Abstract
20S-Hydroxyvitamin D3 [20(OH)D3] is the biologically active major product of the action of CYP11A1 on vitamin D3 and is present in human plasma. 20(OH)D3 displays similar therapeutic properties to 1,25-dihydroxyvitamin D3 [1,25(OH)2D3], but without causing hypercalcaemia and therefore has potential for development as a therapeutic drug. CYP24A1, the kidney mitochondrial P450 involved in inactivation of 1,25(OH)2D3, can hydroxylate 20(OH)D3 at C24 and C25, with the products displaying more potent inhibition of melanoma cell proliferation than 20(OH)D3. CYP3A4 is the major drug-metabolising P450 in liver endoplasmic reticulum and can metabolise other active forms of vitamin D, so we examined its ability to metabolise 20(OH)D3. We found that CYP3A4 metabolises 20(OH)D3 to three major products, 20,24R-dihydroxyvitamin D3 [20,24R(OH)2D3], 20,24S-dihydroxyvitamin D3 [20,24S(OH)2D3] and 20,25-dihydroxyvitamin D3 [20,25(OH)2D3]. 20,24R(OH)2D3 and 20,24S(OH)2D3, but not 20,25(OH)2D3, were further metabolised to trihydroxyvitamin D3 products by CYP3A4 but with low catalytic efficiency. The same three primary products, 20,24R(OH)2D3, 20,24S(OH)2D3 and 20,25(OH)2D3, were observed for the metabolism of 20(OH)D3 by human liver microsomes, in which CYP3A4 is a major CYP isoform present. Addition of CYP3A family-specific inhibitors, troleandomycin and azamulin, almost completely inhibited production of 20,24R(OH)2D3, 20,24S(OH)2D3 and 20,25(OH)2D3 by human liver microsomes, further supporting that CYP3A4 plays the major role in 20(OH)D3 metabolism by microsomes. Since both 20,24R(OH)2D3 and 20,25(OH)2D3 have previously been shown to display enhanced biological activity in inhibiting melanoma cell proliferation, our results show that CYP3A4 further activates, rather than inactivates, 20(OH)D3.
Keywords: 20-Hydroxyvitamin D3, CYP3A4, Hydroxylation, Liver microsomes
Introduction
20S-Hydroxyvitamin D3 [20(OH)D3] is the major product of CYP11A1 action on vitamin D3 (1–4), and acts as a biased agonist on the vitamin D receptor displaying anti-proliferative, anti-inflammatory and pro-differentiative activity (5–11). Unlike 1,25-dihydroxyvitamin D3 [1,25(OH)2D3], it does not induce hypercalcaemia at high doses (up to 60 μg/kg in rodents) (6,12,13). 20(OH)D3 exhibits anti-inflammatory and anti-fibrogenic activity in mice in vivo (8,14), and reduces DNA damage in skin caused by ultraviolet radiation (8,9,15). It therefore has potential as a therapeutic agent via topical or systemic administration. 20(OH)D3 is present in human plasma at a concentration of approximately 3 nM, a concentration higher than that required to observe biological effects in vitro, suggesting it might exert important physiological effects only partially overlapping with those of 1,25(OH)2D3 (16,17).
The kidney mitochondrial P450, CYP24A1, carries out the inactivation of 1,25(OH)2D3, initially hydroxylating it at C24 and then further oxidising the side chain to give the excretory product, calcitroic acid (18–20). CYP24A1 also hydroxylates 20(OH)D3 at C24 giving both enantiomers of 20,24-dihydroxyvitamin D3 [20S,24R(OH)2D3 and 20S,24S(OH)2D3] as well as hydroxylating it at C25 producing 20,25-dihydroxyvitamin D3 [20,25(OH)2D3] (21–23). 20,24R(OH)2D3 and 20,25(OH)2D3 inhibited colony formation by SKMEL-188 melanoma cells more strongly than the parent 20(OH)D3, indicating they have enhanced anti-proliferative activity (21). Therefore, CYP24A1 appears to activate rather than inactivate 20(OH)D3 (21).
Another potential site for the metabolism of 20(OH)D3 is the liver, which is the major tissue involved in drug metabolism in the body. Here, the initial 25-hydroxylation step in the activation of vitamin D3 is carried out by either microsomal CYP2R1, or mitochondrial CYP27A1 (24). CYP27A1 also hydroxylates 20(OH)D3 producing 20,25(OH)2D3 and 20,26-dihydroxyvitamin D3 [20,26(OH)2D3] (25). Human liver microsomes do not contain CYP24A1 or CYP27A1 since these are both mitochondrial P450s (24,26), but do contain CYP3A4, the most abundantly expressed P450 enzyme in the human liver and small intestine (27,28). Besides its well-known role in drug metabolism, CYP3A4 can metabolise various vitamin D analogues. It acts as a vitamin D 24- and 25-hydroxylase on substrates such as 1α-hydroxyvitamin D2, 1α-hydroxyvitamin D3 and vitamin D2, but does not act on vitamin D3 (29,30). The ability of CYP3A4 to hydroxylate 1,25(OH)2D3 has been compared with that of CYP24A1, revealing that both enzymes hydroxylate 1,25(OH)2D3 at the same carbon atoms, C23 and C24, but with opposite product stereoselectivity (31). In addition to this ability to hydroxylate the vitamin D side chain, CYP3A4 has been reported to hydroxylate the A-ring of 25-hydroxyvitamin D3 [25(OH)D3], producing 4α,- and 4β,25-dihydroxyvitamin D3, with these products being detected in the plasma (32).
Mouse liver microsomes can act on 20(OH)D3 and like CYP24A1 in the kidney, produce the biologically active products, 20,24(OH)2D3 and 20,25(OH)2D3 (33). Mouse liver microsomes do not express CYP3A4, but do express six other family 3A isoforms (34). Whether any of these are responsible for 20(OH)D3 metabolism is unknown. Since CYP3A4 is the most abundantly expressed P450 in human liver microsomes and can act on 25(OH)D3 and 1,25(OH)2D3, we investigated its ability to metabolise 20(OH)D3 using both recombinantly expressed enzyme (supersomes) and human liver microsomes.
Materials and Methods
2.1. Materials
A mixed gender 50-donor pool of human liver microsomes, and supersomes made from baculovirus-infected insect cells that express recombinant human CYP3A4 with P450 reductase and cytochrome b5, were purchased from Corning (Corning, NY). NADPH was purchased from Merck (Darmstadt, Germany). Glucose-6-phosphate and azamulin were from Sigma (NSW, Australia), glucose-6-phosphate dehydrogenase was from Roche (NSW, Australia) and 2-hydroxypropyl-β-cyclodextrin (HP-β-CD) was from Cerestar (Hammond, IN). Troleandomycin was from Enzo Life Sciences (Farmingdale, NY). All solvents used were of HPLC grade, and were purchased from Merck (Darmstadt, Germany). 20(OH)D3 was synthesised enzymatically by the action of recombinant bovine CYP11A1 on vitamin D3 and was purified by TLC and reverse-phase HPLC (2,4). 20,25(OH)2D3, 20,24R(OH)2D3 and 20,24S(OH)2D3 were produced by the action of recombinant rat CYP24A1 on 20(OH)D3 (21,22). 20,26(OH)2D3 was produced from the action of recombinant CYP27A1 on 20(OH)D3 (25). The structures of these metabolites including the stereochemistry have been determined previously by NMR (21–23). Concentrations of hydroxyvitamin D standards were determined from their absorbance at 263 nm using an extinction coefficient of 18,000 M−1 cm−1 (35).
2.2. Metabolism by CYP3A4 supersomes
Substrates (see Results for concentrations) solubilised in 0.45% HP-β-CD (3,36) were incubated with CYP3A4 supersomes, at a final CYP3A4 concentration of 60 nM in buffer comprising 50 mM K2HPO4 (pH 7.4), 3.3 mM MgSO4, 500 μM NADPH, 2 mM glucose-6-phosphate and 2 U/mL glucose-6-phosphate dehydrogenase. The typical incubation volume was 0.5 mL. Tubes containing all components except supersomes were preincubated for 8 min at 37°C, then the reaction started by the addition of the CYP3A4 supersomes. Following incubation at 37°C (see Results for times), reactions were stopped by the addition of 2.5 volumes of ice-cold dichloromethane. Tubes were centrifuged (670 × g for 10 min) and the lower organic phase retained. This extraction was repeated twice more, each time with 2.5 volumes dichloromethane. The extracted secosteroids were dried under nitrogen gas at 30°C and dissolved in the solvent required for HPLC analysis (see below), and stored at −20°C.
