Abstract
The major histocompatibility complex class I (MHCI) is a large gene family, with over 30 members in mouse. Some MHCIs are well-known for their critical roles in the immune response. Studies in mice which lack stable cell-surface expression of many MHCI proteins suggest that one or more MHCIs also play unexpected, essential roles in the establishment, function, and modification of neuronal synapses in the central nervous system (CNS). However, there is little information about which genes mediate MHCI’s effects in neurons. In this study, RT-PCR was used to simultaneously assess transcription of many MHCI genes in regions of the central and peripheral nervous system where MHCI has a known or suspected role. In the hippocampus, a part of the CNS where MHCI regulates synapse density, synaptic transmission, and plasticity, we found that more than a dozen MHCI genes are transcribed. Single-cell RT-PCR revealed that individual hippocampal neurons can express more than one MHCI gene, and that the MHCI gene expression profile of CA1 pyramidal neurons differs significantly from that of CA3 pyramidal neurons or granule cells of the dentate gyrus. MHCI gene expression was also assessed at the neuromuscular junction (NMJ), a part of the peripheral nervous system (PNS) where MHCI plays a role in neuronal regeneration, and could potentially influence developmental synapse elimination. Four MHCI genes are expressed at the NMJ at an age when synapse elimination is occurring in three different muscles. Several MHCI mRNA splice variants were detected in hippocampus, but not at the NMJ. Together, these results establish the first profile of MHCI gene expression at the developing NMJ, and demonstrate that MHCI gene expression is under tight spatial and temporal regulation in the nervous system. They also identify more than a dozen MHCIs that could play important roles in synaptic transmission and plasticity in the central and peripheral nervous systems.
Keywords: MHCI, major histocompatibility complex class I, NMJ, alternative splicing, hippocampus, developmental regulation
Introduction
The major histocompatibility complex (MHC) is a group of genes found in vertebrates, where they play a central role in the adaptive immune response. Remarkably, over the past decade, another set of critical functions for MHC have come to light in the healthy vertebrate central nervous system (CNS). These studies have shown that MHCI regulates synapse density, synaptic transmission, and plasticity in the CNS (Datwani et al., 2009; Dixon-Salazar et al., 2014; Edamura et al., 2014; Elmer et al., 2013; Fourgeaud et al., 2010; Glynn et al., 2011; Goddard et al., 2007; Huh et al., 2000; Lee et al., 2014; McConnell et al., 2009; Nelson et al., 2013); reviewed in (Boulanger, 2009; Elmer and McAllister, 2012; Shatz, 2009). However, the MHCI is a large gene family (Fig. 1), and the specific MHCI molecules that mediate these neuronal functions remain largely unknown.
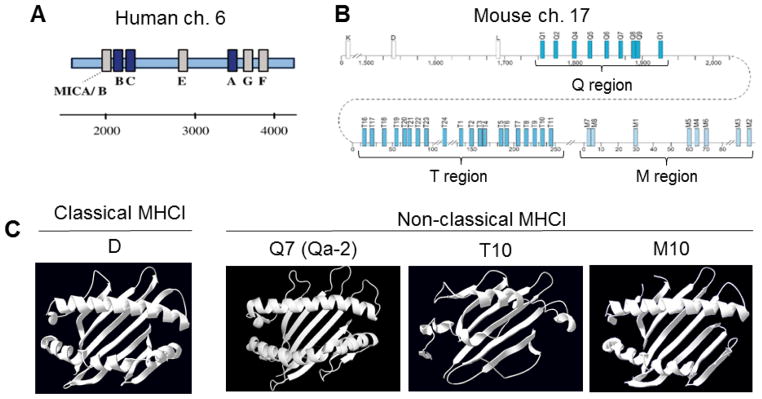
Figure 1. Organization of the MHCI gene family in human and mouse, and structures of classical versus nonclassical MHCI proteins.
A. Human MHCI (HLA) genes are located on chromosome 6, and consist of three classical (blue) and three nonclassical (grey) MHCI genes. MICA/B are MHCI-like genes (adapted from (Cardoso and de Sousa, 2003)).
B. Mouse MHCI genes are located on chromosome 17, and consist of >30 genes, a subset of which are shown here. Three are classical MHCI genes (white). The nonclassical MHCI genes (blue) are further subdivided into Q, T, and M subgroups (adapted from (Boulanger, 2004)).
C. Top-down view of structural renderings of mouse classical and nonclassical MHCI gene products (structural data from RSCB Protein Data Bank (PDB), renderings performed using PyMOL). Only α1 and α2 domains are shown, for clarity. Sources of crystallographic data: D, (Wang et al., 2002); Q7 (Qa-2), (He et al., 2001); T10, (Rudolph et al., 2004); M10, (Olson et al., 2005). In all figures and throughout the text, H2- prefixes were omitted from the names of mouse MHCI genes for clarity.
MHC genes can be divided into three broad categories: class I, II, and III. While there is evidence that members of all three classes of MHC can play important roles in the healthy and/or diseased nervous system (e.g., (Huh et al., 2000; Stevens et al., 2007; Tafti et al., 1996)), the largest number of studies to date support a role for MHCI in the healthy nervous system. The MHCI region is located on chromosome 17 in mice, and the human MHCI (human leukocyte antigen, or HLA) is located on chromosome 6p21.3 (Fig. 1A–B). MHCI genes can be categorized as either classical (MHCIa) or nonclassical (MHCIb). In mice, there are a maximum of three classical MHCI genes: H2-K, H2-D, and H2-L (H2-L is not present in C57Bl/6 mice). In humans, there are also three classical MHCI genes: HLA-A, -B, and –C. Classical MHCI genes are highly polymorphic, are expressed on the surface of most nucleated cells in the body, and present an array of short antigenic peptides for immune surveillance.
The functions of nonclassical MHCI genes, in contrast, are more elusive. Nonclassical MHCI genes consist of members of the H2-Q, -T, and -M gene clusters in mouse, and HLA-E, -F, and –G in humans. Nonclassical MHCI genes show relatively low polymorphism and more restricted expression patterns relative to classical MHCI genes. At least 21 nonclassical MHCI genes are transcribed in C57Bl/6 mice (Guidry and Stroynowski, 2005; Ohtsuka et al., 2008). The mouse and human genomes also contain several MHCI-like genes, which encode proteins that are structurally related to MHCI proteins, but are encoded outside the MHCI gene region. The lack of an antigen-presenting cleft (Fig. 1C) suggests that many nonclassical MHCIs and MHCI-like proteins may be functionally divergent. Despite inroads into understanding the functions of specific nonclassical MHCI proteins (e.g., (Aldrich et al., 1994; Braud et al., 1997; Joyce et al., 1994; Lindahl et al., 1997; Loconto et al., 2003; Wu et al., 1999; Xu et al., 2006)), for most, their function remains unknown.
Although healthy neurons have long been thought to express low or nonexistent levels of MHCI proteins under basal conditions, a growing number of studies have shown that MHCI protein and mRNA are expressed under basal conditions in the healthy developing and adult central nervous system. MHCI mRNA and/or protein has been detected in the CNS in diverse vertebrate species, including mice, rats, cats, marmosets, and humans (Cebrian et al., 2014; Chacon and Boulanger, 2013; Corriveau et al., 1998; Datwani et al., 2009; Edstrom et al., 2004; Huh et al., 2000; Ishii et al., 2003; Ishii and Mombaerts, 2008; Letellier et al., 2008; Lidman et al., 1999; Linda et al., 1998; Linda et al., 1999; Liu et al., 2015; Loconto et al., 2003; Lv et al., 2014; McConnell et al., 2009; Needleman et al., 2010; Ribic et al., 2011; Rolleke et al., 2006; Zhang et al., 2013a; Zhang et al., 2013b; Zohar et al., 2008). Many of these studies used probes or antibodies that do not distinguish specific MHCIs. Studies that used more specific probes and antibodies suggest that both classical and nonclassical MHCIs can be expressed in the nervous system, and that different MHCI genes have distinct expression patterns in the CNS (e.g., (Huh et al., 2000; Liu et al., 2015; Ohtsuka et al., 2008)). However, these studies either examined only a handful of MHCIs in the brain, or examined many MHCIs, but sampled a few large brain regions. As an essential step towards a mechanistic understanding of how MHCI influences neural circuits, it is imperative to determine which of the dozens of mouse MHCI genes are expressed in the relevant tissues. In the current study, we examined the expression of more than a dozen MHCI genes in two specific regions of the nervous system: in the CNS, the hippocampus, and in the peripheral nervous system (PNS), the neuromuscular junction (NMJ).
Results
MHCI gene expression at the developing neuromuscular junction (NMJ)
In the first set of studies, we examined MHCI gene expression at the developing NMJ. MHCI promotes synapse elimination in the developing retino-geniculate projection (Datwani et al., 2009; Huh et al., 2000; Lee et al., 2014), and similar synapse elimination occurs the developing NMJ (Sanes and Lichtman, 1999). MHCI mRNA is expressed in mature motor neurons, as determined using pan-specific probes that detect most MHCI transcripts (Edstrom et al., 2004; Linda et al., 1998; Linda et al., 1999; Thams et al., 2009), and MHCI protein has been detected at the adult NMJ using an antibody that detects the classical MHCI H2-D (Thams et al., 2009). There are also changes in the distribution of synapses in muscles from mice lacking both classical MHCI genes (H2-K and H2-D; (Thams et al., 2009)). However, to date no studies have examined if any MHCI genes are expressed at the developing NMJ, when synapse elimination is occurring.
To assess if any MHCI genes are expressed in a time and place consistent with a role in synapse elimination at the NMJ, cDNA was prepared from microdissected motor end plate band, a region that contains terminal Schwann cells as well as the synaptic connections between motor neurons and muscle fibers. Reverse transcriptase polymerase chain reaction (RT-PCR) was performed on extracts of these samples using primers against 17 MHCI genes, including both of the classical MHCI genes expressed in C57Bl/6 mice (K and D), and members of the Q (Q2, Q4, Q5, Q6, Q7), T (T5, T7, T10, T11, T15, T22, T23, T24), and M (M2, M3) families of nonclassical MHCI genes. Forward primers generally targeted the extracellular α1 domain, one of the most polymorphic regions of MHCI, to maximize primer specificity. Reverse primer sites varied in their location, and included downstream locations in the α2, α3, transmembrane, or cytoplasmic domains (Fig. 2). A complete list of the primers used in these studies, most of which were obtained from (Ohtsuka et al., 2008), can be found in Table 1. As a positive control, RT-PCR was also performed on cDNA from thymus, which expresses most MHCI genes (Ohtsuka et al., 2008). NMJ samples were collected at three different ages: while synapse elimination is ongoing (P7), soon after synapse elimination has concluded (P15), and in more mature animals (P30) (Sanes and Lichtman, 1999). All three ages were profiled in three different muscles: diaphragm (DIA), extensor digitorum longus (EDL), and soleus (SOL). Each experiment was repeated in samples from five different animals. For all experiments, products were verified by matching to predicted size of product, as well as direct sequencing of the RT-PCR amplified fragments, unless otherwise noted.
