Abstract
Biofilm bacteria have developed escape strategies to avoid stresses associated with biofilm growth, respond to changing environmental conditions, and disseminate to new locations. An ever-expanding body of research suggests that cellular release from biofilms is distinct from a simple reversal of attachment and reversion to a planktonic mode of growth, with biofilm dispersion involving sensing of specific cues, regulatory signal transduction, and consequent physiological alterations. However, dispersion is only one of many ways to escape the biofilm mode of growth. The present review is aimed at distinguishing this active and regulated process of dispersion from the passive processes of desorption and detachment by highlighting the regulatory processes and distinct phenotypes specific to dispersed cells.
Graphical Abstract
Biofilms are surface associated communities of microorganisms encased in a self-produced polymeric matrix. The sessile lifestyle affords bacteria multiple protective advantages, allowing bacteria to remain within a favorable environmental niche or host. Compared to free-swimming bacteria, biofilms are better adapted to withstand nutrient deprivation, pH changes, oxygen radicals, biocides, and antimicrobial agents [1]. Adaptation to the sessile lifestyle coincides with altered expression of surface molecules, nutrient utilization, and virulence factors, and resistance to the immune system and antimicrobial agents [2-4]. In fact, bacteria living in biofilms can be up to 1000 times more tolerant to antibacterial compounds than their planktonic counterparts [5,6]. In clinical settings, the extraordinary resistance of biofilms to antimicrobial agents can be devastating, as conventional therapies have proven to be inadequate in the treatment of many if not most chronic biofilm infections. This extraordinary innate resistance to antimicrobial treatments renders biofilms extremely difficult to control in medical settings [7]. It is thus not surprising that biofilms and potential novel strategies to control or eradicate them have received considerable attention. While much research has focused on the development of anti-adhesive surfaces and devices aimed at preventing the formation of biofilms in the first place, recent findings have suggested another promising avenue open for biofilm control: the manipulation of the biofilm lifestyle.
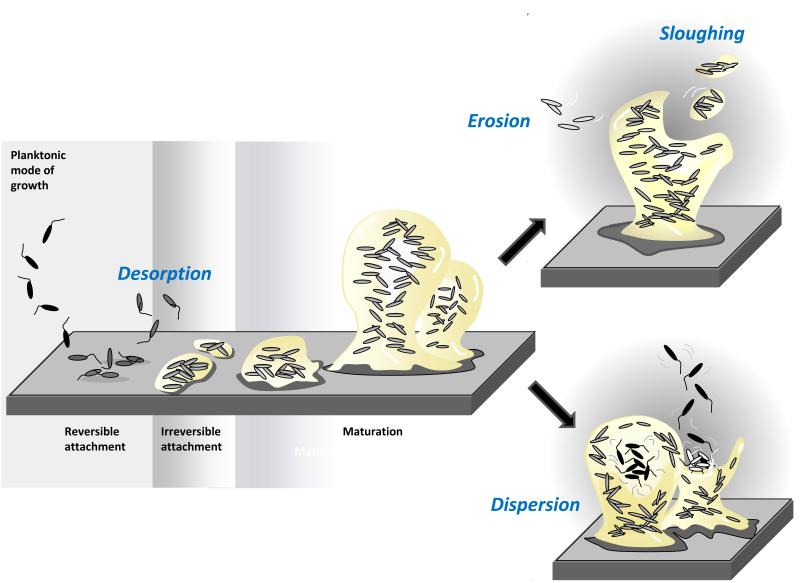
While the regulatory specifics of the biofilm developmental processes of various bacterial species exemplify the remarkable diversity of the microbial world, studies of single-species communities have illustrated some general features characteristic of the biofilm mode of growth. Biofilm formation is initiated with surface attachment by a few planktonic, free-swimming bacteria, which occurs in two stages – reversible and irreversible attachment. The typically unstable reversible attachment is characterized by cells attaching to a surface by a single pole and often returning to the bulk phase (Figure 1). Rod-shaped cells that commit to a more stable surface existence are next seen to attach to the surface via their longitudinal axis. This phenomenon is referred to as ‘irreversible attachment’. Once attached, cells will grow into a more complex multicellular mature form, which in some bacterial species is characterized by the presence of differentiated, mushroom- or pillar-like structures or microcolonies interspersed with fluid-filled channels (Figure 1). The developmental progression leading to a mature biofilm not only coincides with observable phenotypic or architectural changes, but also requires multiple regulatory networks, which translate signals to concerted gene expression changes thereby mediating the spatial and temporal reorganization of the bacterial cells within biofilms [3,8,9]. While these molecular and phenotypic changes provide biofilm-specific benefits, biofilm growth is also associated with certain dangers. As a biofilm grows in size, some cells will become increasingly separated from the bulk liquid interface and essential sources of energy or nutrients. Accumulation of waste products and toxins can present an additional challenge. Being trapped deep within a biofilm can, therefore, threaten cell survival. Thus, biofilm cells have evolved mechanisms to escape the sessile mode of growth as a means of self-preservation and dissemination to new locales [2,9].
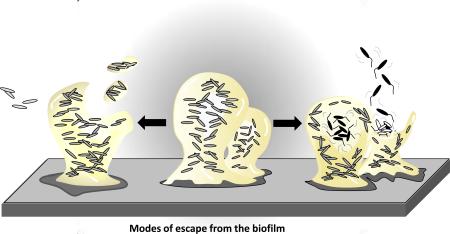
Figure 1. Model of biofilm development and modes of escape.
Based on the analysis of single species biofilms, the formation of biofilms occurs in a stage-specific and progressive manner. The developmental process is initiated by single planktonic cells making contact with the surface. With respect to biofilm formation, several developmental steps are discernable as reversible attachment, irreversible attachment, and biofilm maturation [2,8]. Cells can escape from the biofilm via desorption, detachment, and dispersion. Detachment mechanisms include erosion and sloughing. Erosion describes the continuous removal of biomass while sloughing entails the removal of intact pieces of biofilm or the entire biofilm. In biofilm dispersion, the inside of a biofilm microcolony becomes fluid, and cells within this zone begin to show signs of agitation and movement. Cells escape the biofilm microcolony via a disruption in the microcolony wall through which cells evacuate, entering the bulk liquid as single bacteria.
Not surprisingly, the progression and regulation of the switches enabling bacteria to escape from the biofilm, generally referred to as dispersal or dispersion, have become a major focus of recent research endeavors. Much attention has also been paid to the agents inducing the transition from a sessile to a planktonic, and thus a less protected and more antimicrobial susceptible mode of growth. Studies of bacterial escape from biofilms have ranged from observations of the process naturally occurring in older biofilms to the investigations of the physical and chemical factors triggering the release of cells from the sessile communities. Such factors include but are not limited to shear stress, matrix-degrading enzymes, surfactants, chelating agents, signaling molecules, and environmental cues such as nitric oxide, oxygen levels and variation in carbon and energy source availability. The diversity of biofilm release-inducing factors is matched by the variety of experimental conditions and systems used to study them, which have included microtiter plates, continuous flow reactors and flow cells, microfermentors, slides, drip flow and rotating disc reactors, aggregation in batch culture and colony and pellicle biofilm models. The timing of the experiments have also varied drastically, with induction of cell release tested in time frames ranging from 15 min to 40 hr on bacterial biomass that has been allowed to grow on a surface anywhere from 2 hr to 10 days. Yet despite all these notable differences, majority of these studies refer to the described phenomena as biofilm “dispersion” or “dispersal”.