2.3. Metabolism of 20(OH)D3 by human liver microsomes
The procedure used was similar to the one described previously for mouse liver microsomes (33). Human liver microsomes (1.5 mg/mL) were incubated with 20(OH)D3 solubilised in 0.45% HP-β-CD, in buffer comprising 0.25 M sucrose, 50 mM Hepes (pH 7.4), 20 mM KCl, 5 mM MgSO4, 0.2 mM EDTA, 500 μM NADPH, 2 mM glucose-6-phosphate and 2 U/mL glucose-6-phosphate dehydrogenase. Tubes were preincubated for 5 min at 37°C with all components except the microsomes which were used to start the reaction. Following incubation at 37°C, reactions were stopped with 2.5 volumes ice-cold dichloromethane and extracted as described in section 2.2.
2.4. Reverse-phase HPLC and mass spectrometry
The 20(OH)D3 metabolites produced from the action of CYP3A4 supersomes or human liver microsomes were analysed using a Perkin Elmer HPLC system (Biocompatible Binary Pump 250 or Flexar Binary Pump Series 200) with a UV detector set at 265 nm, equipped with a C18 column (Grace Alltima, 25 cm × 4.6 mm, particle size 5 μm). Metabolites were initially analysed using a gradient of 45 – 100% acetonitrile in water for 30 min, followed by 100% acetonitrile for 35 min, at a flow rate of 0.5 mL/min. As this system was unable to achieve full separation of some metabolites, a second solvent system was used which comprised a gradient of 64 – 100% methanol in water for 20 min, followed by 100% methanol for 25 min or 30 min, at a flow rate of 0.5 mL/min. Quantitation of products was based on the major products having an unaltered conjugated triene structure like 20(OH)D3, as shown by their UV spectrum with a peak at 263 nm (Fig. S2). Therefore, each product gives a proportional response from the UV detector based on their concentration. The amount of each major product could then be calculated from the initial concentration of substrate and the percentage of substrate converted, based on peak integration (3). The kinetic parameters were obtained by fitting the Michaelis-Menten equation to experimental data using Kaleidagraph 4.0 (Synergy Software). For mass spectrometry, metabolites were purified and collected using both acetonitrile-water and methanol-water solvent systems (33,37). The purified metabolites (2 nmol) were reconstituted in 30 μL of 70% methanol + 0.1% formic acid, with 20 μL being injected into the LC/MS. These assays employed electrospray ionization and were performed as a service by UWA Center for Metabolomics, as described previously (37).
2.5. Incubation of microsomes and supersomes with CYP3A4 inhibitors
Incubations were carried out with 20(OH)D3 (10 or 50 μM) solubilised in 0.45% HP-β-CD and buffers as described for CYP3A4 supersomes or human liver microsomes (sections 2.2 or 2.3, respectively). The CYP3A4 inhibitor, troleandomycin (38) was added from an ethanol stock to a final concentration of 250 μM, azamulin (39) was added from a methanol stock to 1 or 10 μM and isoniazid (40) was added from a methanol stock to 1 mM, prior to preincubation (29,30). Reactions were started by the addition of either CYP3A4 supersomes or human liver microsomes and tubes incubated for 20 or 30 min, respectively, at 37°C. Human liver microsomes were also preincubated with troleandomycin (10 μM) and an NADPH regenerating system for 20 min before buffer and 20(OH)D3 (10 μM) were added to start the reaction. Products were extracted as described in section 2.2. Samples were analysed by reverse-phase HPLC (see section 2.4) using a gradient of 45 – 100% acetonitrile in water for 25 min (supersomes) or 30 min (microsomes), then 100% acetonitrile for 35 min, at a flow rate of 0.5 mL/min.
Results
3.1. Metabolism of 20(OH)D3 by CYP3A4
HP-β-CD was used to solubilize 20(OH)D3 for its addition to CYP3A4 supersomes. It has a hydrophobic interior that encapsulates hydrophobic substrates such as 20(OH)D3, and a hydrophilic exterior for favourable interaction with water (3,36,41). Incubation of 20(OH)D3 in HP-β-CD with CYP3A4 supersomes produced 25-fold more products than when the substrate was added from acetonitrile (not shown). Three major products (labelled A, B and C), and another metabolite with a short retention time labelled X (Fig. 1B) were observed, that were not present in the control (Fig. 1A). The three major products were initially identified by comparison of their HPLC retention times to those of authentic standards using an acetonitrile in water gradient on a 25 cm C18 column, as 20,25(OH)2D3, 20,24R(OH)2D3 and 20,24S(OH)2D3 (Fig. 1B). Co-migration of each peak with the respective standard in this solvent system was confirmed by spiking the reaction mixture with each standard (not shown). Each of the major products was collected separately from the acetonitrile-water system and analysed using a methanol-water system (Fig. 1C, E, G). Samples were spiked with an equal amount of standards (Fig. 1D, F, H), confirming that the samples and their respective standards also had identical retention times in this solvent system. We further confirmed that products A, B and C collected from the acetonitrile-water system contained products with retention times identical to 20,25(OH)2D3, 20,24R(OH)2D3 and 20,24S(OH)D3, respectively, on a 15 cm PFP column using an acetonitrile gradient (Fig. S1), further substantiating their identification. Products A, B and C had UV spectra similar to the substrate and typical of vitamin D, indicating an intact triene structure (Fig. S2). These three products were also analysed by mass spectrometry, and all gave major ions at m/z = 399.1 (416.1 + H+ − H2O), m/z = 439.1 (416.1 + Na+) and m/z = 455.1 (416.1 + K+), similar to authentic standards, confirming their classification as species of dihydroxyvitamin D3 (Table 1). Hence, we conclude that product A is 20,25(OH)2D3, product B is 20,24R(OH)2D3 and product C is 20,24S(OH)2D3.
Fig. 1.
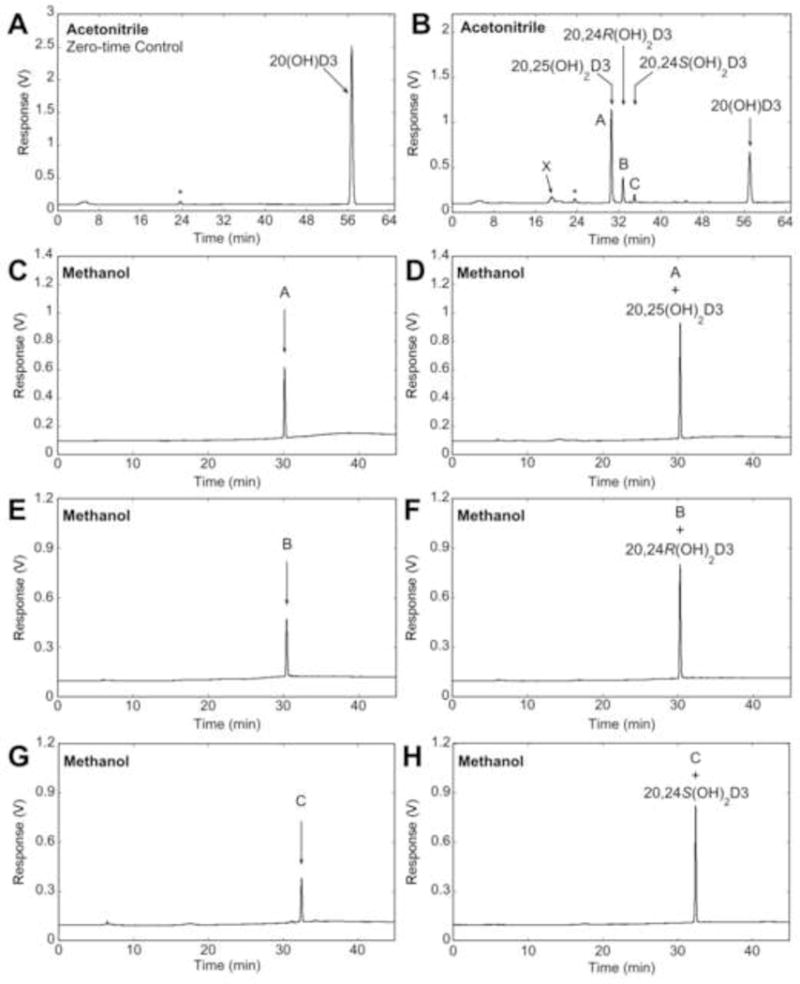
Reverse-phase HPLC analysis and identification of products from 20(OH)D3 metabolism by CYP3A4. 20(OH)D3 (25 μM) in 0.45% HP-β-CD was incubated with 60 nM CYP3A4. (A, B) Products were extracted and analysed by reverse-phase HPLC with a C18 column using an acetonitrile in water gradient. (A) Zero-time control incubation. (B) Test incubation for 60 min, with vertical arrows indicating the retention times of standards. The products with identical retention times to authentic 20,25(OH)2D3, 20,24R(OH)2D3, or 20,24S(OH)2D3 were collected separately and analysed using a methanol in water gradient (C, E, G, respectively) followed by spiking with an equal amount of the corresponding standard (D, F, H, respectively). *UV-absorbing contaminant extracted from supersomes.
Table 1.
