Abstract
Despite the 92% homology of the hematopoietic cell-specific Rac2 to the canonical isoform Rac1, these isoforms have been shown to play non-redundant roles in immune cells. To study isoform-specific dynamics of Rac in live cells we developed a genetically-encoded, single-chain FRET-based biosensor for Rac2. We also made significant improvements to our existing single-chain Rac1 biosensor. We optimized the biosensor constructs for facile expression in hematopoietic cells and performed functional validations in murine macrophage sublines of RAW264.7 cells. Rac2, along with Rac1and Cdc42, have been implicated in the formation of actin-rich protrusions by macrophages, but their individual activation dynamics have not been previously characterized. We found that both Rac1 and Rac2 had similar activation kinetics yet they had very distinct spatial distributions in response to the exogenous stimulus, fMLP. Active Rac1 was mainly localized to the cell periphery, while active Rac2 was distributed throughout the cell with an apparent higher concentration in the perinuclear region. We also performed an extensive morphodynamic analysis of Rac1, Rac2 and Cdc42 activities during the extension of random protrusions. We found that Rac2 appears to play a leading role in the generation of random protrusions, as we observed an initial strong activation of Rac2 in regions distal from the leading edge, followed by the activation of Rac1, a second burst of Rac2 and then Cdc42 immediately behind the leading edge. Overall, isoform-specific biosensors that have been optimized for expression should be valuable for interrogating the coordination of Rho family GTPase activities in living cells.
Introduction
The Rac members of the p21 Rho family of small GTPases include four major isoforms (Paralogs: Rac1, 2, 3 and RhoG) and a splice variant Rac1b (1), and are known to be master regulators of actin-dependent cellular processes (2). Expression patterns vary amongst the isoforms: Rac1 is ubiquitously expressed; Rac3 is found in several tissues but primarily in the brain; while Rac2 is exclusive to hematopoietic cells (3). The relative expression of Rac1 and Rac2 in hematopoietic cells is both cell-type and species-dependent (4). Rac1 and Rac2 share 92% amino acid sequence identity, with the most divergence occurring in their C-terminal polybasic region (4, 5). Importantly, despite their high sequence homology and independent of their relative expression abundance, Rac1 and Rac2 have been shown to play non-redundant roles in leukocyte functions, including development, chemotaxis, phagocytosis and reactive oxygen species (ROS) production for bacterial killing (4, 6). While the two Rac isoforms are known to have identical effector binding domains in their Switch I and II regions, several studies have demonstrated that one basis for their non-redundancy is their subcellular localization that is dictated by their C-terminal polybasic tail (7-9). Rac2 is most-studied for its role in regulating chemotaxis and activation of NADPH oxidase in neutrophils (10, 11). While Rac2 is expressed as the predominant isoform in neutrophils (present at about equal amounts with Rac1 in murine neutrophils, and over 75% in human neutrophils (4, 12)), it is the less abundant isoform in macrophages, where Rac1 was measured to be expressed at approximately 4-fold higher levels (13). Thus in neutrophils and other leukocytes, Rac2 has been shown to have roles different than those driven by its canonical counterpart Rac1 (9, 12-17). Therefore, in addition to their dynamic activation kinetics, insight into the spatial distribution of Rac1 and Rac2 is critical for a complete understanding of the functional roles of these Rac isoforms in leukocytes.
While there are several techniques available to study GTPase dynamics, Forster resonance energy transfer (FRET)-based biosensors have proven to be a powerful means to reveal simultaneously the spatial and temporal activation dynamics of proteins at high-resolution on a single-cell basis, which is otherwise very difficult with more conventional approaches (18). In the case of Rho GTPases, a major focus in the field has been on developing FRET-based biosensors for the canonical members RhoA, Rac1 and Cdc42 (19-25). However, there is increasing awareness that the lesser-studied isoforms, that may be expressed as minor fraction or expressed only in disease states, play different and often critical roles that are specific to such diseased states (26-28). Thus, it is apparent that biosensors for different isoforms of these canonical members are needed to enable their isoform-specific analysis in delineating their non-redundant functional roles.
Previous studies analyzing Rac1 and Rac2 activity in neutrophils or macrophages used bimolecular versions of FRET biosensors (29-31). This approach, while useful, involves cumbersome data analysis due to the non-equimolar distribution of the two separate FRET donor/acceptor components. We have overcome this issue by the development of a fully genetically-encoded, single-chain, FRET-based Rac2 biosensor, which is useful for live-cell imaging of Rac2 activation dynamics in hematopoietic cells. Our design maintains the C-terminal polybasic region of Rac2 and allows for correct intracellular localization and interaction with upstream regulators, including guanosine nucleotide dissociation inhibitor (GDI). In addition, we introduced new optimization strategies to our biosensor expression techniques allowing for facile expression and analyses of Rac2. Moreover, we then extended these optimization strategies to our Rac1 (25) and Cdc42 (24) biosensors, thereby achieving the ability to directly visualize the coordination of several Rho GTPase activities in macrophages. These optimizations should be useful for achieving robust expression and dynamic range of detection in many other cell types.
Materials and Methods
Biosensor construction and fluorometry
Rac1 and Cdc42 biosensor optimization
The previously published single-chain FRET-biosensor for Rac1 (25) was modified to improve the dynamic range of response and the expression stability in living cells. Briefly, the original Rac1 biosensor contained a monomeric Cerulean1 (mCer1) as the FRET donor, two tandem p21 binding domains (PBD) of p21 activated kinase 1 (PAK1, amino acid residues 70-149) to achieve autoinhibitory regulation, separated by a structurally optimized linker (GSGGPPGSGGSG), monomeric Venus (mVen) as FRET acceptor, and a full-length wild-type (WT) Rac1 (25). The second PBD contained H83D, H86D point mutations to render it unable to bind to active GTPase. To further optimize the stability and response of this system, the FRET acceptor mVen was replaced by monomeric circularly permutated (cp) versions cp49, cp157, cp173, cp195 or cp229Ven (32), PCR amplified using the primer pairs: 5'-cataagaatgcggccgcaatgaccggcaagctgcccg-3' and 5'-gggggaattccttggtgcagatcaacttcagggtc-3' (cp49); 5'-gcgtaccatggatgcagaagaacggcatcaaggc-3' and 5'-ggccgaattccttcttgtcggcggtgatatagac-3' (cp157); 5'-cataagaatgcggccgcaatggacggcggcgtgcagc-3' and 5'-ggaattccttctcgatgttgtggcggatcttgaagtttgc-3' (cp173); 5'-cataagaatgcggccgcaatgctgcccgacaaccactacc-3' and 5' -ccggaattccttcagcacggggccgtcgccgatg-3' (cp195); and 5'-cataagaatgcggccgcaatgatcactctcggcatggacgag-3' and 5'-ggaattccttcccggcggcggtcacgaactc-3' (cp229). The PCR-amplified fragments were subcloned into the biosensor backbone using the NotI and EcoRI restriction sites (Supplemental Fig. 1A). In order to achieve the required expression stability needed for the production of stable-inducible cell lines, the codon usages of the mCer1 and the second PBD were synonymously modified (33) in Rac1 (25) and Cdc42 (24) biosensors (Supplemental Fig. 1B, C). Furthermore, the final version of the optimized Rac1 biosensor construct also contained additional linker optimization applied to monomeric cp229Ven (mcp229Ven), by replacing the first 16 amino acids with the flexible, structureless linker that is resistant to protease cleavage (34) Briefly, mcp229Ven was PCR amplified using a primer pair 5'-gatatatatgcggccgcaatgggcagcaccagcggcagcggcaaaccgggcagcggcgaaggcagcatggtgagcaagggcgaggagctg-3' (optimized linker sequence underlined (34)) and 5'-taaataaataaagaattccccggcggcggtcacgaactccag-3', and subcloned into the biosensor backbone using NotI and EcoRI (Supplemental Fig. 1A) restriction sites. This version of the biosensor did not yield different fluorometric responses from the original mcp229Ven version of the sensor without this modification (data not shown).
Rac2 Biosensor construction
To create the Rac2 biosensor, Rac1 was replaced by full-length WT Rac2 in the mVen and mcp229Ven versions of the Rac1 sensor. WT Rac2 was PCR amplified using the primer pair 5'-gtatatatatatatgaattcatgcaggccatcaagtgtgtg-3' and 5'-ccaattaattaattaactcgagctagaggaggctgcaggcgcg-3', and was subcloned into the biosensor backbone using the EcoRI and XhoI restriction sites (Supplemental Fig. 1A). The synonymous modifications (Supplemental Fig. 1B, C) were applied to the mCer1 and the second PBD as in Rac1 biosensor. To optimize the mcp229Ven version of the Rac2 biosensor, the linker was also optimized by replacing the first 16 amino acids within the mcp229Ven to confer protease resistance and stability, as in the optimized Rac1 sensor.
Fluorometric validation
The biosensor cDNA cassettes were subcloned into pTriEX-4 (Novagen) for transient expression. Characterization of biosensor response was performed in HEK293T cells by overexpressing WT or mutant versions of the biosensor, with or without the appropriate upstream regulators as previously described (35). Briefly, HEK293T cells were plated overnight at 1.2×106 cells/well of 6-well plates coated with poly-L-lysine, and transfected the next day using Lipofectamine2000 (Invitrogen) following the manufacturer's protocols. Biosensors were co-transfected at ratios of 1:3 (Rac1) and 1:4 (Rac2) with GDI or GAP, or 1:0.5 – 10 for co-transfection with GEFs (+/− GDI) as indicated. Adherent cells were washed in PBS and fixed using 3.7% formaldehyde 48 h following the transfection, and fluorescence emission spectra was measured by spectrofluorometer using a plate reader (Horiba-Jobin-Yvon Fluorolog-3MF2 with MicroMax plate reader). The spectra were obtained by exciting with 433 nm light, with emission scanned from 450 – 600 nm. The fluorescence reading of cells with empty vector (pCDNA3.1) was used to measure light scatter and autofluorescence, which were subtracted from the data. The resulting spectra were normalized to the peak mCer1 emission intensity at 474 nm to generate the final ratiometric spectra.
Production of stable-inducible biosensor systems
The second generation tetracycline-inducible system was used to produce stable-inducible cell lines. Tet-OFF tetracycline Trans-Activator (tTA) was restriction digested out of a pRetroX-advanced tet-OFF backbone (Clontech) using the BamHI/EcoRI sites flanking the tTA gene cassette. This fragment was then ligated into the new pQEXIN retroviral vector at its multiple cloning sites. The pQEXIN vector was produced by modifying the pQCXIN (Clontech) retroviral vector, originally containing the CMV promoter, a multiple cloning site, an internal ribosomal entry site, and a neomycin resistance gene cassette. The CMV promoter was digested out using the BglII/NotI sites flanking the promoter, and a PCR-amplified EF1α promoter containing BamHI/NotI sites was ligated into the backbone (amplified using the primer pair: 5’-gtcgacattattgactagatggatccgcgtgaggctccggtgcccgtcagtg-3’ and 5’-gctagcgcatatgcttaattgcggccgctatattcctcacgacacctgaaatggaag-3’). This produced the pQEXIN backbone system, resistant to gene silencing in hematopoietic/stem cells.
The expression cassettes for the optimized Rac1, Rac2 and Cdc42 biosensors were placed into pRetro-X inducible retroviral backbone system (Clontech), modified to contain the Gateway recombination cassette (Invitrogen) and an antibiotic resistance for Zeocin (pRetro-X-Zeo-DEST). Briefly, Rac1, Rac2 and Cdc42 biosensor cassettes were subcloned into the Gateway pENTR-4 entry vector using the NcoI/XhoI restriction sites. The Gateway recombination reaction was performed following the manufacturer's protocols (Invitrogen), producing pRetro-X-Zeo-Rac1, pRetro-X-Zeo-Rac2 and pRetro-X-Zeo-Cdc42.