Ways to leave a biofilm
Given the wide variety of agents and conditions attributed to cellular release from the biofilm, however, one has to wonder whether the nature of the “escape” from the attached biomass is indeed the same. This is particularly important considering that there is more than one way for bacteria to leave the biofilm. In fact, there are at least three types of “escapes” or ways for bacteria to leave the biofilm: desorption, detachment, and dispersion (reviewed in [10]). The transfer of bacteria directly from a substratum to the bulk liquid is known as desorption. Desorption may be observed at early stages of biofilm development, when the first cell contact with the surface is initiated. Thus, desorption is in many ways a reversion of the bacterial attachment process or factors involved in surface contact (Figure 1, Table 1). Considering that attachment has been demonstrated in many bacterial species to be a regulated process, desorption may likewise be an active process. Detachment has long been considered the primary process that limits biofilm accumulation [11] and occurs when external forces, such as shear stress (e.g. due to air bubbles), become sufficiently high or alternatively too low to maintain the biofilm structure. In 1988, Bryers [12] categorized four distinct mechanisms by which bacteria may passively detach from a biofilm. These are abrasion, grazing, erosion, and sloughing (Figure 1). Abrasion is the release of cells from a biofilm as a result of collisions with particles from the bulk liquid, while grazing is the act of removal of biofilm cells by the feeding activity of eukaryotic organisms. Erosion is the continuous loss of small portions of the biofilm due to fluid shear in a flowing system. Organisms that are closest to the bulk water interface of a biofilm are the only cells susceptible to this form of detachment. Cells not enmeshed within the biofilm matrix and daughter cells that are produced at the interface are particularly prone to loss by erosion. While similar to erosion [13], sloughing refers to the removal of intact pieces of biofilm or the entire biofilm by fluid frictional forces. In addition to shear forces, the rate and degree of detachment from a biofilm will be impacted by any modification of the biofilm structure, such as exogenously induced degradation of the biofilm matrix by chemical or enzymatic means. Considering that biofilms are encased in an extracellular polymeric matrix composed of polysaccharides, proteins, and DNA, it is not surprising that most detachment agents have degradative (Table 1), chelating [14], or detergent-like functions [15,16]. An example of a matrix degrading detachment agent is the hydrolase Dispersin B from Aggregatibacter actinomycetemcomitans that catalyzes the hydrolysis of the linear polymers of N-acetyl-D-glucosamines. Exogenously added Dispersin B has been shown to coincide with the loss of the structural integrity of the biofilm structure of the oral pathogen A. actinomycetemcomitans [17,18]. As N-acetyl-D-glucosamines is present in the matrix of several Gram-positive and Gram-negative biofilm-forming bacteria, it is not surprising that Dispersin B is capable of inducing detachment in a large variety of biofilm forming species including Staphylococcal species [18,19]. Detachment has also been demonstrated for various bacterial species upon treatment with periodate, cellulose, and proteinase K (Table 1). DNAse has been used in the treatment of pulmonary diseases since the early 90s, primarily to reduce the viscoelasticity of the sputum from cystic fibrosis patients [20,21]. Since then, extracellular DNA has been recognized as one of the major matrix components of bacterial biofilms [22]. It was shown that DNAse treatment led to the detachment of young, but not mature, flow-chamber-grown P. aeruginosa biofilms [22], probably due to mature biofilms harboring increasing amounts of matrix material other than extracellular DNA. Treatment with DNAse coinciding with detachment has been described in a large number of other biofilm forming species including P. putida, S. aureus, Shewanella oneidensis, and Bacillus licheniformis [23-26].
Table 1.
Cues, signals and agents linked to the different modes of escapes from the biofilm. PDE, phosphodiesterase; DGC, diguanylate cyclase; batch-grown biofilms, biofilms grown in microtiter plates, petri dishes or other containers without medium being replaced over the course of biofilm growth; CV staining, biofilm biomass was stained with crystal violet; CFU, viability count based on colony forming units; SNP, sodium nitroprusside.
| Cue, Signal, factors | Species | Biofilm growth method/ Biofilm age and treatment | Effector Regulatory System | Source |
|---|---|---|---|---|
| Conditions linked to induction of biofilm dispersion | ||||
| Oxygen depletion | Pseudomonas aeruginosa | Semi-batch growth system using polystyrene surface; 2d biofilms; detection of biomass reduction by CV staining | PDE RbdA | [59] |
| Oxygen depletion, cessation of flow | Shewanella oneidensis | Flow cell 16 hr biofilms; dispersion noted by microscopy within 5 min post cessation of flow | Transcriptional regulators ArcA and CRP | [42,74] |
| Hydrogen peroxide | a. actinomycetemco mitans | 6 hr microtiter dish biofilms or 4 -hr colony biofilms, and murine abscess infection model | Up-regulation of dspB expression encoding Dispersin B | [70] |
| Starvation, cessation of flow | P. putida | Flow cell 4d biofilms; detachment observed 15 min post flow cessation | [53] | |
| Starvation, cessation of flow |
P. aeruginosa
P. fluorescens |
Drip flow or capillary flow biofilm reactors; 4 d biofilms; dispersion assessed 3 d post flow cessation by spectroscopy | [75-78] | |
| Sudden step-increase of carbon source concentration (glucose, glutamate, succinate, citrate) | P. aeruginosa | Continuous flow reactors (tube reactors, flow cells); 4-5 d biofilms; monitoring biofilm by CFU and microscopy for up to 60 min, with release of cells from the surface observable within 10 min Acute and chronic murine virulence model, using dispersion-deficient mutants |
• Phosphorylation-dependent signaling (response inhibited with phosphatase inhibitor) • Increased cellular PDE activity • Decreased cellular c-di-GMP levels • Chemotaxis transducer BdlA • Heme-associated BdlA may be involved in redox/energy state sensing; BdlA cleavage and activation modulated by c-di-GMP • PDE DipA • DGC GcbA • Sensory protein NicD, DGC activity • Sensory protein NbdA, PDE activity • Release of matrix degrading enzymes • Virulence gene expression • Dispersion phenotype • Transcriptomic analysis |
[27,38,39,44,49-52,79] |
| Sudden step-increase of carbon source concentration | Acinetobacter sp | Continuous-flow slide culture; dispersion response monitored by microcopy | [80] | |
| Sudden step-increase of carbon source concentration | S. pneumoniae | 72 hr biofilms grown on human respiratory epithelial cells (HRECs); 2 hr treatment | [54] | |
| Ammonium chloride | P. aeruginosa | Continuous flow reactors (tube reactors, flow cells); 4-5 d biofilms; monitoring biofilm by CFU, microscopy, and biofilm effluent measurements for up to 60 min, with release of cells from the surface observable within 10 min | • Phosphorylation-dependent signaling (response inhibited with phosphatase inhibitor) • Chemotaxis transducer BdlA • PDE DipA |
[44,51,52] |
| Heavy metals (mercury chloride, sodium arsenate), silver nitrate | P. aeruginosa | Continuous flow reactors (tube reactors, flow cells); 4-5 d biofilms; monitoring biofilm by CFU, microscopy, and biofilm effluent measurements for up to 60 min, with release of cells from the surface observable within 10 min | • Chemotaxis transducer BdlA • PDE DipA • DGC GcbA |
[50-52] |
| Nitric oxide (via SNP) | P. aeruginosa | Batch-grown 24 hr biofilms attached to slides in petri dishes; biofilms exposed to NO for 24 hr; SYTO staining and microscopy | • Increased cellular PDE activity • Decreased cellular c-di-GMP levels • Chemotaxis transducer BdlA • Transcriptomic analysis |
[40] |
| Nitric oxide (via SNP) | P. aeruginosa | Continuous flow reactors; 6 d biofilms; dispersion assessed via effluent measurement and microscopy under flowing conditions | • Sensory protein NbdA, PDE activity • MucR, dual activity (PDE, DGC) • Chemotaxis transducer BdlA • Release of matrix degrading enzymes • Virulence gene expression • Dispersion phenotype |
[27,39] |
| Nitric oxide (via SNP) | Nitrosomonas europaea Ammonia oxidizer | Batch-grown 24 hr biofilms attached to wells of microtiter plate; biofilms treated for additional 24 hr; biomass detected by CV staining | [81] | |
| Cis-2 decenoic acid (fatty acid signaling molecule) |
P. aeruginosa
E. coli K. pneumonia B.subtilis P. mirabilis S. aureus S. pyogenes C.albicans |