Mass spectrometry of primary and secondary products from 20(OH)D3 metabolism by CYP3A4. Mass spectra were recorded as described in the Experimental section. Authentic standards of 20,25(OH)2D3, 20,24R(OH)2D3 and 20,24S(OH)2D3 gave the same parent and daughter ions as the corresponding identified CYP3A4 products.
| Product | [M + Na]+ | [M + K]+ | [M + H – H2O]+ | [M + H – 2H2O]+ | Identification |
|---|---|---|---|---|---|
| 20,25(OH)2D3 | 439.1 | 455.1 | 399.1 | 381.1 | Dihydroxyvitamin D3 |
| 20,24R(OH)2D3 | 439.1 | 455.1 | 399.1 | 381.1 | Dihydroxyvitamin D3 |
| 20,24S(OH)2D3 | 439.1 | 455.0 | 399.1 | 381.1 | Dihydroxyvitamin D3 |
| 1 (from 20,24R(OH)2D3) | 455.4 | – | 415.4 | 397.4 | Trihydroxyvitamin D3 |
| a (from 20,24S(OH)2D3) | 455.4 | – | 415.4 | 397.4 | Trihydroxyvitamin D3 |
A time course for the metabolism of 20(OH)D3 showing products greater than 0.1% of total extracted secosteroid, revealed that the rate of formation of 20,25(OH)2D3, 20,24R(OH)2D3 and 20,24S(OH)2D3 declined throughout the incubation (Fig. S3A), most likely due to CYP3A4 inactivation since only 18% of the substrate was consumed by the end of the incubation. Formation of product X displayed a lag, not being detectable until 5 min, suggesting that it is a secondary metabolite (Fig. S3A). CYP3A4 has previously been reported to metabolise 25(OH)D3 (32), therefore we compared its ability to metabolise this substrate to that of 20(OH)D3. The time course for formation of total products from each substrate showed that 20(OH)D3 was metabolised approximately 30 times faster than 25(OH)D3 (Fig. S3B). The products from 25(OH)D3 were not identified since authentic standards were not available, but according to Wang et al. (32), the major products are 4β,25(OH)2D3 and 4α,25(OH)2D3.
The presence of product X (Fig. 1B) with a shorter retention time than the major products suggested that CYP3A4 may be able to further metabolise the major products to secondary products. This was investigated by incubating 20,25(OH)2D3, 20,24R(OH)2D3 and 20,24S(OH)2D3 with CYP3A4. 20,25(OH)2D3 was poorly metabolised by CYP3A4, with essentially no products formed after incubation with the enzyme for one hour (Fig. 2A). In contrast, incubation of 20,24R(OH)2D3 or 20,24S(OH)2D3 with CYP3A4 resulted in three products from each of these substrates, products 1 – 3 and products a – c, respectively (Fig. 2B, C). Products 3 and b had identical retention times, suggesting they are the same compound, which can only be 20,24R,24S-trihydroxyvitamin D3. The retention time of product 1 was identical to that of product X produced directly from 20(OH)D3 (Fig. 1B), confirming that product X is a secondary metabolite and that it arises from 20,24R(OH)2D3. For both 20,24R(OH)2D3 and 20,24S(OH)2D3, the major product (product 1 and a, respectively) was produced in sufficient amounts to collect for analysis by mass spectrometry. Both gave major ions at m/z = 397.4 (432.4 + H+ − 2H2O), m/z = 415.4 (432.4 + H+ − H2O) and m/z = 455.4 (432.4 + Na+), resulting in their classification as species of trihydroxyvitamin D3 (Table 1). The time course for 20,24R(OH)2D3 metabolism showed that all products were produced without a lag, with the rate being approximately linear for the first 5 min (Fig. S4). Insufficient 20,24S(OH)2D3 was available to carry out a time course since enzymatic synthesis using CYP24A1 is inefficient (21).
Fig. 2.
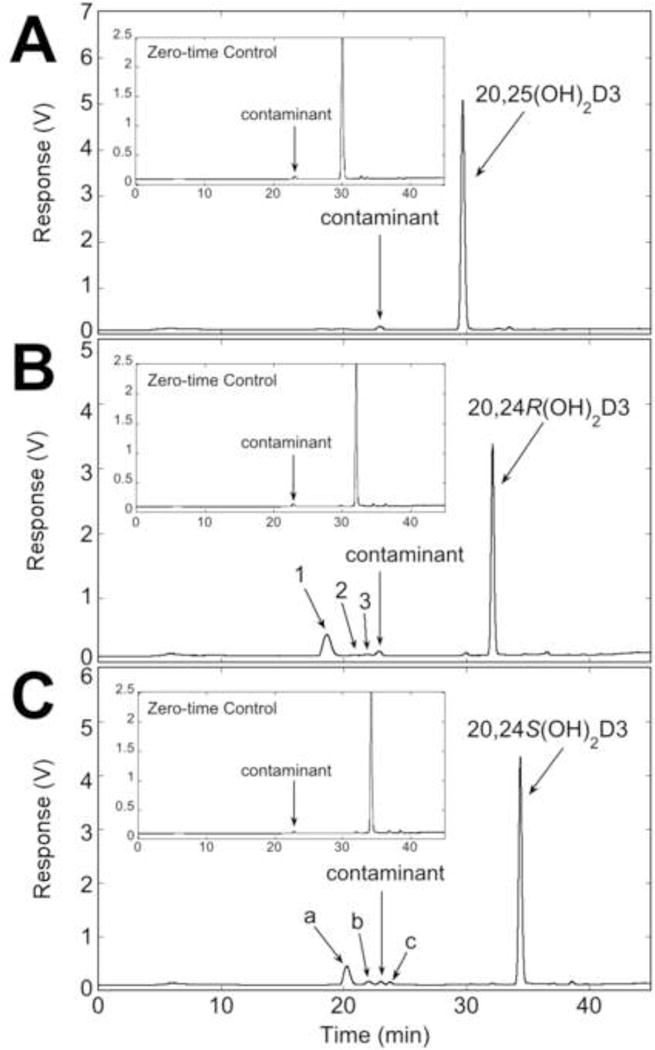
Further metabolism of 20,25(OH)2D3, 20,24R(OH)2D3 and 20,24S(OH)2D3 by CYP3A4. Substrates (25 μM) in 0.45% HP-β-CD were incubated with 60 nM CYP3A4. Products were extracted and analysed by reverse-phase HPLC using an acetonitrile in water gradient. (A) Incubation of 20,25(OH)2D3 with CYP3A4. (B) Formation of products from 20,24R(OH)2D3. (C) Formation of products from 20,24S(OH)2D3.
The kinetic parameters for the metabolism of 20(OH)D3, 20,24R(OH)2D3 and 20,24S(OH)2D3 were determined from the initial rates of formation of all products, by HPLC. We used a simple model where the Michaelis-Menten equation gave a good fit to the data, as shown in Fig. 3, providing confidence in the parameter estimates. The resulting data are summarised in Table 2. 20(OH)D3 metabolism by CYP3A4 gave the lowest Km and the highest kcat/Km value amongst the three substrates (Table 2), indicating that 20(OH)D3 is a better substrate for CYP3A4 being metabolised more efficiently than either 20,24R(OH)2D3 or 20,24S(OH)2D3. The data indicates that there should be little production of the trihydroxyvitamin D3 secondary metabolites from either 20,24R(OH)2D3 or 20,24S(OH)2D3 in the presence of 20(OH)D3, as observed experimentally (Fig. 1).
Fig. 3.
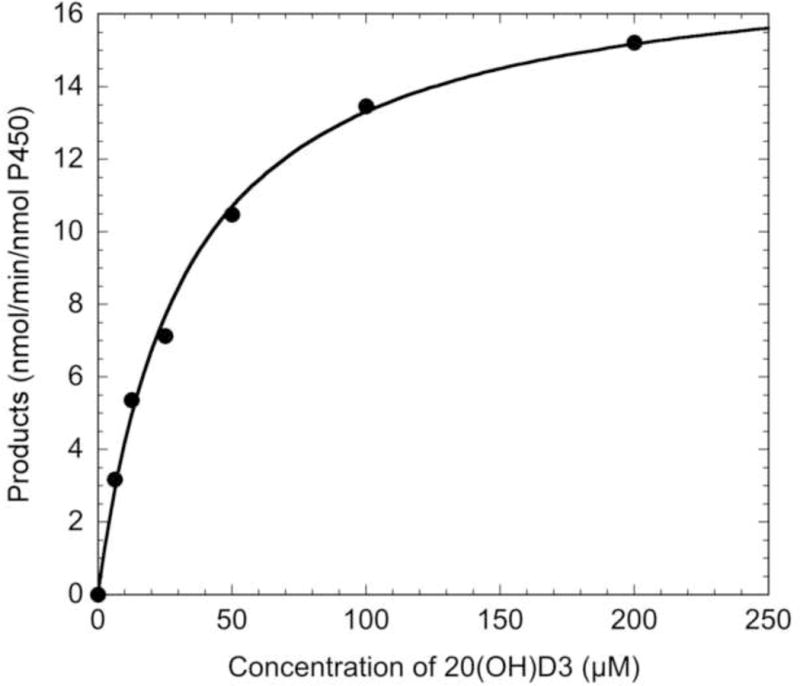
Analysis of the kinetics of the conversion of 20(OH)D3 to total products by CYP3A4. Various concentrations of 20(OH)D3 in 0.45% HP-β-CD were incubated with 60 nM CYP3A4 for 2.5 min, with the products being extracted and analysed by reverse-phase HPLC. The Michaelis-Menten equation was fitted to experimental data, with an R value of 0.998.
