Abstract
The RNA chaperone protein Hfq is critical to the function of small, base pairing RNAs in many bacteria. In the past few years, structures and modeling of wild type Hfq and assays of various mutants have documented that the homohexameric Hfq ring can contact RNA at four sites (proximal face, distal face, rim and C-terminal tail) and that different RNAs bind to these sites in various configurations. These studies together with novel in vitro and in vivo experimental approaches are beginning to give mechanistic insights into how Hfq acts to promote small RNA-mRNA pairing and indicate that flexibility is integral to the Hfq role in RNA matchmaking.
Introduction
In many bacteria, the RNA chaperone protein Hfq, which was identified nearly 50 years ago as an essential host factor for bacteriophage Qβ RNA replication in Escherichia coli [1], is required for the function of small, stress-induced regulatory RNAs (sRNAs) that act by limited base pairing. Hfq stabilizes the sRNAs and promotes their interactions with mRNAs leading to altered stability and/or translation of these targets (reviewed in [2]). This broad Hfq role in facilitating base pairing sRNA function is reflected in the pleiotropic phenotypes of hfq deletion strains observed for a variety of bacteria, including stress sensitivity and reduced virulence (reviewed in [3]). Given the central role played by Hfq in sRNA-mediated gene regulation in many bacteria, the protein has been the focus of extensive study.
Initial crystal structures of the apoprotein showed that Hfq adopts a homohexameric toroid of roughly 65 Å diameter (reviewed in [4-6]). Each subunit is composed of an amino (N)-terminal α-helix followed by five highly twisted and curved antiparallel β-strands, terminating in an unstructured carboxy (C)-terminal region. The surface of the toroid with the N-terminal α-helices is commonly referred to as the ‘proximal’ face, the opposite face the ‘distal’ face, and the outer ring the ‘rim’ or ‘lateral’ face. The sequences and structures of Hfq unambiguously show that the protein is a member of the Sm and Sm-like (Lsm) family of proteins that are involved in various aspects of RNA metabolism in virtually all eukaryotes and archaea (reviewed in [7,8]).
Two comprehensive reviews about Hfq were published in 2011 and 2012 [2,3]. However, given the interest in the chaperone, much has been learned about the protein and its mechanisms of promoting RNA base pairing in the past three years. Here we discuss the findings from a wide range of approaches focusing on the Hfq role in promoting RNA base pairing, particularly in Gram-negative bacteria (summarized in Figure 1).
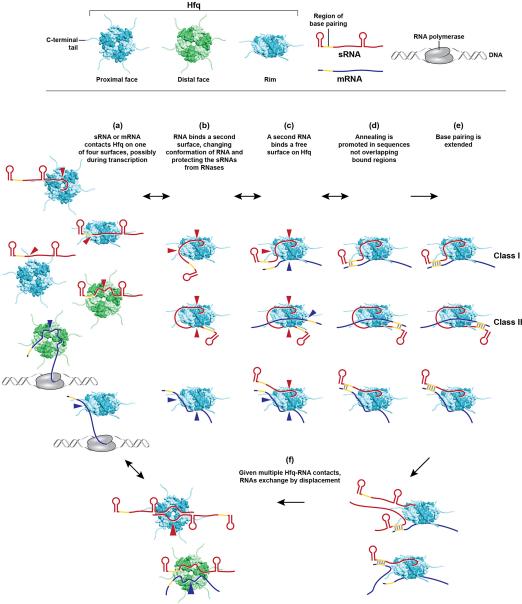
Figure 1.
