989;44:483–491. doi: 10.1016/0024-3205(89)90464-5. [DOI] [PubMed] [Google Scholar]
- Godderz D, Heinen C, Marchese FP, Kurz T, Acs K, Dantuma NP. Cdc48-independent proteasomal degradation coincides with a reduced need for ubiquitylation. Scientific reports. 2015;5:7615. doi: 10.1038/srep07615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenblatt EJ, Olzmann JA, Kopito RR. Derlin-1 is a rhomboid pseudoprotease required for the dislocation of mutant alpha-1 antitrypsin from the endoplasmic reticulum. Nature structural & molecular biology. 2011;18:1147–1152. doi: 10.1038/nsmb.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halawani D, LeBlanc AC, Rouiller I, Michnick SW, Servant MJ, Latterich M. Hereditary inclusion body myopathy-linked p97/VCP mutations in the NH2 domain and the D1 ring modulate p97/VCP ATPase activity and D2 ring conformation. Molecular and cellular biology. 2009;29:4484–4494. doi: 10.1128/MCB.00252-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann P, Buchberger A, Schindelin H. Hierarchical binding of cofactors to the AAA ATPase p97. Structure. 2011;19:833–843. doi: 10.1016/j.str.2011.03.018. [DOI] [PubMed] [Google Scholar]
- Hanzelmann P, Schindelin H. The structural and functional basis of the p97/valosin-containing protein (VCP)-interacting motif (VIM): mutually exclusive binding of cofactors to the N-terminal domain of p97. The Journal of biological chemistry. 2011;286:38679–38690. doi: 10.1074/jbc.M111.274506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzelmann P, Schindelin H. Characterization of an Additional Binding Surface on the p97 N-Terminal Domain Involved in Bipartite Cofactor Interactions. Structure. 2016a;24:140–147. doi: 10.1016/j.str.2015.10.027. [DOI] [PubMed] [Google Scholar]
- Hanzelmann P, Schindelin H. Structural Basis of ATP Hydrolysis and Intersubunit Signaling in the AAA+ ATPase p97. Structure. 2016b;24:127–139. doi: 10.1016/j.str.2015.10.026. [DOI] [PubMed] [Google Scholar]
- Hemion C, Flammer J, Neutzner A. Quality control of oxidatively damaged mitochondrial proteins is mediated by p97 and the proteasome. Free radical biology & medicine. 2014;75:121–128. doi: 10.1016/j.freeradbiomed.2014.07.016. [DOI] [PubMed] [Google Scholar]
- Heo JM, Livnat-Levanon N, Taylor EB, Jones KT, Dephoure N, Ring J, Xie J, Brodsky JL, Madeo F, Gygi SP, Ashrafi K, Glickman MH, Rutter J. A stress-responsive system for mitochondrial protein degradation. Molecular cell. 2010;40:465–480. doi: 10.1016/j.molcel.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitchcock AL, Krebber H, Frietze S, Lin A, Latterich M, Silver PA. The conserved npl4 protein complex mediates proteasome-dependent membrane-bound transcription factor activation. Molecular biology of the cell. 2001;12:3226–3241. doi: 10.1091/mbc.12.10.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huyton T, V, Pye E, Briggs LC, Flynn TC, Beuron F, Kondo H, Ma J, Zhang X, Freemont PS. The crystal structure of murine p97/VCP at 3.6A. Journal of structural biology. 2003;144:337–348. doi: 10.1016/j.jsb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- Isaacson RL, V, Pye E, Simpson P, Meyer HH, Zhang X, Freemont PS, Matthews S. Detailed structural insights into the p97-Npl4-Ufd1 interface. The Journal of biological chemistry. 2007;282:21361–21369. doi: 10.1074/jbc.M610069200. [DOI] [PubMed] [Google Scholar]
- Jarosch E, Taxis C, Volkwein C, Bordallo J, Finley D, Wolf DH, Sommer T. Protein dislocation from the ER requires polyubiquitination and the AAA-ATPase Cdc48. Nature cell biology. 2002;4:134–139. doi: 10.1038/ncb746. [DOI] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A. A proteolytic pathway that recognizes ubiquitin as a degradation signal. The Journal of biological chemistry. 1995;270:17442–17456. doi: 10.1074/jbc.270.29.17442. [DOI] [PubMed] [Google Scholar]