Table 2.
Kinetic parameters for metabolism of 20(OH)D3 and its primary products by CYP3A4. Data for Km and kcat are presented as mean ± standard error of the curve fit.
| Substrate | Km (μM) | kcat (min−1) | kcat / Km (μM−1 min−1) |
|---|---|---|---|
| 20(OH)D3 | 32.6 ± 2.8 | 17.7 ± 0.5 | 0.54 |
| 20,24R(OH)2D3 | 331.8 ± 193.9 | 65.0 ± 28.5 | 0.20 |
| 20,24S(OH)2D3 | 86.9 ± 44.3 | 16.5 ± 4.3 | 0.19 |
3.2. Metabolism of 20(OH)D3 by human liver microsomes
To determine whether CYP3A4 plays a major role in the metabolism of 20(OH)D3 by human liver microsomes, 20(OH)D3 was solubilised in HP-β-CD and incubated with human liver microsomes, with the metabolites being extracted and analysed by reverse-phase HPLC (Fig. 4). Six metabolites were initially observed when products were analysed using an acetonitrile in water gradient and each represented greater than 0.2% of total extracted secosteroids (Fig. 4B). Three products were tentatively identified as 20,25(OH)2D3, 20,24S(OH)2D3, and either 20,24R(OH)2D3 or 20,26(OH)2D3 which have an identical retention time in the acetonitrile-water system (33). Baseline separation could not be achieved for products labelled H2/H3 or H4/H5 so the overlapping peaks were collected and analysed using a methanol in water gradient (Fig. 4D, E). The product labelled H1 was also collected from the initial chromatography and analysed on a C18 Alltima column in this methanol-water system (Fig. 4C), as well as on a 15 cm PFP column with an acetonitrile in water gradient (Fig. S1G). Its retention time matched that of 20,25(OH)2D3 standard in both systems. The overlapping products labelled H2/H3 from the acetonitrile gradient separated into three peaks in the methanol-water system, with two of them being identified as 20,24R(OH)2D3 and 20,26(OH)2D3 by comparison of their retention times to those of standards (Fig. 4D). In the methanol-water system, the combined H4/H5 peaks from acetonitrile clearly separated into two major peaks, one of which had an identical retention time to 20,24S(OH)2D3 (Fig. 4E). The peaks corresponding to H2/H3 and H4/H5 were collected from the acetonitrile-water system and further analysed using a PFP column with an acetonitrile in water gradient, which also showed they contained products with identical retention times to 20,24R(OH)2D3, 20,24S(OH)2D3 and 20,26(OH)2D3 (Fig. S1H, I). Thus the four products identified from human liver microsomes are 20,25(OH)2D3, 20,24R(OH)2D3, 20,24S(OH)2D3 and 20,26(OH)2D3, with three being the same as the three major products seen for 20(OH)D3 metabolism by CYP3A4, and 20,26(OH)2D3 being the additional product seen with microsomes. The retention time of H6 did not align with any available standards, including 20,23-dihydroxyvitamin D3, and is as yet unidentified. The time course for formation of both identified (Fig. S5A) and unidentified products (Fig. S5B) from 20(OH)D3 metabolism by human liver microsomes showed that formation of all 20(OH)D3 metabolites was approximately linear for 15 min with no lags evident, suggesting that none of the products are secondary metabolites. Further support for CYP3A4 being responsible for the formation of 20,25(OH)2D3, 20,24R(OH)2D3 and 20,24S(OH)2D3 in human liver microsomes is provided by comparing the ratios of products to those for CYP3A4. For microsomes, the proportion of 20,25(OH)2D3:20,24R(OH)2D3:20,24S(OH)2D3 at the end of the time course (Fig. S5A) was 1:0.47:0.16, close to the ratios seen for CYP3A4 (1:0.41:0.14) (Fig. S3A).
Fig. 4.
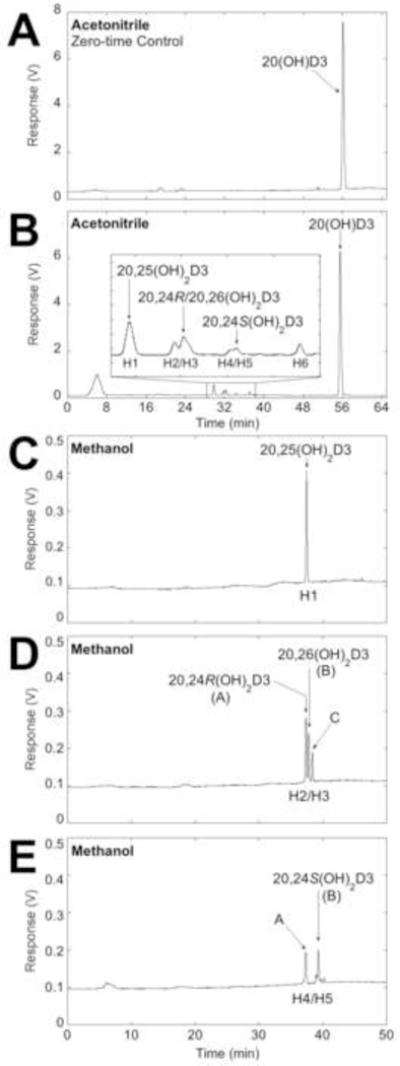
Analysis of the metabolism of 20(OH)D3 by human liver microsomes. 20(OH)D3 (50 μM) in 0.45% HP-β-CD was incubated with microsomes (1.5 mg/mL). (A, B) Products were extracted and analysed by reverse-phase HPLC with a C18 column using an acetonitrile in water gradient. (A) Zero-time control incubation. (B) Test incubation for 45 min. (C – E) The products in the major peaks were collected and analysed using a methanol in water gradient. (C) Peak labelled H1 with identical retention time to 20,25(OH)2D3. (D) Overlapping peaks labelled H2/H3 separated into three peaks, A – C. (E) Overlapping peaks labelled H4/H5 separated into two products, A and B. Arrows indicate retention times of standards.
The kinetic parameters for formation of products from 20(OH)D3 metabolism by human liver microsomes were obtained by measuring product conversion at a range of substrate concentrations. Kinetic parameters were measured separately for each product since they may arise from different microsomal enzymes (Table 3). Both acetonitrile-water and methanol-water systems were used to obtain baseline separation of all products. The Km values for formation of each product were reasonably similar, within the range of 114 – 217 μM, with the Km calculated for total product formation being 165 μM. The similarity of the Km values for 20,25(OH)2D3, 20,24R(OH)2D3 and 20,24S(OH)2D3 (Fig. 5) suggests that they could be produced by a single P450 species, namely CYP3A4 as the product profile would suggest, but the participation of other P450s cannot be excluded. The Vmax values were within the range of 14 – 56 pmol/min/mg protein, except for formation of the major product, 20,25(OH)2D3, which had a Vmax of 134.2 ± 36.0 pmol/min/mg protein.
Table 3.
Kinetic parameters for production of 20(OH)D3 metabolites by human liver microsomes. Data for Km and Vmax are presented as mean ± standard error of the curve fit. Similar data was obtained in two other experiments.
| Product | Km (μM) | Vmax (min−1) | Km/Vmax(μM−1min−1) |
|---|---|---|---|
| H1 (20,25(OH)2D3) | 217 ± 88 | 134.2 ± 36.0 | 1.6 |
| H2/H3 A (20,24R(OH)2D3) | 138 ± 49 | 56.1 ± 11.5 | 2.5 |
| H2/H3 B (20,26(OH)2D3) | 155 ± 42 | 34.5 ± 5.5 | 4.5 |
| H2/H3 C | 163 ± 51 | 24.4 ± 4.6 | 6.7 |
| H4/H5 A | 149 ± 32 | 14.1 ± 1.8 | 10.6 |
| H4/H5 B (20,24S(OH)2D3) | 114 ± 31 | 15.2 ± 2.2 | 7.5 |
| H6 | 139 ± 74 | 21.0 ± 6.5 | 6.6 |
| Total Products | 165 ± 56 | 292.6 ± 60.3 | 0.6 |
Fig. 5.