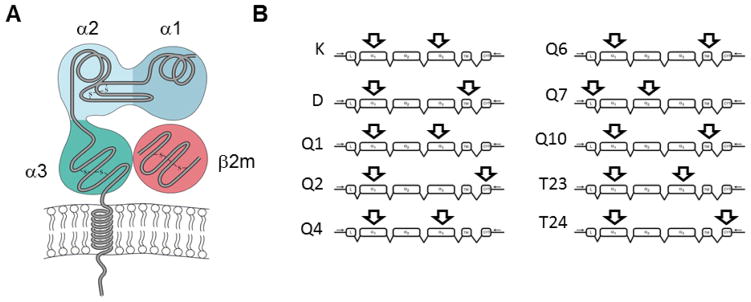
Figure 2. Representative forward and reverse primer binding sites.
A. Schematic of the structure of a typical MHCI molecule (adapted from (Goldsby and Goldsby, 2003)).
B. Examples show approximate locations of predicted forward and reverse primer binding on specific MHCI cDNAs. The locations of primer binding determine the size of the predicted product, and also define the sequence area within which alternative splicing events will be detected. MHCI transcript skeleton adapted from (Renthal et al., 2011).
Table 1. Primer sets used for RT-PCR amplification of MHCI genes.
MHCI primers were obtained from (Ohtsuka et al., 2008). Primers for controls and MHCI-like genes are predicted to recognize unique regions of the nucleotide sequence for each gene. Size of the predicted RT-PCR product is given in base pairs. Multiplex RT-PCR primers were designed to amplify most transcribed MHCI genes, using a degenerate forward primer and an AL1 poly(dT) reverse primer. Y, pyrimidine (C or T); R, purine (A or G).
| Gene | Forward Primer | Reverse Primer | |
|---|---|---|---|
| MHCI | K | GGGAGCCCCGGTACATGGAA | GGTGACTTTATCTTCAGGTCTGCT |
| D | TCGGCTATGTGGACAACAAGG | GGCCATAGCTCCAAGGACAC | |
| Q1 | CTGCGGTATTTCGAGACCTCG | GGTATCTGTGGAGCCACATCAG | |
| Q2 | ACACACAGGTCTCCAAGGAA | TGGATCTTGAGCGTAGTCTCTTA | |
| Q4 | CTTGCTGAGTTATTTCTACACCT | ACCGTCAGATCTGTGGTGACAT | |
| Q5 | GGGAGCCCCGGTTCATCATC | CAGGGTGACAGCATCATAAGATA | |
| Q6 | GTATTTCCACACTGCTGTGTCCT | AAGGACAACCAGAATAGCTACGT | |
| Q7 | CGGGCCAACACTCGCTGCAA | GTATCTGCGGAGCGACTGCAT | |
| Q10 | CACACTCCATGAGGTATTTCGAA | CAGATCAGCAATGTGTGACATGATA | |
| T5 | GGTGGTGTTGCAGAGACGCT | CTGCTCTTCAACACAAAAGG | |
| T7 | CTTCACACGTTCCAGCTGTTGTT | AGGCCTGGTCTCCACAAGCTCT | |
| T9 | ACCGCCCTGGCCCCGACCCGA | CATCCGTGCATATCCTGGATA | |
| T10 | CCCTTTGGGTTCACACTCGCTT | CCTGGTCTCCACAAACTCCACTTCT | |
| T11 | CGGTATTTCCACACCGTCGTA | TAGAGATATGCGAGGCTAAGTTG | |
| T15 | ACCGCCCTGGCCCCGACCCAA | CATCCGTGCATATCCTGGATT | |
| T22 | CTGGAGCAGGAGGAAGCAGATA | CAAATGATGAACAAAATGAAAACCA | |
| T23 | AGTATTGGGAGCGGGAGACTT | AGCACCTCAGGGTGACTTCAT | |
| T24 | ATGCACAGTACTTCACTCATG | CCCCTAGCATATACTCCTGTCG | |
| M2 | GAGGAGACCCACTACATGACTGTT | GAAAATGAAAGACTGAGGAGGTCTAC | |
| M3 | CAGCGCTGTGATAGCATTGA | ACAACAATAGTGATCACACCT | |
| MHCI-like | CD1d | AAAAACCCCAGAGCCTTTGTGTACC | TGTCTGCCAGGACGTTCCCCA |
| FcRn | CCTCAGTGCCCACGGCTGTG | CTGCGCATCCTGCCCCACAA | |
| HFE | CCGGTGGACCCAGCTGAGGA | ACGGCCAATGTCAGCCAGCC | |
| MR1 | TCCGATCCTGGTCCCGTCGTC | TTCCCGAGGGGCCTCCAGAAC | |
| ZAG | GGTGCCTGTCCTGCTGTCCC | AGGTATGCCTTGGCTCGCTGC | |
| Positive controls | Arf | ATGGGGAATATCTTTGCAAACCTC | CTTCTGGTTCCGGAGCTG |
| ODC | TGGAGTGAGAATCATAGCTG | TTGGCCTCTGGAACCCATTG | |
| Multiplex PCR | Pan-MHCI/AL1 poly dT | ACYYTGARRTGYTGGGCYYT | ATTGGATCCAGGCCGCTCTGGACAAAATATGAATTCT(24) |
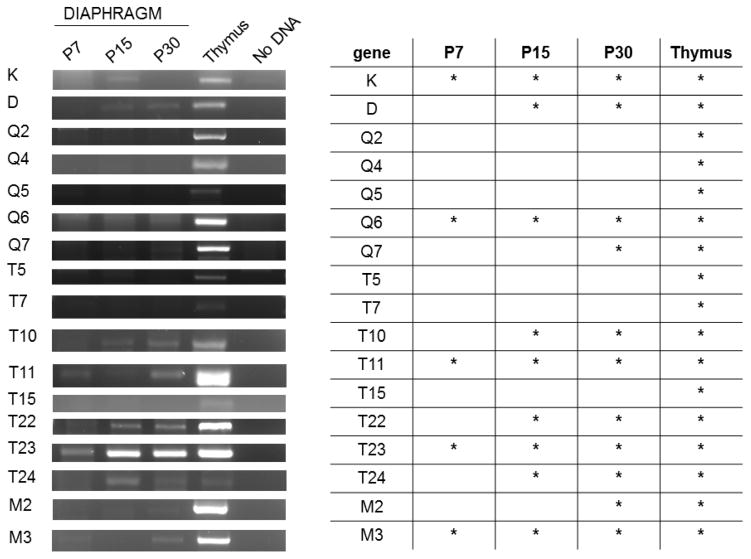
Using this RT-PCR-based approach, expression of many members of both the classical and nonclassical MHCI gene families were detected at the developing NMJ. At least five MHCI genes are expressed in the diaphragm at P7, when synapse elimination is occurring (K, Q6, T11, T23, and M3; Fig. 3). By P15, when synapse elimination in the diaphragm is complete (Sanes and Lichtman, 1999), more MHCI genes--about half of the MHCI genes assessed--are detected, and by P30, two-thirds of the profiled MHCI genes are expressed at the NMJ. All of the profiled MHCI genes were detectable in thymus, confirming that the lack of some MHCI genes in diaphragm was not due to inability of the primers to detect their target.
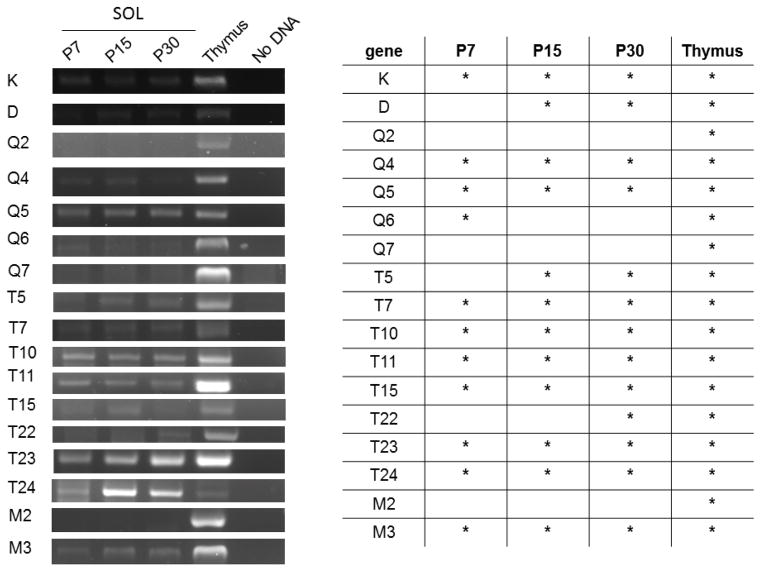
Figure 3. Detection of MHCI mRNA in the end plate band of diaphragm muscle at three different ages.
Left, Example gels show products amplified from the diaphragm end plate band of one animal per age. Band brightness can be influenced by many variables, including levels of transcript, efficiency of primer sets, and the size of product, and should not be considered a reliable measure of transcript levels. Five animals per age were tested.
Right, summary table. Asterisks denote positive detection of the correct product, verified by sequencing, in at least 3/5 of the animals tested at that age (most commonly, 5/5).
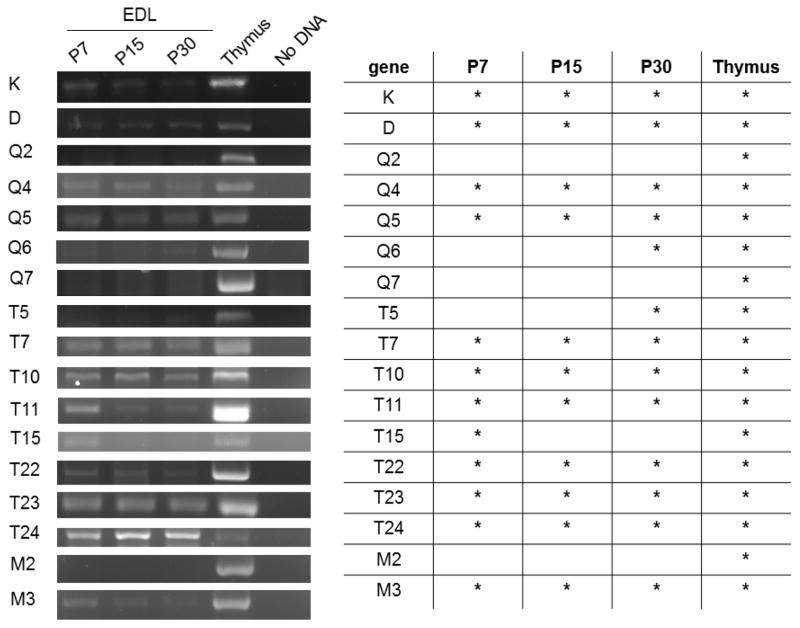
In order to determine if MHCI gene expression is a unique feature of diaphragm, or is a general feature the NMJ, we examined two hind-limb muscles, the EDL and SOL. Both undergo developmental synapse elimination on roughly the same timetable as diaphragm (reviewed in (Sanes and Lichtman, 1999)). In the P7 EDL, more than twice as many MHCIs were detected compared to P7 diaphragm. Expression of 12 MHCI genes (K, D, Q4, Q5, T7, T10, T11, T15, T22, T23, T24, and M3) is detectable in the EDL at P7, when synapse elimination is ongoing. At P15 in EDL, T15 is no longer detected. By P30, however, even more MHCI genes--three-quarters of the profiled MHCI genes--are expressed at the NMJ in EDL (Fig. 4). Interestingly, of the 12 MHCI transcripts we detected in P7 EDL, only four-- K, T11, T23, and M3--are also expressed in diaphragm muscle at the same age. These same four were also expressed at all ages examined in both muscles, suggesting they are consistently expressed at the NMJ.