- Johnson JO, Mandrioli J, Benatar M, Abramzon Y, Van Deerlin VM, Trojanowski JQ, Gibbs JR, Brunetti M, Gronka S, Wuu J, Ding J, McCluskey L, Martinez-Lage M, Falcone D, Hernandez DG, Arepalli S, Chong S, Schymick JC, Rothstein J, Landi F, Wang YD, Calvo A, Mora G, Sabatelli M, Monsurro MR, Battistini S, Salvi F, Spataro R, Sola P, Borghero G, Galassi G, Scholz SW, Taylor JP, Restagno G, Chio A, Traynor BJ Consortium I. Exome sequencing reveals VCP mutations as a cause of familial ALS. Neuron. 2010;68:857–864. doi: 10.1016/j.neuron.2010.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Fuentealba RA, Miller SE, Jackson E, Piwnica-Worms D, Baloh RH, Weihl CC. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. The Journal of cell biology. 2009;187:875–888. doi: 10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JS, Weihl CC. p97/VCP at the intersection of the autophagy and the ubiquitin proteasome system. Autophagy. 2010;6:283–285. doi: 10.4161/auto.6.2.11063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, Kang W, Suh SW, Yang JK. Crystal structure of FAF1 UBX domain in complex with p97/VCP N domain reveals a conformational change in the conserved FcisP touch-turn motif of UBX domain. Proteins. 2011;79:2583–2587. doi: 10.1002/prot.23073. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Kim EE. Crystallization and preliminary X-ray crystallographic analysis of the complex between the N-D1 domain of VCP from Homo sapiens and the N domain of OTU1 from Saccharomyces cerevisiae. Acta crystallographica Section F, Structural biology communications. 2014;70:1087–1089. doi: 10.1107/S2053230X14013351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimonis VE, Kovach MJ, Waggoner B, Leal S, Salam A, Rimer L, Davis K, Khardori R, Gelber D. Clinical and molecular studies in a unique family with autosomal dominant limb-girdle muscular dystrophy and Paget disease of bone. Genetics in medicine : official journal of the American College of Medical Genetics. 2000;2:232–241. doi: 10.1097/00125817-200007000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimonis VE, Watts GD. Autosomal dominant inclusion body myopathy, Paget disease of bone, and frontotemporal dementia. Alzheimer Dis Assoc Disord. 2005;19(Suppl 1):S44–47. doi: 10.1097/01.wad.0000183081.76820.5a. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Fukushi J, Hori S, Matsuda N, Okatsu K, Kakiyama Y, Kawawaki J, Kakizuka A, Tanaka K. Different dynamic movements of wild-type and pathogenic VCPs and their cofactors to damaged mitochondria in a Parkin-mediated mitochondrial quality control system. Genes Cells. 2013;18:1131–1143. doi: 10.1111/gtc.12103. [DOI] [PubMed] [Google Scholar]
- Kirchner P, Bug M, Meyer H. Ubiquitination of the N-terminal region of caveolin-1 regulates endosomal sorting by the VCP/p97 AAA-ATPase. The Journal of biological chemistry. 2013;288:7363–7372. doi: 10.1074/jbc.M112.429076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koller KJ, Brownstein MJ. Use of a cDNA clone to identify a supposed precursor protein containing valosin. Nature. 1987;325:542–545. doi: 10.1038/325542a0. [DOI] [PubMed] [Google Scholar]
- Kondo H, Rabouille C, Newman R, Levine TP, Pappin D, Freemont P, Warren G. p47 is a cofactor for p97-mediated membrane fusion. Nature. 1997;388:75–78. doi: 10.1038/40411. [DOI] [PubMed] [Google Scholar]
- Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen EL, Thumm M. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. The Journal of cell biology. 2010;190:965–973. doi: 10.1083/jcb.201002075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latterich M, Frohlich KU, Schekman R. Membrane fusion and the cell cycle: Cdc48p participates in the fusion of ER membranes. Cell. 1995;82:885–893. doi: 10.1016/0092-8674(95)90268-6. [DOI] [PubMed] [Google Scholar]