Cell lines, transfection and induction
RAW/LR5 cells, a derivative of monocyte/macrophage RAW 264.7 cells, and FMLPR.2 (RAW/LR5 expressing full length human formyl-methionyl-leucyl-phenylalanine (fMLP) receptor) (36) were cultured in complete RAW medium (RPMI 1640 supplemented with 10% newborn calf serum, 100 U/mL penicillin and 100 μg/mL streptomycin) and maintained at 37°C and 5% CO2 atmosphere. For transient expression of synonymously-modified WT or mutant versions of the biosensor used for the fixed cell experiments, cells were transfected with biosensor construct in pTriEX-4 vector using FugeneHD (Fisher Scientific) according to the manufacturer's protocols: cells were plated in a 12-well plate the day before transfection; next day cells (70-90% confluent) were incubated in the transfection mix containing 1 μg DNA (1:3 DNA:FugeneHD ratio, prepared in Opti-MEM) for 2 h, then replated onto 12-mm round glass coverslips in a 24-well plate and incubated overnight at 37°C and 5% CO2 atmosphere. Stable macrophage cell line with constitutive expression of EGFP was produced by viral transduction of pQEXIN-EGFP into RAW/LR5 cells and selected in 2 mg/mL G418 (Teknova). Stable macrophage cell lines with inducible expression of synonymously-modified Rac1, Rac2 or Cdc42 biosensors used for live cell imaging were produced by two-step viral transduction of RAW/LR5 cells: first, cells were transduced with pQEXIN-tetOFF and selected in G418 up to 2 mg/mL, gradually increasing concentration following infection; the resulting cell line with stable expression of tTA was transduced with pRetro-X-Zeo inducible retroviral backbone containing biosensor cassettes and selected in Zeocin (Invitrogen) up to 1 mg/mL, in gradual increments. The retroviral production, concentration and infection of pQEXIN-EGFP, pQEXIN-tetOFF, pRetro-X-Zeo-Rac1, pRetro-X-Zeo-Rac2 and pRetro-X-Zeo-Cdc42 were performed following the manufacturer's protocols (Clontech). Successful infection with biosensor was monitored by microscopy before repression with doxycycline. Furthermore, stable Rac1 and Rac2 inducible RAW/LR5 cell lines were transfected with full-length human fMLP receptor subcloned into pApuro (construct originally described in (36)) and selected in puromycin (R&D Systems), up to 7 μg/mL in gradual increments, to generate cell lines responsive to fMLP. To repress the biosensor expression during normal culture, cells were maintained in complete RAW medium supplemented with 2 μg/mL doxycycline (MP Biomedicals). To induce biosensor expression, doxycycline was removed 48 h prior to imaging by a brief trypsinization and replating at a low density; 24 h later cells were detached again by a brief trypsinization and replated at a low density onto 25-mm round coverslips in 6 well dishes. For optimal induction of biosensor expression, cells were maintained in complete RAW medium containing FBS, certified to be free of tetracycline.
RNA-mediated interference
Reduction of Rac1 and Rac2 expression in RAW/LR5 cells was achieved by short hairpin RNA directed against their respective mRNA. For Rac1, coding DNA targeting shRNA sequence 5’-agacggagctgttggtaaa-3’ was generated using siDESIGN (GE Dharmacon) and inserted into pSuper.retro.puro retroviral vector (Oligoengine) via BglII and HindIII restriction sites; empty pSuper.retro.puro vector was used as control (shCtrl1). For Rac2, shRNA sequence 5'-caaagggagagatgtggaa-3' (targeting 3’UTR) in pGIPZ lentiviral vector or empty pGIPZ vector (Dharmacon) as control (shCtrl2) were obtained from Einstein shRNA Core Facility. Control and shRNA plasmids were transfected into GP2-293 packaging cell lines (Clontech) using Lipofectamine2000 (Invitrogen) according to manufacturer's instructions. Viral supernatants were concentrated using Retro-X concentrator (Clontech) following the manufacturer's protocol and the concentrated viral stocks were used to infect RAW/LR5 cells. Transduced cells were selected in puromycin, up to 7 μg/mL in gradual increments, to achieve stable genomic integration. Reduction of protein expression was determined by western blotting.
Western blotting
Whole cell lysate were prepared by lysing cells in ice-cold Buffer A (25mM Tris, 137mM NaCl, 1% Triton X-100, 2 mM EDTA, 1 mM orthovanadate, 1 mM benzamidine, 10 μg/ml aprotinin, 10 μg/ml leupeptin, pH 7.4; protease inhibitors from Sigma). Lysates were resolved by SDS-PAGE and transferred to PVDF membrane (BioRad). After blocking, blots were incubated in primary antibody (monoclonal anti-GFP (13.1/7.1), Roche; monoclonal anti-Rac1 (23A8), Millipore; polyclonal anti-Rac2 (07-604), Millipore; polyclonal anti-Cdc42 (P1), Santa Cruz; monoclonal anti-β-actin (AC15), Santa Cruz) overnight at 4°C, followed by secondary antibody conjugated with Licor DX800 dye (LiCor). Blots were analyzed using the Odyssey Infrared Imaging System (LI-COR, Lincoln, NE). For quantification, the integrated values of background-corrected band densities were measured with Metamorph software.
FACS sorting
Stably transduced, inducible biosensor cell lines were FACS sorted to enrich for near-100% populations of cells expressing the biosensors. FACS sorting was performed at Albert Einstein College of Medicine Flow Cytometry Facility, by gating for the CFP/YFP double-positive population, as previously described (33).
Fixed-cell imaging
For ratiometric imaging RAW/LR5 or FMLPR.2 cells were transiently transfected with Rac1 or Rac2 biosensor. Cells mounted on 12-mm round coverslips were serum-starved in RPMI for at least 1 h prior to stimulation with 50 ng/mL CX3CL1 (R&D Systems) or 100 nM fMLP (Sigma) in Buffer with Divalent (BWD = 125 mM NaCl, 5 mM KCl, 1 mM KH2PO4, 5 mM glucose, 10 mM NaHCO3, 1 mM MgCl2, 1 mM CaCl2, and 20 mM Hepes) for indicated times at 37°C before fixation. Cells were fixed in 3.7% formaldehyde in BWD, permeabilized with 0.2% Triton X-100 in BWD for 10 min, stained for F-actin with Alexa Flour 568-phalloidin (1:400) (Invitrogen), and mounted in 50% glycerol in PBS. Microscope imaging at 60X magnification and image processing was performed as previously described in detail (24, 37, 38). Whole cell level of average GTPase activity was determined by thresholding the whole cell area in the FRET/donor ratiometric image using Metamorph Software. To measure GTPase activity at cell periphery we performed unbiased edge erosion measurements of ratio intensities as described below.
Measurements of biosensor activity as a function of the radial distance
To measure average ratio intensity per unit area as a function of the radial distance away from the edge of a cell, a new Matlab algorithm was constructed (“erodeEdgeStack.m”). This program tracks the edge position of a cell, makes edge erosion in the radial direction by a user-defined distance, then it makes a binary mask spanning the distance between the original edge position and the newly created edge position through erosion. This is repeated until the entire radial distance of a cell is traversed by the erosion process to produce successively smaller circumferential binary masks. These binary masks are next applied to the raw ratio image, and the average ratio intensity per unit area is calculated at every radial position and plotted to represent the radial scan of the ratio intensity values of every cell. The resulting average ratio intensity distribution per each cell is then normalized to the minimal ratio value within the distribution, normally corresponding to the central, nuclear region of the cell.
Live-cell imaging
The stable-inducible cell system was used for live-cell imaging. Biosensor induction was performed as described above and cells were plated onto 25-mm round coverslips. The coverslip was mounted in a custom-made imaging chamber, described in (38), and maintained at 37°C during imaging. Imaging was performed in BWD to achieve optimal signal-to-noise ratio. To image random protrusion events, cells were mounted in a closed chamber configuration, BWD was supplemented with 5% FBS and images were acquired at 10 s intervals for 10 min at 60X magnification. To image stimulation with fMLP, cells were mounted in an open chamber configuration in 500 μL BWD. Following temperature stabilization, images were acquired at every 10 s for 15 min, where cells were imaged in BWD without the stimulant for the first 24 frames to establish baseline activity, followed by an addition of fMLP prior to exposure at the 25th frame in 500 μL at 2x concentration (final concentration 100 nM). At every time point, FRET and mCer1 emission channels were acquired simultaneously using two side-mounted cameras in order to eliminate motion artifacts (38), followed by an acquisition of the differential interference contrast (DIC) channel at the third camera mounted on the bottom port of the microscope. mVen emission was captured to confirm biosensor expression at first frame only to minimize photobleaching during imaging session. Detailed descriptions of live-cell imaging, microscope settings and image processing are provided in (37, 38). To quantitatively analyze the leading edge dynamics of GTPase activity we applied the Morphodynamic-mapping and Cross-correlation analysis as described previously (39). Whole cell level of average GTPase activity was determined by thresholding the whole area of cell in the FRET/donor ratiometric image at every frame using Metamorph Software.
Morphodynamic-mapping and Cross-correlation analysis
Morphodynamic-mapping and cross-correlation methods were described previously (39). We measured Rho GTPase activity and cell edge velocity in sampling windows of 0.65 × 1.3 μm constructed along the leading edge of the cells (Supplemental Fig. 2A) and tracked cell edge motion during complete protrusion/retraction cycles. Sampling windows were moved away from the leading edge in 0.65 μm increments to measure GTPase activities at successively distal regions away from the edge. The extent of coupling between changes in Rho GTPase activity and edge velocity was determined using cross-correlation function xcov in Matlab and Pearson's correlation coefficient was used to determine the strength of coupling of protrusive edge motion to changes in GTPase activities. A positive cross-correlation coefficient indicates increasing Rho GTPase activity, while negative values indicate reducing activity during the corresponding protrusive edge motion. In addition to determining the spatial coupling, this approach also measures the temporal relationship between Rho GTPase activation and protrusion onset. As with previous analyses using this approach (24, 27, 39), individual windows were assumed to be independent measurement entities and thus n=733 windows from 25 cells for Rac1, n=778 windows from 29 cells for Rac2, and n=520 windows from 19 cells for Cdc42 were measured, compiled and analyzed using the 2000x bootstrapping of the smooth-spline fit functions from the individual windows to determine the 95% confidence intervals. The autocorrelation function of the leading edge protrusion velocities were used as a measure to indicate the periodicity of the intrinsic protrusion/retraction cycling (39). This analysis indicated that in all cases analyzed for Rac1, Rac2 and Cdc42, the expressions of these biosensors did not alter the basic protrusion cycling periodicity (Supplemental Fig. 2B).
Measurement of cell area change
To quantify the differences in cellular protrusive activity, we focused on the instantaneous rate of change in area (ΔA/Δt) and reasoned that cells undergoing more “robust” protrusion/retraction cycling are likely experiencing greater absolute change in area as a function of time. We analyzed the “cell area change index”, determined from the live cell DIC movie stack of cells undergoing random protrusions. We manually traced the outlines of each cell in the field of view at every 10-frame intervals and measured the area of such regions of interest and calculated the absolute values of the differences between each successive 10-frame intervals to determine the average square pixels of cell area change.
Statistical analysis
All results are presented as the mean +/− SEM. Data were analyzed using two-tailed unpaired Student's t test unless noted otherwise, and differences with a p value < 0.05 were considered as significant.