Figure 4. Detection of MHCI mRNA in the end plate band of EDL muscle at three different ages.
Left, Example gels show products amplified from the EDL end plate band of one animal per age. Five animals per age were tested.
Right, summary table. Asterisks denote positive detection of the correct product, verified by sequencing, in at least 3/5 of the animals tested at that age.
In the SOL, mRNA encoded by at least 11 MHCI genes (K, Q4, Q5, Q6, T7, T10, T11, T15, T23, T24, and M3) is expressed at P7. By P15, 12 of the 17 profiled MHCI genes are expressed, and the number of MHCI genes expressed is highest at P30 (Fig. 5). In both SOL and EDL, some MHCI genes are detected when synapse elimination is occurring, but not at other ages (Q6 and T15, respectively). In both diaphragm and SOL, K is the only classical MHCI expressed at P7, while both K and D are expressed in EDL during synapse elimination.
Figure 5. Detection of MHCI mRNA in the end plate band of SOL muscle at three different ages.
Left, Example gels show products amplified from the SOL end plate band of one animal per age. Five animals per age were tested.
Right, summary table. Asterisks denote positive detection of the correct product, verified by sequencing, in at least 3/5 of the animals tested at that age.
One trend across all three muscles is that the number of MHCI transcripts that are detectable increases with age, with the fewest MHCI genes expressed at P7, a trend that is most striking in diaphragm. In addition, the overlap in profiled gene expression at P7 is significantly higher between SOL and EDL (59% overlap) than between either of these hind limb muscles and the diaphragm (SOL and diaphragm, 29% overlap; EDL and diaphragm, 24% overlap). These results together show that a large number of both classical and nonclassical MHCI genes are expressed at the developing NMJ, and that their expression is dynamic over development, and may be specialized for different muscles. They also identify a set of four MHCI genes (K, T11, T23, and M3) that are expressed in the right time and place to play a role in developmental synapse elimination in multiple muscles.
MHCI protein expression at the developing NMJ
The above results demonstrate that many MHCI genes are transcribed into mRNA at the developing NMJ. However, presence of mRNA does not guarantee that the mRNA will be translated into protein. Previous studies have shown that MHCI protein is expressed at the NMJ in adults (Thams et al., 2009). To our knowledge, however, no one has examined if MHCI protein is expressed at the developing NMJ, as synapse elimination is occurring. Therefore, MHCI protein expression was examined in diaphragm muscle at three ages: while synapse elimination is ongoing (P7), just after synapse elimination has concluded (P15), and more mature animals (P30) (Sanes and Lichtman, 1999). Whole-mount diaphragm muscles were immunolabeled with antibodies against MHCI or an IgG isotype control primary antibody, as well as an antibody against the presynaptic marker synaptophysin. Postsynaptic nicotinic acetylcholine receptors (nAChRs) were labeled using fluorescently-conjugated alpha bungarotoxin (α-btx; see Methods).
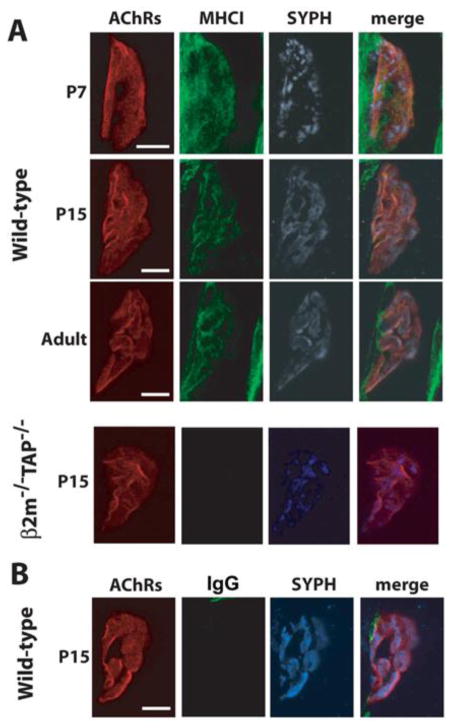
Immunostaining revealed that specific MHCI protein labeling is present at the NMJ at all three ages (Fig. 6). This labeling is largely abolished in samples from β2m−/−TAP−/− mice, which lack stable cell-surface expression of most MHCI proteins (see Methods). MHCI protein is not expressed uniformly throughout the tissue, but is enriched at the NMJ. There is also MHCI labeling that extends beyond the NMJ, but this appears to be nonspecific, as it is still present when the anti-MHCI primary antibody is omitted and an IgG control primary antibody is used (Fig, 6B). These results show that one or more MHCI mRNAs are translated into protein at the NMJ at a wide range of ages. They are also the first demonstration that MHCI protein is expressed at the developing NMJ.
Figure 6. Detection of MHCI protein at the NMJ in diaphragm at multiple ages.
(A) Representative confocal micrographs (maximum projections of Z-stacks) of triple-labeled NMJs from WT (top) or MHCI-deficient β2m−/−TAP−/− (bottom) mouse diaphragm at the indicated ages. MHCI immunoreactivity (green); nAChRs (red); synaptophysin (SYPH, blue). As expected, β2m−/−TAP−/− mice lack specific MHCI labeling.
(B) IgG1 isotype control for MHCI immunostaining in P15 WT diaphragm. Representative images of triple-labeling of the diaphragm using α-btx (nAChRs, red), isotype-specific control antibody for the anti-MHCI antibody (IgG1, green) and anti-synaptophysin antibody (SYPH, blue). Experiments in (B) were performed on hemidiaphragms from the same WT P15 animal as in (A), imaged on the same day, with identical illumination and exposure parameters. Scales, 10mm.
MHCI gene expression in the hippocampus
In the hippocampus, one or more MHCIs are necessary for normal synapse density (Dixon-Salazar et al., 2014), synaptic transmission (Fourgeaud et al., 2010), and synaptic plasticity (Goddard et al., 2007; Huh et al., 2000; Nelson et al., 2013), and for some hippocampus-dependent forms of learning and memory (Nelson et al., 2013). Importantly, these studies were all conducted in β2m−/−TAP−/− mice, which lack stable cell-surface expression of many classical and nonclassical MHCIs as well as some MHCI-like proteins (Bhatt et al., 2007; Brutkiewicz et al., 1995; Ljunggren et al., 1995; Praetor and Hunziker, 2002; Yamaguchi and Hashimoto, 2002). While multiple studies have detected MHCI protein in hippocampal neurons (e.g., (Dixon-Salazar et al., 2014; Goddard et al., 2007)), these studies did not resolve which MHCI genes are expressed. As an essential step towards a mechanistic understanding of how MHCI regulates hippocampal synaptic transmission and plasticity, it is imperative to determine which of the dozens of mouse MHCI genes are expressed in the hippocampus.
To characterize MHCI gene expression in the hippocampus, primer sets for twenty classical and nonclassical MHCI genes and five MHCI-like genes were used to probe samples from P14 mice. P14 was chosen because NMDAR-mediated synaptic transmission is enhanced in hippocampal slices from P13-P16 mice (Fourgeaud et al., 2010). MHCI-like genes were included because some MHCI-like proteins, including HFE, FcRn, MR1, and CD1d1, normally associate with β2m, and therefore their function may also be disrupted in β2m−/−TAP−/− mice (Bhatt et al., 2007; Brutkiewicz et al., 1995; Praetor and Hunziker, 2002; Yamaguchi and Hashimoto, 2002). Liver and spleen samples from the same animals were used as a positive control.
These twenty-five primer pairs yielded RT-PCR products corresponding to a total of fourteen distinct MHCI and MHCI-like genes from P14 mouse hippocampus. Both of the classical MHCI genes in this mouse strain, K and D, were expressed, as were multiple nonclassical MHCI genes of each subtype (Fig. 7 and Table 2). These included the four MHCI genes that were also detected in all muscles at all ages (K, T11, T23, and M3). Thus these four MHCI genes appear to be expressed in the CNS as well as in the PNS at a variety of ages. Three MHCI-like genes, FcRn, Cd1d1, and MR1, were also detected in hippocampus (Fig. 7), while two other MHCI-like genes, HFE and ZAG, were not detected.
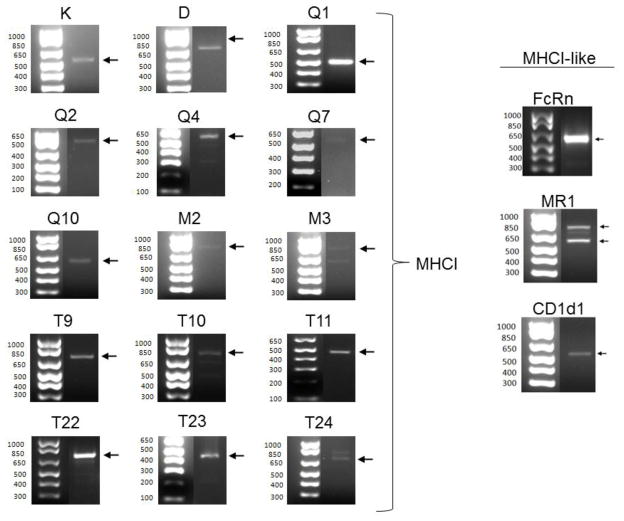
Figure 7. RT-PCR products encoding 13 different MHCI and 3 MHCI-like genes detected in hippocampal extracts.
Arrows indicate primary RT-PCR product. All products but Q2, Q10 and T9 ran at the expected size; Q2 and T9 also could not be verified by sequencing, and therefore are omitted from the list of transcribed genes. The smaller-than-expected Q10 product was verified as a splice variant of Q10 by sequencing (see Fig. 8). All other products’ identities were also verified by sequencing.