- Lee JJ, Park JK, Jeong J, Jeon H, Yoon JB, Kim EE, Lee KJ. Complex of Fas-associated factor 1 (FAF1) with valosin-containing protein (VCP)-Npl4-Ufd1 and polyubiquitinated proteins promotes endoplasmic reticulum-associated degradation (ERAD) The Journal of biological chemistry. 2013;288:6998–7011. doi: 10.1074/jbc.M112.417576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley BN, Ploegh HL. A membrane protein required for dislocation of misfolded proteins from the ER. Nature. 2004;429:834–840. doi: 10.1038/nature02592. [DOI] [PubMed] [Google Scholar]
- Madeo F, Schlauer J, Zischka H, Mecke D, Frohlich KU. Tyrosine phosphorylation regulates cell cycle-dependent nuclear localization of Cdc48p. Molecular biology of the cell. 1998;9:131–141. doi: 10.1091/mbc.9.1.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnaghi P, D’Alessio R, Valsasina B, Avanzi N, Rizzi S, Asa D, Gasparri F, Cozzi L, Cucchi U, Orrenius C, Polucci P, Ballinari D, Perrera C, Leone A, Cervi G, Casale E, Xiao Y, Wong C, Anderson DJ, Galvani A, Donati D, O’Brien T, Jackson PK, Isacchi A. Covalent and allosteric inhibitors of the ATPase VCP/p97 induce cancer cell death. Nature chemical biology. 2013;9:548–556. doi: 10.1038/nchembio.1313. [DOI] [PubMed] [Google Scholar]
- Maric M, Maculins T, De Piccoli G, Labib K. Cdc48 and a ubiquitin ligase drive disassembly of the CMG helicase at the end of DNA replication. Science. 2014;346:1253596. doi: 10.1126/science.1253596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta SG, Khare M, Ramani R, Watts GD, Simon M, Osann KE, Donkervoort S, Dec E, Nalbandian A, Platt J, Pasquali M, Wang A, Mozaffar T, Smith CD, Kimonis VE. Genotype-phenotype studies of VCP-associated inclusion body myopathy with Paget disease of bone and/or frontotemporal dementia. Clinical genetics. 2013;83:422–431. doi: 10.1111/cge.12000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H, Bug M, Bremer S. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nature cell biology. 2012;14:117–123. doi: 10.1038/ncb2407. [DOI] [PubMed] [Google Scholar]
- Meyer H, Weihl CC. The VCP/p97 system at a glance: connecting cellular function to disease pathogenesis. Journal of cell science. 2014;127:3877–3883. doi: 10.1242/jcs.093831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Kondo H, Warren G. The p47 co-factor regulates the ATPase activity of the membrane fusion protein, p97. FEBS letters. 1998;437:255–257. doi: 10.1016/s0014-5793(98)01232-0. [DOI] [PubMed] [Google Scholar]
- Meyer HH, Shorter JG, Seemann J, Pappin D, Warren G. A complex of mammalian ufd1 and npl4 links the AAA-ATPase, p97, to ubiquitin and nuclear transport pathways. The EMBO journal. 2000;19:2181–2192. doi: 10.1093/emboj/19.10.2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer HH, Wang Y, Warren G. Direct binding of ubiquitin conjugates by the mammalian p97 adaptor complexes, p47 and Ufd1-Npl4. The EMBO journal. 2002;21:5645–5652. doi: 10.1093/emboj/cdf579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir D, Stewart SE, Osmond BC, Botstein D. Cold-sensitive cell-division-cycle mutants of yeast: isolation, properties, and pseudoreversion studies. Genetics. 1982;100:547–563. doi: 10.1093/genetics/100.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno SP, Bailey R, Campion N, Herron S, Gambus A. Polyubiquitylation drives replisome disassembly at the termination of DNA replication. Science. 2014;346:477–481. doi: 10.1126/science.1253585. [DOI] [PubMed] [Google Scholar]
- Mosbech A, Gibbs-Seymour I, Kagias K, Thorslund T, Beli P, Povlsen L, Nielsen SV, Smedegaard S, Sedgwick G, Lukas C, Hartmann-Petersen R, Lukas J, Choudhary C, Pocock R, Bekker-Jensen S, Mailand N. DVC1 (C1orf124) is a DNA damage-targeting p97 adaptor that promotes ubiquitin-dependent responses to replication blocks. Nature structural & molecular biology. 2012;19:1084–1092. doi: 10.1038/nsmb.2395. [DOI] [PubMed] [Google Scholar]
- Murayama Y, Ogura T, Yamanaka K. Characterization of C-terminal adaptors, UFD-2 and UFD-3, of CDC-48 on the polyglutamine aggregation in C. elegans. Biochemical and biophysical research communications. 2015;459:154–160. doi: 10.1016/j.bbrc.2015.02.088. [DOI] [PubMed] [Google Scholar]