Results
Rac1 biosensor optimization and generation of Rac2 biosensor
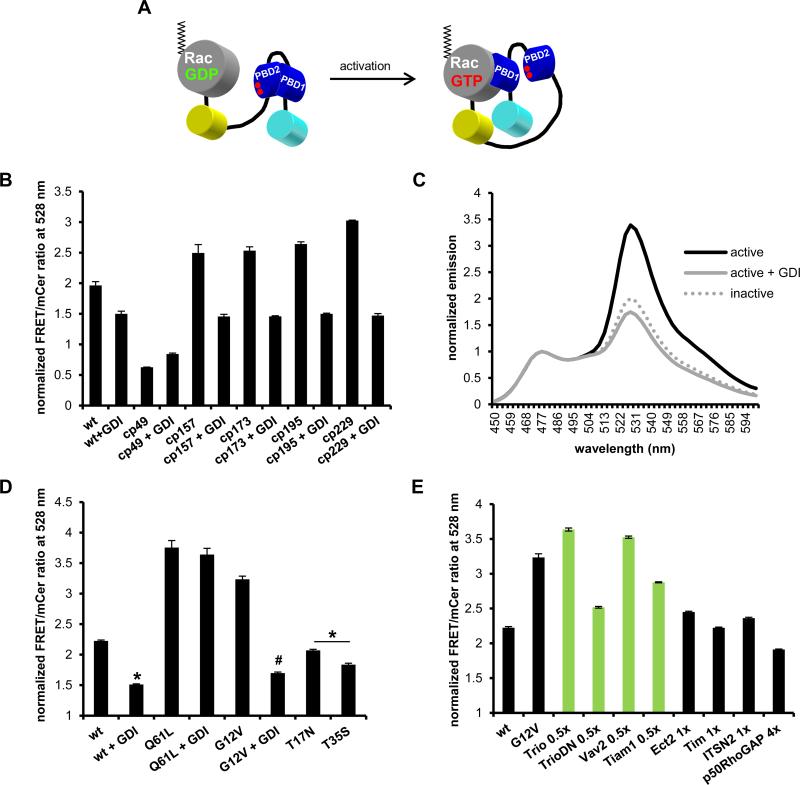
Our laboratory recently developed a fully genetically-encoded, single-chain, FRET-based Rac1 biosensor using mCer1 and mVen fluorescent proteins (FP) as FRET pair (25). Briefly, this biosensor is composed of mCer1 at the N-terminus, followed by two tandem PBD of PAK1 with a structurally optimized linker connecting them, followed by mVen and the full-length, WT Rac1 at the C-terminus (Fig. 1A; Supplemental Fig. 1A). Importantly, the C-terminal incorporation of the full-length GTPase allows for proper membrane localization of the active biosensor and negative regulation by the native GDI. FRET response is produced by the affinity-driven interaction between PBD1 and the built-in GTPase in the GTP-loaded ON state, while PBD2 serves to auto-inhibit PBD1 to minimize FRET in the GDP-loaded OFF state of the biosensor. PBD2 contains a pair of GTPase-binding deficient mutations (H83D, H86D) to prevent interaction with the built-in or other endogenous GTPases, and to restrict its function to the auto-inhibition of PBD1. The original configuration of the biosensor reported an approximate 80% change in FRET levels between ON vs OFF state (25). As the linker lengths had already been fully optimized, we attempted to further increase the dynamic range by optimizing dipole coupling of the FRET FP pair by testing the available circular permutations of mVen (32). We characterized the resulting biosensor responses as routinely done in our laboratory (23-25) by overexpressing the biosensor constructs in HEK293 cells and measuring the fluorescence emission spectra between 450 – 600 nm in fixed adherent cells with excitation at 433 nm. Using a constitutively active version (G12V) of the biosensor, with or without negative regulator GDI, we found that except for mcp49Ven, all circular permutations improved the FRET response compared to the original mVen. Mcp229Ven produced the best improvement, increasing the FRET/donor ratio from GDI-inhibited to active state by 146%, an 82.5% improvement over the original configuration (Fig. 1B). The FRET/donor ratio remained at similar levels in the GDI-inhibited OFF state for both mVen and mcp229Ven versions, indicating that the measured increase in FRET resulted from an improved dipole coupling in the ON state. To confirm that introducing mcp229Ven into Rac1 biosensor did not affect its overall functionality compared to the published mVen version, we repeated the characterization of biosensor response as previously performed (25) by introducing GTPase mutations that either activate (constitutively active G12V, Q61L) or inactivate (dominant negative T17N, effector-binding deficient T35S) the biosensor. We found that the mcp229Ven version of the biosensor behaved similarly to the original version, with G12V and Q61L mutations increasing FRET/donor ratio, while remaining low in case of T17N and T35S mutations (Fig.1C, D). Furthermore, we re-tested FRET response to an array of upstream negative (GDI, GTPase Activating Protein (GAP)) and positive (Guanine nucleotide exchange factor (GEF)) regulators. As expected, in the presence of excess GDI, FRET levels were reduced for the WT and G12V GTPase mutant, but not in the case of Q61L mutant, as it is unable to bind GDI (40). In co-expression of WT biosensor with p50RhoGAP or non-Rac1 GEFs (Ect2, Tim, ITSN2) FRET/donor ratios remained low, while Rac1 GEFs (Trio, Vav2, Tiam1) increased FRET to similar levels as the G12V activating mutation (Fig. 1E). These fluorometry measurements confirm that the mcp229Ven version of Rac1 biosensor retains its original functionality, but with a much improved dynamic range.
FIGURE 1.
Rac1 biosensor optimization. A) Single-chain Rac biosensor design showing inactive low FRET (left) versus active high FRET states (right). Red dots in PBD2 domain indicate GTPase-binding mutations H83D, H86D. B) Optimization of WT Rac1 biosensor with circular permutations cp49, cp157, cp173, cp195 and cp229 of mVen with or without excess GDI. C) Emission spectra of constitutively active (G12V) with or without excess GDI and dominant negative (T17N) Rac1 biosensor with mcp229Ven, normalized to mCer1 emission peak at 474 nm. D) WT or mutant GTPase versions of mcp229Ven Rac1 biosensor in the presence or absence of excess GDI. E) WT GTPase version of mcp229Ven Rac1 biosensor co-expressed with Rac1-targeting (in green) or non-targeting GEFs (no GDI) and Rac1-targeting p50RhoGAP. Fluorometry data, performed in HEK293 cells, are the mean +/− SEM of 3 independent experiments performed in triplicates. * p < 0.0001 vs WT alone; # p < 0.00001 vs G12V alone.
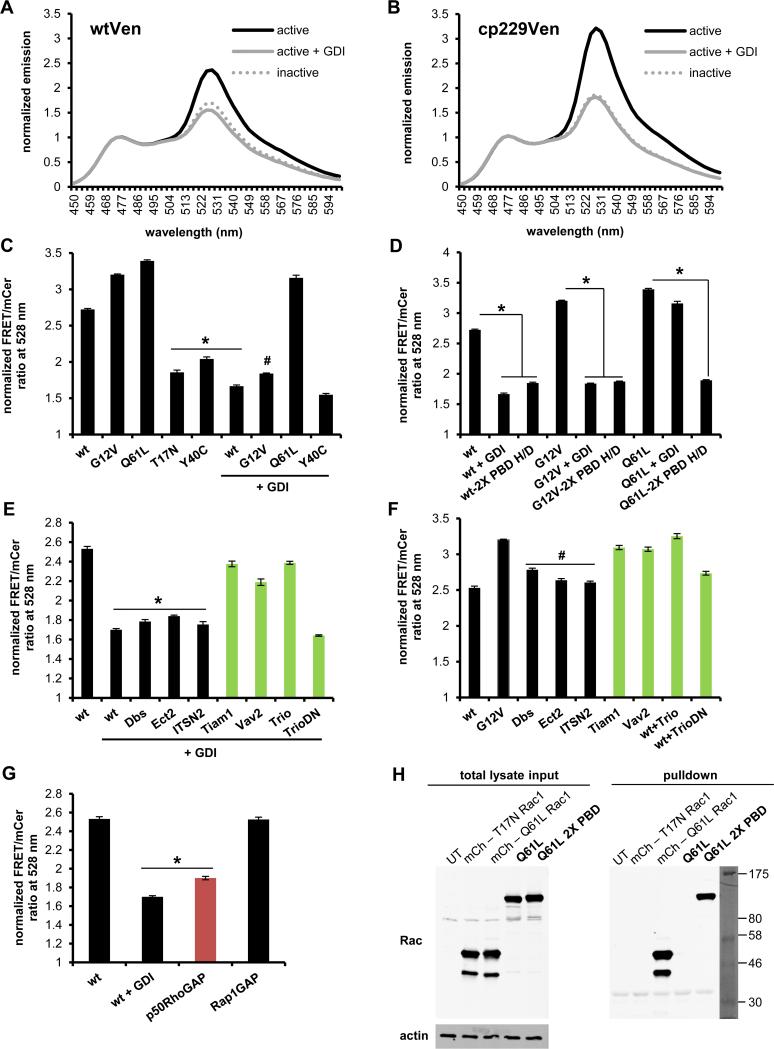
Based on the high homology (~92%) between Rac1 and Rac2, we also generated a Rac2 biosensor by replacing WT Rac1 with full-length WT Rac2 in the mVen and mcp229Ven versions of the Rac1 biosensor. Similarly to Rac1 biosensor, the mcp229Ven version exhibited a significantly improved dynamic range, 119% change in FRET levels between OFF versus ON states, compared to 65% in case of the original mVen version (Fig. 2A, B), therefore we proceeded to fully characterize the mcp229Ven version. WT Rac2 biosensor or versions containing the activating G12V and Q61L GTPase mutations showed high FRET/donor ratios, while the ratios remained low when containing the inactivating T17N or effector-binding-deficient Y40C mutations, as expected. Co-expression of WT and G12V versions with a 4-fold excess of GDI reduced FRET to similar levels as the inactivating mutations (Fig. 2C). Furthermore, introducing GTPase-binding deficient H83D, H86D mutations into PBD1 reduced FRET levels for WT and G12V mutants similar to GDI-inhibited levels, as well as for Q61L mutant (Fig. 2D). Next, we tested Rac2 biosensor modulation in response to GEFs and GAPs. GDI-mediated inhibition of the WT Rac2 biosensor was rescued by Rac GEFs (Tiam, Vav2, Trio), while non-Rac GEFs (Dbs, Ect2, ITSN2) had no effect (Fig. 2E) In addition, when the biosensor was co-expressed with GEFs in the absence GDI, only Rac GEFs were able to further activate the biosensor to levels similar to the constitutively active G12V mutation (Fig. 2F). As an internal control, a dominant negative (DN) mutation in TrioGEF fails to activate the biosensor as expected (Fig. 2E, F). In testing regulation by GAPs, p50RhoGAP reduced FRET similar to GDI-inhibited levels, while non-targeting Rap1GAP had no effect (Fig. 2G).
FIGURE 2.
Characterization of new Rac2 biosensor. Normalized emission spectra of constitutively active (G12V) with or without excess GDI and dominant negative (T17N) Rac2 sensor containing A) original mVen B) mcp229Ven, normalized to mCer1 donor emission peak at 474 nm. C) WT or mutant GTPase versions of mcp229Ven Rac2 biosensor in the presence or absence of excess GDI. D) WT and mutant versions of Rac2 biosensor with or without the GTPase-binding deficient H83D, H86D mutations in PBD1 (2X PBD H/D = both PBD1 and PBD2 are GTPase-binding deficient) in the presence or absence of excess GDI. E) WT GTPase version of mcp229Ven biosensor co-expressed with Rac2-targeting (in green) or non-targeting GEFs in the presence or F) absence of excess GDI. G) WT GTPase version of mcp229Ven biosensor co-expressed with excess of targeting (in red) or non-targeting GAPs. H) Western blot of GST-PAK pulldown of mCherry-tagged Q16L and T17N Rac1 for assay control, and Q61L constitutively active version of the Rac2 biosensor with functional or GTPase-binding-deficient mutations in the PBD1 domain, overexpressed in HEK293 cells. Total lysates and actin was used as a loading control. Fluorometry data, performed in HEK293 cells, are the mean +/− SEM of 3 independent experiments performed in triplicates. * p < 0.00001 vs WT alone; # p < 0.00001 vs G12V alone.