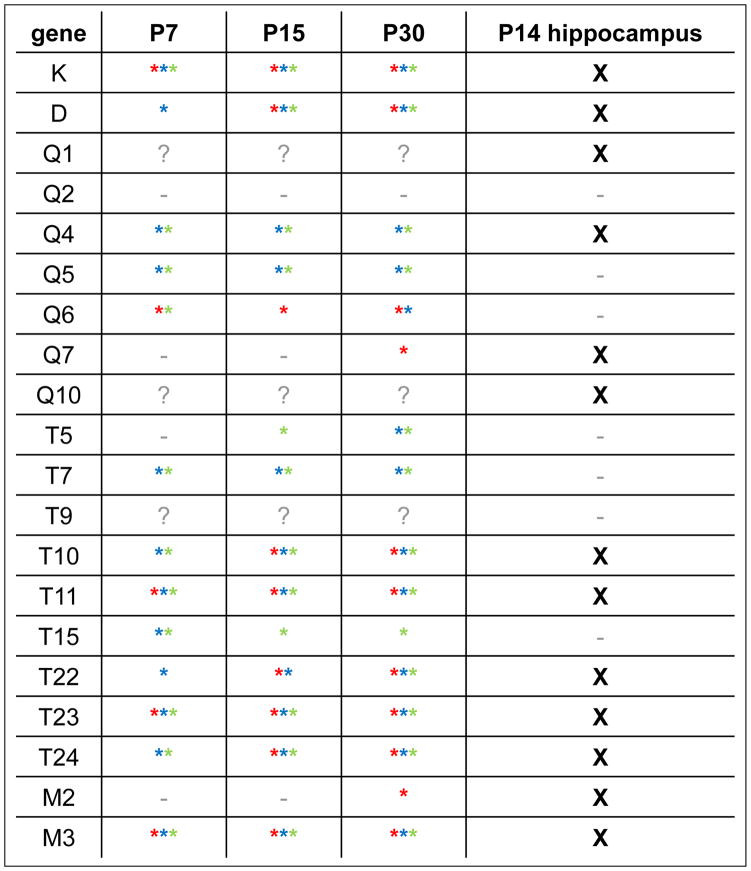
Table 2. Summary table of MHCI gene expression in three different developing muscles at three different ages, and in P14 hippocampus.
Left, asterisks indicate verified (sequenced) products detected in at least 3/5 animals in diaphragm (DIA; red), extensor digitorum longus (EDL; blue), or soleus (SOL; green). “?” indicates primer pairs that were not attempted in muscle. Far right, X indicates verified products detected in P14 hippocampus extract. “-“ on both sides indicates products that were never detected in at least five independent experiments.
MHCI gene expression in individual hippocampal neurons
The above results demonstrate that many MHCI transcripts can be detected in extracts of whole hippocampus. Importantly, MHCI’s roles in the hippocampus appear to be restricted to specific regions and cell types. For example, synapse density is elevated in area CA3, but not area CA1, of the adult hippocampus in MHCI-deficient β2m−/−TAP−/− mice (Dixon-Salazar et al., 2014), while synaptic transmission and plasticity are altered at synapses made onto CA1 neurons in the same MHCI-deficient mice (Fourgeaud et al., 2010; Huh et al., 2000; Nelson et al., 2013). Thus it is important to determine which MHCIs are present in the particular cell types where these phenotypes are observed. To date, however, most studies of MHCI mRNA and protein expression in the hippocampus have been performed in hippocampal extracts, or in hippocampal neurons in culture, where cell types are mixed. In situ hybridization studies have been performed in brain slices for a handful of MHCI mRNAs (e.g., (Huh et al., 2000; Liu et al., 2015)). These studies hint at within-hippocampus differences in MHCI gene expression, but their spatial resolution is not sufficient to identify specific cell types.
To definitively assess MHCI gene expression differences within the hippocampus, we performed single-cell RT-PCR. Whole-cell patch pipettes were used to form a gigaohm seal onto individual neurons in acute hippocampal slices from P14–18 mice, and the cell contents were aspirated into the pipette. The tip of the pipette was then broken into a reaction tube, and RT-PCR was performed. A multiplex RT-PCR strategy was used to first amplify all MHCI mRNAs, using pan-specific primers, followed by a second round of amplification using gene-specific primers (see Methods). Three types of neurons were sampled: CA1 pyramidal neurons, CA3 pyramidal neurons, and granule cells of the dentate gyrus. RT-PCR was attempted for both classical MHCIs (K and D), as well as members of the Q (Q1, Q2), T (T9, T11, T23, T24) and M (M3) families which were detected in our whole-hippocampus samples at the same age (Table 2), suggesting they are expressed in one or more cell types in hippocampus.
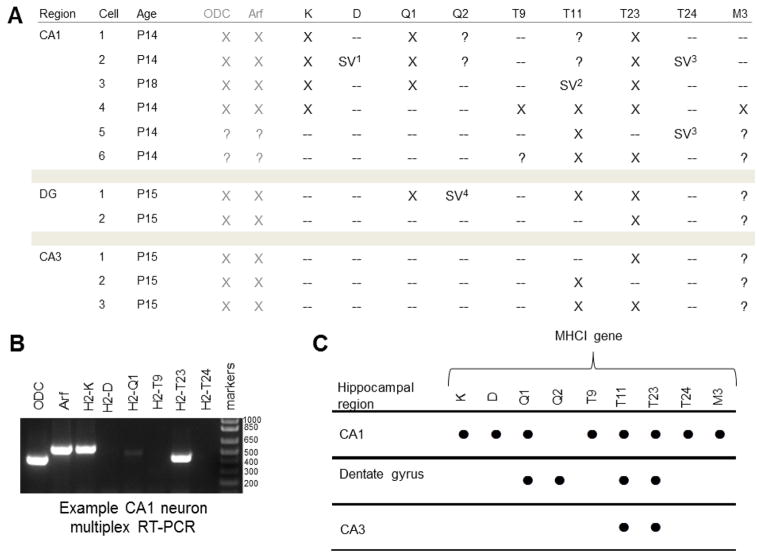
Neurons isolated from area CA1 consistently expressed the largest number of MHCI genes, of the three neuron types examined. MHCI mRNAs detected in CA1 neurons included T23 (present in 5/6 neurons sampled), K (4/6), Q1 (3/6) and T24 (2/6). Other MHCI genes appear to be more restricted in their expression: D, T9, and M3 were each detected in only one CA1 neuron. However, this method could have a high false-negative rate, given the low quantities of mRNA involved. To assess the false-negative rate, we included two positive control probe sets for transcripts that are expressed at moderate levels in neurons (Dolter and Braman, 2001; Koirala and Corfas, 2010): ornithine decarboxylase (ODC), a key enzyme in polyamine biosynthesis (Slotkin and Bartolome, 1986), and ADP-ribosylation factor 1 (Arf), a central component of both COPI and clathrin coat assemblies (Haynes et al., 2005). Both yielded products in every neuron examined (Fig. 7), suggesting that the false-negative rate for moderately-expressed gene products is low. However, the false-negative rate for MHCI may be higher if its abundance is lower. Therefore it is impossible to conclude that MHCIs that are not detected are truly absent. The likelihood of a false positive result, in contrast, is small, particularly since RT-PCR products were verified by sequencing. Thus the MHCI genes detected here represent the minimum set of MHCI genes that are expressed in these hippocampal neuron types.
Cells isolated from other regions of the hippocampus had strikingly and consistently different MHCI gene expression patterns. Unlike CA1 pyramidal neurons, which in total expressed eight different MHCI genes, only two MHCI mRNAs were detected in CA3 pyramidal neurons (T11 and T23). Granule cells of the dentate gyrus also expressed T11 and T23, as well as Q1 and Q2 (Fig 8). Larger sample sizes will ultimately be required to fully define the entire pool of MHCI genes expressed in these cell types. However, the current studies suggest that there are significant, cell-type-specific differences in MHCI gene expression within the hippocampus. In addition, they demonstrate, for the first time, that individual neurons can express more than one MHCI, and can simultaneously express both classical and nonclassical MHCI genes.
Figure 8. MHCI gene expression in individual P14–18 hippocampal neurons.
A. Table summarizing single-cell RT-PCR data for individual neurons from specific regions of mouse hippocampus.
B. Example gel showing multiplex RT-PCR of cDNA extracted from a single CA1 neuron. Summary. mRNA transcripts for eight MHCI genes were observed in individual CA1 neurons (indicated with ●), while four were observed in granule cells of the dentate gyrus and two were observed in neurons of the CA3 region. Number of individual neurons sampled: CA1, n = 6; Dentate gyrus, n = 2; CA3, n = 3. DG, dentate gyrus; ODC, ornithine decarboxylase (positive control); Arf, ADP-ribosylation factor 1 (positive control); SV, splice variant. “?” indicates RT-PCR not performed on a given sample.
SV1 D splice variant: 818 base pairs (bp) predicted product, 548bp observed, no α1.
SV2 T11 splice variant: 415bp predicted product, 659bp observed, retained second intron.
SV3 T24 splice variant: 736bp predicted product, 184 bp observed, no α2 or α3.
SV 4 Q2 splice variant: 785bp predicted product, 509bp observed, no α3.
Alternative splicing of MHCI in the CNS but not PNS
Interestingly, several MHCI primer pairs generated RT-PCR products in whole hippocampus samples which did not match the predicted size, either as the sole product, or in addition to a product of the predicted size. Four of these alternatively-sized products were confirmed by sequencing to be splice variants of MHCI genes (Fig. 9). Some of these putative splice variants contained one or more introns, which is also a feature of unspliced genomic sequences. To ensure removal of any possible contaminating genomic DNA, experiments were repeated in the presence of DNase. In addition, in all experiments, cDNA was generated using primers against the polyA tail, which is characteristic of mature mRNA. Consistent with the idea that these procedures successfully eliminated genomic contamination, we never detected sequences containing all introns, indicative of genomic sequences. Furthermore, retained introns or other splice variants were never detected in samples from the NMJ at any age in any of the three muscles examined.
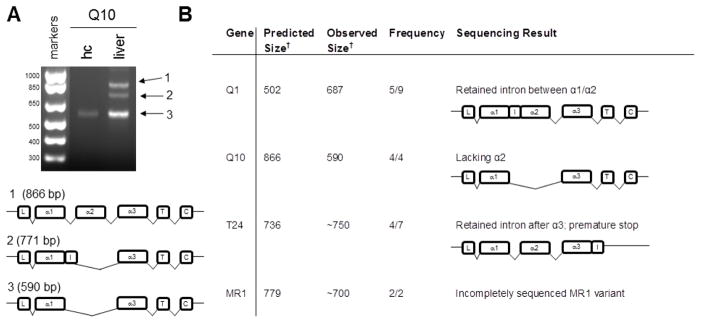
Figure 9. Alternative splicing of MHCI genes in whole hippocampus.
A. Products of smaller-than-expected size were detected using primers against the nonclassical MHCI Q10. The MHCI heavy chain is organized into eight exons, each of which encodes a functional domain. Exon 1 encodes the signal peptide, and exons 2, 3, and 4 encode the α1, α2, and α3 domains, respectively. The transmembrane domain is encoded in exon 5, and the short cytoplasmic tail is encoded by exons 6, 7, and 8. Sequencing confirmed that the products of unexpected size were splice variants of Q10 that both lacked the α2 domain, while one, which was detected in liver, included a retained intron (I). The α2-deficient transcript (3) was the only form of Q10 detected in hippocampus (hc). L, leader sequence; T, transmembrane domain; C, cytoplasmic domain, I, retained intron.