- Nalbandian A, Donkervoort S, Dec E, Badadani M, Katheria V, Rana P, Nguyen C, Mukherjee J, Caiozzo V, Martin B, Watts GD, Vesa J, Smith C, Kimonis VE. The multiple faces of valosin-containing protein-associated diseases: inclusion body myopathy with Paget’s disease of bone, frontotemporal dementia, and amyotrophic lateral sclerosis. Journal of molecular neuroscience : MN. 2011;45:522–531. doi: 10.1007/s12031-011-9627-y. [DOI] [PubMed] [Google Scholar]
- Neuber O, Jarosch E, Volkwein C, Walter J, Sommer T. Ubx2 links the Cdc48 complex to ER-associated protein degradation. Nature cell biology. 2005;7:993–998. doi: 10.1038/ncb1298. [DOI] [PubMed] [Google Scholar]
- Nishikori S, Esaki M, Yamanaka K, Sugimoto S, Ogura T. Positive cooperativity of the p97 AAA ATPase is critical for essential functions. The Journal of biological chemistry. 2011;286:15815–15820. doi: 10.1074/jbc.M110.201400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Ewens CA, Tsang C, Yeung HO, Zhang X, Freemont PS. The role of the N-domain in the ATPase activity of the mammalian AAA ATPase p97/VCP. The Journal of biological chemistry. 2012;287:8561–8570. doi: 10.1074/jbc.M111.302778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura T, Wilkinson AJ. AAA+ superfamily ATPases: common structure--diverse function. Genes Cells. 2001;6:575–597. doi: 10.1046/j.1365-2443.2001.00447.x. [DOI] [PubMed] [Google Scholar]
- Peters JM, Walsh MJ, Franke WW. An abundant and ubiquitous homo-oligomeric ring-shaped ATPase particle related to the putative vesicle fusion proteins Sec18p and NSF. The EMBO journal. 1990;9:1757–1767. doi: 10.1002/j.1460-2075.1990.tb08300.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puumalainen MR, Lessel D, Ruthemann P, Kaczmarek N, Bachmann K, Ramadan K, Naegeli H. Chromatin retention of DNA damage sensors DDB2 and XPC through loss of p97 segregase causes genotoxicity. Nature communications. 2014;5:3695. doi: 10.1038/ncomms4695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye VE, Beuron F, Keetch CA, McKeown C, Robinson CV, Meyer HH, Zhang X, Freemont PS. Structural insights into the p97-Ufd1-Npl4 complex. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:467–472. doi: 10.1073/pnas.0603408104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pye VE, Dreveny I, Briggs LC, Sands C, Beuron F, Zhang X, Freemont PS. Going through the motions: the ATPase cycle of p97. Journal of structural biology. 2006;156:12–28. doi: 10.1016/j.jsb.2006.03.003. [DOI] [PubMed] [Google Scholar]
- Qiu L, Pashkova N, Walker JR, Winistorfer S, Allali-Hassani A, Akutsu M, Piper R, Dhe-Paganon S. Structure and function of the PLAA/Ufd3-p97/Cdc48 complex. The Journal of biological chemistry. 2010;285:365–372. doi: 10.1074/jbc.M109.044685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinovich E, Kerem A, Frohlich KU, Diamant N, Bar-Nun S. AAA-ATPase p97/Cdc48p, a cytosolic chaperone required for endoplasmic reticulum-associated protein degradation. Molecular and cellular biology. 2002;22:626–634. doi: 10.1128/MCB.22.2.626-634.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabouille C, Levine TP, Peters JM, Warren G. An NSF-like ATPase, p97, and NSF mediate cisternal regrowth from mitotic Golgi fragments. Cell. 1995;82:905–914. doi: 10.1016/0092-8674(95)90270-8. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan SK, den Besten W, Deshaies RJ. p97-dependent retrotranslocation and proteolytic processing govern formation of active Nrf1 upon proteasome inhibition. eLife. 2014;3:e01856. doi: 10.7554/eLife.01856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan K, Bruderer R, Spiga FM, Popp O, Baur T, Gotta M, Meyer HH. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature. 2007;450:1258–1262. doi: 10.1038/nature06388. [DOI] [PubMed] [Google Scholar]