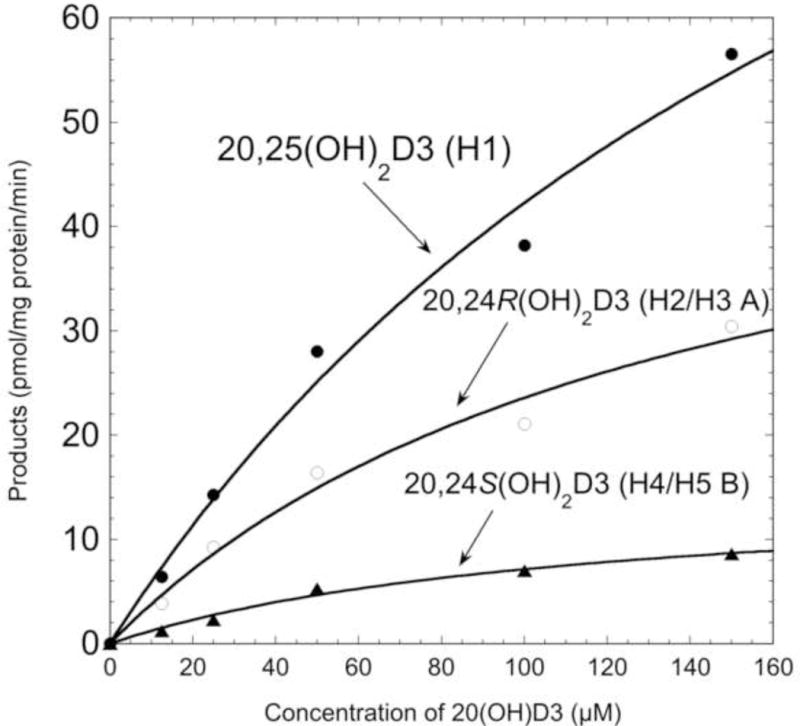
Kinetics of the conversion of 20(OH)D3 to 20,25(OH)2D3, 20,24R(OH)2D3 and 20,24S(OH)2D3 by human liver microsomes. Various concentrations of 20(OH)D3 in 0.45% HP-β-CD were incubated with human liver microsomes (1.5 mg/mL) for 10 min at 37°C. Products were extracted and analysed by reverse-phase HPLC, as described in Materials and Methods. The Michaelis-Menten equation was fitted to experimental data, with R values of 0.994, 0.992 and 0.994 for 20,25(OH)2D3, 20,24R(OH)2D3 and 20,24S(OH)2D3, respectively.
3.3. Effect of CYP3A family-specific inhibitors on metabolism of 20(OH)D3
To further define the role of CYP3A4 in human liver microsomes to metabolise 20(OH)D3, troleandomycin and azamulin, known inhibitors of the CYP3A family (38,39), were added to the incubations of 20(OH)D3 with human liver microsomes. As a control, the effect of troleandomycin on 20(OH)D3 metabolism by CYP3A4 was also tested. The metabolism of 20(OH)D3 by either CYP3A4 or human liver microsomes, both in the presence and absence of troleandomycin, was analysed by reverse-phase HPLC (Fig. 6). In the absence of troleandomycin, 20(OH)D3 was metabolised by CYP3A4 to products previously identified as 20,25(OH)2D3, 20,24R(OH)2D3 and 20,24S(OH)2D3 (see section 3.1) (Fig. 6A). However, when troleandomycin was added to the reaction at the same time as substrate, no products were observed indicating that troleandomycin completely inhibited CYP3A4 activity (Fig. 6B), as expected (38). Metabolism of 20(OH)D3 by human liver microsomes in the absence of troleandomycin was similar to that described earlier, with six products being observed, four of which were identified as 20,25(OH)2D3, 20,24R(OH)2D3, 20,24S(OH)2D3 and 20,26(OH)2D3 (Fig. 6C, E). In the presence of 250 μM troleandomycin, total product formation was reduced by 72.6%. The peak corresponding to 20,25(OH)2D3 was reduced by this treatment, as was the combined peak corresponding to 20,24R(OH)2D3/20,26(OH)2D3 and the peak containing 20,24S(OH)2D3 (Fig. 6C, D). Since troleandomycin is a mechanism-based inhibitor which forms an inactive P450-Fe(II)-metabolite complex with CYP3A family members (42–44), we also tested the inhibition of 20(OH)D3 metabolism where microsomes were preincubated with a low concentration of troleandomycin (10 μM) for 20 min prior to starting the reaction with substrate (10 μM). Under these conditions, total product formation was decreased by 63.7% and again decreases were seen in the peaks corresponding to 20,25(OH)2D3, 20,24R(OH)2D3/20,26(OH)2D3 and 20,24S(OH)2D3 (Fig. 6E, F). The only product of microsomes whose production was not markedly reduced by troleandomycin under either conditions, was H6 (Fig. 6D, F).
Fig. 6.
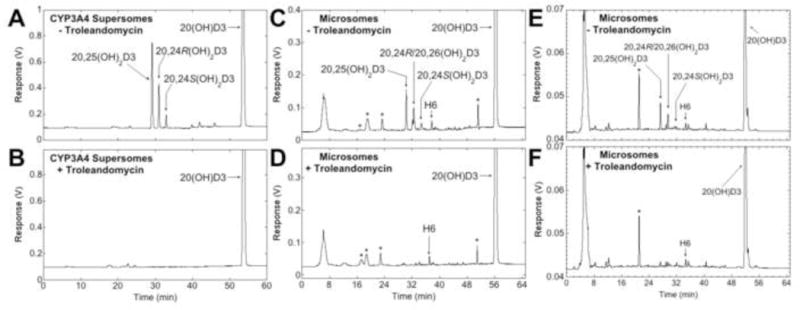
Effects of the CYP3A family inhibitor, troleandomycin, on metabolism of 20(OH)D3 by CYP3A4 and human liver microsomes. (A – D) 20(OH)D3 (50 μM) in 0.45% HP-β-CD was incubated in the presence (B, D) or absence (A, C) of 250 μM troleandomycin with CYP3A4 for 20 min (A, B) or human liver microsomes for 30 min (C, D), where the inhibitor and substrate were added before starting the reaction with enzyme. (E, F) Human liver microsomes were preincubated with 10 μM troleandomycin (F) or without (E) for 20 min then the reaction initiated by the addition of 20(OH)D3 (10 μM) in 0.45% HP-β-CD. *Denotes contaminant peaks present in the no-substrate control.
In the presence of azamulin (1 μM or 10 μM), total products from microsomes were decreased by 60.0% and 75.9%, respectively. The peaks corresponding to 20,25(OH)2D3, 20,24R(OH)2D3/20,26(OH)2D3 and 20,24S(OH)2D3, but not H6, were reduced (Fig. 7C, D). Furthermore, isoniazid, reported to inhibit CYP3A family members as well as CYP1A2, CYP2A6, CYP2C19 (40), also markedly decreased peaks corresponding to 20,25(OH)2D3, 20,24R(OH)2D3/20,26(OH)2D3 and 20,24S(OH)2D3, as well as H6 (Fig. S6).
Fig. 7.
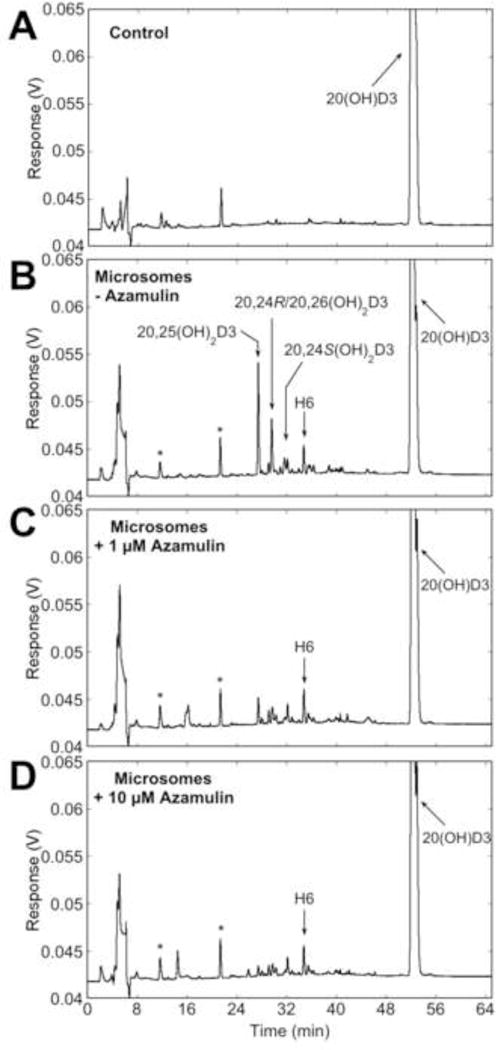
Effects of the CYP3A family inhibitor, azamulin, on metabolism of 20(OH)D3 by human liver microsomes. 20(OH)D3 (50 μM) in 0.45% HP-β-CD was incubated with human liver microsomes for 30 min in the absence (B) or presence of 1 μM (C) or 10 μM (D) azamulin. *Denotes contaminant peaks present in the control.