B. RT-PCR products larger or smaller than the predicted size were observed with twelve primer pairs. Frequency reported as number of times a band of that size observed in experiments using the same sample type where any band was detected with that primer set. Sequencing of several products confirmed their identity as an MHCI splice variant. Sequencing was inconclusive for experiments indicated with a “-“. †Size in base pairs.
For Q10, the only product ever observed in whole hippocampus extracts (4/4 experiments) was smaller than predicted. Sequencing data show that this short product is a splice variant which completely lacks the α2 domain. This splicing event would likely eliminate the capacity of the protein to present antigen. In contrast, full-length Q10 was readily detected in liver, alongside two α2-free forms (Fig. 9). Alternatively-spliced forms of the MHCI-like gene MR1 were also detected in hippocampus (Fig. 7 & 9). We frequently observed a retained second intron (between α1 and α2) in Q1 from hippocampus (5/9 experiments). A T24 variant observed in hippocampal tissue retained the fourth intron, between the α3 and transmembrane domains, as confirmed by sequencing, effectively eliminating the transmembrane and intracellular domains, and is predicted to encode a soluble protein (Fig. 9). MHCI splice variants for D, Q2, T11 and T24 were also detected in hippocampal single-cell samples (Fig. 8). Intriguingly, while most of the same primers were used to detect MHCI at the NMJ, using the same sample concentrations, over an even greater number of experiments, no splice variants were ever detected in NMJ samples. These alternatively-spliced transcripts suggest that MHCI proteins could be produced in the hippocampus with divergent structures, and altered or absent immune functions.
Discussion
A growing literature supports important roles for MHCI in the development, function, and plasticity of the vertebrate nervous system (reviewed in (Boulanger, 2009; Elmer and McAllister, 2012; Shatz, 2009)). However, little is known about the specific MHCI genes that mediate the newly-discovered neuronal functions of MHCI. Of the roughly twenty-one nonclassical MHCI genes that are known to be transcribed (Guidry and Stroynowski, 2005; Ohtsuka et al., 2008), we detected sixteen in the nervous system (Q1, Q4, Q5, Q6, Q7, Q10, T5, T7, T10, T11, T15, T22, T23, T24, M2, and M3). We also detected both classical MHCI genes found in C57Bl/6 mice (K and D), as well as three MHCI-like genes (FcRn, MR1, and Cd1d1). This list includes eight MHCI transcripts that were not detected in the nervous system via RT-PCR in previous studies (Q5, Q6, Q10, T5, T7, T9, T10, and M2; (Guidry and Stroynowski, 2005; Ohtsuka et al., 2008)), which examined different regions of the CNS, at different ages. Our results at the NMJ demonstrate that MHCI gene expression can change rapidly during early postnatal development, and our single-cell studies in hippocampus show that MHCI gene expression can differ even in different cell types within a single brain region. This remarkably dynamic and spatially specialized expression of different MHCI genes in the nervous system may have contributed to the long-standing belief that MHCI is not expressed in the nervous system under basal conditions.
Hippocampal synapse density and hippocampal synaptic transmission and plasticity are altered in β2m−/−TAP−/− mice, which lack stable cell-surface expression of many MHCI proteins (Dixon-Salazar et al., 2014; Fourgeaud et al., 2010; Huh et al., 2000; Nelson et al., 2013). The current study identified a pool of MHCI genes and MHCI-like genes that are expressed in the hippocampus. The protein products of several of these MHCI genes are likely disrupted in β2m−/−TAP−/− mice, because they require β2m and/or TAP for their stable cell-surface expression (Chiu et al., 1999; Crowley et al., 1997; Ke and Warner, 2000; Ljunggren et al., 1995; Robinson et al., 1998; Zappacosta et al., 2000). The list of MHCI genes that are expressed in the hippocampus could be used to guide targeted manipulations of the expression and function of specific MHCI genes in future studies.
This study also identified significant cell-type-specific differences in MHCI gene expression within the hippocampus. These differences may be relevant to phenotypes that are restricted to certain regions of the hippocampus in systemically MHCI-deficient mice, such as increased synapse number, which is seen in area CA3, but not area CA1 (Dixon-Salazar et al., 2014), and synaptic transmission and plasticity, which are altered at synapses made onto CA1 pyramidal cells by CA3 pyramidal cells (Fourgeaud et al., 2010; Huh et al., 2000; Nelson et al., 2013). The current studies identified nine MHCI genes which could be involved in the regulation of synaptic transmission and plasticity at the CA1/CA3 synapse, because they are expressed in pre- and/or post-synaptic cells at that synapse. These single-cell studies also demonstrate that individual neurons can express more than one MHCI.
Studies in MHCI-deficient β2m−/−TAP−/ −, β2m−/− (Huh et al., 2000), and Kb−/−Db−/− (Datwani et al., 2009; Lee et al., 2014) mice all support the idea that MHCI is involved in synapse elimination in the developing visual system. In the current study, we examined MHCI gene expression at another site of well-documented developmental synapse elimination, the NMJ. In adults, MHCI is expressed in presynaptic motor neurons, and has been implicated in responses to motor neuron injury (Edstrom et al., 2004; Linda et al., 1998; Linda et al., 1999; Oliveira et al., 2004; Thams et al., 2009). This is the first study to demonstrate that MHCI genes are expressed at the developing NMJ, as synapse elimination is occurring. We detected both classical and nonclassical MHCI mRNAs at the NMJ in three different developing muscles. While there were muscle-specific differences in gene expression, in all three muscles the fewest MHCI genes were expressed at the youngest ages. Furthermore, four MHCI genes—K, T11, T23 and M3—were expressed in all three NMJs at P7, when synapse elimination is ongoing. The current study was designed to sample mRNAs from all cell types at the NMJ, including muscle cells, motor neurons, and Schwann cells. In future studies, it will be of interest to determine which of the four MHCI genes expressed at the developing NMJ are expressed in each cell type. The fact that MHCI is dynamically expressed during synapse elimination at the NMJ raises the possibility that MHCI could be involved in synapse elimination at the NMJ, as it is in the developing visual system. The current studies pave the way for targeted experiments manipulating the expression of specific MHCI genes at the NMJ during early development.
Immunohistochemistry in developing (Fig. 6) and adult (Thams et al., 2009) animals suggest that MHCI protein is also present at the NMJ. There are two important technical limitations of any current study of MHCI protein expression. First, the specificity of most MHCI antibodies has not been fully characterized for the dozens of members of the MHCI family. Second, there are currently no antibodies to detect most protein products of the MHCI gene family. Therefore we focused on an mRNA-based approach, which allowed us to reliably discriminate among products of the different MHCI genes. An important subsequent step will be to validate translation of each of the mRNAs we detected, by generating and validating new anti-MHCI antibodies against the predicted protein products.
While the functional significance of MHCI splicing remains largely unknown, alternative splicing is more frequent in the MHCI than in the rest of the genome in humans (Vandiedonck et al., 2011). Splice isoforms of Q5 appear to be enriched in brain (Renthal et al., 2011). Intriguingly, alternative splicing of MHCI genes was detected in hippocampus, but not at the NMJ. The specific splicing events that we detected in the hippocampus have also been observed outside the CNS (Ohtsuka et al., 2008; Tajima et al., 2003). When considering the splicing events that have been identified to date, it is important to keep in mind that two classes of splicing events will be missed: (1) those that fall outside the area between the two primers, and (2) those that excise one or both sites targeted by the primers. An important part of understanding the functional significance of these splicing events in the brain will be to determine if the MHCI transcript isoforms observed here are translated into stable proteins.
Taken together, the current results significantly expand our understanding of the potential role of specific MHCI genes in the developing and adult nervous system. They define a pool of MHCI genes that could underlie the changes in hippocampal synapse number, synaptic function and plasticity seen in β2m−/−TAP−/− mice (Dixon-Salazar et al., 2014; Fourgeaud et al., 2010; Huh et al., 2000; Nelson et al., 2013). They also show that individual neurons can express multiple MHCI genes, and suggest that alternative splicing may further expand the functional diversity of CNS-expressed MHCI genes. Finally, these studies provide the first evidence that MHCI is expressed at the developing NMJ, and identify specific members of the large MHCI family that could potentially play a role in development of the NMJ.
Methods
Experimental Animals
All mice were housed and handled in accordance with the Princeton Institutional Animal Care and Use Committee (IACUC) and Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC)-approved procedures and standards. C57BL/6J mice, which are MHCI haplotype b, were obtained from Jackson Labs. Mice were housed in a 12 hour light-dark cycle with food and water access ad libitum. For studies of MHCI in the peripheral nervous system, diaphragm, EDL, and SOL muscle were collected from developing (P7 and P15) and mature (P30) mice (see below). Five animals (male and female) were examined per age group. To identify genes expressed in the hippocampus at an age when synaptic transmission is known to be regulated by MHCI (Fourgeaud et al., 2010), P14 mice were examined. For immunolabeling, we used P7, P15, and P30 WT mice, and P15 β2m−/−TAP−/− mice. These knockout mice lack two key components of the MHCI expression pathway: β2-microglobulin (β2m), the invariant light chain of the MHCI protein, and the transporter associated with antigen processing 1 (TAP1), an endoplasmic reticulum-expressed transporter required to load antigenic peptides onto MHCI. β2m−/−TAP−/− mice have reduced stable cell-surface expression of most of the over 50 MHCI proteins found in mouse (Dorfman et al., 1997; Ljunggren et al., 1995). MHCI-deficient β2m−/−TAP−/− mice were obtained from D. Raulet and C. Shatz, and were backcrossed more than 8 times to C57Bl/6.