- Raman M, Havens CG, Walter JC, Harper JW. A genome wide screen identifies p97 as an essential regulator of DNA damage-dependent CDT1 destruction. Molecular cell. 2011 doi: 10.1016/j.molcel.2011.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan HN, Ye Y. The p97 ATPase associates with EEA1 to regulate the size of early endosomes. Cell Res. 2012;22:346–359. doi: 10.1038/cr.2011.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rape M, Hoppe T, Gorr I, Kalocay M, Richly H, Jentsch S. Mobilization of processed, membrane-tethered SPT23 transcription factor by CDC48(UFD1/NPL4), a ubiquitin-selective chaperone. Cell. 2001;107:667–677. doi: 10.1016/s0092-8674(01)00595-5. [DOI] [PubMed] [Google Scholar]
- Ritz D, Vuk M, Kirchner P, Bug M, Schutz S, Hayer A, Bremer S, Lusk C, Baloh RH, Lee H, Glatter T, Gstaiger M, Aebersold R, Weihl CC, Meyer H. Endolysosomal sorting of ubiquitylated caveolin-1 is regulated by VCP and UBXD1 and impaired by VCP disease mutations. Nature cell biology. 2011;13:1116–1123. doi: 10.1038/ncb2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothballer A, Tzvetkov N, Zwickl P. Mutations in p97/VCP induce unfolding activity. FEBS letters. 2007;581:1197–1201. doi: 10.1016/j.febslet.2007.02.031. [DOI] [PubMed] [Google Scholar]
- Rouiller I, V, Butel M, Latterich M, Milligan RA, Wilson-Kubalek EM. A major conformational change in p97 AAA ATPase upon ATP binding. Molecular cell. 2000;6:1485–1490. doi: 10.1016/s1097-2765(00)00144-1. [DOI] [PubMed] [Google Scholar]
- Rouiller I, DeLaBarre B, May AP, Weis WI, Brunger AT, Milligan RA, Wilson-Kubalek EM. Conformational changes of the multifunction p97 AAA ATPase during its ATPase cycle. Nature structural biology. 2002;9:950–957. doi: 10.1038/nsb872. [DOI] [PubMed] [Google Scholar]
- Ruggiano A, Foresti O, Carvalho P. Quality control: ER-associated degradation: protein quality control and beyond. The Journal of cell biology. 2014;204:869–879. doi: 10.1083/jcb.201312042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumpf S, Jentsch S. Functional division of substrate processing cofactors of the ubiquitin-selective Cdc48 chaperone. Molecular cell. 2006;21:261–269. doi: 10.1016/j.molcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- Schaeffer V, Akutsu M, Olma MH, Gomes LC, Kawasaki M, Dikic I. Binding of OTULIN to the PUB domain of HOIP controls NF-kappaB signaling. Molecular cell. 2014;54:349–361. doi: 10.1016/j.molcel.2014.03.016. [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. Membrane-bound Ubx2 recruits Cdc48 to ubiquitin ligases and their substrates to ensure efficient ER-associated protein degradation. Nature cell biology. 2005;7:999–1006. doi: 10.1038/ncb1299. [DOI] [PubMed] [Google Scholar]
- Schuberth C, Buchberger A. UBX domain proteins: major regulators of the AAA ATPase Cdc48/p97. Cell Mol Life Sci. 2008;65:2360–2371. doi: 10.1007/s00018-008-8072-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuller JM, Beck F, Lossl P, Heck AJ, Forster F. Nucleotide-dependent conformational changes of the AAA+ ATPase p97 revisited. FEBS letters. 2016 doi: 10.1002/1873-3468.12091. [DOI] [PubMed] [Google Scholar]
- Shcherbik N, Haines DS. Cdc48p(Npl4p/Ufd1p) binds and segregates membrane-anchored/tethered complexes via a polyubiquitin signal present on the anchors. Molecular cell. 2007;25:385–397. doi: 10.1016/j.molcel.2007.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Grimaud R, Hoskins JR, Wickner S, Maurizi MR. Unfolding and internalization of proteins by the ATP-dependent proteases ClpXP and ClpAP. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:8898–8903. doi: 10.1073/pnas.97.16.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C, Wang Q, Li CC. ATPase activity of p97-valosin-containing protein (VCP). D2 mediates the major enzyme activity, and D1 contributes to the heat-induced activity. The Journal of biological chemistry. 2003;278:3648–3655. doi: 10.1074/jbc.M208422200. [DOI] [PubMed] [Google Scholar]