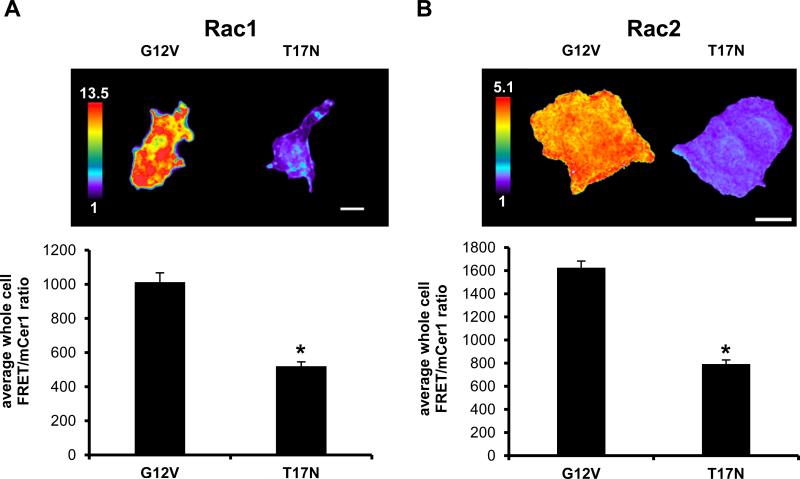
A possible concern is that expression of our biosensors may cause overexpression artifacts in cells due to competitively binding to and activating endogenous downstream effectors. To exclude this possibility we probed for extraneous effector binding by performing a GST-PAK pulldown assay using constitutively active Q61L Rac2 biosensors containing either functional or GTPase-binding-deficient (H83D, H86D) PBD1. Only the biosensor with the non-functional PBD1 was found in the pulldown fraction, demonstrating that the GTPase within the biosensor interacts exclusively with its built-in effector PBD1 (Fig. 2H). Lastly, we tested whether the Rac1 and Rac2 biosensors would retain their dynamic range in an appropriate immune cell type, macrophages. We transiently overexpressed G12V or T17N mutants in a murine monocytic/macrophage cell line (RAW/LR5), and measured approximately 2-fold difference in whole-cell FRET/donor ratios between the active (G12V) and inactive (T17N) states (Fig. 3A, B) similar to what was observed in the fluorometry experiments in HEK293 cells (Figures 1, 2). This data supports the usefulness of the newly developed and improved biosensors.
FIGURE 3.
Validation of Rac1 and Rac2 biosensors in macrophages. A) Ratiometric images of transiently overexpressed constitutively active (G12V) or dominant negative (T17N) Rac1 and B) Rac2 biosensor (with mcp229Ven) in RAW/LR5 cells. Scale bar = 10 μm. Bar graphs are quantitation of whole cell levels of Rac activity, n = at least 15 cells/condition, mean +/− SEM, p < 0.00001.
Differential patterns of Rac1 and Rac2 activation in macrophages in response to fMLP
To further validate proper functionality of the improved Rac1 and the new Rac2 biosensors, we next tested the responsiveness of the WT version of the biosensors in macrophages in a biological process known to require both Rac1and Rac2 activities. While the role of Rac2 has been well-studied in neutrophils, little is known about its isoform-specific function in macrophages. In neutrophils both Rac1 and Rac2 have been demonstrated to be required for superoxide production and chemotaxis in response to fMLP (41, 42) and Rac2 has been found to be required for superoxide production in response to Phorbol 12-myristate 13-acetate (PMA) in macrophages (13). Therefore, we chose to use fMLP as a stimulant to elicit a receptor-mediated response as a means to validate the newly developed Rac2 biosensor, as well as for testing isoform-specific reporting in parallel with the Rac1 biosensor.
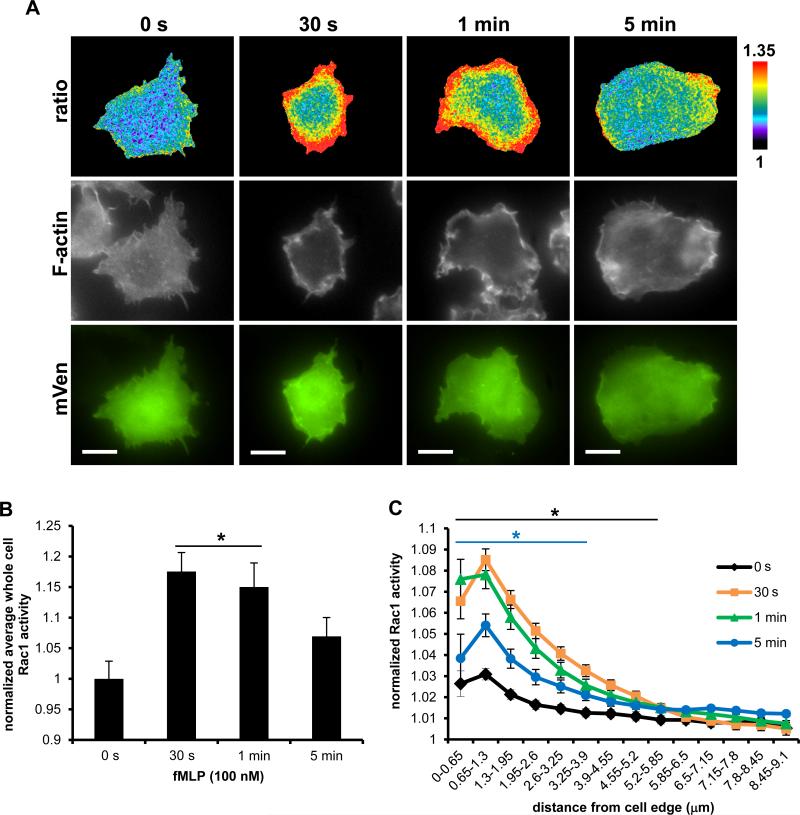
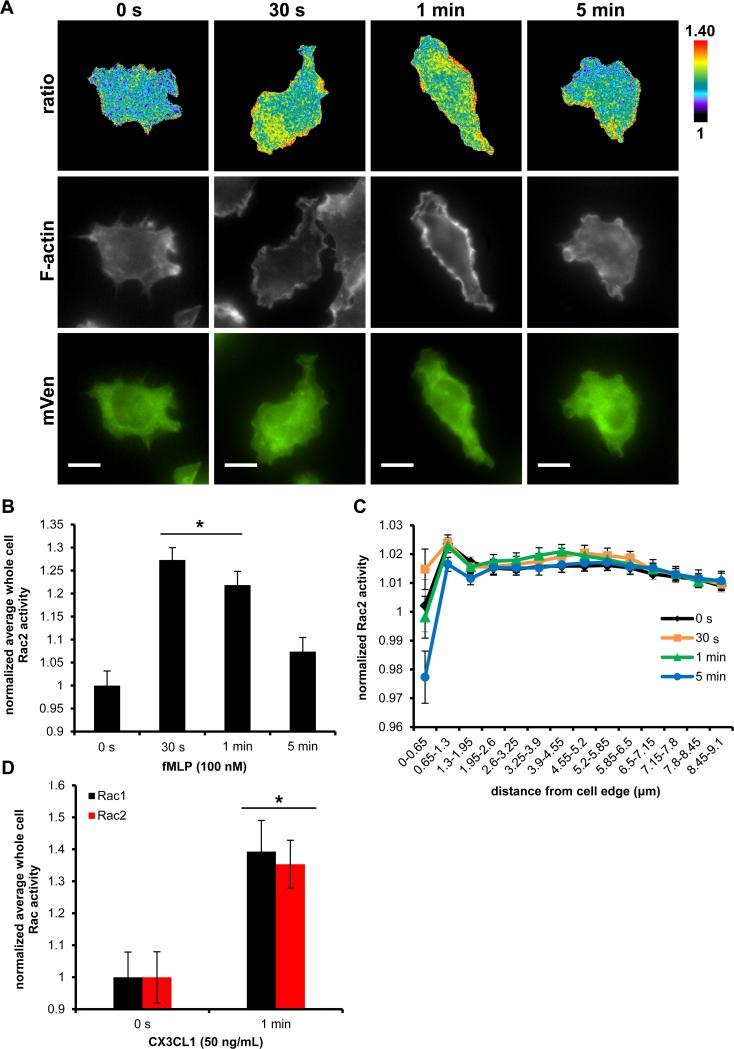
Using a dominant-negative approach Rac1 has been previously shown by us to be important for the generation of actin rich protrusion in response to fMLP in a subline of RAW264.7 (RAW/LR5) cells that express the fMLP receptor (FMLPR.2) (36) and both Rac1 and Rac2 are important for fMLP generated free actin barbed ends in neutrophils (41). We transiently overexpressed WT Rac1 biosensor in FMLPR.2 cells and stimulated with 100 nM fMLP for up to 5 min. Rac1 exhibited a robust and highly polarized activation at the plasma membrane at 30 s and 1 min, which dissipated by 5 min, coinciding with regions of increased F-actin content in space and time (Fig. 4A). The kinetics of whole cell level of Rac1 activity, significantly increased at 30 s (18%), 1 min (15%), and began to decline at 5 min (Fig. 4B). In addition, we took an unbiased approach to measure Rac1 activity at the cell periphery. Using an edge erosion algorithm that samples biosensor activity from the cell edge radially towards the cell center, we measured a significant increase in Rac1 activity at the cell periphery compared to the central cell area at 30 s and 1 min; at 5 min the edge activity was still significantly elevated, although returning to baseline levels (Fig. 4C). These results are consistent with the reported role for Rac1 in the generation of actin protrusions at the cell periphery in response to fMLP.
FIGURE 4.
Rac1 activation dynamics during fMLP response by macrophages. A) Representative ratiometric images (top panel) of FMLPR.2 cells transiently expressing WT Rac1 biosensor stimulated with 100 nM fMLP for indicated times before fixation, stained for F-actin (middle panel) and mVen emission to confirm biosensor expression (bottom panel); scale bar = 10 μm. B) Quantitation of whole cell levels of Rac1 activity; * p < 0.0001 vs 0 s. C) Quantitation of Rac1 activity at the cell edge by radial edge-erosion analysis, where activity was normalized to the lowest value (center region of the cell) within each time point to show changes in relative edge activity. Black * p < 0.0001 for 30 s and 1 min vs 0 s in 0 – 5.85 μm region; blue * p < 0.02 for 5 min vs 0 s in 0 – 3.9 μm. Data are the mean +/− SEM from 3 independent experiments, ≥ 15 cells/time point/experiment.
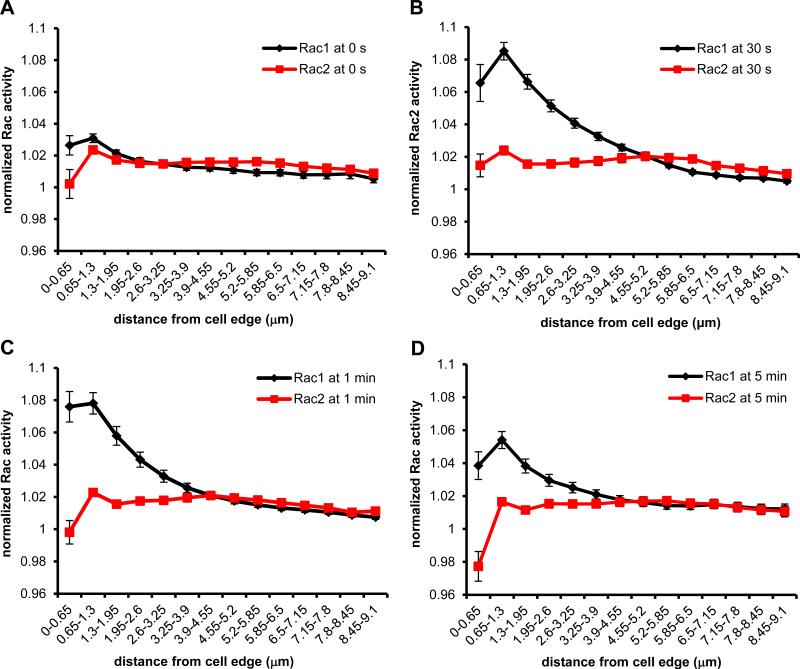
To evaluate isoform-specific reporting by the biosensors, we compared the activation dynamics between Rac1 and Rac2. In addition to the possible role for Rac2 in actin polymerization (41), Yamauchi et al. reported that macrophages derived from Rac2−/− mice produce significantly reduced levels of reactive oxygen species in response to PMA compared to WT cells, suggesting that Rac2 plays a role during respiratory burst in murine macrophages (13). Studies in human neutrophils demonstrated that during stimulation with PMA or fMLP, production of superoxide anion began within 1 min (43), therefore we reasoned we can expect to detect measurable Rac2 activity early during stimulation of macrophages. Figure 5A shows representative ratiometric images of localized Rac2 activity in response to fMLP. As actin polymerization normally occurs at the cell periphery and the superoxide-generating NADPH oxidase complex is known to translocate to plasma membrane (11), we expected Rac2 activity to localize at the cell periphery. However, while we observed an overall increase in Rac2 activity, the Rac2 activity was not dramatically increased at the plasma membrane. The bulk of activity was mainly localized in the cell body in a perinuclear region, reaching maximal response at 30 s and 1 min. We also quantified whole cell levels of Rac2 activity and measured significant increase peaking at 30 s (27%) and 1 min (21%), and leveling off by 5 min (Fig. 5B), similar to the kinetics observed in neutrophils (44). In contrast to Rac1, there was no significant increase in edge activity detected for Rac2 (Fig. 5C). To directly compare the edge activity of Rac1 and Rac2, we overlaid their measured activities (from Fig. 4C and 5C) at every time point, shown in Figure 6. Overall, these findings demonstrated that there were spatially distinct activation patterns of Rac1 versus Rac2 in response to fMLP. To confirm these results using a more physiologically relevant stimulant to macrophages, we used CX3CL1 that signals to Rac1 and Rac2 (45) via a G protein-coupled receptor, similarly as fMLP (46). WT Rac1 or Rac2 biosensors were transiently expressed in RAW/LR5 cells and then stimulated for 1 min, the time of peak F-actin content in macrophages (47). As expected, whole cell levels of Rac1 and Rac2 activities were significantly elevated, at similar levels for both isoforms (Fig. 5D). These results suggest that the Rac biosensors are functional reporters of activity in response to multiple stimuli.