Discussion
In this study, we have shown that liver microsomal CYP3A4 metabolises 20(OH)D3 to 20,24R(OH)2D3, 20,24S(OH)2D3 and 20,25(OH)2D3 (Fig. 8). Two of the products, 20,25(OH)2D3 and 20,24R(OH)2D3, are more potent at inhibiting colony formation by melanoma cells than 20(OH)D3 (21,22), indicating CYP3A4 is further activating, rather than inactivating, 20(OH)D3, at least with respect to cell proliferation. Both 20,24R(OH)2D3 and 20,24S(OH)2D3 are active on the immune system, including modulating INFγ production by splenocytes (23), however this activity has not been directly compared to that of 20(OH)D3. As mentioned in the introduction, because 20(OH)D3 lacks calcaemic activity (6,12,13) but possesses the ability to inhibit proliferation, stimulate differentiation, reduce inflammation and inhibit collagen synthesis (5–8,10,14), it has the potential to be used as a drug to treat a range of disorders. Thus if used therapeutically, metabolism of 20(OH)D3 by CYP3A4 or human liver microsomes should not terminate its activity, but rather, may enhance it.
Fig. 8.
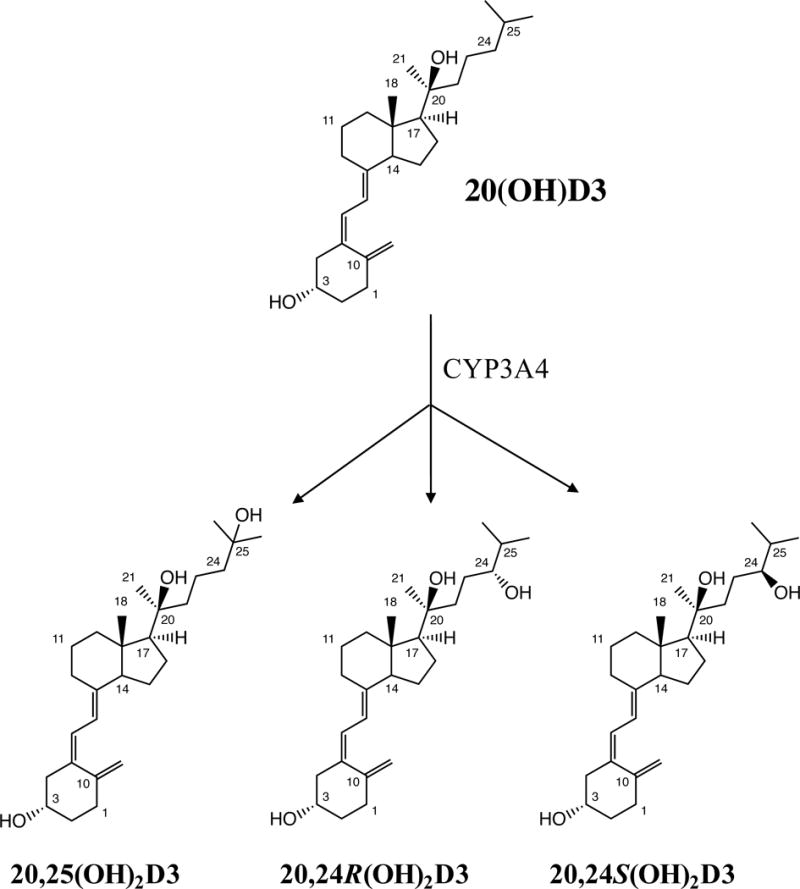
Metabolism of 20(OH)D3 by CYP3A4.
In the current study we used HP-β-CD to solubilise 20(OH)D3 for addition to both supersomes and microsomes. This encapsulating agent enables hydrophobic substrates to be held in solution above their normal solubility limit in aqueous solution (36,41) and is particularly useful for membrane bound-enzymes that display a high Km for substrate (3,16). At low final concentrations of HP-β-CD, as used in this study, there is relatively weak binding of the substrate to the cyclodextrin and it is readily available to the membrane-bound P450 (3,16,25,36). Ishikawa et al (45) reported that the addition of HP-β-CD caused a decrease in the rate of 7′-hydroxylation of 7-benzoyl-4-trifluoromethylcoumarin by CYP3A4, but in these experiments substrate was not pre-dissolved in the HP-β-CD. In contrast, we observed a markedly higher rate of 20(OH)D3 metabolism by CYP3A4 supersomes when the substrate was added from a HP-β-CD solution than when added from an acetonitrile stock.
The ability of CYP3A4 to hydroxylate 20(OH)D3 at C24 and C25 found in our study is consistent with previous reports that it can hydroxylate vitamin D2, 1-hydroxyvitamin D2, 1-hydroxyvitamin D3 at these positions (29,30). CYP3A4 also displays 24-hydroxylase activity towards 1,25(OH)2D3, as well as the ability to hydroxylate it at C23 (31). A more recent study reported that CYP3A4 can hydroxylate 25(OH)D3 at C4, producing 4β,25-dihydroxyvitamin D3 which was found at concentrations comparable to 1,25(OH)2D3 in human plasma (32). We found no evidence for hydroxylation of 20(OH)D3 at the 4β-position in our study.
The reported catalytic efficiency for metabolism of 25(OH)D3 by CYP3A4 (1.32 min−1 μM−1) (32) is higher than we observed for 20(OH)D3 (0.54 min−1μM−1), with 25(OH)D3 displaying a lower Km but comparable kcat. However, under our assay conditions 25(OH)D3 appeared to be a much poorer substrate for CYP3A4 compared to 20(OH)D3. Using 50 μM substrate, which is above the reported Km values for both substrates, 20(OH)D3 was metabolised approximately 30 times faster than 25(OH)D3. In contrast, the reported metabolism of 1,25(OH)2D3 by CYP3A4 (31) is of similar efficiency (0.70 min−1μM−1) to that of 20(OH)D3. The plasma concentration of 20(OH)D3 has been reported to be 3 nM (17) and assuming a similar concentration in the liver, this is well below the Km observed for 20(OH)D3 (32.6 ± 2.8 μM), indicating low rates of metabolism per P450 molecule. However, as CYP3A4 is expressed at high levels in liver as a high capacity and low affinity enzyme (27,28,46,47), the total rate of 20(OH)D3 metabolism may be of physiological significance. Its high capacity to metabolise 20(OH)D3 may also be of pharmacological importance in therapeutic use, as mentioned earlier.
Interestingly, the same products of 20(OH)D3 metabolism we have identified for CYP3A4, 20,24R(OH)2D3, 20,24S(OH)2D3 and 20,25(OH)2D3, have previously been identified as the major products of metabolism of 20(OH)D3 by the kidney mitochondrial CYP24A1 despite it having a smaller substrate binding pocket than CYP3A4 and a very much more selective active site (47,48). The CYP24A1 studies were carried out with the 20(OH)D3 substrate incorporated into phospholipid vesicles and substrate concentrations were expressed as a ratio to the phospholipid solvent (24), making direct comparison of their kinetic parameters difficult. Nevertheless, recalculation of the kinetic parameters for CYP24A1 concentrations in terms of aqueous molarity gives a catalytic efficiency (kcat/Km) of approximately 1 μM−1 min−1. This is double that for metabolism of 20(OH)D3 by CYP3A4 (0.54 μM−1 min−1) found in the current study, so it is likely that both enzymes contribute to the metabolism of 20(OH)D3 in vivo. Supporting that metabolism of 20(OH)D3 by CYP3A4 and/or CYP24A1 does occur in vivo, both 20,24R(OH)2D3 and 20,25(OH)2D3 have been detected in human serum (17). Which enzyme is the major metaboliser of 20(OH)D3 will largely depend on the relative concentration of each enzyme. In the case of CYP24A1 this will be determined by the 1,25(OH)2D3 concentration as this is a potent stimulator of CYP24 expression (21), and for CYP3A4 will depend on glucocorticoid levels and possible induction by other drugs (27,28).
Knockdown of 20,25(OH)2D3, 20,24R(OH)2D3 and 20,24S(OH)2D3 formation from 20(OH)D3 by troleandomycin and azamulin in human liver microsomes supports that a CYP3A family member is responsible for this metabolism of 20(OH)D3. Given that the ratio of these three hydroxylated metabolites produced by the microsomes is almost identical to that seen with CYP3A4 supersomes, and that CYP3A4 is the major CYP3A family member expressed in liver, we conclude that CYP3A4 is primarily responsible for 20(OH)D3 metabolism in liver microsomes. However, it would appear not to be the only P450 responsible for 20(OH)D3 metabolism in liver microsomes, since 20,26(OH)2D3 was also identified as a product, as well as H6. Furthermore, H6 was formed in the presence of both CYP3A family-specific inhibitors, indicating that there is at least one other P450, not of family 3A, involved in its formation. Inhibition of its formation by isoniazid suggests the P450 responsible may be CYP1A2, CYP2A6 or CYP2C19 (40), as these are known targets of isoniazid as well as CYP3A4. H6 is also synthesised from 20(OH)D3 by mouse liver microsomes and in that study was characterised as a dehydro-dihydroxyvitamin D3 species (M = 414) (33). Production of 20,25(OH)2D3 was not completely blocked by the CYP3A family inhibitors, leaving the possibility that other microsomal P450s may contribute to some degree to its formation with a prime candidate being CYP2R1, known to hydroxylate vitamin D3 at C25 (49,50).