- Stapf C, Cartwright E, Bycroft M, Hofmann K, Buchberger A. The general definition of the p97/valosin-containing protein (VCP)-interacting motif (VIM) delineates a new family of p97 cofactors. The Journal of biological chemistry. 2011;286:38670–38678. doi: 10.1074/jbc.M111.274472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka A, Cleland MM, Xu S, Narendra DP, Suen DF, Karbowski M, Youle RJ. Proteasome and p97 mediate mitophagy and degradation of mitofusins induced by Parkin. The Journal of cell biology. 2010;191:1367–1380. doi: 10.1083/jcb.201007013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WK, Li D, Li CC, Esser L, Dai R, Guo L, Xia D. A novel ATP-dependent conformation in p97 N-D1 fragment revealed by crystal structures of disease-related mutants. The EMBO journal. 2010;29:2217–2229. doi: 10.1038/emboj.2010.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WK, Xia D. Structural and functional deviations in disease-associated p97 mutants. Journal of structural biology. 2012;179:83–92. doi: 10.1016/j.jsb.2012.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WK, Xia D. Altered intersubunit communication is the molecular basis for functional defects of pathogenic p97 mutants. The Journal of biological chemistry. 2013;288:36624–36635. doi: 10.1074/jbc.M113.488924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang WK, Xia D. Role of the D1–D2 Linker of Human VCP/p97 in the Asymmetry and ATPase Activity of the D1-domain. Scientific reports. 2016;6:20037. doi: 10.1038/srep20037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Jokitalo E, Kano F, Murata M, Zhang X, Canas B, Newman R, Rabouille C, Pappin D, Freemont P, Kondo H. VCIP135, a novel essential factor for p97/p47-mediated membrane fusion, is required for Golgi and ER assembly in vivo. The Journal of cell biology. 2002;159:855–866. doi: 10.1083/jcb.200208112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama K, Kondo H. p97/p47-Mediated biogenesis of Golgi and ER. J Biochem (Tokyo) 2005;137:115–119. doi: 10.1093/jb/mvi028. [DOI] [PubMed] [Google Scholar]
- Uchiyama K, Totsukawa G, Puhka M, Kaneko Y, Jokitalo E, Dreveny I, Beuron F, Zhang X, Freemont P, Kondo H. p37 is a p97 adaptor required for Golgi and ER biogenesis in interphase and at the end of mitosis. Dev Cell. 2006;11:803–816. doi: 10.1016/j.devcel.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Verma R, Oania R, Fang R, Smith GT, Deshaies RJ. Cdc48/p97 mediates UV-dependent turnover of RNA Pol II. Molecular cell. 2011;41:82–92. doi: 10.1016/j.molcel.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma R, Oania RS, Kolawa NJ, Deshaies RJ. Cdc48/p97 promotes degradation of aberrant nascent polypeptides bound to the ribosome. eLife. 2013;2:e00308. doi: 10.7554/eLife.00308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Li L, Ye Y. Inhibition of p97-dependent protein degradation by Eeyarestatin I. The Journal of biological chemistry. 2008 doi: 10.1074/jbc.M708347200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Mora-Jensen H, Weniger MA, Perez-Galan P, Wolford C, Hai T, Ron D, Chen W, Trenkle W, Wiestner A, Ye Y. ERAD inhibitors integrate ER stress with an epigenetic mechanism to activate BH3-only protein NOXA in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2200–2205. doi: 10.1073/pnas.0807611106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Shinkre BA, Lee JG, Weniger MA, Liu Y, Chen W, Wiestner A, Trenkle WC, Ye Y. The ERAD Inhibitor Eeyarestatin I Is a Bifunctional Compound with a Membrane-Binding Domain and a p97/VCP Inhibitory Group. PloS one. 2010;5:e15479. doi: 10.1371/journal.pone.0015479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Song C, Yang X, Li CC. D1 ring is stable and nucleotide-independent, whereas D2 ring undergoes major conformational changes during the ATPase cycle of p97-VCP. The Journal of biological chemistry. 2003;278:32784–32793. doi: 10.1074/jbc.M303869200. [DOI] [PubMed] [Google Scholar]