FIGURE 5.
Rac2 activation dynamics during fMLP response by macrophages. A) Representative ratiometric images (top panel) of FMLPR.2 cells transiently expressing WT Rac2 biosensor stimulated with 100 nM fMLP for indicated times, stained for F-actin (middle panel) and mVen emission to confirm biosensor expression (bottom panel); scale bar = 10 μm. B) Quantitation of whole cell levels of Rac2 activity; * p < 0.0001 vs 0 s. C) Quantitation of Rac2 activity at the cell edge by radial edge-erosion analysis, where activity was normalized to the lowest value (center region of the cell) within each time point to show relative edge activity. Data are the mean +/− SEM from 3 independent experiments, ≥ 15 cells/time point/experiment. D) Quantitation of whole cell levels of Rac1 and Rac2 activity in RAW/LR5 cells transiently expressing Rac1 or Rac2 biosensor and stimulated with 50 ng/mL CX3CL1 for 1 min. Data mean +/− SEM of n = 12 cells, p < 0.005.
FIGURE 6.
Rac1 versus Rac2 activity at the cell periphery during fMLP response. Rac1 and Rac2 activity at the cell edge measured by radial edge-erosion analysis (from Fig. 4C and 5C) overlaid at A) 0 s B) 30 s C) 1 min and D) 5 min.
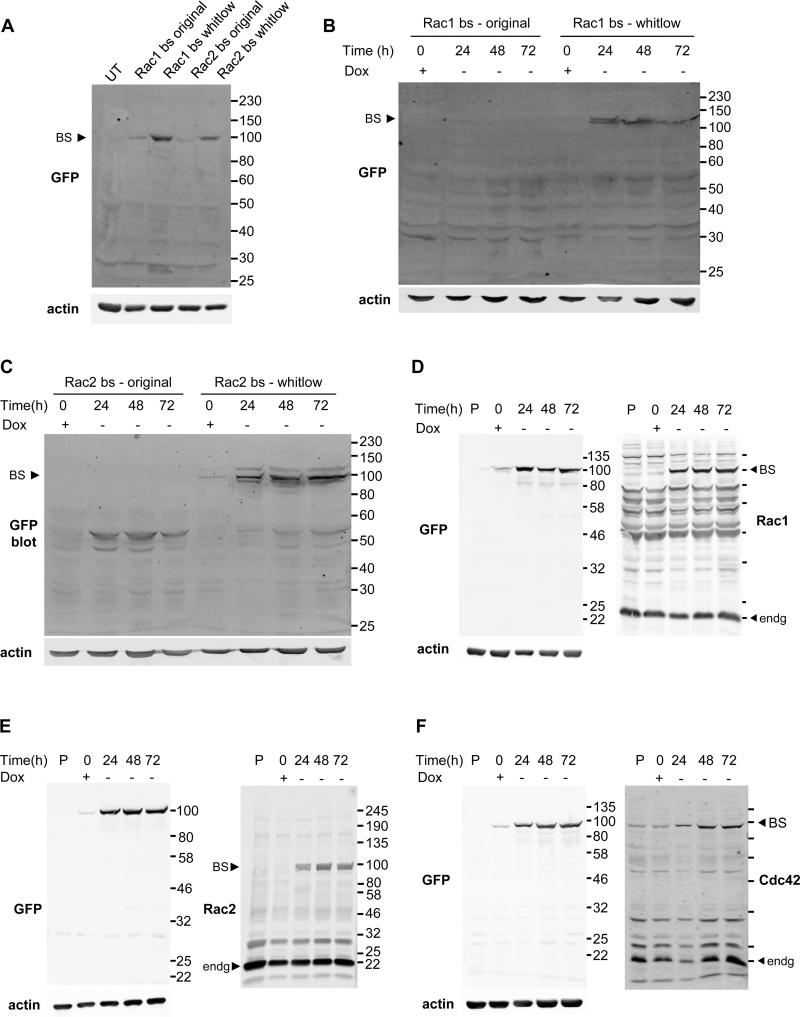
To further explore the kinetics of Rac1 and Rac2 activation in greater detail and to demonstrate usefulness of the biosensors in live-cell conditions, we also performed live-cell imaging of fMLP stimulation. To do this we generated additional cell lines with stable expression of the fMLP receptor (36) as well as stable-inducible expression of either Rac1 or Rac2 biosensors. In order to incorporate the biosensor gene expression cassette stably and inducibly into macrophages, additional optimizations were required. Macrophages are exceptionally challenging for controlled expression due to their highly-proteolytic nature and their tendency to tightly regulate exogenous gene expression through promoter silencing (48, 49). Initially, cell lines with stable, constitutive expression of Rac biosensors were generated; however this resulted in massive biosensor truncation, with little expression of full-length biosensor (data not shown). We identified a potential non-specific lysine-protease cleavage site within the linker structure originally used to produce the mcp229Ven (32) and mutated this region to confer resistance against proteolytic cleavage using partial fragment of a protease resistant linker structure reported by Whitlow et al. (34). In addition, to further minimize negative effects of biosensor overexpression, we opted to generate a system with inducible expression of the biosensor. Whitlow-modification of Rac1 and Rac2 biosensors substantially improved the full length expression of the Rac biosensors, during both transient and inducible expression (Fig. 7A-C). However, this resulted in only a partial elimination of truncated biosensor expression and required further optimization. To fully overcome this issue, we applied the synonymous codon modification approach. We recently reported the use of this approach for the stable expression of probes containing tandem sequence motifs that have high levels of sequence homology, as these are highly susceptible to deletions via homologous recombination due to the inherent nature of retroviruses (33). The codon usages of the mCer1 and one of the tandem PBD domains in the Rac2 biosensor were modified to reduce the base sequence homology within the biosensor cassette, assuring a full-length genetic incorporation of the biosensor cassette during retroviral transduction. As demonstrated in detail in (33), ensuring full length expression of the biosensors is critical for proper data interpretation and without the synonymous modification single-color biosensor fragments were still detected by FACS analysis, resulting in little or no Rac1 activity in random protrusion of macrophages. This modification was also applied to our optimized Rac1 biosensor and our recently developed Cdc42 biosensor (24). Lastly, the second generation tTA was virally transduced under an EF1α promoter to confer increased resistance against promoter silencing, as the CMV promoter is subject to silencing in hematopoietic and stem cells (48, 49). In fact, tTA expression under the control of CMV promoter was silenced in a relatively short amount of time, approximately 2-3 weeks in case of Rac2 biosensor, longer for Rac1 (personal observations), in which case we could no longer induce Rac2 biosensor expression. Only after switching to EF1α promoter were we able to achieve efficient induction long term. Collectively, with these modifications and improvements in expression strategies, we were able to achieve stable, inducible expression of our biosensors in macrophages, as shown by western blotting analysis in Fig. 7D-F.
FIGURE 7.
Optimization of biosensor expression in macrophages. Western blot analysis of A) original (non-optimized) or whitlow-modified Rac1 and Rac2 biosensors transiently expressed overnight. B) Tet-OFF inducible expression of original or whitlow-modified Rac1 and C) Rac2 biosensor. D) Tet-OFF induction of fully optimized, FACS-sorted Rac1, E) Rac2 and F) Cdc42 biosensors. BS = biosensor, Dox = doxycycline at 2 μg/mL, endg = endogenous, P = untransduced parental RAW/LR5 lysate, UT = untransfected.
Next, we FACS sorted the stable, inducible cells to obtain near-100% inducible-expressor populations of Rac1, Rac2 and Cdc42 biosensor cell lines to better characterize the biosensor expression levels in these cells. We confirmed that these cell populations isolated retained the ability to induce biosensor expression at similar efficiencies following sorting and a freeze-thaw cycle (data not shown). Using these, we determined biosensor expression levels in comparison to the endogenous GTPases (Rac1 biosensor 65.2% at 24 h, 62.1% at 48 h and 61.3% at 72 h of endogenous Rac1; Rac2 biosensor 22.3% at 24 h, 28.6% at 48 h and 23.5% at 72h of endogenous Rac2; Cdc42 biosensor 33.4% at 24h, 26.2% at 48 h and 7.3% at 72 h of endogenous Cdc42; Fig. 7D-F). Compared to the previously reported guideline for the biosensor expression to be within approximately 20-30% of the endogenous proteins (35, 50), the Rac1 biosensor expression was higher. Therefore, to ensure that cell protrusive edge dynamics driven by actin cytoskeleton was not aberrantly impacted by the higher level of Rac1 biosensor expression, we compared the edge protrusion dynamics to the parental RAW/LR5 stably expressing EGFP and measured the edge cycling periodicity. We did not observe statistically significant differences in the periodicity of the edge protrusion dynamics between the biosensor stable cell lines versus the EGFP control (Supplemental Fig. 2B).
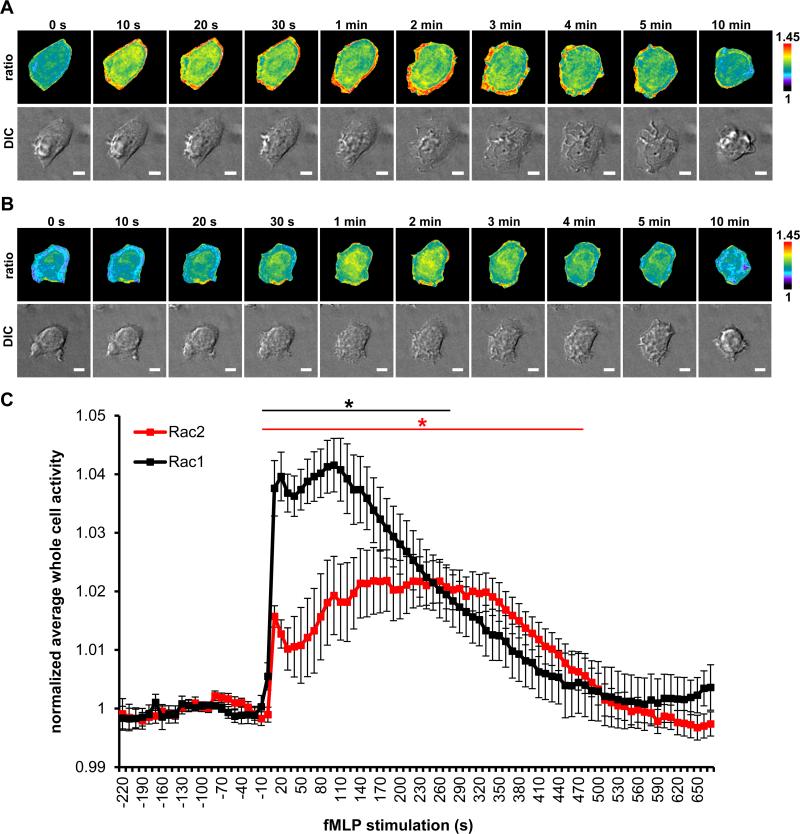
Stimulation with fMLP elicited changes in cell morphology within 20 s and displayed dynamic dorsal ruffling by 1 min that lasted for ~ 8 min (movie S1, S2) in these stable-inducible cell lines. Live-cell image analysis of Rac1 and Rac2 activation recapitulated our observations from fixed-cell images (compare Figure. 8A and B with Figures 4A and 5A). Ratiometric analysis revealed robust Rac1 activation at the cell edge in addition to the measurable increase at the whole cell levels (Fig. 8A; movie S1) and Rac2 displayed activation that was more localized to the perinuclear region of the cell (Fig. 8B; movie S2). Furthermore, measurements of whole cell levels of Rac activity revealed an earlier spike in the activation of Rac1 and Rac2 than seen with the fixed cell data, with both isoforms peaking within 10 s of fMLP addition (Fig. 8C). The increased activation of Rac1 was of shorter duration and activity began to decrease within 3 min of activation, however, Rac2 activation was sustained longer, approximately 6 min, before beginning to decrease (Fig. 8C). Overall, our Rac biosensors yielded appropriate and isoform-specific measurements during fMLP responses based on previously published results. Furthermore, we have shown that these biosensors report consistent ratiometric measurements between fixed versus live-imaging experiments, demonstrating usefulness under both settings.