In conclusion, CYP3A4 metabolises 20(OH)D3 to the major products 20,24R(OH)2D3, 20,24S(OH)2D3 and 20,25(OH)2D3, which display enhanced biological activity on melanoma cells. CYP3A4 is the major, but not only, P450 involved in metabolism of 20(OH)D3 by human liver microsomes. The further activation of 20(OH)D3 rather than inactivation by CYP3A4 in liver microsomes supports the potential for 20(OH)D3 to be developed as a therapeutic drug.
Supplementary Material
Highlights.
-
-
Human CYP3A4 metabolises 20-hydroxyvitamin D3 to three major and one minor product.
-
-
Major products were 20,24R/S-dihydroxyvitamins D3 and 20,25-dihydroxyvitamin D3.
-
-
CYP3A4 plays the major role in 20-hydroxyvitamin D3 metabolism by liver microsomes.
-
-
Hydroxyvitamin D3 products of CYP3A4 action display enhanced biological activity.
Footnotes
Conflict of Interest:
The authors declare no conflict of interest
References
- 1.Guryev O, Carvalho RA, Usanov S, Gilep A, Estabrook RW. A pathway for the metabolism of vitamin D3: unique hydroxylated metabolites formed during catalysis with cytochrome P450scc (CYP11A1) Proc Natl Acad Sci U S A. 2003;100:14754–14759. doi: 10.1073/pnas.2336107100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Slominski A, Semak I, Zjawiony J, Wortsman J, Li W, Szczesniewski A, Tuckey RC. The cytochrome P450scc system opens an alternate pathway of vitamin D3 metabolism. FEBS J. 2005;272:4080–4090. doi: 10.1111/j.1742-4658.2005.04819.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuckey RC, Nguyen MN, Slominski A. Kinetics of vitamin D3 metabolism by cytochrome P450scc (CYP11A1) in phospholipid vesicles and cyclodextrin. Int J Biochem Cell Biol. 2008;40:2619–2626. doi: 10.1016/j.biocel.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tuckey RC, Li W, Shehabi HZ, Janjetovic Z, Nguyen MN, Kim TK, Chen J, Howell DE, Benson HAE, Sweatman T, Baldisseri DM, Slominski A. Production of 22-hydroxy metabolites of vitamin D3 by cytochrome P450scc (CYP11A1) and analysis of their biological activities on skin cells. Drug Metab Dispos. 2011;39:1577–1588. doi: 10.1124/dmd.111.040071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janjetovic Z, Zmijewski MA, Tuckey RC, DeLeon DA, Nguyen MN, Pfeffer LM, Slominski AT. 20-Hydroxycholecalciferol, product of vitamin D3 hydroxylation by P450scc, decreases NF-κB activity by increasing IκBα levels in human keratinocytes. PLoS ONE. 2009;4:e5988. doi: 10.1371/journal.pone.0005988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slominski AT, Janjetovic Z, Fuller BE, Zmijewski MA, Tuckey RC, Nguyen MN, Sweatman T, Li W, Zjawiony J, Miller D, Chen TC, Lozanski G, Holick MF. Products of vitamin D3 or 7-dehydrocholesterol metabolism by cytochrome P450scc show anti-leukemia effects, having low or absent calcemic activity. PLoS ONE. 2010;5:e9907. doi: 10.1371/journal.pone.0009907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zbytek B, Janjetovic Z, Tuckey RC, Zmijewski MA, Sweatman TW, Jones E, Nguyen MN, Slominski AT. 20-Hydroxyvitamin D3, a product of vitamin D3 hydroxylation by cytochrome P450scc, stimulates keratinocyte differentiation. J Invest Dermatol. 2008;128:2271–2280. doi: 10.1038/jid.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slominski AT, Kim TK, Li W, Yi AK, Postlethwaite A, Tuckey RC. The role of CYP11A1 in the production of vitamin D metabolites and their role in the regulation of epidermal functions. J Steroid Biochem Mol Biol. 2014;144(Pt A):28–39. doi: 10.1016/j.jsbmb.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slominski AT, Janjetovic Z, Kim TK, Wasilewski P, Rosas S, Hanna S, Sayre RM, Dowdy JC, Li W, Tuckey RC. Novel non-calcemic secosteroids that are produced by human epidermal keratinocytes protect against solar radiation. J Steroid Biochem Mol Biol. 2015;148:52–63. doi: 10.1016/j.jsbmb.2015.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Slominski AT, Li W, Kim TK, Semak I, Wang J, Zjawiony JK, Tuckey RC. Novel activities of CYP11A1 and their potential physiological significance. J Steroid Biochem Mol Biol. 2015;151:25–37. doi: 10.1016/j.jsbmb.2014.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slominski AT, Manna PR, Tuckey RC. On the role of skin in the regulation of local and systemic steroidogenic activities. Steroids. 2015;103:72–88. doi: 10.1016/j.steroids.2015.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang J, Slominski A, Tuckey RC, Janjetovic Z, Kulkarni A, Chen J, Postlethwaite AE, Miller D, Li W. 20-Hydroxyvitamin D3 inhibits proliferation of cancer cells with high efficacy while being non-toxic. Anticancer Res. 2012;32:739–746. [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J, Wang J, Kim TK, Tieu EW, Tang EK, Lin Z, Kovacic D, Miller DD, Postlethwaite A, Tuckey RC, Slominski AT, Li W. Novel vitamin D analogs as potential therapeutics: metabolism, toxicity profiling, and antiproliferative activity. Anticancer Res. 2014;34:2153–2163. [PMC free article] [PubMed] [Google Scholar]
- 14.Slominski A, Janjetovic Z, Tuckey RC, Nguyen MN, Bhattacharya KG, Wang J, Li W, Jiao Y, Gu W, Brown M, Postlethwaite AE. 20S-Hydroxyvitamin D3, noncalcemic product of CYP11A1 action on vitamin D3, exhibits potent antifibrogenic activity in vivo. J Clin Endocr Metab. 2013;98:E298–E303. doi: 10.1210/jc.2012-3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tongkao-On W, Carter S, Reeve VE, Dixon KM, Gordon-Thomson C, Halliday GM, Tuckey RC, Mason RS. CYP11A1 in skin: an alternative route to photoprotection by vitamin D compounds. J Steroid Biochem Mol Biol. 2015;148:72–78. doi: 10.1016/j.jsbmb.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Slominski AT, Kim TK, Shehabi HZ, Semak I, Tang EKY, Nguyen MN, Benson HAE, Korik E, Janjetovic Z, Chen J, Yates CR, Postlethwaite A, Li W, Tuckey RC. In vivo evidence for a novel pathway of vitamin D3 metabolism initiated by P450scc and modified by CYP27B1. FASEB J. 2012;26:3901–3915. doi: 10.1096/fj.12-208975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Slominski AT, Kim TK, Li W, Postlethwaite A, Tieu EW, Tang EK, Tuckey RC. Detection of novel CYP11A1-derived secosteroids in the human epidermis and serum and pig adrenal gland. Sci Rep. 2015;5:14875. doi: 10.1038/srep14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tieu EW, Tang EK, Tuckey RC. Kinetic analysis of human CYP24A1 metabolism of vitamin D via the C24-oxidation pathway. FEBS J. 2014;281:3280–3296. doi: 10.1111/febs.12862. [DOI] [PubMed] [Google Scholar]
- 19.Jones G, Prosser DE, Kaufmann M. 25-Hydroxyvitamin D-24-hydroxylase (CYP24A1): its important role in the degradation of vitamin D. Arch Biochem Biophys. 2012;523:9–18. doi: 10.1016/j.abb.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 20.Sakaki T, Sawada N, Nonaka Y, Ohyama Y, Inouye K. Metabolic studies using recombinant escherichia coli cells producing rat mitochondrial CYP24 CYP24 can convert 1alpha,25-dihydroxyvitamin D3 to calcitroic acid. Eur J Biochem. 1999;262:43–48. doi: 10.1046/j.1432-1327.1999.00375.x. [DOI] [PubMed] [Google Scholar]