- Watts GD, Wymer J, Kovach MJ, Mehta SG, Mumm S, Darvish D, Pestronk A, Whyte MP, Kimonis VE. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- Weihl CC, Dalal S, Pestronk A, Hanson PI. Inclusion body myopathy-associated mutations in p97/VCP impair endoplasmic reticulum-associated degradation. Human molecular genetics. 2006;15:189–199. doi: 10.1093/hmg/ddi426. [DOI] [PubMed] [Google Scholar]
- Weihl CC, Temiz P, Miller SE, Watts G, Smith C, Forman M, Hanson PI, Kimonis V, Pestronk A. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. Journal of neurology, neurosurgery, and psychiatry. 2008;79:1186–1189. doi: 10.1136/jnnp.2007.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox AJ, Laney JD. A ubiquitin-selective AAA-ATPase mediates transcriptional switching by remodelling a repressor-promoter DNA complex. Nature cell biology. 2009;11:1481–1486. doi: 10.1038/ncb1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S, Peng G, Wang Y, Fang S, Karbowski M. The AAA-ATPase p97 is essential for outer mitochondrial membrane protein turnover. Molecular biology of the cell. 2011;22:291–300. doi: 10.1091/mbc.E10-09-0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K, Sasagawa Y, Ogura T. Recent advances in p97/VCP/Cdc48 cellular functions. Biochimica et biophysica acta. 2012;1823:130–137. doi: 10.1016/j.bbamcr.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Ye Y. Diverse functions with a common regulator: ubiquitin takes command of an AAA ATPase. Journal of structural biology. 2006;156:29–40. doi: 10.1016/j.jsb.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature. 2001;414:652–656. doi: 10.1038/414652a. [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. The Journal of cell biology. 2003;162:71–84. doi: 10.1083/jcb.200302169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. doi: 10.1038/nature02656. [DOI] [PubMed] [Google Scholar]
- Zhang L, Ashendel CL, Becker GW, Morre DJ. Isolation and characterization of the principal ATPase associated with transitional endoplasmic reticulum of rat liver. The Journal of cell biology. 1994;127:1871–1883. doi: 10.1083/jcb.127.6.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Ye Y. The final moments of misfolded proteins en route to the proteasome. DNA and cell biology. 2014;33:477–483. doi: 10.1089/dna.2014.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Gui L, Zhang X, Bulfer SL, Sanghez V, Wong DE, Lee Y, Lehmann L, Lee JS, Shih PY, Lin HJ, Iacovino M, Weihl CC, Arkin MR, Wang Y, Chou TF. Altered cofactor regulation with disease-associated p97/VCP mutations. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:E1705–1714. doi: 10.1073/pnas.1418820112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Shaw A, Bates PA, Newman RH, Gowen B, Orlova E, Gorman MA, Kondo H, Dokurno P, Lally J, Leonard G, Meyer H, van Heel M, Freemont PS. Structure of the AAA ATPase p97. Molecular cell. 2000;6:1473–1484. doi: 10.1016/s1097-2765(00)00143-x. [DOI] [PubMed] [Google Scholar]
- Zhao G, Zhou X, Wang L, Li G, Schindelin H, Lennarz WJ. Studies on peptide:N-glycanase-p97 interaction suggest that p97 phosphorylation modulates endoplasmic reticulum-associated degradation. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:8785–8790. doi: 10.1073/pnas.0702966104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Shen Y, Ballar P, Apostolou A, Agami R, Fang S. AAA ATPase p97/valosin-containing protein interacts with gp78, a ubiquitin ligase for endoplasmic reticulum-associated degradation. The Journal of biological chemistry. 2004;279:45676–45684. doi: 10.1074/jbc.M409034200. [DOI] [PubMed] [Google Scholar]
- Zhou HJ, Wang J, Yao B, Wong S, Djakovic S, Kumar B, Rice J, Valle E, Soriano F, Menon MK, Madriaga A, Kissvonsoly S, Kumar A, Parlati F, Yakes MF, Shawver L, Le Moigne R, Anderson DJ, Rolfe M, Wustrow D. Discovery of a First-in-Class, Potent, Selective and Orally Bioavailable Inhibitor of the p97 AAA ATPase (CB-5083) Journal of medicinal chemistry. 2015 doi: 10.1021/acs.jmedchem.5b01346. [DOI] [PubMed] [Google Scholar]