FIGURE 8.
Live-cell imaging of Rac1 and Rac2 activation dynamics during fMLP response by macrophages. Time lapse ratiometric image (top panel) and DIC (bottom panel) of RAW/LR5 cell line with inducible expression of WT A) Rac1 B) Rac2 biosensor and fMLP-R stably incorporated, stimulated with 100 nM fMLP; scale bar = 5 μm. C) Quantitation of whole cell levels of Rac1 and Rac2 activity, normalized to average whole cell activity before stimulation (baseline activity during the first 4 min prior to stimulation). Data are the mean +/− SEM of 11 cells for Rac1 and 10 cells for Rac2; black * p < 0.05, 10-270 s vs 0 s (Rac1); red * p < 0.05, 10–480 s vs 0 s (Rac2). Statistical significance was determined using one-tailed, paired student t test.
Coordination of Rho GTPase activity in random protrusions of macrophages
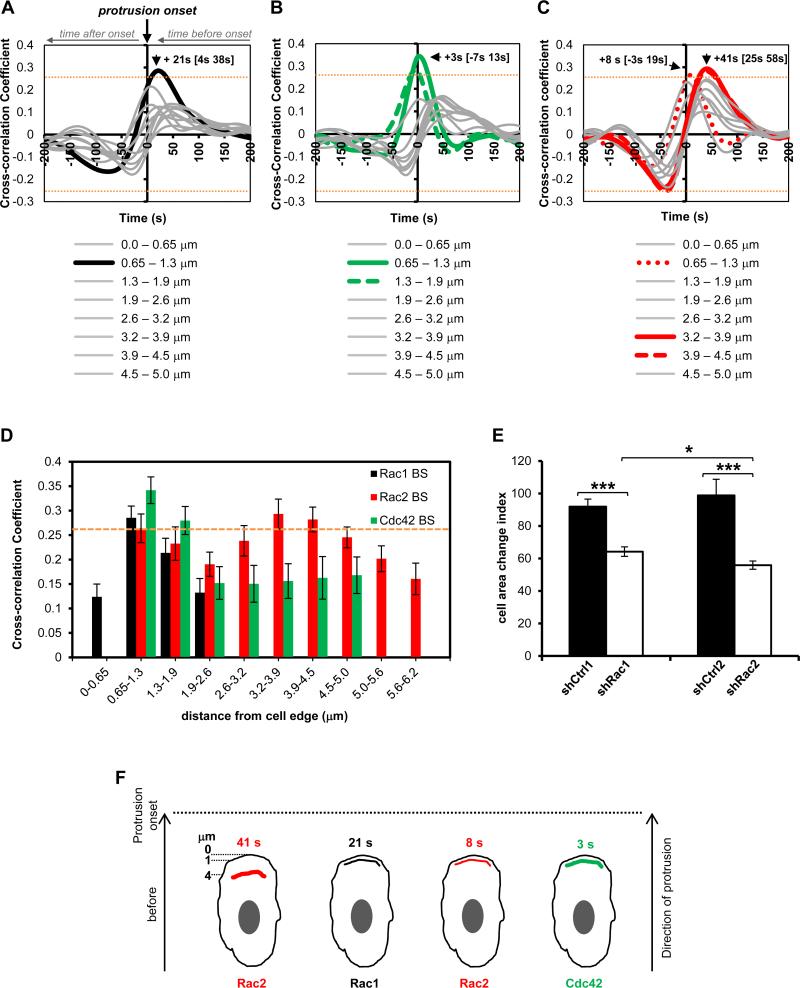
Rho GTPases are master regulators of actin cytoskeletal dynamics in cells (2, 51). In macrophages Rac2, along with its closely related isoform Rac1, has been shown to play a role in actin remodeling necessary for changes in cell morphology. Macrophages from mice with genetic deletions of Rac1, Rac2 or both isoforms displayed significantly reduced dorsal ruffling in response to CSF-1 in parallel with significant decrease in the formation of F-actin (52, 53). Clearly, the Rac isoforms play roles in F-actin and morphological changes in cells during protrusion and ruffling, yet no information exists on the spatiotemporal dynamics of these proteins in macrophages. Furthermore, Cdc42, another member of the Rho GTPase family, is required for F-actin polymerization in macrophages, and reducing Cdc42 protein levels is associated with a defective ability to extend protrusions (54). Here, we sought to use our biosensors to determine the coordination of Rac1, Rac2 and Cdc42 activities during macrophage protrusion using a previously described computational approach (39). All three Rho GTPases displayed complex activation patterns during random protrusion/retraction cycles in RAW/LR5 cells grown in serum (movies S3-S5 and Supplemental Fig. 2C-E; Rac1, Rac2, and Cdc42 respectively). Rac1 displayed a strong positive cross-correlation at the 0.65-1.3 μm region immediately behind the leading edge, while an even stronger coupling occurred with Cdc42 in slightly wider, 0.65-1.9 μm region (Fig. 9A, B, D). Unlike Rac1 and Cdc42, Rac2 appears to exhibit two distinct peak locations of positive cross-correlation in space, at 0.65 – 1.3 μm and 3.2 – 4.5 μm from the edge (Fig. 9C, D). The frontal Rac2 peak, at the 0.65 – 1.3 μm from the edge (+8s[−3s 19s]; peak of cross-correlation function in time [+/− 95% confidence interval in time], Fig. 9C; Supplemental Fig. 3A), is indistinguishable in time from the peaks of Rac1 (+21s[4s 38s], Fig. 9A) and Cdc42 (+3s[−7s 13s], Fig. 9B) at this location and it coincides with protrusion (p=0.2605 for Rac1 versus Rac2; p=0.3683 for Cdc42 versus Rac2; 95% confidence intervals for both Cdc42 and Rac2 peaks at this location span zero time lag to indicate coincident with the protrusion onset). The second, more pronounced peak at a distal location of 3.2 – 3.9 μm from the edge with approximately + 41s lead time (+41s[24.5s 57.5s], Fig. 9C), is significantly different in time compared to the front-associated Rac2 activity (p=0.048), as well as to the Cdc42 activation timing (p=0.024), but indistinguishable from Rac1 activation timing due to Rac1 trending more to the leading time shift (p=0.1993). Interestingly, regions even further away from this position maintain nearly identical temporal lead times, though the correlation peak values drop below the p=0.05 threshold by 4.5 – 4.9 μm away from the edge. This trend is measurable up to 6.21 μm away from the edge, suggesting that Rac2 activation at these distal regions is not a function of the retrograde diffusive flux of activated material from the frontal portion, but rather activation of Rac2 specifically at those distal locations (Supplemental Fig. 3B).
FIGURE 9.
Morphodynamic analysis of Rac1, Rac2 and Cdc42 activation dynamics during random protrusions of macrophages. Temporal cross-correlation of GTPase activation relative to edge velocity, arrows showing peak activation times for A) Rac1 B) Cdc42 and C) Rac2. Data from n=733 windows from 25 cells for Rac1, n=520 windows from 19 cells for Cdc42, and n=778 windows from 29 cells for Rac2. Orange dashed line indicates that cross-correlations above 0.2552 or below −0.2552 are significant with p < 0.05. Gray lines indicate non-significant correlations. See text for additional statistical values for morphodynamic analysis. D) Spatial dependence of temporal cross-correlation of GTPase activation as a function of distance from the edge of the protrusion (data extracted from panels A-C). E) Cell area change index for shRac1 and shRac2 macrophages and their respective controls, shCtrl1 and shCtrl2; data mean +/− SEM, * p = 0.0352, *** p < 2.5 × 10−6, shCtrl1 = 127 cells, shRac1 = 119 cells, shCtrl2 = 69 cells and shRac2 cells = 101 cells from 3 independent experiments and at least 5 individual fields/experiments. F) Model of Rho GTPase activation dynamics in random protrusions of macrophages showing that Rac2 is activated first in a 3.2-4.5 μm region 41 s before protrusion onset, followed a second pool of Rac2 activation right behind the leading edge, in parallel with Rac1 and Cdc42, at indicated times before protrusion onset.
Overall, these results indicate that Rac2 is activated first in the rear of the protrusion, followed by Rac1, Rac2 and Cdc42 activations immediately behind the leading edge at the onset of protrusion. Furthermore, these measurements reveal the existence of two distinct fractions of Rac2 activity, separable both in space and time. The + 41s lead time of Rac2 activation distal to the protrusion onset suggests it supersedes the front-associated Rac1 and Cdc42 activity, and loss of Rac2 activity could impede extension of a protrusion. To test this hypothesis, we reduced endogenous Rac2 via shRNA-mediated interference and performed time-lapse imaging of random protrusions, similarly to the biosensor imaging conditions used for the morphodynamic analysis. Reducing Rac2 levels (~80%) did not affect protein expression levels of Rac1 and Cdc42 (Supplemental Fig. 3D). Time lapse images show that macrophages with reduced Rac2 were defective in their ability to extend protrusions. The majority of shRac2 cells failed to extend a protrusion or was only able to produce weak protrusions (movie S6), indicating a strong role for Rac2 in driving robust edge protrusions. We quantified the difference between control (shCtrl2) and shRac2 cells by measuring the average cell area change at every 10 frames over 61 frames and saw a 50% decrease in area change in shRac2 macrophages (Fig. 9E). However, the decrease in cell area change was only 30% in shRac1 macrophages compared to its control (shCtrl1) (movie S7; Rac1 levels reduced by ~70% without affecting expression levels of Rac2 and Cdc42, Supplemental Fig. 3C). The differences in area change between shRac1 and shRac2 were also significant while the respective controls for shRac1 and shRac2 were not significantly different from each other or the parental RAW/LR5 cell line (Supplemental Fig. 3E). These results are consistent with our morphodynamic measurements in random protrusions in which Rac2 is shown to be activated first followed by Rac1 activation, suggesting a potential hierarchical order of activation of Rac GTPases. Collectively, this differential spatiotemporal coupling of Rac1, Rac2 and Cdc42 activation to the leading edge are suggestive of different roles during the generation of protrusions, with Rac2 playing a critical role in macrophages. In addition, it further demonstrates and reinforces the usefulness for isoform-specific reporting of our single-chain Rac biosensors.