Continuous flow reactors and microtiter plate biofilms; 4-7 d biofilms; endogenous and exogenous addition of cis-2 decenoic acid; direct microscopic evaluation of dispersion response under flowing conditions and microtiter plate dispersion assays | Fatty acid synthase DspI | [32] |
| Iron | P. aeruginosa | Batch-grown 48 hr biofilms in microtiter plate; co-treatment for 48 hrs; biomass evaluation by CV staining Continuous flow reactors (tube reactors, flow cells); 4-5 d biofilms; monitoring biofilm by microscopy and biofilm effluent measurements for up to 60 min, with release of cells from the surface observable within 10 min |
[37,82] | |
| Bile salt taurocholate | Vibrio cholerae | Prevents 24 hr biofilm formation in glass tubes when present in growth medium; Causes cell detachment from 24 hr glass tube biofilms within 1-2 hr; CV staining and microscopy | [55] | |
| Factors linked to biofilm detachment (Enzymatic degradation of matrix components) | ||||
| Cellulase | P. putida | 2-3 d biofilms;1hr treatment | Cellulose-like matrix component | [41] |
| Proteinase K | P. putida | Batch-grown 2-3 d pellicle biofilms;1hr treatment | Proteolysis of adhesin LapA | [41] |
| Proteinase K |
S. lugdunensis
S. aureus Listeria monocytogenes |
Batch-grown 24-72 hr biofilms in microtiter plates or petri dish systems; 2-6 hr treatment | Biofilm-associated protein Bap | [19,83,84] |
| Periodate | P. putida | Batch-grown 2-3 d pellicle biofilms; 1hr treatment | Cellulose-like matrix component | [41] |
| Periodate | S. epidermidis | Batch-grown 24 hr biofilms in microtiter plates or petri dish systems; 2 hr treatment | Cellulose-like matrix component | [19] |
| Dispersin B |
A. actinomycetemco mitans
A. pleuropneumoniae S. epidermidis |
Batch grown 24 hr biofilms attached to polystyrene; 2hr treatment Batch grown 18-24 hr biofilms in microtiter plate; 2hr treatment |
linear polymers of N-acetyl-D-glucosamines, matrix component | [17-19] |
| DNase | L. monocytogenes | 72-hr polystyrene peg biofilms; 24 hr treatment | [84] | |
| Alginate lyase | P. aeruginosa | Biofilms grown in petri dish system | [85] | |
| Biosurfactant (viscosin) | P. fluorescens | 1-d flow cell-grown biofilms; 7.5-17.5-hr microtiter biofilms; 9.5 hr treatment | [16] | |
| Biosurfactant (rhamnolipids) | S. epidermidis | 18-hr flow cell biofilms; 1 hr treatment | [16] | |
| Conditions linked to desorption | ||||
| Proteinase K | P. putida | Batch-grown 2-3 d pellicle biofilms; 1hr treatment | Adhesion LapA | [41] |
| Nitric oxide (via SNP) |
Serratia marcescens
V. cholerae E. coli Bacillus licheniformis S. epidermidis Fusobacterium nucleatum Candida albicans |
Cell grown anaerobically in Schaedler Broth to an OD600 of 0.1; SNP added and cells allowed to attach for 4 h on a sterile glass slide; Slides washed and stained with CV; Microscopy for number of cells | [86] | |
| Cis-2 decenoic acid (fatty acid signaling molecule) | P. aeruginosa | Continuous flow reactors; Microscopy under flowing conditions | [32] | |
| Conditions linked to dispersion or biofilm architectural collapse | ||||
| Reduction of c-di-GMP | P. aeruginosa | Overexpression of rcsB impairs formation of biofilms grown in microfermentors for 4 days. Cells attached to 24 well plates after 6 hr, induction of rscB, and dispersion (reduction in CV biomass) apparent within 45 min, with experiment taken out to 18 hr for complete biofilm removal |
• Response regulator RcsB • Induction of PDE PvrR |
[67] |
| Reduction of c-di-GMP | P. aeruginosa | 4d drip flow or capillary flow biofilm reactors; dispersion assessed 3d post flow cessation | [77] | |
| Reduction of c-di-GMP | P. putida | Batch-grown 10-24 hr biofilms, in microtiter plates; biofilm biomass evaluation by CV staining | • PDE YhjH • Adhesion LapA • Protease LapG |
[41] |
| Cell-to-cell signaling molecules, mechanism of escape unclear | ||||
| Quorum sensing: Acylated homoserine lactones (AHLs) |
Yersinia pseudotuberculosis
Rhodobacter sphaeroides |
Aggregation in batch culture | Mutant of LuxR homolog YpsR are hyperaggreagative Inactivation of cerI, encoding a 7,8-cis-N-(tetradecenoyl)homoserine lactone synthase, results in mucoid colony phenotype and hyperaggregation in liquid |
[87] [88] |
| Quorum sensing, Agr | S. aureus | Flow cells; 2-day old biofilms; loss of biofilm biomass monitored 1-2 d post glucose depletion | No biofilm biomass loss noted in agr mutant post 2 days of glucose depletion | [89] |
| DSF | Xanthamonas campestris | Aggregates in batch medium after overnight growth of rpfF mutants; Enzyme causes disaggregation within 30 min; DSF can prevent aggregate formation or cause complete disaggregation within 3 hr Aggregates in batch medium after overnight growth; DSF causes disaggregation within 3 hr |
DSF and Rpf genes positively control synthesis of manA-encoded endo-β-1,4-mannanase which degrades the matrix | [35] |
| AI-2 | V. vulnificus | 12 hr biofilms in microtiter plates; CV staining to quantify attached cells; CFU counts to quantify detached cells | Host cells increase smcR (LuxR homolog) expression; SmcR downregulates genes associated with biofilm formation and up-regulates those associated with detachment including vvpE encoding an elastolytic protease | [71] |
The third mode of cellular release from biofilms, the process of dispersion, is distinct from the passive release of cells occurring during desorption and detachment in that it is characterized by an active phenotypic switch, which allows the bacterial cells to leave a biofilm. Specifically, dispersion involves the sensing of certain signals or cues and their transduction through regulatory networks to enable physiological changes that facilitate cellular release from biofilm communities. Similarly to detachment, dispersion relies on factors to weaken the biofilm matrix. However, in contrast to the exogenous addition of such factors during detachment, dispersion requires the direct production by the biofilm bacteria of matrix-degrading enzymes, such as alginate lyase, the glycosyl hydrolase PslG, or β-N-acetylglucosaminidase [17,27-30]. The source of the triggering signals distinguishes between two classes of dispersion. While native dispersion occurs upon sensing of self-synthesized signaling molecules, environmentally induced dispersion is triggered by factors and changes in the external environment. Native dispersion, which is generally characterized as the terminal stage in biofilm development, has also been referred to as seeding dispersal [31], as it is assumed to lead to the translocation of bacteria to new sites for colonization. Native dispersion has been observed in various biofilm forming species (Table 2) to occur from within microcolonies. Following differentiation of the interior of the microcolony into a motile phenotype and the periphery into a non-motile surrounding ‘wall phenotype’ [31], cells coordinately evacuate the microcolony from local break out points and enter the bulk liquid [2,31,32]. However, dispersion rarely involves the entire biofilm. Typically, only selected microcolonies or areas within a biofilm will undergo a dispersion event at any particular time, in a manner often dependent on microcolony diameter [31]. The evacuation of bacteria from within a microcolony, first described in Davies in 1999 [33] and expanded on by Tolker-Nielsen et al. [34], Sauer et al. [2], and Purevdorj-Gage et al. [31], is illustrated in Figure 1. The factor responsible for native dispersion in Pseudomonas aeruginosa biofilms has been identified as the fatty acid signaling molecule cis-2-decenoic acid (cis-DA) [32]. Similarly, disaggregation of flocs by Xanthomonas campesitris in liquid has been linked to the cis-11-methyl-2-dodecenoic acid (DSF) [35]. Dispersion in response to cis-unsaturated fatty acids is fairly conserved, as cis-DA has been shown to induce the dispersion of Escherichia coli, Klebsiella pneumoniae, Proteus mirabilis, Streptococcus pyogenes, Bacillus subtilis, Staphylococcus aureus, and the yeast Candida albicans biofilms [32], while the Burkholderia cenocepatia cis-2-dodecenoic acid BDSF has been shown to trigger dispersion of Francisella novicida biofilms [36]. In contrast, environmentally induced dispersion occurs following the sensing of external conditions including starvation, oxidative or nitrosative stress, host factors such as bile salts, and availability of oxygen, iron, and carbon sources (Table 1) [37-43]. Induced dispersion coincides with 80% or more of the biofilm biomass being removed [44], with the evacuating bacteria having been ascribed phenotypes similar to those derived from native dispersion events [31,45,46]. These findings have suggested that native and environmentally induced dispersion are subjects to overlapping mechanisms that are likely conserved across kingdoms and domains [26,32].