RT-PCR
The presence of MHCI genes was assessed by reverse transcription (RT)-PCR of cDNA, which allows rapid, unambiguous discrimination among the many members of the MHCI gene family. Mice were deeply anesthetized with isoflurane inhalation anesthesia to a surgical plane, and rapidly decapitated prior to dissection. Liver, thymus, diaphragm (DIA), extensor digitorium longus (EDL), soleus (SOL) and/or brain were then removed (see below for individual dissections). Tissues were dissected under 10X magnification in a binocular dissecting microscope, flash frozen on dry ice, and stored at −80 degrees C until processing. Dissections were performed as follows. Diaphragm: The body was placed in a supine position on a Sylgard- (Dow Corning) lined dissecting tray and pinned through all four paws with the limbs extended. Identical horizontal incisions (right and left) were made at the level of the 4th rib, proximal to the diaphragm, beginning at the manubrium and moving posteriorly to the spinal cord. Similar incisions were made distal to the diaphragm. After severing the spinal cord, the diaphragm was removed while still attached to the adjacent ribs and connective tissue. The diaphragm and associated ribs and connective tissue was pinned onto a Sylgard- (Dow Corning) lined glass petri dish. A narrow (~1mm wide) strip of muscle including the end plate band (identified by position) was removed. EDL/SOL: The lower limb was transected at the mid-point of the femur, skinned, and pinned supine to a Sylgard-lined glass petri dish (5″ diameter). The tibialis anterior muscle was first removed by exposing and cutting its distal tendon, and then using the tendon as a guide to peel away the overlying muscle, exposing the EDL underneath. EDL was then microdissected, beginning at the distal tendon in the foot, and flash frozen. Once the EDL was removed, the leg was turned into the prone position and re-pinned. An incision detaching all of the tendons beginning at the calcaneus was made, and the tendons peeled upward together, exposing the proximal tendon of the soleus. The soleus was then removed by snipping at the origin and insertion. Liver, spleen, and thymus: gross dissections to remove the liver, spleen, and thymus were performed using a mouse anatomical atlas as a guide. Liver and thymus were used for primer design and optimization, and as positive controls, since many MHCI genes are expressed in these tissues (Ohtsuka et al., 2008). Hippocampus: a midline incision through the skin was made from the back of the neck to the nose using surgical scissors. The skin was peeled away from the skull from caudal to rostral. A second midline incision was made through the skull using fine surgical scissors, keeping the scissors tips pointing up towards the skull, and making small snips. The skull was flapped down from the midline toward the base of the skull. The brain was blocked using a razor blade to cut straight down, removing the olfactory bulbs (rostrally) and the cerebellum (caudally). The blocked brain was scooped from the skull using a small flat-bottomed weighing scoop, and placed on a Sylgard-lined petri dish. The brain was bisected down the midline using a razor blade, and the cerebral cortex teased away gently using two pairs of forceps, exposing the hippocampus, which was removed using forceps and flash-frozen.
Total RNA was extracted from the resulting samples using an RNAeasy Mini Prep kit (QIAGEN), following manufacturer’s instructions. mRNA was reverse transcribed to cDNA using qScript cDNA synthesis KIT (Quanta Biosciences), which uses oligo-dT adapter primers to preferentially reverse-transcribe mature mRNAs. RT-PCR was performed in a 20 μl total reaction volume under the following Thermocycler conditions: 95°C for 5 min (to denature cDNA), followed by 35 cycles of amplification (95°C for 45 s, 51–64°C for 30 s [based on primer melting temperature and optimization experiments] and 72°C for 1 min) and 5 min at 72°C. MHCI gene-specific primer sets were previously described (Ohtsuka et al., 2008) (for these and other primers, see Table 1). RT-PCR products were subjected to electrophoresis on 1.5% w/v agarose gels and visualized using an ethidium bromide stain. RT-PCR products were excised from gels and isolated using a Zymoclean Gel DNA Recovery Kit (Zymo Research). Resulting DNA samples were submitted with appropriate forward and reverse primers to GeneWiz for sequencing. Sequenced products were compared to known MHCI sequences on the NCBI Gene database (http://www.ncbi.nlm.nih.gov/gene) to confirm product identity.
MHCI and synaptophysin immunohistochemistry in diaphragm
Dissected diaphragm muscle (see above) was drop-fixed with 4% PFA (Electron Microscopy Systems) in phosphate buffered saline (PBS, Sigma) for 30 min. All subsequent steps were performed at 4° C. Muscle was rinsed briefly in PBS, and nonspecific labeling was reduced by incubating in a blocking solution containing 5% bovine serum albumin, 0.01% Triton X-100 and 0.1% sodium azide (Fischer) for one hour. Muscles were treated with primary antibodies against synaptophysin (SYPH, Santa Cruz, 1:100) and MHCI (OX18, AbdSerotec, 1:100) in block overnight. OX18 is a mouse monoclonal antibody that recognizes a monomorphic epitope in the α3 region of the rat MHCI RT-1A (Fukumoto et al., 1982), and reacts with mouse MHCI in Western blots and immunohistochemistry (Corriveau et al., 1998; Datwani et al., 2009; Dixon-Salazar et al., 2014; Goddard et al., 2007; Huh et al., 2000). The suspected mouse orthologue of rat RT-1A, H-2K1, is 84.9% identical and 89.2% similar to RT-1A at the amino acid level in the α3 region, the site of the OX-18 epitope. Several lines of evidence support OX-18’s ability to bind to MHCI in the mouse nervous system. First, genetically deleting the MHCI light chain, β2m, reduces the amount of MHCI that reaches the cell surface (Dorfman et al., 1997; Zijlstra et al., 1989), and cell surface OX-18 immunofluorescence is greatly attenuated in both β2m−/− (Needleman et al., 2010) and β2m−/−TAP−/− (Goddard et al., 2007) neurons in vitro. Second, OX-18 recognizes proteins of the expected molecular weight in western blots of adult mouse brain (Corriveau et al., 1998; Dixon-Salazar et al., 2014; Huh et al., 2000) and similar labeling is seen in rat brain using a rabbit polyclonal antibody that recognizes a distinct epitope of MHCI (Needleman et al., 2010). The secondary antibodies we used to detect MHCI cause some nonspecific background labeling, which can be clearly distinguish hed from specific labeling in two ways: (1) it is still present when labeling with an isotype-control antibody, and (2) it is not abolished in MHCI-deficient animals (Fig. 6). Taken together, these results provide strong support for the specificity of OX-18 in recognizing MHCI at the mouse NMJ.
Incubation in primary antibody was followed by 3x 40 min rinses in PBS, and a four hour treatment with a cocktail of Alexa-488-conjugated donkey anti-mouse IgG (Invitrogen, 1:1000) and Cy5-conjugated donkey anti-goat IgG (Jackson, 1:100) in block. After rinsing 3x 40 min, muscles were treated for 30 min with rhodamine-conjugated α-bungarotoxon (α-btx; Molecular Probes; diluted to 0.1μg/μl in ddH2O and frozen in 100 μl aliquots, then diluted 1:10 in ddH2O immediately before use, for a final concentration of 0.01 μg/μl) to label postsynaptic nicotinic acetylcholine receptors. Samples were rinsed in 3×20 min in PBS and mounted on slides using fluorescence-stabilizing mounting media (Vectashield hard set mount). Coverslipped slides were sandwiched between two pieces of paper towel and compressed under a lead brick overnight, and stored at 4° C until imaging. Z-serial images (0.5 μm per step, up to 20μm total depth per junction) were collected from intact muscles using a Leica (Wetzlar, Germany) DMI6000 inverted microscope outfitted with a Yokogawa (Tokyo, Japan) spinning disk confocal head and an Orca ER high resolution B&W cooled CCD camera (Hamamatsu, Sewickley, PA) under a 63x oil immersion lens at 1.6x magnification. WT and β2m−/−TAP−/− samples were imaged under identical conditions during the same microscopy session.
Single-neuron mRNA extraction and RT-PCR
mRNA from the cellular contents of individual neurons was isolated by glass pipette as described previously (Eberwine et al., 1992; Koirala and Corfas, 2010; Toledo-Rodriguez et al., 2004). Briefly, acute coronal hippocampal slices were obtained from P13 to P16 C57Bl/6 mice. Following anesthesia to a surgical plane via isoflurane inhalation, mice were decapitated and the brains were removed and submersed in ice cold ACSF (in mM: 126 NaCl, 2.5 KCl, 1.25 NaH2PO4, 2 MgCl2, 2.5 CaCl2, 26 NaHCO3, and 10 glucose (Sigma)), equilibrated with 95% O2/5% CO2, and sectioned to 350 μM with a vibratome. Slices were equilibrated in oxygenated ACSF at RT for at least 15 min before being transferred to the recording chamber. Visualized whole-cell patch-clamp experiments of CA1, CA3, or dentate gyrus neurons were performed at RT. Patch electrodes (3–5 mΩ) were filled with an internal solution (in mM: 108 cesium gluconate, 20 HEPES, 0.4 EGTA, 2.8 NaCl, 5 TEACl, 4 MgATP, 0.3 NaGTP, and 10 phosphocreatine (Sigma), adjusted to pH7.2 with CsOH (290 mosM)). A gigaohm seal was made on the surface of the cell, using an Axopatch 200B amplifier (Axon Instruments), signals digitized at 10 or 20 kHz with a Digidata 1322A (Axon Instruments), and filtered at 2 kHz with Clampex 9.2. After break-in, the contents of the cell were aspirated into the recording pipette under visual control. The pipette tip was broken off in a sterile reaction tube, and RT-PCR was performed on the pipette contents. One cell was collected per animal. A multiplex RT-PCR strategy was used for increased sensitivity, as previously described (Chamberlain et al., 1988), to amplify multiple MHCI genes from a single-cell sample. An initial round of RT-PCR was performed using a degenerate pan-MHCI forward primer and an AL1-poly(dT) reverse primer to amplify most transcribed MHCI genes. Primer sets used for identification of specific MHCI genes were the same as those used for analysis of whole hippocampus (Table 1). Each sample was divided in two, and MHCI-specific primers were grouped according to their melting temperature.
Structural renderings of crystallized proteins and structural predictions of splice variants
For proteins that have been crystallized, structural data was obtained from the RSCB Protein Data Bank (PDB; http://www.rcsb.org/pdb/home/home.do), and structural renderings were generated in PyMOL Molecular Graphics System, version 1.3.0 (Schrödinger, LLC) (https://www.pymol.org/). For splice variants which have not been crystallized, structural predictions were performed for the translation products of MHCI splice variants isolated from hippocampus, using the I-TASSER server (http://zhanglab.ccmb.med.umich.edu/I-TASSER; (Roy et al., 2010; Zhang, 2008). The I-TASSER server produces five weighted structural predictions for each input sequence. Only the highest-probability structural prediction for each input sequence were considered. All structures were rendered and manipulated in PyMOL.
Highlights.
More than a dozen MHCI genes are expressed in the adult mouse hippocampus.
MHCI gene expression is specialized in different cell types in hippocampus.
Individual hippocampal neurons can express more than one MHCI gene.
MHCI mRNAs undergo alternative splicing in the central nervous system.
MHCI mRNA and protein are expressed at the developing NMJ.
Acknowledgments
Supported by a Ruth L. Kirschstein National Research Service Award (NRSA) Individual Predoctoral MD/PhD Award 1F30AG046044-01A1 (M.T) and a Princeton Neuroscience Institute Innovation Award (L.M.B).