- 21.Tieu EW, Tang EKY, Chen J, Li W, Nguyen MN, Janjetovic Z, Slominski A, Tuckey RC. Rat CYP24A1 acts on 20-hydroxyvitamin D3 producing hydroxylated products with increased biological activity. Biochem Pharmacol. 2012;84:1696–1704. doi: 10.1016/j.bcp.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tieu EW, Li W, Chen J, Kim TK, Ma D, Slominski AT, Tuckey RC. Metabolism of 20-hydroxyvitamin D3 and 20,23-dihydroxyvitamin D3 by rat and human CYP24A1. J Steroid Biochem Mol Biol. 2015;149:153–165. doi: 10.1016/j.jsbmb.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin Z, Marepally SR, Ma D, Myers LK, Postlethwaite AE, Tuckey RC, Cheng CY, Kim TK, Yue J, Slominski AT, Miller DD, Li W. Chemical Synthesis and Biological Activities of 20S,24S/R-Dihydroxyvitamin D3 Epimers and Their 1alpha-Hydroxyl Derivatives. J Med Chem. 2015;58:7881–7887. doi: 10.1021/acs.jmedchem.5b00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuster I. Cytochromes P450 are essential players in the vitamin D signaling system. Biochim Biophys Acta. 2011;1814:186–199. doi: 10.1016/j.bbapap.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 25.Tieu EW, Li W, Chen J, Baldisseri DM, Slominski AT, Tuckey RC. Metabolism of cholesterol, vitamin D3 and 20-hydroxyvitamin D3 incorporated into phospholipid vesicles by human CYP27A1. J Steroid Biochem. 2012;129:163–171. doi: 10.1016/j.jsbmb.2011.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prosser DE, Jones G. Enzymes involved in the activation and inactivation of vitamin D. Trends Biochem Sci. 2004;29:664–673. doi: 10.1016/j.tibs.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Watkins PB, Wrighton SA, Schuetz EG, Molowa DT, Guzelian PS. Identification of glucocorticoid-inducible cytochromes P-450 in the intestinal mucosa of rats and man. J Clin Invest. 1987;80:1029–1036. doi: 10.1172/JCI113156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thummel KE, Wilkinson GR. In vitro and in vivo drug interactions involving human CYP3A. Annu Rev Pharmacol Toxicol. 1998;38:389–430. doi: 10.1146/annurev.pharmtox.38.1.389. [DOI] [PubMed] [Google Scholar]
- 29.Gupta RP, Hollis BW, Patel SB, Patrick KS, Bell NH. CYP3A4 is a human microsomal vitamin D 25-hydroxylase. J Bone Miner Res. 2004;19:680–688. doi: 10.1359/JBMR.0301257. [DOI] [PubMed] [Google Scholar]
- 30.Gupta RP, He YA, Patrick KS, Halpert JR, Bell NH. CYP3A4 as a vitamin D-24- and 25-hydroxylase: analysis of structure function by site-directed mutagenesis. J Clin Endocr Metab. 2005;90:1210–1219. doi: 10.1210/jc.2004-0966. [DOI] [PubMed] [Google Scholar]
- 31.Xu Y, Hashizume T, Shuhart MC, Davis CL, Nelson WL, Sakaki T, Kalhorn TF, Watkins PB, Schuetz EG, Thummel KE. Intestinal and hepatic CYP3A4 catalyze hydroxylation of 1alpha,25-dihydroxyvitamin D(3): implications for drug-induced osteomalacia. Mol Pharmacol. 2006;69:56–65. doi: 10.1124/mol.105.017392. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z, Lin YS, Zheng XE, Senn T, Hashizume T, Scian M, Dickmann LJ, Nelson SD, Baillie TA, Hebert MF, Blough D, Davis CL, Thummel KE. An inducible cytochrome P450 3A4-dependent vitamin D catabolic pathway. Mol Pharmacol. 2012;81:498–509. doi: 10.1124/mol.111.076356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheng CY, Slominski AT, Tuckey RC. Metabolism of 20-hydroxyvitamin D3 by mouse liver microsomes. J Steroid Biochem Mol Biol. 2014;144:286–293. doi: 10.1016/j.jsbmb.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deb S, Pandey M, Adomat H, Guns ES. Cytochrome P450 3A-mediated microsomal biotransformation of 1alpha,25-dihydroxyvitamin D3 in mouse and human liver: drug-related induction and inhibition of catabolism. Drug Metab Dispos. 2012;40:907–918. doi: 10.1124/dmd.111.041681. [DOI] [PubMed] [Google Scholar]
- 35.Hiwatashi A, Nishii Y, Ichikawa Y. Purification of cytochrome P-450D1 alpha (25-hydroxyvitamin D3–1 alpha-hydroxylase) of bovine kidney mitochondria. Biochem Biophys Res Commun. 1982;105:320–327. doi: 10.1016/s0006-291x(82)80047-8. [DOI] [PubMed] [Google Scholar]
- 36.De Caprio J, Yun J, Javitt NB. Bile acid and sterol solubilization in 2-hydroxypropyl-beta-cyclodextrin. J Lipid Res. 1992;33:441–443. [PubMed] [Google Scholar]
- 37.Clarke M, Tuckey R, Gorman S, Holt B, Hart P. Optimized 25-hydroxyvitamin D analysis using liquid–liquid extraction with 2D separation with LC/MS/MS detection, provides superior precision compared to conventional assays. Metabolomics. 2013;9:1031–1040. [Google Scholar]
- 38.Khojasteh SC, Prabhu S, Kenny JR, Halladay JS, Lu AY. Chemical inhibitors of cytochrome P450 isoforms in human liver microsomes: a re-evaluation of P450 isoform selectivity. Eur J Drug Metab Pharmacokinet. 2011;36:1–16. doi: 10.1007/s13318-011-0024-2. [DOI] [PubMed] [Google Scholar]
- 39.Stresser DM, Broudy MI, Ho T, Cargill CE, Blanchard AP, Sharma R, Dandeneau AA, Goodwin JJ, Turner SD, Erve JC, Patten CJ, Dehal SS, Crespi CL. Highly selective inhibition of human CYP3Aa in vitro by azamulin and evidence that inhibition is irreversible. Drug Metab Dispos. 2004;32:105–112. doi: 10.1124/dmd.32.1.105. [DOI] [PubMed] [Google Scholar]
- 40.Wen X, Wang JS, Neuvonen PJ, Backman JT. Isoniazid is a mechanism-based inhibitor of cytochrome P450 1A2, 2A6, 2C19 and 3A4 isoforms in human liver microsomes. Eur J Clin Pharmacol. 2002;57:799–804. doi: 10.1007/s00228-001-0396-3. [DOI] [PubMed] [Google Scholar]
- 41.Wallimann P, Marti T, Furer A, Diederich F. Steroids in Molecular Recognition. Chem Rev. 1997;97:1567–1608. doi: 10.1021/cr960373b. [DOI] [PubMed] [Google Scholar]
- 42.Ghosal A, Satoh H, Thomas PE, Bush E, Moore D. Inhibition and kinetics of cytochrome P4503A activity in microsomes from rat, human, and cdna-expressed human cytochrome P450. Drug Metab Dispos. 1996;24:940–947. [PubMed] [Google Scholar]
- 43.Ma B, Prueksaritanont T, Lin JH. Drug interactions with calcium channel blockers: possible involvement of metabolite-intermediate complexation with CYP3A. Drug Metab Dispos. 2000;28:125–130. [PubMed] [Google Scholar]
- 44.Lim HK, Duczak N, Jr, Brougham L, Elliot M, Patel K, Chan K. Automated screening with confirmation of mechanism-based inactivation of CYP3A4, CYP2C9, CYP2C19, CYP2D6, and CYP1A2 in pooled human liver microsomes. Drug Metab Dispos. 2005;33:1211–1219. doi: 10.1124/dmd.104.003475. [DOI] [PubMed] [Google Scholar]
- 45.Ishikawa M, Yoshii H, Furuta T. Interaction of modified cyclodextrins with cytochrome P-450. Biosci Biotechnol Biochem. 2005;69:246–248. doi: 10.1271/bbb.69.246. [DOI] [PubMed] [Google Scholar]
- 46.Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci U S A. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J Biol Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 48.Annalora AJ, Goodin DB, Hong WX, Zhang Q, Johnson EF, Stout CD. Crystal structure of CYP24A1, a mitochondrial cytochrome P450 involved in vitamin D metabolism. J Mol Biol. 2010;396:441–451. doi: 10.1016/j.jmb.2009.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shinkyo R, Sakaki T, Kamakura M, Ohta M, Inouye K. Metabolism of vitamin D by human microsomal CYP2R1. Biochem Bioph Res Co. 2004;324:451–457. doi: 10.1016/j.bbrc.2004.09.073. [DOI] [PubMed] [Google Scholar]
- 50.Cheng JB, Motola DL, Mangelsdorf DJ, Russell DW. De-orphanization of cytochrome P450 2R1: a microsomal vitamin D 25-hydroxylase. J Biol Chem. 2003;278:38084–38093. doi: 10.1074/jbc.M307028200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