Table 2.
Dispersion events confirmed by direct microscopic examination, with dispersion being part of biofilm developmental life cycle.
| Species | Growth conditions and observations | Source |
|---|---|---|
| Pseudomonas aeruginosa | Biofilms grown under flowing conditions, dispersion and hollowing noted in > 6 d biofilms | [2,15,31,91] |
| Candida albicans | Biofilms grown under flowing conditions for 24 hr; Dispersion of biomass noted by cell enumeration and hemocytometer. | [92] |
| Neisseria subflava Actinobacillus actinomycetemcomitans Haemophilus aphrophilus Streptococcus mitis oral Streptococcus sp. | Batch polystyrene petri dish surface; 24hr growth; cell release apparent within 8hr | [93] |
| P. fluorescens | Cells attached on glass slides, grown in Batch culture system; Dispersion/loss of biomass noted within 40 hrs; mediated by an EPS lyase | [94] |
All biofilm escapes are not created equal
Considering these diverse findings, the question arises of whether the manner by which bacterial cells escape from the biofilm matters. The answer is yes, as the way bacterial cells escape from the biofilm affects the physiology and phenotype of the respective cells. In contrast to detachment or desorption, which involve the passive release of cells from a biofilm structure, native and induced dispersion involves the active release of bacteria from a biofilm as a physiologically regulated response to internal or external stimuli, with dispersing cells experiencing a change in their behavior. While cells freed via detachment or desorption may ultimately undergo a phenotypic change as a result of their release from the biofilm environment, signal transduction and phenotypic changes are required for cells to be released from a biofilm via dispersion. This is supported by the findings that dispersed cells differ significantly from biofilm cells with respect to gene expression, protein production, and posttranslational modification [2,9,44,47-49]. Compared to biofilm cells, dispersed cells are furthermore characterized by decreased levels of the intracellular signaling molecule c-di-GMP and increased production of matrix degrading enzymes [27,38-40,50,51]. While dispersion has often been described to coincide with increased motility of the escaping bacteria, several studies demonstrated that motility is not required for dispersion [42,44,52,53], suggesting that motility may be an indicator of dispersed cells returning to the planktonic mode of growth, but is not part of the mechanism enabling dispersion. Findings such as these have suggested that the process of dispersion may be a simple reversal of the motile-sessile transition and the switch of biofilm cells to planktonic cells. However, dispersed cells are distinct from planktonic cells with respect to gene expression, release of matrix-degrading enzymes, and pathogenicity [27,29,54,55], with dispersed cells being highly virulent against macrophages and Caenorhabditis elegans compared to planktonic cells [29] and contributing to the severity of both acute and chronic lung infections [27]. Moreover, gene expression and protein production profiles of dispersed cells are also markedly different form those of the biofilm cells from which they are derived [2,27,29,44,54]. In contrast, very little to no difference has been noted between intact and detached biofilm cells. Fux et al. [46] demonstrated cell aggregates detached from S. aureus biofilms to be highly tolerant to the antibiotic oxacillin compared to exponential-phase planktonic cultures. The same study, however, demonstrated that cells dispersed from S. aureus biofilms due to native dispersion events were as susceptible as exponential-phase planktonic cultures to oxacillin [46]. In a manner similar to native dispersion, induced dispersion has been linked to released cells being rendered significantly more susceptible to antimicrobial agents compared to biofilm cells [44,45,52]. Synergistic activity was noted upon exposure of P. aeruginosa biofilms to citrate with amikacin disulphate, colistin methanesulphonate or erythromycin, succinic acid with colistin methanesulphonate [45], and various carbon sources with hydrogen peroxide [44], but not nitric oxide with colistin[56].
A tale of c-di-GMP modulation
While desorption and detachment are dependent on exogenous cellular release factors and may lead to eventual phenotypic switches, physiological changes in biofilm cells and production of the release factors by biofilm cells appear to be a prerequisite for dispersion to occur. But what are the regulatory events underlying and driving these phenotypic transitions? The dispersion response, induced upon sensing cues such as cis-unsaturated fatty acids, NO, or changes in carbon availability via cue-specific membrane-bound sensory proteins such as RfpCG, NicD, and NbdA [35,38,39,57], requires specific regulatory events including phosphotransfers and coincides with coordinated changes in protein phosphorylation and gene expression [40,44,49].
One of the most noticeable regulatory consequences of induction of dispersion is the modulation of the intracellular signaling molecule cyclic di-GMP (c-di-GMP), high levels of which promote sessile growth, while low levels correlate with planktonic existence [58]. Levels of c-di-GMP are enzymatically modulated by diguanylate cyclases (DGCs), proteins containing a GGDEF domain, and phosphodiesterases (PDEs) harboring either an EAL or HD-GYP domain. In P. aeruginosa, dispersion upon exposure to NO and elevated nutrient concentrations has been linked to the reduction of the cellular c-di-GMP levels, requiring the PDEs DipA, NbdA, and RbdA [40,51,52,59]. The finding of dispersed cells being characterized by reduced c-di-GMP levels has led to the hypothesis that dispersed cells can be generated by reducing the intracellular c-di-GMP content through modulation of PDEs [60]. In P. putida and P. fluorescens, the large adhesive outer-membrane protein LapA mediates attachment to surfaces and matrix components and is required for the transition from reversible to irreversible attachment [41,61-63]. Elevated c-di-GMP levels contribute to the localization of LapA to the cell surface, while low c-di-GMP levels, as in the case of phosphate limitation, result in LapA being released from the outer membrane via cleavage by the periplasmic cysteine protease LapG and consequently in impaired attachment [62]. It is thus not surprising that lowering the intracellular c-di-GMP level via induction of yhjH, an E. coli-derived PDE, led to the disaggregation of P. putida pellicles. Considering that no disaggregation was noted in ΔlapG mutant pellicles, the finding suggested that LapG exerts its activity on LapA in response to a decrease in the intracellular c-di-GMP level [41]. The adhesin controlled by LapG in P. aeruginosa, a strain lacking LapA, has recently been demonstrated to be CdrA [63,64]. Disaggregation of P. putida pellicles was furthermore accomplished upon treatment with proteinase K, which the authors attributed to the proteolytic degradation of LapA [41]. While both disaggregation events coincide with LapA being released from the cell surface and thus, cells being released from the substratum, the mechanisms by which this is achieved likely differ. While the phenotype of the escaped bacterial populations was not investigated, it is likely that protease treatment induces detachment or sloughing rather than dispersion events, while modulation of the c-di-GMP level may induce dispersion. However, it is likely that reduced c-di-GMP levels coincide with escaped cells being rendered susceptible to antimicrobial agents. This is supported by the finding that in P. aeruginosa, reducing cellular c-di-GMP levels of biofilm cells from >75 to ≤40 pmol/mg correlated with increased susceptibility to antimicrobial agents [65]. Regardless of the mechanism, the contribution of LapA to the escape from the biofilm raises the question of whether conditions that prevent biofilm formation can induce dispersion. In P. fluorescens, phosphate limitation functions as an inhibitor of biofilm formation. This is likely due to reduced c-di-GMP levels, as activation of the Pho regulon, the major pathway for adaptation to phosphate limitation, results in expression of the PDE RapA, with rapA overexpression inhibiting biofilm formation in a manner similar to Pi limitation [62,66]. Interestingly, the same conditions that prevent P. fluorescens biofilm formation also inhibit LapA secretion [62]. These conditions are in stark contrast to the rapid changes in the carbon concentration that trigger dispersion but are unlikely to prevent biofilm formation and development (Table 1).