Abbreviations
- Arf
ADP-ribosylation factor 1
- CNS
central nervous system
- DG
dentate gyrus
- DIA
diaphragm
- EDL
extensor digitorum longus
- HLA
human leukocyte antigen
- MHCI
major histocompatibility complex class I
- mRNA
messenger ribonucleic acid
- nAChRs
nicotinic acetylcholine receptors
- NMJ
neuromuscular junction
- ODC
ornithine decarboxylase
- RT-PCR
reverse-transcriptione polymerase chain reaction
- PNS
peripheral nervous system
- SOL
soleus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldrich CJ, DeCloux A, Woods AS, Cotter RJ, Soloski MJ, Forman J. Identification of a Tap-dependent leader peptide recognized by alloreactive T cells specific for a class Ib antigen. Cell. 1994;79:649–658. doi: 10.1016/0092-8674(94)90550-9. [DOI] [PubMed] [Google Scholar]
- Bhatt L, Horgan CP, Walsh M, McCaffrey MW. The hereditary hemochromatosis protein HFE and its chaperone beta2-microglobulin localise predominantly to the endosomal-recycling compartment. Biochem Biophys Res Commun. 2007;359:277–284. doi: 10.1016/j.bbrc.2007.05.100. [DOI] [PubMed] [Google Scholar]
- Boulanger LM. MHC class I in activity-dependent structural and functional plasticity. Neuron Glia Biol. 2004;1:283–289. doi: 10.1017/S1740925X05000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulanger LM. Immune proteins in brain development and synaptic plasticity. Neuron. 2009;64:93–109. doi: 10.1016/j.neuron.2009.09.001. [DOI] [PubMed] [Google Scholar]
- Braud V, Jones EY, McMichael A. The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur J Immunol. 1997;27:1164–1169. doi: 10.1002/eji.1830270517. [DOI] [PubMed] [Google Scholar]
- Brutkiewicz RR, Bennink JR, Yewdell JW, Bendelac A. TAP-independent, beta 2-microglobulin-dependent surface expression of functional mouse CD1.1. J Exp Med. 1995;182:1913–1919. doi: 10.1084/jem.182.6.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso CS, de Sousa M. HFE, the MHC and hemochromatosis: paradigm for an extended function for MHC class I. Tissue Antigens. 2003;61:263–275. doi: 10.1034/j.1399-0039.2003.00065.x. [DOI] [PubMed] [Google Scholar]
- Cebrian C, Zucca FA, Mauri P, Steinbeck JA, Studer L, Scherzer CR, Kanter E, Budhu S, Mandelbaum J, Vonsattel JP, et al. MHC-I expression renders catecholaminergic neurons susceptible to T-cell-mediated degeneration. Nat Commun. 2014;5:3633. doi: 10.1038/ncomms4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacon MA, Boulanger LM. MHC class I protein is expressed by neurons and neural progenitors in mid-gestation mouse brain. Mol Cell Neurosci. 2013;52:117–127. doi: 10.1016/j.mcn.2012.11.004. [DOI] [PubMed] [Google Scholar]
- Chamberlain JS, Gibbs RA, Ranier JE, Nguyen PN, Caskey CT. Deletion screening of the Duchenne muscular dystrophy locus via multiplex DNA amplification. Nucleic Acids Res. 1988;16:11141–11156. doi: 10.1093/nar/16.23.11141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu NM, Chun T, Fay M, Mandal M, Wang CR. The majority of H2-M3 is retained intracellularly in a peptide-receptive state and traffics to the cell surface in the presence of N-formylated peptides. J Exp Med. 1999;190:423–434. doi: 10.1084/jem.190.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron. 1998;21:505–520. doi: 10.1016/s0896-6273(00)80562-0. [DOI] [PubMed] [Google Scholar]
- Crowley MP, Reich Z, Mavaddat N, Altman JD, Chien Y. The recognition of the nonclassical major histocompatibility complex (MHC) class I molecule, T10, by the gammadelta T cell, G8. J Exp Med. 1997;185:1223–1230. doi: 10.1084/jem.185.7.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datwani A, McConnell MJ, Kanold PO, Micheva KD, Busse B, Shamloo M, Smith SJ, Shatz CJ. Classical MHCI molecules regulate retinogeniculate refinement and limit ocular dominance plasticity. Neuron. 2009;64:463–470. doi: 10.1016/j.neuron.2009.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon-Salazar TJ, Fourgeaud L, Tyler CM, Poole JR, Park JJ, Boulanger LM. MHC class I limits hippocampal synapse density by inhibiting neuronal insulin receptor signaling. J Neurosci. 2014;34:11844–11856. doi: 10.1523/JNEUROSCI.4642-12.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolter KE, Braman JC. Small-sample total RNA purification: laser capture microdissection and cultured cell applications. Biotechniques. 2001;30:1358–1361. doi: 10.2144/01306pf01. [DOI] [PubMed] [Google Scholar]
- Dorfman JR, Zerrahn J, Coles MC, Raulet DH. The basis for self-tolerance of natural killer cells in beta2-microglobulin- and TAP-1- mice. J Immunol. 1997;159:5219–5225. [PubMed] [Google Scholar]
- Eberwine J, Yeh H, Miyashiro K, Cao Y, Nair S, Finnell R, Zettel M, Coleman P. Analysis of gene expression in single live neurons. Proc Natl Acad Sci U S A. 1992;89:3010–3014. doi: 10.1073/pnas.89.7.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edamura M, Murakami G, Meng H, Itakura M, Shigemoto R, Fukuda A, Nakahara D. Functional deficiency of MHC class I enhances LTP and abolishes LTD in the nucleus accumbens of mice. PLoS One. 2014;9:e107099. doi: 10.1371/journal.pone.0107099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edstrom E, Kullberg S, Ming Y, Zheng H, Ulfhake B. MHC class I, beta2 microglobulin, and the INF-gamma receptor are upregulated in aged motoneurons. J Neurosci Res. 2004;78:892–900. doi: 10.1002/jnr.20341. [DOI] [PubMed] [Google Scholar]
- Elmer BM, Estes ML, Barrow SL, McAllister AK. MHCI requires MEF2 transcription factors to negatively regulate synapse density during development and in disease. J Neurosci. 2013;33:13791–13804. doi: 10.1523/JNEUROSCI.2366-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmer BM, McAllister AK. Major histocompatibility complex class I proteins in brain development and plasticity. Trends Neurosci. 2012 doi: 10.1016/j.tins.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourgeaud L, Davenport CM, Tyler CM, Cheng TT, Spencer MB, Boulanger LM. MHC class I modulates NMDA receptor function and AMPA receptor trafficking. Proc Natl Acad Sci U S A. 2010;107:22278–22283. doi: 10.1073/pnas.0914064107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumoto T, McMaster WR, Williams AF. Mouse monoclonal antibodies against rat major histocompatibility antigens. Two Ia antigens and expression of Ia and class I antigens in rat thymus. Eur J Immunol. 1982;12:237–243. doi: 10.1002/eji.1830120313. [DOI] [PubMed] [Google Scholar]
- Glynn MW, Elmer BM, Garay PA, Liu XB, Needleman LA, El-Sabeawy F, McAllister AK. MHCI negatively regulates synapse density during the establishment of cortical connections. Nat Neurosci. 2011;14:442–451. doi: 10.1038/nn.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goddard CA, Butts DA, Shatz CJ. Regulation of CNS synapses by neuronal MHC class I. Proc Natl Acad Sci U S A. 2007;104:6828–6833. doi: 10.1073/pnas.0702023104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsby RA, Goldsby RA. Immunology. 5. New York: W.H. Freeman; 2003. [Google Scholar]
- Guidry PA, Stroynowski I. The murine family of gut-restricted class Ib MHC includes alternatively spliced isoforms of the proposed HLA-G homolog, “blastocyst MHC”. J Immunol. 2005;175:5248–5259. doi: 10.4049/jimmunol.175.8.5248. [DOI] [PubMed] [Google Scholar]
- Haynes LP, Thomas GM, Burgoyne RD. Interaction of neuronal calcium sensor-1 and ADP-ribosylation factor 1 allows bidirectional control of phosphatidylinositol 4-kinase beta and trans-Golgi network-plasma membrane traffic. J Biol Chem. 2005;280:6047–6054. doi: 10.1074/jbc.M413090200. [DOI] [PubMed] [Google Scholar]
- He X, Tabaczewski P, Ho J, Stroynowski I, Garcia KC. Promiscuous antigen presentation by the nonclassical MHC Ib Qa-2 is enabled by a shallow, hydrophobic groove and self-stabilized peptide conformation. Structure. 2001;9:1213–1224. doi: 10.1016/s0969-2126(01)00689-x. [DOI] [PubMed] [Google Scholar]
- Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, Shatz CJ. Functional requirement for class I MHC in CNS development and plasticity. Science. 2000;290:2155–2159. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii T, Hirota J, Mombaerts P. Combinatorial coexpression of neural and immune multigene families in mouse vomeronasal sensory neurons. Curr Biol. 2003;13:394–400. doi: 10.1016/s0960-9822(03)00092-7. [DOI] [PubMed] [Google Scholar]
- Ishii T, Mombaerts P. Expression of nonclassical class I major histocompatibility genes defines a tripartite organization of the mouse vomeronasal system. J Neurosci. 2008;28:2332–2341. doi: 10.1523/JNEUROSCI.4807-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce S, Tabaczewski P, Angeletti RH, Nathenson SG, Stroynowski I. A nonpolymorphic major histocompatibility complex class Ib molecule binds a large array of diverse self-peptides. J Exp Med. 1994;179:579–588. doi: 10.1084/jem.179.2.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke X, Warner CM. Regulation of Ped gene expression by TAP protein. J Reprod Immunol. 2000;46:1–15. doi: 10.1016/s0165-0378(99)00053-4. [DOI] [PubMed] [Google Scholar]
- Koirala S, Corfas G. Identification of novel glial genes by single-cell transcriptional profiling of Bergmann glial cells from mouse cerebellum. PLoS One. 2010;5:e9198. doi: 10.1371/journal.pone.0009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Brott BK, Kirkby LA, Adelson JD, Cheng S, Feller MB, Datwani A, Shatz CJ. Synapse elimination and learning rules co-regulated by MHC class I H2-D. Nature. 2014;509:195–200. doi: 10.1038/nature13154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letellier M, Willson ML, Gautheron V, Mariani J, Lohof AM. Normal adult climbing fiber monoinnervation of cerebellar Purkinje cells in mice lacking MHC class I molecules. Dev Neurobiol. 2008;68:997–1006. doi: 10.1002/dneu.20639. [DOI] [PubMed] [Google Scholar]
- Lidman O, Olsson T, Piehl F. Expression of nonclassical MHC class I (RT1-U) in certain neuronal populations of the central nervous system. Eur J Neurosci. 1999;11:4468–4472. doi: 10.1046/j.1460-9568.1999.00904.x. [DOI] [PubMed] [Google Scholar]