Discussion
In this report we present the development and optimization of a fully genetically-encoded, single-chain FRET-based Rac2 biosensor, using monomeric Cerulean1 and monomeric circularly permutated Venus. Importantly, this design incorporates the full-length Rac2 with the C-terminal end of the molecule to maintain the native hypervariable region to allow for post-translational modifications for correct isoform specific membrane localization. In addition, our biosensor is able to interact with endogenous upstream regulators, including GDI. Furthermore, the single-chain design ensures equimolar distribution of FRET donor and acceptor, making data analysis and interpretation more straightforward. We employed the combination of three distinct protein engineering and expression strategies to ensure the stable, full-length expression of the biosensor in hematopoietic cells: 1) synonymous modification (33) ensures the incorporation of full-length biosensor gene cassette during the production of stable inducible system, critical for proper data interpretation; 2) to minimize proteolytic cleavage post-translationally, we optimized the original linker region in mcp229Ven by replacing it with a protease-resistant linker (34); and 3) rational selection of promoter system where tTA is expressed under the control of EF1α promoter to confer increased resistance against promoter silencing (48). We then further extended these new optimization strategies to our previous biosensors for Rac1 and Cdc42 GTPase (24, 25). The approaches presented here should be widely useful for achieving robust biosensor expression with a dynamic range of detection in many cell types, and are not limited to hematopoietic cell types. One of our immediate goals is the expression of these biosensors in primary cells and we are in the process of such optimization. Primary cells are often not very transfectable, slow-growing or non-dividing, and require transduction by lentiviral system that integrates only one or two copies of the gene of interest, or carefully titrated transduction by adenovirus to avoid overexpression. The main factor in these approaches is achieving high enough expression of the biosensor where the signal-to-noise ratio is suitable for imaging but not so much as to cause overexpression artifacts. This is dependent on the titrated transduction (adenovirus) or the strength of the promoter that is driving the expression of the biosensor (lentivirus). At the same time, one must consider the gene silencing tendencies of hematopoietic cells that may also interfere with biosensor expression. Thus, it is important to test for robust and ready expression using multiple promoters (including CMV, EF1α, UbC (Ubiquitin-C), PGK (Mouse phosphoglycerate kinase 1) and viral 5’-LTR, etc.) and a reporter construct such as EGFP, to determine which system would produce the best transduction and expression in the target primary cell of choice.
In addition to our extensive validation and characterization in HEK293 cells, we performed functional validations in RAW/LR5 cells, a murine monocyte/macrophage cell line, to further demonstrate appropriate isoform-specific reporting by the Rac2 biosensor based on previously published observations. Consistent with data showing that Rac2 plays a role in NADPH oxidase activation in macrophages (13), we observed a significant increase in whole-cell activation levels of Rac2, as well as Rac1, within the first minute of stimulation, similar to that seen in neutrophils (44). Interestingly, while we showed that Rac2 activation was primarily perinuclear during fMLP stimulation, Rac1 activity dramatically translocated and accumulated at the cell periphery during its peak activity. This result appears to be more in line with reports of Rac1 as the isoform that interacts with the NADPH complex at the plasma membrane following stimulation, as observed previously in human monocytes (55). Our observation of active Rac2 localization is consistent with previously reported Rac2 localization in the endosomal compartment (40). However, it does not exclude this isoform from having a role, in addition to but distinct from Rac1, during ROS production. Indeed, we observed reduced ROS production in murine macrophages either with reduced Rac1 or Rac2 levels (data not shown), thus determining their individual roles in murine macrophages requires further investigation.
We also performed morphodynamic mapping of Rac2 activity in parallel with Rac1 and Cdc42 during random protrusions of macrophages. These GTPases have been implicated in the generation of protrusions in macrophages to a variety of stimuli (36, 52-54), however their spatiotemporal dynamics have not been previously characterized in this particular context. Cross-correlation analysis of GTPase activation and cell edge movement revealed distinct spatial and temporal activation patterns for Rac1, Rac2 and Cdc42. Unexpectedly, we detected a tight coupling of Rac2 activity to protrusion farther away from the leading edge, in the distal 3.2-4.5 μm region, in addition to a weak but significant coupling to protrusion at the 0.65-1.3 μm region. Interestingly, Rac2 is activated first in this distal region with a significant lead time from the onset of protrusion, followed by activation of Rac1, Rac2 and Cdc42 immediately behind the leading edge coinciding with the onset of edge protrusions. This significant lead-time in the primary peak activity of Rac2 at a distal location from the edge suggested to us that perhaps Rac2 is acting as the master orchestrator of the leading edge protrusive cycling in these cells. Indeed, depletion of Rac2 had a more potent effect on the cell's ability to form random protrusions in comparison to depletion of Rac1, corroborating a potentially leading role for Rac2 in driving the protrusive machinery in macrophages during serum-mediated random protrusions.
The spatial coupling of GTPase activation to the dynamics of protrusion is exquisitely dependent on the balance and activities of their respective, upstream regulators (18, 56). Indeed, our observations indicate an extremely tight spatiotemporal coordination of GTPase activation dynamics in macrophages. Unlike previous observations in fibroblasts in which significant and broad regions of diffusive flux of activated Rac1/Cdc42 were noted (39), in macrophages Rac1 and Cdc42 activities are more tightly defined in spatiotemporal localization. The peaks of the cross-correlation at 0.65 – 1.3 μm appear to rapidly decay and result in insignificant correlations for Rac1 (Supplemental Fig. 3F) and much reduced but still significant correlations for Cdc42 (Supplemental Fig. 3G) within already the adjacent measurement window at 1.3 – 1.9 μm away from the leading edge. The temporal positions of the decaying peaks move in the negative-“lag” direction (Rac1: +21s[4s 38s] at 0.65 – 1.3 μm and +7s[−9s 23s] at 1.3 – 1.9 μm; Cdc42: +3s[−7s 13s] at 0.65 – 1.3 μm and −4s[−14s 6s] at 1.3 – 1.9 μm). This suggests a diffusive flux of activated material from the 0.65 – 1.3 μm location moving backwards in space to this shifted position (Supplemental Fig. 3F, G), similar to what was observed previously for these two GTPases in fibroblasts (39). However unlike fibroblasts, these trends only last within this single adjacent window in macrophages (Supplemental Fig. 3F, G). The edge-population of Rac2 also goes through a rapid diffusive flux and decay in that region, similar to Rac1 and Cdc42, reaching non-significance already by the adjacent window position, and the peak position in time shifts also in the negative-“lag” direction (Rac2: from +8s[−3s 19s] at 0.65 – 1.3 μm to +2s[−10s 14s] at 1.3 – 1.9 μm) (Supplemental Fig. 3A). In contrast, Rac2 activity at the distal region from the edge does not appear to shift in time lag as a function of distance away from the edge (Supplemental Fig. 3B), indicating that Rac2 activation is occurring in relatively broad regions away from the edge and is not susceptible to diffusive flux at these distal regions suggesting differential regulatory dynamics. These observations suggest that the spatiotemporal couplings of Rac1, Rac2 and Cdc42 activities to the protrusive edge motion in macrophages at the leading edge are much tighter compared to fibroblasts, where broader spatial and temporal decay patterns were observed (39). Thus, it is interesting to speculate that, through evolutionary mechanisms, macrophages that are required to mobilize quickly and home-in on signals to migrate and clear immunological threats, have evolved a much tighter spatiotemporal regulation of these GTPases compared to other cell types including the fibroblasts.
In previous studies using other cell types, we routinely expressed genetically encoded biosensors at 20-30% of the endogenous protein levels to minimize the possibility of overexpression effects (23, 25, 57). Here, we established stable, inducible cell lines for Rac1, Rac2, and Cdc42 biosensors in RAW/LR5, at 20-30% (Rac2 and Cdc42), and up to 60% (Rac1) of the endogenous protein levels. In order to ascertain that the moderately elevated level of Rac1 biosensor expression compared to endogenous proteins did not impact cell protrusion dynamics, we quantified the edge protrusion rates compared to the parental RAW/LR5 expressing EGFP as a control (Supplemental Fig.2B), and showed no significant difference in the measured protrusive periodicity. Furthermore, to ensure that different levels of biosensors in different cells do not impact the measured morphodynamic cross-correlations, we parsed our cross-correlation data sets for Rac1, Rac2 and Cdc42 as functions of the biosensor expression levels in individual cells analyzed (Supplemental Fig.4). There was no observable correlation between the absolute biosensor expression levels in individual cells to the resulting edge cross-correlation peak positions measured from those cells (Supplemental Fig.4). The range of biosensor expression levels amongst individual cells (between 4-7 fold difference in intensity levels, comparing the lowest to highest expressors analyzed; Fig.4A) was similar to previously published work where it also showed no specific dependence of morphodynamic correlation measurements on expression level variability (39).
In conclusion, we introduce a new Rac2 biosensor that is useful for the high-resolution spatiotemporal analysis of the activation dynamics of this hematopoietic-cell-specific Rac isoform in both fixed and live-cell imaging. Our observations reveal for the first time, the spatiotemporal coupling and coordination of Rho family GTPase activation dynamics during random edge protrusions of macrophages (Fig. 9F). Moreover, this is the first direct observation of two distinct and significantly separable pools of Rac2 being recruited during protrusions of macrophages. The definitive separation both in space and time suggests that they play distinct roles and likely involve regulation by distinct set of upstream regulators. The temporal order of GTPase activation suggests that Rac2 plays an upstream role that is supported by our observations of significantly inhibited formation of protrusions in macrophages with reduced Rac2 levels. A recent study demonstrated that family members may compensate by relocalization of activity (58), thus this remains an area for future studies. Further investigation is needed to determine if both pools of Rac2 activity are required for the extension of a protrusion, and to uncover the individual molecular roles of Rac1, Rac2 and Cdc42 in macrophage motility.
Supplementary Material
Acknowledgments
This work is in partial fulfillment of the PhD requirements for V.M. We thank Drs. Jeffrey E. Segall, Robert A. Coleman and Sara K. Donnelly for their constructive feedback on this manuscript.
This work was supported by National Institutes of Health Grants GM093121 (to L.H.) GM071828 (to D.C.) and T32GM007491 (to V.M.).