Similar to Gjermansen et al. [41], Mikkelsen et al. [67] linked modulation of c-di-GMP to the disaggregation of P. aeruginosa biofilms, a phenotype noted upon overexpression of the response regulator RcsB. RcsB has previously been described to activate the expression of genes involved in the Chaperone Usher Pathway (Cup), which leads to the production of CupD fimbriae and increased attachment [68]. The authors linked the dispersal phenotype upon overexpression of RcsB to the increased expression of pvrR encoding a PDE, with PvrR having previously been demonstrated to counteract that activity of RcsB on cupD gene expression [67,68]. The findings suggest that the reversion of mechanisms and factors involved in attachment contribute to dispersion. However, the effect of c-di-GMP modulation on attachment factors can also be seen in a context different from that of biofilm development.
While most developmental processes are governed by hierarchically organized genetic networks that establish checkpoints in the developmental process, which once passed, commit cells to a specific fate [3], there is an increasing body of literature indicating that the formation of biofilms is reversible. In P. aeruginosa, at least four two-component regulatory systems, namely SagS, BfiRS, BfmRS, and MifRS, are required to coordinate the progression of P. aeruginosa biofilm development in a stage-specific manner. Together, these systems form a coordinated signaling network that regulates three committed steps of the P. aeruginosa biofilm life cycle, in particular the transition to three later biofilm developmental stages following initial attachment [8]. Despite the presence of this coordinated signaling network, loss of expression of the respective regulatory systems coincided with biofilm architecture collapse, with the remaining biofilm cells demonstrating phenotypic characteristics of earlier biofilm stages [8,69]. Such findings suggest the likelihood of a 4th mode of escape from the biofilm lifestyle, that of collapse. However, additional studies will be required to determine the manner by which cells escape during biofilm architectural collapse and whether modulation of c-di-GMP levels contribute to collapse or dispersion of biofilms.
Conclusions
Various modes of bacterial escape from biofilm communities differ in the means of cell release, as well as in the distinct phenotypic manifestations and properties of the released cells. Thus, it is essential to consider the conditions and characteristics of the respective processes in both basic research and transitional applications targeting biofilms. It is now becoming clear that biofilm disruption approaches have to consider interspecies and interkingdom signaling in polymicrobial biofilms [36,54,70], as well as the effect of the host environment on bacterial biofilms [54,55,71]. Eliciting the release of cells from a biofilm represents a promising strategy for controlling biofilms in industrial and medical settings. Yet, it is important to consider the manner by which cells are released, as this will determine their physiology, particularly with respect to differences in susceptibility to antimicrobial agents and their pathogenicity. For instance, while detachment agents have proven beneficial in eliminating pathogenic biofilms from the oral cavity or from medical equipment [72], induction of detachment and release of large cellular aggregates may be undesirable in the treatment of chronic infections as it may lead to complications such as emboli [46]. Similarly, bacterial dissemination from the peritoneal cavity of mice to the spleen was noted upon overproduction of the PDE YhjH [73]. In order to promote such transitional research and proper practical applications for biofilm control, there is an increasing need from the basic research side to integrate the findings from the various biofilm escape studies to map complete dispersion signaling networks and gain a better understanding of the phenotypic switches associated with the release of biofilm cells. While dispersion pathways have begun to be mapped for such species as P. aeruginosa, P. putida, P. fluorescens, and V. vulnificus [38,39,41,49,71], the bridging of the dispersion-inducing cues with the detachment factors via specific regulatory networks has remained limited and represents an important focus of the biofilm escape research.
Supplementary Material
Highlights.
At least three types of “escapes” or ways for bacteria to leave the biofilm are known; desorption, detachment, and dispersion
Dispersion is a highly regulated, active process, coinciding with a physiological change in the dispersing population
Dispersed cells are characterized by altered susceptibility to antimicrobial agents and virulence compared to planktonic and biofilm cells
The degree of detachment from a biofilm will be impacted by shear forces and modification of the biofilm matrix
Collapse of the biofilm architecture may be linked to reversion of biofilm developmental regulatory networks
Acknowledgement
This work was supported by grants from the National Institutes of Health (R01AI080710, RO1A10752570) and F. Hoffmann-La Roche Ltd.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu. Rev. Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 2.Sauer K, Camper AK, Ehrlich GD, Costerton JW, Davies DG. Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 2002;184:1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Monds RD, O'Toole GA. The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 2009;17:73–87. doi: 10.1016/j.tim.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Jensen PØ, Givskov M, Bjarnsholt T, Moser C. The immune system vs. Pseudomonas aeruginosa biofilms. FEMS Immunology & Medical Microbiology. 2010;59:292–305. doi: 10.1111/j.1574-695X.2010.00706.x. [DOI] [PubMed] [Google Scholar]
- 5.Høiby N, Bjarnsholt T, Givskov M, Molin S, Ciofu O. Antibiotic resistance of bacterial biofilms. International journal of antimicrobial agents. 2010;35:322–332. doi: 10.1016/j.ijantimicag.2009.12.011. [DOI] [PubMed] [Google Scholar]
- 6.Van Acker H, Van Dijck P, Coenye T. Molecular mechanisms of antimicrobial tolerance and resistance in bacterial and fungal biofilms. Trends in Microbiology. 2014;22:326–333. doi: 10.1016/j.tim.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Costerton J. Introduction to biofilm. Int J Antimicrob Agents. 1999;11:217–221. doi: 10.1016/s0924-8579(99)00018-7. [DOI] [PubMed] [Google Scholar]
- 8.Petrova OE, Sauer K. A novel signaling network essential for regulating Pseudomonas aeruginosa biofilm development. PLoS Pathog. 2009;5:e1000668. doi: 10.1371/journal.ppat.1000668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stoodley P, Sauer K, Davies DG, Costerton JW. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 2002;56:187–209. doi: 10.1146/annurev.micro.56.012302.160705. [DOI] [PubMed] [Google Scholar]
- 10.Davies DG. Biofilm Highlights. 1-28. Springer; Berlin: 2011. Biofilm Dispersion. [Google Scholar]
- 11.van Loosdrecht MCM, Picioreanu C, Heijnen JJ. A more unifying hypothesis for the structure of microbial biofilms. FEMS Microbial Ecology. 1997;24:181–183. [Google Scholar]
- 12.Breyers JD. Modeling biofilm accumulation. In: Bazin MJ, Prosser JI, editors. Physiology Models in Microbiology. Vol. 2. Boca Raton, FL: 1988. pp. 109–144. [Google Scholar]
- 13.Stewart PS. A model of biofilm detachment. Biotechnology and Bioengineering. 1993;41:111–117. doi: 10.1002/bit.260410115. [DOI] [PubMed] [Google Scholar]
- 14.Banin E, Brady KM, Greenberg EP. Chelator-induced dispersal and killing of Pseudomonas aeruginosa cells in a biofilm. Applied and Environmental Microbiology. 2006;72:2064–2069. doi: 10.1128/AEM.72.3.2064-2069.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boles BR, Thoendel M, Singh PK. Rhamnolipids mediate detachment of Pseudomonas aeruginosa from biofilms. Mol. Microbiol. 2005;57:1210–1223. doi: 10.1111/j.1365-2958.2005.04743.x. [DOI] [PubMed] [Google Scholar]