- Linda H, Hammarberg H, Cullheim S, Levinovitz A, Khademi M, Olsson T. Expression of MHC class I and beta2-microglobulin in rat spinal motoneurons: regulatory influences by IFN-gamma and axotomy. Exp Neurol. 1998;150:282–295. doi: 10.1006/exnr.1997.6768. [DOI] [PubMed] [Google Scholar]
- Linda H, Hammarberg H, Piehl F, Khademi M, Olsson T. Expression of MHC class I heavy chain and beta2-microglobulin in rat brainstem motoneurons and nigral dopaminergic neurons. J Neuroimmunol. 1999;101:76–86. doi: 10.1016/s0165-5728(99)00135-6. [DOI] [PubMed] [Google Scholar]
- Lindahl KF, Byers DE, Dabhi VM, Hovik R, Jones EP, Smith GP, Wang CR, Xiao H, Yoshino M. H2-M3, a full-service class Ib histocompatibility antigen. Annu Rev Immunol. 1997;15:851–879. doi: 10.1146/annurev.immunol.15.1.851. [DOI] [PubMed] [Google Scholar]
- Liu J, Shen Y, Li M, Lv D, Zhang A, Peng Y, Miao F, Zhang J. Spatial-Temporal Expression of Non-classical MHC Class I Molecules in the C57 Mouse Brain. Neurochem Res. 2015;40:1487–1496. doi: 10.1007/s11064-015-1620-3. [DOI] [PubMed] [Google Scholar]
- Ljunggren HG, Van Kaer L, Sabatine MS, Auchincloss H, Jr, Tonegawa S, Ploegh HL. MHC class I expression and CD8+ T cell development in TAP1/beta 2-microglobulin double mutant mice. Int Immunol. 1995;7:975–984. doi: 10.1093/intimm/7.6.975. [DOI] [PubMed] [Google Scholar]
- Loconto J, Papes F, Chang E, Stowers L, Jones EP, Takada T, Kumanovics A, Fischer Lindahl K, Dulac C. Functional expression of murine V2R pheromone receptors involves selective association with the M10 and M1 families of MHC class Ib molecules. Cell. 2003;112:607–618. doi: 10.1016/s0092-8674(03)00153-3. [DOI] [PubMed] [Google Scholar]
- Lv D, Shi Q, Liu J, Zhang A, Miao F, He Y, Shen Y, Zhang J. The similar expression pattern of MHC class I molecules in human and mouse cerebellar cortex. Neurochem Res. 2014;39:180–186. doi: 10.1007/s11064-013-1204-z. [DOI] [PubMed] [Google Scholar]
- McConnell MJ, Huang YH, Datwani A, Shatz CJ. H2-K(b) and H2-D(b) regulate cerebellar long-term depression and limit motor learning. Proc Natl Acad Sci U S A. 2009;106:6784–6789. doi: 10.1073/pnas.0902018106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman LA, Liu XB, El-Sabeawy F, Jones EG, McAllister AK. MHC class I molecules are present both pre- and postsynaptically in the visual cortex during postnatal development and in adulthood. Proc Natl Acad Sci U S A. 2010;107:16999–17004. doi: 10.1073/pnas.1006087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PA, Sage JR, Wood SC, Davenport CM, Anagnostaras SG, Boulanger LM. MHC class I immune proteins are critical for hippocampus-dependent memory and gate NMDAR-dependent hippocampal long-term depression. Learning and memory. 2013;20:505–517. doi: 10.1101/lm.031351.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuka M, Inoko H, Kulski JK, Yoshimura S. Major histocompatibility complex (Mhc) class Ib gene duplications, organization and expression patterns in mouse strain C57BL/6. BMC Genomics. 2008;9:178. doi: 10.1186/1471-2164-9-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira AL, Thams S, Lidman O, Piehl F, Hokfelt T, Karre K, Linda H, Cullheim S. A role for MHC class I molecules in synaptic plasticity and regeneration of neurons after axotomy. Proc Natl Acad Sci U S A. 2004;101:17843–17848. doi: 10.1073/pnas.0408154101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson R, Huey-Tubman KE, Dulac C, Bjorkman PJ. Structure of a pheromone receptor-associated MHC molecule with an open and empty groove. PLoS Biol. 2005;3:e257. doi: 10.1371/journal.pbio.0030257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praetor A, Hunziker W. beta(2)-Microglobulin is important for cell surface expression and pH-dependent IgG binding of human FcRn. J Cell Sci. 2002;115:2389–2397. doi: 10.1242/jcs.115.11.2389. [DOI] [PubMed] [Google Scholar]
- Renthal NE, Guidry PA, Shanmuganad S, Renthal W, Stroynowski I. Isoforms of the nonclassical class I MHC antigen H2-Q5 are enriched in brain and encode Qdm peptide. Immunogenetics. 2011;63:57–64. doi: 10.1007/s00251-010-0488-2. [DOI] [PubMed] [Google Scholar]
- Ribic A, Flugge G, Schlumbohm C, Matz-Rensing K, Walter L, Fuchs E. Activity-dependent regulation of MHC class I expression in the developing primary visual cortex of the common marmoset monkey. Behav Brain Funct. 2011;7:1. doi: 10.1186/1744-9081-7-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PJ, Travers PJ, Stackpoole A, Flaherty L, Djaballah H. Maturation of Qa-1b class I molecules requires beta 2-microglobulin but is TAP independent. J Immunol. 1998;160:3217–3224. [PubMed] [Google Scholar]
- Rolleke U, Flugge G, Plehm S, Schlumbohm C, Armstrong VW, Dressel R, Uchanska-Ziegler B, Ziegler A, Fuchs E, Czeh B, et al. Differential expression of major histocompatibility complex class I molecules in the brain of a New World monkey, the common marmoset (Callithrix jacchus) J Neuroimmunol. 2006;176:39–50. doi: 10.1016/j.jneuroim.2006.04.015. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph MG, Wingren C, Crowley MP, Chien YH, Wilson IA. Combined pseudo-merohedral twinning, non-crystallographic symmetry and pseudo-translation in a monoclinic crystal form of the gammadelta T-cell ligand T10. Acta crystallographica Section D, Biological crystallography. 2004;60:656–664. doi: 10.1107/S0907444904002239. [DOI] [PubMed] [Google Scholar]
- Sanes JR, Lichtman JW. Development of the vertebrate neuromuscular junction. Annu Rev Neurosci. 1999;22:389–442. doi: 10.1146/annurev.neuro.22.1.389. [DOI] [PubMed] [Google Scholar]
- Shatz CJ. MHC class I: an unexpected role in neuronal plasticity. Neuron. 2009;64:40–45. doi: 10.1016/j.neuron.2009.09.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotkin TA, Bartolome J. Role of ornithine decarboxylase and the polyamines in nervous system development: a review. Brain Res Bull. 1986;17:307–320. doi: 10.1016/0361-9230(86)90236-4. [DOI] [PubMed] [Google Scholar]
- Stevens B, Allen NJ, Vazquez LE, Howell GR, Christopherson KS, Nouri N, Micheva KD, Mehalow AK, Huberman AD, Stafford B, et al. The classical complement cascade mediates CNS synapse elimination. Cell. 2007;131:1164–1178. doi: 10.1016/j.cell.2007.10.036. [DOI] [PubMed] [Google Scholar]
- Tafti M, Nishino S, Aldrich MS, Liao W, Dement WC, Mignot E. Major histocompatibility class II molecules in the CNS: increased microglial expression at the onset of narcolepsy in canine model. J Neurosci. 1996;16:4588–4595. doi: 10.1523/JNEUROSCI.16-15-04588.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima A, Tanaka T, Ebata T, Takeda K, Kawasaki A, Kelly JM, Darcy PK, Vance RE, Raulet DH, Kinoshita K, et al. Blastocyst MHC, a putative murine homologue of HLA-G, protects TAP-deficient tumor cells from natural killer cell-mediated rejection in vivo. J Immunol. 2003;171:1715–1721. doi: 10.4049/jimmunol.171.4.1715. [DOI] [PubMed] [Google Scholar]
- Thams S, Brodin P, Plantman S, Saxelin R, Karre K, Cullheim S. Classical major histocompatibility complex class I molecules in motoneurons: new actors at the neuromuscular junction. J Neurosci. 2009;29:13503–13515. doi: 10.1523/JNEUROSCI.0981-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo-Rodriguez M, Blumenfeld B, Wu C, Luo J, Attali B, Goodman P, Markram H. Correlation maps allow neuronal electrical properties to be predicted from single-cell gene expression profiles in rat neocortex. Cereb Cortex. 2004;14:1310–1327. doi: 10.1093/cercor/bhh092. [DOI] [PubMed] [Google Scholar]
- Vandiedonck C, Taylor MS, Lockstone HE, Plant K, Taylor JM, Durrant C, Broxholme J, Fairfax BP, Knight JC. Pervasive haplotypic variation in the spliceo-transcriptome of the human major histocompatibility complex. Genome Res. 2011;21:1042–1054. doi: 10.1101/gr.116681.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Sharma A, Maile R, Saad M, Collins EJ, Frelinger JA. Peptidic termini play a significant role in TCR recognition. J Immunol. 2002;169:3137–3145. doi: 10.4049/jimmunol.169.6.3137. [DOI] [PubMed] [Google Scholar]
- Wu L, Feng H, Warner CM. Identification of two major histocompatibility complex class Ib genes, Q7 and Q9, as the Ped gene in the mouse. Biol Reprod. 1999;60:1114–1119. doi: 10.1095/biolreprod60.5.1114. [DOI] [PubMed] [Google Scholar]
- Xu H, Chun T, Choi HJ, Wang B, Wang CR. Impaired response to Listeria in H2-M3-deficient mice reveals a nonredundant role of MHC class Ib-specific T cells in host defense. J Exp Med. 2006;203:449–459. doi: 10.1084/jem.20051866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H, Hashimoto K. Association of MR1 protein, an MHC class I-related molecule, with beta(2)-microglobulin. Biochem Biophys Res Commun. 2002;290:722–729. doi: 10.1006/bbrc.2001.6277. [DOI] [PubMed] [Google Scholar]
- Zappacosta F, Tabaczewski P, Parker KC, Coligan JE, Stroynowski I. The murine liver-specific nonclassical MHC class I molecule Q10 binds a classical peptide repertoire. J Immunol. 2000;164:1906–1915. doi: 10.4049/jimmunol.164.4.1906. [DOI] [PubMed] [Google Scholar]
- Zhang A, Yu H, He Y, Shen Y, Pan N, Liu J, Fu B, Miao F, Zhang J. The spatio-temporal expression of MHC class I molecules during human hippocampal formation development. Brain Res. 2013a;1529:26–38. doi: 10.1016/j.brainres.2013.07.001. [DOI] [PubMed] [Google Scholar]
- Zhang A, Yu H, Shen Y, Liu J, He Y, Shi Q, Fu B, Miao F, Zhang J. The expression patterns of MHC class I molecules in the developmental human visual system. Neurochem Res. 2013b;38:273–281. doi: 10.1007/s11064-012-0916-9. [DOI] [PubMed] [Google Scholar]
- Zhang Y. I-TASSER server for protein 3D structure prediction. BMC Bioinformatics. 2008;9:40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlstra M, Li E, Sajjadi F, Subramani S, Jaenisch R. Germ-line transmission of a disrupted beta 2-microglobulin gene produced by homologous recombination in embryonic stem cells. Nature. 1989;342:435–438. doi: 10.1038/342435a0. [DOI] [PubMed] [Google Scholar]
- Zohar O, Reiter Y, Bennink JR, Lev A, Cavallaro S, Paratore S, Pick CG, Brooker G, Yewdell JW. Cutting edge: MHC class I-ly49 interaction regulates neuronal function. J Immunol. 2008;180:6447–6451. doi: 10.4049/jimmunol.180.10.6447. [DOI] [PMC free article] [PubMed] [Google Scholar]