Abbreviations used in this article
- BWD
Buffer with Divalent
- cp
circular permutation
- DIC
Differential Interference Contrast
- EF1α
Elongation Factor 1α
- fMLP
formyl-Methionyl-Leucyl-Phenylalanine
- FRET
Forster Resonance Energy Transfer
- FP
Fluorescent Protein
- GAP
GTPase Activating Protein
- GDI
Guanine Nucleotide Dissociation Inhibitor
- GEF
Guanine Nucleotide Exchange Factor
- mCer1
monomeric Cerulean1
- mVen
monomeric Venus
- mcp229Ven
monomeric circular permutation 229 of mVenus
- PAK1
p21 Activated Kinase 1
- PBD
p21 Binding Domain
- PMA
Phorbol 12-myristate 13-acetate
- ROS
Reactive Oxygen Species
- tTA
tetracycline Trans-Activator
- WT
wild-type
Footnotes
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Boureux A, Vignal E, Faure S, Fort P. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol Biol Evol. 2007;24:203–216. doi: 10.1093/molbev/msl145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Etienne-Manneville S, Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- 3.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 4.Bokoch GM. Regulation of innate immunity by Rho GTPases. Trends Cell Biol. 2005;15:163–171. doi: 10.1016/j.tcb.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 5.Didsbury J, Weber RF, Bokoch GM, Evans T, Snyderman R. rac, a novel ras-related family of proteins that are botulinum toxin substrates. J Biol Chem. 1989;264:16378–16382. [PubMed] [Google Scholar]
- 6.Pai SY, Kim C, Williams DA. Rac GTPases in human diseases. Disease markers. 2010;29:177–187. doi: 10.3233/DMA-2010-0738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tao W, Filippi MD, Bailey JR, Atkinson SJ, Connors B, Evan A, Williams DA. The TRQQKRP motif located near the C-terminus of Rac2 is essential for Rac2 biologic functions and intracellular localization. Blood. 2002;100:1679–1688. [PubMed] [Google Scholar]
- 8.Filippi MD, Harris CE, Meller J, Gu Y, Zheng Y, Williams DA. Localization of Rac2 via the C terminus and aspartic acid 150 specifies superoxide generation, actin polarity and chemotaxis in neutrophils. Nature immunology. 2004;5:744–751. doi: 10.1038/ni1081. [DOI] [PubMed] [Google Scholar]
- 9.Yamauchi A, Marchal CC, Molitoris J, Pech N, Knaus U, Towe J, Atkinson SJ, Dinauer MC. Rac GTPase isoform-specific regulation of NADPH oxidase and chemotaxis in murine neutrophils in vivo. Role of the C-terminal polybasic domain. J Biol Chem. 2005;280:953–964. doi: 10.1074/jbc.M408820200. [DOI] [PubMed] [Google Scholar]
- 10.Shelef MA, Tauzin S, Huttenlocher A. Neutrophil migration: moving from zebrafish models to human autoimmunity. Immunol Rev. 2013;256:269–281. doi: 10.1111/imr.12124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pick E. Role of the Rho GTPase Rac in the activation of the phagocyte NADPH oxidase: outsourcing a key task. Small GTPases. 2014;5:e27952. doi: 10.4161/sgtp.27952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, Yamauchi A, Marchal CC, Molitoris JK, Quilliam LA, Dinauer MC. Chemoattractant-stimulated Rac activation in wild-type and Rac2-deficient murine neutrophils: preferential activation of Rac2 and Rac2 gene dosage effect on neutrophil functions. J Immunol. 2002;169:5043–5051. doi: 10.4049/jimmunol.169.9.5043. [DOI] [PubMed] [Google Scholar]
- 13.Yamauchi A, Kim C, Li S, Marchal CC, Towe J, Atkinson SJ, Dinauer MC. Rac2-deficient murine macrophages have selective defects in superoxide production and phagocytosis of opsonized particles. J Immunol. 2004;173:5971–5979. doi: 10.4049/jimmunol.173.10.5971. [DOI] [PubMed] [Google Scholar]
- 14.Lacy P, Mahmudi-Azer S, Bablitz B, Gilchrist M, Fitzharris P, Cheng D, Man SF, Bokoch GM, Moqbel R. Expression and translocation of Rac2 in eosinophils during superoxide generation. Immunology. 1999;98:244–252. doi: 10.1046/j.1365-2567.1999.00873.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glogauer M, Marchal CC, Zhu F, Worku A, Clausen BE, Foerster I, Marks P, Downey GP, Dinauer M, Kwiatkowski DJ. Rac1 deletion in mouse neutrophils has selective effects on neutrophil functions. J Immunol. 2003;170:5652–5657. doi: 10.4049/jimmunol.170.11.5652. [DOI] [PubMed] [Google Scholar]
- 16.Abdel-Latif D, Steward M, Macdonald DL, Francis GA, Dinauer MC, Lacy P. Rac2 is critical for neutrophil primary granule exocytosis. Blood. 2004;104:832–839. doi: 10.1182/blood-2003-07-2624. [DOI] [PubMed] [Google Scholar]
- 17.Baier A, Ndoh VN, Lacy P, Eitzen G. Rac1 and Rac2 control distinct events during antigen-stimulated mast cell exocytosis. J Leukoc Biol. 2014 doi: 10.1189/jlb.0513281. [DOI] [PubMed] [Google Scholar]
- 18.Pertz O. Spatio-temporal Rho GTPase signaling - where are we now? J Cell Sci. 2010;123:1841–1850. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- 19.Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- 20.Itoh RE, Kurokawa K, Ohba Y, Yoshizaki H, Mochizuki N, Matsuda M. Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol Cell Biol. 2002;22:6582–6591. doi: 10.1128/MCB.22.18.6582-6591.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seth A, Otomo T, Yin HL, Rosen MK. Rational design of genetically encoded fluorescence resonance energy transfer-based sensors of cellular Cdc42 signaling. Biochemistry. 2003;42:3997–4008. doi: 10.1021/bi026881z. [DOI] [PubMed] [Google Scholar]
- 22.Nalbant P, Hodgson L, Kraynov V, Toutchkine A, Hahn KM. Activation of endogenous Cdc42 visualized in living cells. Science. 2004;305:1615–1619. doi: 10.1126/science.1100367. [DOI] [PubMed] [Google Scholar]
- 23.Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- 24.Hanna S, Miskolci V, Cox D, Hodgson L. A New Genetically Encoded Single-Chain Biosensor for Cdc42 Based on FRET, Useful for Live-Cell Imaging. PLoS One. 2014;9:e96469. doi: 10.1371/journal.pone.0096469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moshfegh Y, Bravo-Cordero JJ, Miskolci V, Condeelis J, Hodgson L. A Trio-Rac1-Pak1 signalling axis drives invadopodia disassembly. Nat Cell Biol. 2014;16:574–586. doi: 10.1038/ncb2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bravo-Cordero JJ, Oser M, Chen X, Eddy R, Hodgson L, Condeelis J. A Novel Spatiotemporal RhoC Activation Pathway Locally Regulates Cofilin Activity at Invadopodia. Curr Biol. 2011;21:635–644. doi: 10.1016/j.cub.2011.03.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zawistowski JS, Sabouri-Ghomi M, Danuser G, Hahn KM, Hodgson L. A RhoC biosensor reveals differences in the activation kinetics of RhoA and RhoC in migrating cells. PLoS One. 2013;8:e79877. doi: 10.1371/journal.pone.0079877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donnelly SK, Bravo-Cordero JJ, Hodgson L. Rho GTPase isoforms in cell motility: Don't fret, we have FRET. Cell Adh Migr. 2014;8:526–534. doi: 10.4161/cam.29712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gardiner EM, Pestonjamasp KN, Bohl BP, Chamberlain C, Hahn KM, Bokoch GM. Spatial and temporal analysis of Rac activation during live neutrophil chemotaxis. Curr Biol. 2002;12:2029–2034. doi: 10.1016/s0960-9822(02)01334-9. [DOI] [PubMed] [Google Scholar]
- 30.Hoppe AD, Swanson JA. Cdc42, Rac1, and Rac2 display distinct patterns of activation during phagocytosis. Mol Biol Cell. 2004;15:3509–3519. doi: 10.1091/mbc.E03-11-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yoshida S, Hoppe AD, Araki N, Swanson JA. Sequential signaling in the plasma-membrane domains during macropinosome formation in macrophages. Journal of Cell Science. 2009;122:3250–3261. doi: 10.1242/jcs.053207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagai T, Yamada S, Tominaga T, Ichikawa M, Miyawaki A. Expanded dynamic range of fluorescent indicators for Ca(2+) by circularly permuted yellow fluorescent proteins. Proc Natl Acad Sci U S A. 2004;101:10554–10559. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu B, Miskolci V, Sato H, Tutucci E, Kenworthy CA, Donnelly SK, Yoon YJ, Cox D, Singer RH, Hodgson L. Synonymous modification results in high-fidelity gene expression of repetitive protein and nucleotide sequences. Genes Dev. 2015;29:876–886. doi: 10.1101/gad.259358.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Whitlow M, Bell BA, Feng SL, Filpula D, Hardman KD, Hubert SL, Rollence ML, Wood JF, Schott ME, Milenic DE, Yokota T, Schlom J. An improved linker for single-chain Fv with reduced aggregation and enhanced proteolytic stability. Protein Eng. 1993;6:989–995. doi: 10.1093/protein/6.8.989. [DOI] [PubMed] [Google Scholar]
- 35.Hodgson L, Pertz O, Hahn KM. Design and optimization of genetically encoded fluorescent biosensors: GTPase biosensors. Methods Cell Biol. 2008;85:63–81. doi: 10.1016/S0091-679X(08)85004-2. [DOI] [PubMed] [Google Scholar]
- 36.Cox D, Chang P, Zhang Q, Reddy PG, Bokoch GM, Greenberg S. Requirements for both Rac1 and Cdc42 in membrane ruffling and phagocytosis in leukocytes. J Exp Med. 1997;186:1487–1494. doi: 10.1084/jem.186.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spiering D, Bravo-Cordero JJ, Moshfegh Y, Miskolci V, Hodgson L. Quantitative Ratiometric Imaging of FRET-Biosensors in Living Cells. Methods Cell Biol. 2013;114:593–609. doi: 10.1016/B978-0-12-407761-4.00025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spiering D, Hodgson L. Multiplex imaging of Rho family GTPase activities in living cells. Methods Mol Biol. 2012;827:215–234. doi: 10.1007/978-1-61779-442-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Machacek M, Hodgson L, Welch C, Elliott H, Pertz O, Nalbant P, Abell A, Johnson GL, Hahn KM, Danuser G. Coordination of Rho GTPase activities during cell protrusion. Nature. 2009;461:99–103. doi: 10.1038/nature08242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Michaelson D, Silletti J, Murphy G, D'Eustachio P, Rush M, Philips MR. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J Cell Biol. 2001;152:111–126. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun CX, Magalhaes MA, Glogauer M. Rac1 and Rac2 differentially regulate actin free barbed end formation downstream of the fMLP receptor. J Cell Biol. 2007;179:239–245. doi: 10.1083/jcb.200705122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H, Sun C, Glogauer M, Bokoch GM. Human neutrophils coordinate chemotaxis by differential activation of Rac1 and Rac2. J Immunol. 2009;183:2718–2728. doi: 10.4049/jimmunol.0900849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Quinn MT, Evans T, Loetterle LR, Jesaitis AJ, Bokoch GM. Translocation of Rac correlates with NADPH oxidase activation. Evidence for equimolar translocation of oxidase components. J Biol Chem. 1993;268:20983–20987. [PubMed] [Google Scholar]
- 44.Ming W, Li S, Billadeau DD, Quilliam LA, Dinauer MC. The Rac effector p67phox regulates phagocyte NADPH oxidase by stimulating Vav1 guanine nucleotide exchange activity. Mol Cell Biol. 2007;27:312–323. doi: 10.1128/MCB.00985-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park H, Cox D. Syk regulates multiple signaling pathways leading to CX3CL1 chemotaxis in macrophages. J Biol Chem. 2011;286:14762–14769. doi: 10.1074/jbc.M110.185181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Migeotte I, Communi D, Parmentier M. Formyl peptide receptors: a promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine & growth factor reviews. 2006;17:501–519. doi: 10.1016/j.cytogfr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 47.Gevrey JC, Isaac BM, Cox D. Syk is required for monocyte/macrophage chemotaxis to CX3CL1 (Fractalkine). J Immunol. 2005;175:3737–3745. doi: 10.4049/jimmunol.175.6.3737. [DOI] [PubMed] [Google Scholar]
- 48.Qin JY, Zhang L, Clift KL, Hulur I, Xiang AP, Ren BZ, Lahn BT. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One. 2010;5:e10611. doi: 10.1371/journal.pone.0010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Teschendorf C, Warrington KH, Jr., Siemann DW, Muzyczka N. Comparison of the EF-1 alpha and the CMV promoter for engineering stable tumor cell lines using recombinant adeno-associated virus. Anticancer research. 2002;22:3325–3330. [PubMed] [Google Scholar]
- 50.Hodgson L, Shen F, Hahn K. Biosensors for characterizing the dynamics of rho family GTPases in living cells. Curr Protoc Cell Biol. 2010 doi: 10.1002/0471143030.cb1411s46. Chapter 14: Unit 14 11 11-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 52.Wells CM, Walmsley M, Ooi S, Tybulewicz V, Ridley AJ. Rac1-deficient macrophages exhibit defects in cell spreading and membrane ruffling but not migration. J Cell Sci. 2004;117:1259–1268. doi: 10.1242/jcs.00997. [DOI] [PubMed] [Google Scholar]
- 53.Wheeler AP, Wells CM, Smith SD, Vega FM, Henderson RB, Tybulewicz VL, Ridley AJ. Rac1 and Rac2 regulate macrophage morphology but are not essential for migration. J Cell Sci. 2006;119:2749–2757. doi: 10.1242/jcs.03024. [DOI] [PubMed] [Google Scholar]
- 54.Park H, Cox D. Cdc42 regulates Fc gamma receptor-mediated phagocytosis through the activation and phosphorylation of Wiskott-Aldrich syndrome protein (WASP) and neural-WASP. Mol Biol Cell. 2009;20:4500–4508. doi: 10.1091/mbc.E09-03-0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao X, Carnevale KA, Cathcart MK. Human monocytes use Rac1, not Rac2, in the NADPH oxidase complex. J Biol Chem. 2003;278:40788–40792. doi: 10.1074/jbc.M302208200. [DOI] [PubMed] [Google Scholar]
- 56.Bravo-Cordero JJ, Sharma VP, Roh-Johnson M, Chen X, Eddy R, Condeelis J, Hodgson L. Spatial regulation of RhoC activity defines protrusion formation in migrating cells. J Cell Sci. 2013 doi: 10.1242/jcs.123547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bravo-Cordero JJ, Moshfegh Y, Condeelis J, Hodgson L. Live Cell Imaging of RhoGTPase Biosensors in Tumor Cells. Methods Mol Biol. 2013;1046:359–370. doi: 10.1007/978-1-62703-538-5_22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Veltman DM, King JS, Machesky LM, Insall RH. SCAR knockouts in Dictyostelium: WASP assumes SCAR's position and upstream regulators in pseudopods. J Cell Biol. 2012;198:501–508. doi: 10.1083/jcb.201205058. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.