- 16.Bonnichsen L, Svenningsen NB, Rybtke M, de Bruijn I, Raaijmakers JM, Tolker-Nielsen T, Nybroe O. The lipopeptide biosurfactant viscosin enhances dispersal of Pseudomonas fluorescens SBW25 biofilms. Microbiology. 2015 doi: 10.1099/mic.0.000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan JB, Ragunath C, Ramasubbu N, Fine DH. Detachment of Actinobacillus actinomycetemcomitans biofilm cells by an endogenous β-hexosaminidase activity. Journal of Bacteriology. 2003;185:4693–4698. doi: 10.1128/JB.185.16.4693-4698.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaplan JB, Ragunath C, Velliyagounder K, Fine DH, Ramasubbu N. Enzymatic detachment of Staphylococcus epidermidis biofilms. Antimicrob. Agents Chemother. 2004;48:2633–2636. doi: 10.1128/AAC.48.7.2633-2636.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chaignon P, Sadovskaya I, Ragunah C, Ramasubbu N, Kaplan JB, Jabbouri S. Susceptibility of staphylococcal biofilms to enzymatic treatments depends on their chemical composition. Applied Microbiology and Biotechnology. 2007;75:125–132. doi: 10.1007/s00253-006-0790-y. [DOI] [PubMed] [Google Scholar]
- 20.Shah P, Bush A, Canny G, Colin A, Fuchs H, Geddes D, Johnson C, Light M, Scott S, Tullis D. Recombinant human DNase I in cystic fibrosis patients with severe pulmonary disease: a short-term, double-blind study followed by six months open-label treatment. European Respiratory Journal. 1995;8:954–958. [PubMed] [Google Scholar]
- 21.Fuchs HJ, Borowitz DS, Christiansen DH, Morris EM, Nash ML, Ramsey BW, Rosenstein BJ, Smith AL, Wohl ME. Effect of aerosolized recombinant human DNase on exacerbations of respiratory symptoms and on pulmonary function in patients with cystic fibrosis. New England Journal of Medicine. 1994;331:637–642. doi: 10.1056/NEJM199409083311003. [DOI] [PubMed] [Google Scholar]
- 22.Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- 23.Nijland R, Hall MJ, Burgess JG. Dispersal of biofilms by secreted, matrix degrading, bacterial DNase. PLoS ONE. 2010;5:e15668. doi: 10.1371/journal.pone.0015668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Boles BR, Horswill AR. Staphylococcal biofilm disassembly. Trends in Microbiology. 2011;19:449–455. doi: 10.1016/j.tim.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gödeke J, Heun M, Bubendorfer S, Paul K, Thormann KM. Roles of two Shewanella oneidensis MR-1 extracellular endonucleases. Applied and Environmental Microbiology. 2011;77:5342–5351. doi: 10.1128/AEM.00643-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat. Rev. Micro. 2012;10:39–50. doi: 10.1038/nrmicro2695. [DOI] [PubMed] [Google Scholar]
- 27*.Li Y, Petrova OE, Su S, Lau GW, Panmanee W, Na R, Hassett DJ, Davies DG, Sauer K. BdlA, DipA and induced dispersion contribute to acute virulence and chronic persistence of Pseudomonas aeruginosa. PLoS Pathog. 2014;10:e1004168. doi: 10.1371/journal.ppat.1004168. [This work demonstrated that dispersed cells have a phenotype that is distinct from biofilm and planktonic cells with respect to gene expression, release of matrix-degrading enzymes, and pathogenicity. It also suggested that environmentally induced dispersion plays a role in the progression of both chronic and acute infections and potentially the switch between the two.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaplan JB. Biofilm dispersal: mechanisms, clinical Implications, and potential therapeutic uses. J. Dent. Res. 2010;89:205–218. doi: 10.1177/0022034509359403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29*.Chua SL, Liu Y, Yam JKH, Chen Y, Vejborg RM, Tan BGC, Kjelleberg S, Tolker-Nielsen T, Givskov M, Yang L. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat Commun. 2014;5 doi: 10.1038/ncomms5462. [This study demonstrated that the transcriptome of dispersed cells is distinct from that of biofilm and planktonic cells. It also provided evidence that dispersed cells exhibit enhanced virulence against macrophages and Caenorhabditis elegans relative to planktonic cells.] [DOI] [PubMed] [Google Scholar]
- 30*.Yu S, Su T, Wu H, Liu S, Wang D, Zhao T, Jin Z, Du W, Zhu M-J, Chua SL, et al. PslG, a self-produced glycosyl hydrolase, triggers biofilm disassembly by disrupting exopolysaccharide matrix. Cell Res. 2015;25:1352–1367. doi: 10.1038/cr.2015.129. [This study provided the first evidence that the P. aeruginosa Psl exopolysaccharide biofilm matrix can be disrupted by the self-synthesized glycosyl hydrolase PslG. The work also demonstrated that biofilm susceptibility to antibiotic agents and macrophages could be increased as a result of PslG treatment.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purevdorj-Gage B, Costerton WJ, Stoodley P. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology. 2005;151:1569–1576. doi: 10.1099/mic.0.27536-0. [DOI] [PubMed] [Google Scholar]
- 32.Davies DG, Marques CNH. A fatty acid messenger is responsible for inducing dispersion in microbial biofilms. J. Bacteriol. 2009;191:1393–1403. doi: 10.1128/JB.01214-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davies DG. Regulation of matrix polymer in biofilm formation and dispersion. In: Wingender J, Neu TR, Flemming H-C, editors. Microbial extrapolymeric substances, characterization, structure and function. 93-112. Springer-Verlag; Berlin: 1999. [Google Scholar]
- 34.Tolker-Nielsen T, Brinch UC, Ragas PC, Andersen JB, Jacobsen CS, Molin S. Development and dynamics of Pseudomonas sp. biofilms. J Bacteriol. 2000;182:6482–6489. doi: 10.1128/jb.182.22.6482-6489.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dow JM, Crossman L, Findlay K, He Y-Q, Feng J-X, Tang J-L. Biofilm dispersal in Xanthomonas campestris is controlled by cell-cell signaling and is required for full virulence to plants. Proc.Natl. Acad. Sci. 2003;100:10995–11000. doi: 10.1073/pnas.1833360100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dean SN, Chung M-C, Hoek ML. Burkholderia diffusible signal factor signals to Francisella novicida to disperse biofilm and increase siderophore production. Applied and Environmental Microbiology. 2015;81:7057–7066. doi: 10.1128/AEM.02165-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37*.Lanter BB, Sauer K, Davies DG. Bacteria present in carotid arterial plaques are found as biofilm deposits which may contribute to enhanced risk of plaque rupture. mBio. 2014;5:01206–01214. doi: 10.1128/mBio.01206-14. [This work demonstrated that biofilms of P. acnes, which can colonize the carotid arteries of patients with atherosclerosis, undergo dispersion in respose to physiologically relevant levels of norepinephrine in the presence of iron-bound transferrin or with free iron, with the dispersion event correlating with an increase in degradative enzyme levels.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38*.Basu Roy A, Sauer K. Diguanylate cyclase NicD-based signalling mechanism of nutrient-induced dispersion by Pseudomonas aeruginosa. Mol. Microbiol. 2014;94:771–793. doi: 10.1111/mmi.12802. [The results of this study established an outside-in signalling mechanism model for the process of glutamate-induced dispersion, in which the seven transmembrane receptor (7TMR) diguanylate cyclase NicD senses the disperion-inducing cues and regulates c-di-GMP levels to activate the dispersion regulatory network consisting of BdlA and the PDE DipA.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39*.Li Y, Heine S, Entian M, Sauer K, Frankenberg-Dinkel N. NO-induced biofilm dispersion in Pseudomonas aeruginosa is mediated by a MHYT-domain coupled phosphodiesterase. J. Bacteriol. 2013;195:3531–3542. doi: 10.1128/JB.01156-12. [This work provided the first evidence of a PDE, the membrane-bound MHYT-containing NbdA, that involved in regulating c-di-GMP levels specifically during dispersion in response to NO.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barraud N, Schleheck D, Klebensberger J, Webb JS, Hassett DJ, Rice SA, Kjelleberg S. Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J. Bacteriol. 2009;191:7333–7342. doi: 10.1128/JB.00975-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gjermansen M, Nilsson M, Yang L, Tolker-Nielsen T. Characterization of starvation-induced dispersion in Pseudomonas putida biofilms: genetic elements and molecular mechanisms. Mol. Microbiol. 2010;75:815–826. doi: 10.1111/j.1365-2958.2009.06793.x. [DOI] [PubMed] [Google Scholar]
- 42.Thormann KM, Saville RM, Shukla S, Spormann AM. Induction of rapid detachment in Shewanella oneidensis MR-1 biofilms. J. Bacteriol. 2005;187:1014–1021. doi: 10.1128/JB.187.3.1014-1021.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Applegate DH, Bryers JD. Effects on carbon and oxygen limitations and calcium concentrations on biofilm removal processes Biotechnol. Bioeng. 1991;37:17–25. doi: 10.1002/bit.260370105. [DOI] [PubMed] [Google Scholar]
- 44.Sauer K, Cullen MC, Rickard AH, Zeef LAH, Davies DG, Gilbert P. Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 2004;186:7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ross SS, Fiegel J. Nutrient dispersion enhances conventional antibiotic activity against Pseudomonas aeruginosa biofilms. International journal of antimicrobial agents. 2012;40:177–181. doi: 10.1016/j.ijantimicag.2012.04.015. [DOI] [PubMed] [Google Scholar]
- 46.Fux CA, Wilson S, Stoodley P. Detachment characteristics and oxacillin resistance of Staphyloccocus aureus biofilm emboli in an in vitro catheter infection model. J. Bacteriol. 2004;186:4486–4491. doi: 10.1128/JB.186.14.4486-4491.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hindré T, Brüggemann H, Buchrieser C, Héchard Y. Transcriptional profiling of Legionella pneumophila biofilm cells and the influence of iron on biofilm formation. Microbiology. 2008;154:30–41. doi: 10.1099/mic.0.2007/008698-0. [DOI] [PubMed] [Google Scholar]
- 48.Barraud N, Hassett DJ, Hwang S-H, Rice SA, Kjelleberg S, Webb JS. Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 2006;188:7344–7353. doi: 10.1128/JB.00779-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Petrova OE, Sauer K. Dispersion by Pseudomonas aeruginosa requires an unusual posttranslational modification of BdlA. Proc. National Acad. Sci. 2012;109:16690–16695. doi: 10.1073/pnas.1207832109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Petrova OE, Cherny KE, Sauer K. The diguanylate cyclase GcbA facilitates Pseudomonas aeruginosa biofilm dispersion by activating BdlA. Journal of Bacteriology. 2015;197.1:174–187. doi: 10.1128/JB.02244-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Basu Roy A, Petrova OE, Sauer K. The phosphodiesterase DipA (PA5017) is essential for Pseudomonas aeruginosa biofilm dispersion. J. Bacteriol. 2012;194:2904–2915. doi: 10.1128/JB.05346-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morgan R, Kohn S, Hwang S-H, Hassett DJ, Sauer K. BdlA, a chemotaxis regulator essential for biofilm dispersion in Pseudomonas aeruginosa. J. Bacteriol. 2006;188:7335–7343. doi: 10.1128/JB.00599-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gjermansen M, Ragas P, Sternberg C, Molin S, Tolker-Nielsen T. Characterization of starvation- induced dispersion in Pseudomonas putida biofilms. Environ. Microbiol. 2005;7:894–904. doi: 10.1111/j.1462-2920.2005.00775.x. [DOI] [PubMed] [Google Scholar]
- 54.Marks LR, Davidson BA, Knight PR, Hakansson AP. Interkingdom signaling induces Streptococcus pneumoniae biofilm dispersion and transition from asymptomatic colonization to disease. mBio. 2013;4:e00438–00413. doi: 10.1128/mBio.00438-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55*.Hay AJ, Zhu J. Host intestinal signal-promoted biofilm dispersal induces Vibrio cholerae colonization. Infection and Immunity. 2015;83:317–323. doi: 10.1128/IAI.02617-14. [This work demonstrated that the dispersion of Vibrio cholerae biofilms upon entry into the host is likely mediated by bile salt taurocholate, which also contributes to the activation of virulece post-dispersion.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56*.Chua SL, Tan SY-Y, Rybtke MT, Chen Y, Rice SA, Kjelleberg S, Tolker-Nielsen T, Yang L, Givskov M. Bis-(3′-5′)-cyclic dimeric GMP regulates antimicrobial peptide resistance in Pseudomonas aeruginosa. Antimicrobial Agents and Chemotherapy. 2013;57:2066–2075.. doi: 10.1128/AAC.02499-12. [This work proposed utilizing the manipulation of c-di-GMP levels in P. aeruginosa, via inducible expression of the E. coli PDE-encoding gene yhjH, as an approach to model biofilm disperison in vitro and in vivo.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ryan RP, McCarthy Y, Andrade M, Farah CS, Armitage JP, Dow JM. Cell–cell signal-dependent dynamic interactions between HD-GYP and GGDEF domain proteins mediate virulence in Xanthomonas campestris. Proc. National Acad. Sci. 2010;107:5989–5994. doi: 10.1073/pnas.0912839107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Boyd CD, O'Toole GA. Second messenger regulation of biofilm formation: breakthroughs in understanding c-di-GMP effector systems. Annual review of cell and developmental biology. 2012;28:439–462. doi: 10.1146/annurev-cellbio-101011-155705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.An S, Wu Je, Zhang L-H. Modulation of Pseudomonas aeruginosa biofilm dispersal by a cyclic-di- GMP phosphodiesterase with a putative hypoxia-sensing fomain. Appl. Environ. Microbiol. 2010;76:8160–8173. doi: 10.1128/AEM.01233-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chua SL, Hultqvist LD, Yuan M, Rybtke M, Nielsen TE, Givskov M, Tolker-Nielsen T, Yang L. In vitro and in vivo generation and characterization of Pseudomonas aeruginosa biofilm-dispersed cells via c-di-GMP manipulation. Nat. Protocols. 2015;10:1165–1180. doi: 10.1038/nprot.2015.067. [DOI] [PubMed] [Google Scholar]
- 61.Hinsa SM, Espinosa-Urgel M, Ramos JL, O'Toole GA. Transition from reversible to irreversible attachment during biofilm formation by Pseudomonas fluorescens WCS365 requires an ABC transporter and a large secreted protein. Mol. Microbiol. 2003;49:905–918. doi: 10.1046/j.1365-2958.2003.03615.x. [DOI] [PubMed] [Google Scholar]
- 62.Monds RD, Newell PD, Gross RH, O'Toole GA. Phosphate-dependent modulation of c-di-GMP levels regulates Pseudomonas fluorescens Pf0-1 biofilm formation by controlling secretion of the adhesin LapA. Mol. Microbiol. 2007;63:656–679. doi: 10.1111/j.1365-2958.2006.05539.x. [DOI] [PubMed] [Google Scholar]
- 63.Rybtke M, Berthelsen J, Yang L, Høiby N, Givskov M, Tolker-Nielsen T. The LapG protein plays a role in Pseudomonas aeruginosa biofilm formation by controlling the presence of the CdrA adhesin on the cell surface. MicrobiologyOpen. 2015 doi: 10.1002/mbo3.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Borlee BR, Goldman AD, Murakami K, Samudrala R, Wozniak DJ, Parsek MR. Pseudomonas aeruginosa uses a cyclic-di-GMP-regulated adhesin to reinforce the biofilm extracellular matrix. Molecular Microbiology. 2010;75:827–842. doi: 10.1111/j.1365-2958.2009.06991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gupta K, Liao J, Petrova OE, Cherny KE, Sauer K. Elevated levels of the second messenger c-di-GMP contribute to antimicrobial resistance of Pseudomonas aeruginosa. Mol. Microbiol. 2014;92:488–506. doi: 10.1111/mmi.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Monds RD, Silby MW, Mahanty HK. Expression of the Pho regulon negatively regulates biofilm formation by Pseudomonas aureofaciens PA147-2. Mol. Microbiol. 2001;42:415–426. doi: 10.1046/j.1365-2958.2001.02641.x. [DOI] [PubMed] [Google Scholar]
- 67.Mikkelsen H, Hui K, Barraud N, Filloux A. The pathogenicity island encoded PvrSR/RcsCB regulatory network controls biofilm formation and dispersal in Pseudomonas aeruginosa PA14. Molecular Microbiology. 2013;89:450–463. doi: 10.1111/mmi.12287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68*.Mikkelsen H, Ball G, Giraud C, Filloux A. Expression of Pseudomonas aeruginosa cupD fimbrial genes is antagonistically controlled by RcsB and the EAL-containing PvrR response regulators. PLoS ONE. 2009;4:e6018. doi: 10.1371/journal.pone.0006018. [This work demonstrated that the PvrSR/RcsCB regulatory network that modulates biofilm formation via regulation of cupD genes can also contribute to biofilm dispersal via the action of the RcsB- regulated PDE PvrR.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Petrova OE, Sauer K. SagS contributes to the motile-sessile switch and acts in concert with BfiSR to enable Pseudomonas aeruginosa biofilm formation. J. Bacteriol. 2011;193:6614–6628. doi: 10.1128/JB.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Stacy A, Everett J, Jorth P, Trivedi U, Rumbaugh KP, Whiteley M. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proceedings of the National Academy of Sciences. 2014;111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71*.Kim SM, Park JH, Lee HS, Kim WB, Ryu JM, Han HJ, Choi SH. LuxR homologue SmcR is essential for Vibrio vulnificus pathogenesis and biofilm detachment, and its expression is induced by host cells. Infection and Immunity. 2013;81:3721–3730. doi: 10.1128/IAI.00561-13. [This work provided a clinically relevant model of V. vulnificus biofilm dispersion, in which exposure of biofilms to host cells activates quorum sensing signaling via SmcR, leading to biofilm dispersion via up-regulation of vvpE, which encodes an elastolytic protease capable of disaggregating established V. vulnificus biofilms.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marion K, Freney J, James G, Bergeron E, Renaud F, Costerton J. Using an efficient biofilm detaching agent: an essential step for the improvement of endoscope reprocessing protocols. Journal of Hospital Infection. 2006;64:136–142. doi: 10.1016/j.jhin.2006.06.011. [DOI] [PubMed] [Google Scholar]
- 73.Christensen LD, van Gennip M, Rybtke MT, Wu H, Chiang W-C, Alhede M, Høiby N, Nielsen TE, Givskov M, Tolker-Nielsen T. Clearance of Pseudomonas aeruginosa foreign-body biofilm infections through reduction of the cyclic di-GMP level in the bacteria. Infect. Immun. 2013;81:2705–2713. doi: 10.1128/IAI.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thormann KM, Duttler S, Saville RM, Hyodo M, Shukla S, Hayakawa Y, Spormann AM. Control of formation and cellular detachment from Shewanella oneidensis MR-1 biofilms by cyclic di-GMP. J. Bacteriol. 2006;188:2681–2691. doi: 10.1128/JB.188.7.2681-2691.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Delaquis PJ, Caldwell DE, Lawrence JR, McCurdy AR. Detachment of Pseudomonas fluorescens from biofilms on glass surfaces in response to nutrient stress. Microbial ecology. 1989;18:199–210. doi: 10.1007/BF02075808. [DOI] [PubMed] [Google Scholar]
- 76.Delille A, Quiles F, Humbert F. In situ monitoring of the nascent Pseudomonas fluorescens biofilm response to variations in the dissolved organic carbon level in low-nutrient water by attenuated total reflectance-Fourier transform infrared spectroscopy. Applied and Environmental Microbiology. 2007;73:5782–5788. doi: 10.1128/AEM.00838-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schleheck D, Barraud N, Klebensberger J, Webb JS, McDougald D, Rice SA, Kjelleberg S. Pseudomonas aeruginosa PAO1 preferentially grows as aggregates in liquid batch cultures and disperses upon starvation. 2009 doi: 10.1371/journal.pone.0005513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hunt SM, Werner EM, Huang B, Hamilton MA, Stewart PS. Hypothesis for the role of nutrient starvation in biofilm detachment. Applied and Environmental Microbiology. 2004;70:7418–7425. doi: 10.1128/AEM.70.12.7418-7425.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Petrova OE, Sauer K. PAS domain residues and prosthetic group involved in BdlA-dependent dispersion response by Pseudomonas aeruginosa biofilms. J. Bacteriol. 2012;194:5817–5828. doi: 10.1128/JB.00780-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.James GA, Korber DR, Caldwell DE, Costerton JW. Digital image analysis of growth and starvation responses of a surface-colonizing Acinetobacter sp. J. Bacteriol. 1995;177:907–915. doi: 10.1128/jb.177.4.907-915.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Schmidt I, Steenbakkers PJ, op den Camp HJ, Schmidt K, Jetten MS. Physiologic and proteomic evidence for a role of nitric oxide in biofilm formation by Nitrosomonas europaea and other ammonia oxidizers. Journal of Bacteriology. 2004;186:2781–2788. doi: 10.1128/JB.186.9.2781-2788.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Musk DJ, Banko DA, Hergenrother PJ. Iron salts perturb biofilm formation and disrupt existing biofilms of Pseudomonas aeruginosa. Chemistry & biology. 2005;12:789–796. doi: 10.1016/j.chembiol.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 83.Kumar Shukla S, Rao TS. Dispersal of Bap-mediated Staphylococcus aureus biofilm by proteinase K. J Antibiot. 2012 doi: 10.1038/ja.2012.98. [DOI] [PubMed] [Google Scholar]
- 84.Nguyen UT, Burrows LL. DNase I and proteinase K impair Listeria monocytogenes biofilm formation and induce dispersal of pre-existing biofilms. International Journal of Food Microbiology. 2014;187:26–32. doi: 10.1016/j.ijfoodmicro.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 85.Boyd A, Chakrabarty Aá. Role of alginate lyase in cell detachment of Pseudomonas aeruginosa. Applied and Environmental Microbiology. 1994;60:2355–2359. doi: 10.1128/aem.60.7.2355-2359.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barraud N, Storey MV, Moore ZP, Webb JS, Rice SA, Kjelleberg S. Nitric oxide-mediated dispersal in single-and multi-species biofilms of clinically and industrially relevant microorganisms. Microbial biotechnology. 2009;2:370–378. doi: 10.1111/j.1751-7915.2009.00098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Atkinson S, Throup JP, Stewart GS, Williams P. A hierarchical quorum-sensing system in Yersinia pseudotuberculosis is involved in the regulation of motility and clumping. Molecular Microbiology. 1999;33:1267–1277. doi: 10.1046/j.1365-2958.1999.01578.x. [DOI] [PubMed] [Google Scholar]
- 88.Puskas A, Greenberg Eá, Kaplan S, Schaefer Aá. A quorum-sensing system in the free-living photosynthetic bacterium Rhodobacter sphaeroides. Journal of Bacteriology. 1997;179:7530–7537. doi: 10.1128/jb.179.23.7530-7537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Boles BR, Horswill AR. agr mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathogens. 2008;4:e1000052. doi: 10.1371/journal.ppat.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tao F, Swarup S, Zhang LH. Quorum sensing modulation of a putative glycosyltransferase gene cluster essential for Xanthomonas campestris biofilm formation. Environmental Microbiology. 2010;12:3159–3170. doi: 10.1111/j.1462-2920.2010.02288.x. [DOI] [PubMed] [Google Scholar]
- 91.Allison D, Brown M, Evans D, Gilbert P. Surface hydrophobicity and dispersal of Pseudomonas aeruginosa from biofilms. FEMS Microbiology Letters. 1990;71:101–104. doi: 10.1016/0378-1097(90)90039-s. [DOI] [PubMed] [Google Scholar]
- 92.Uppuluri P, Chaturvedi AK, Srinivasan A, Banerjee M, Ramasubramaniam AK, Köhler JR, Kadosh D, Lopez-Ribot JL. Dispersion as an important step in the Candida albicans biofilm developmental cycle. PLoS Pathog. 2010;6:e1000828. doi: 10.1371/journal.ppat.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kaplan JB, Fine DH. Biofilm dispersal of Neisseria subflava and other phylogenetically diverse oral bacteria. Applied and Environmental Microbiology. 2002;68:4943–4950. doi: 10.1128/AEM.68.10.4943-4950.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Allison DG, Ruiz B, SanJose C, Jaspe A, Gilbert P. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiology Letters. 1998;167:179–184. doi: 10.1111/j.1574-6968.1998.tb13225.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.