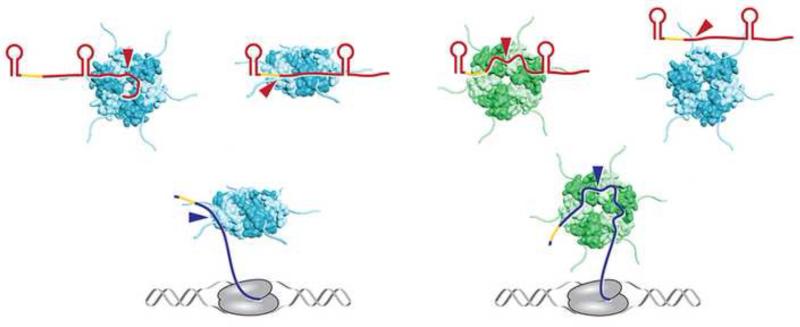
Recent reports have added new details to the model of how Hfq binds to sRNAs and mRNAs and stimulates their interactions. (a) E. coli Hfq (teal for the proximal face and rim views and green for the distal face view) employs four solvent exposed surfaces to interact with RNA; sRNAs (red) have been found to contact the proximal and distal faces, rim and C-terminus, and mRNAs (blue) have been shown to contact the distal face, rim and C-terminus (not shown). Red and blue arrows denote sRNA and mRNA binding to Hfq, respectively. Some of the interactions occur concurrent with transcription of the RNAs, although Hfq binding to Rho-independent terminator of sRNAs most likely occurs after transcription termination, when the U-rich end of the terminator is accessible. Hfq binding to mRNAs and sRNAs is proposed to be in random order. Initial binding is likely to involve only a subset of subunits allowing for rapid displacement by other RNAs. (b) Many RNAs bind multiple surfaces, resulting in changes in RNA secondary structure and protection against RNase degradation, particularly for sRNAs. (c) The free surface(s) of Hfq not already bound to RNA interacts with the cognate RNA partner, positioning the unbound seed region of the sRNA in close proximity to the unbound complementary region of the mRNA. (d) Basic residues on the Hfq rim surface neutralize the negative charge of the RNAs and help to catalyze initial nucleation. (e) Hfq also promotes base pairing of the remaining complementary region. (f) Lower affinity of duplex RNA for Hfq causes free RNAs to compete off sRNA-mRNA pairs, allowing Hfq to serve as a matchmaker for another pair of RNAs. Lists of Class I and Class II sRNAs can be found in [25].
Binding of sRNAs and mRNAs
Hfq has four solvent exposed regions, the proximal face, distal face, rim and C-terminal tail, which possess unique architectures and electrostatic surfaces and are surprisingly varied from one homolog to another (reviewed in [3]). Early crystal structures of Staphylococcus aureus Hfq in complex with short oligonucleotides showed that the proximal face binds polyU sequences such that the uridines are stacked in pockets between neighboring monomers around the central pore [9]. Similar studies of E. coli Hfq revealed that the distal face binds A-rich oligonucleotides with three nucleotides per subunit of Hfq [10]. These triplet sequences were first defined as A-R-N motifs where the adenosine (A-site) and purine (R-site) nucleotides stack into the solvent exposed pockets while the third nucleotide (N) lacks protein contacts. More recent work has led to further definition of these binding sites as well as appreciation that the rim and C-terminal tails also contribute to RNA binding, and that binding to sRNAs and mRNAs is more varied than perhaps initially thought (Figure 1a).
Proximal face binding to polyU sequences in Rho-independent terminators of sRNAs
The importance of the proximal face in binding polyU sequences, particularly those comprising the Rho-independent terminators found on all Hfq binding sRNAs characterized thus far [11,12], has been affirmed in multiple studies. Co-crystal structures of E. coli [13,14] and Listeria monocytogenes [15] Hfq bound to U-rich oligonucleotides showed geometries very similar to what was found for the S. aureus protein [9] with uridines stacked adjacent to phenylalanine in each Hfq monomer [13,14]. The proximal-face uridine-binding pocket thus is conserved in Hfq from Gram-negative and Gram-positive bacteria. Nonetheless, a study that examined sRNA accumulation and target mRNA regulation in E. coli strains expressing various Hfq mutants from the chromosome [16], showed that while mutations in some conserved proximal face residues (such as Q8, F42 and K56) affected most sRNAs analyzed, mutations in other proximal face residues (such as D9 and F39) had varying effects toward different sRNAs. It also was recently reported that certain acidic residues (D9, E18 and E37) on the proximal face of the E. coli protein are important for the discrimination of sRNAs from other cellular RNAs; mutations of these residues allowed for more non-discriminant Hfq binding to RNA while reducing the efficiency of the sRNA-mRNA annealing [17]. Generally, although sRNAs appear to be similarly anchored to the proximal face of Hfq via the polyU tail of the Rho-independent terminator, there are some differences in the way individual sRNAs contact this face.
Distal face binding to varied A-rich sequences in mRNAs and sRNAs
Further binding studies have revealed that RNA contacts on the distal face are more varied than those on the proximal face, though Hfq hexamers from both Gram-negative and Gram-positive organisms contain R-site stacking interactions with adenosine on the distal side. Tryptophan fluorescence quenching assays led to further definition of the preferred distal face binding sequence to (A-A-N)n for the E. coli protein and (A-L)n for the S. aureus protein, where L is a linker nucleotide [18]. Co-crystal structures of E. coli Hfq bound to the A-rich region of the OxyS sRNA [19], S. aureus Hfq bound to an (A-A)3-A oligonucleotide [20] and Bacillus subtilis Hfq bound to an (A-G)3-A oligonucleotide [21] similarly show refinement of the canonical A-R-N motif as well as differences in the binding configuration.
Regardless of the exact sequence, several studies have demonstrated that for mRNAs, the position of the A-rich motif relative to the base-pairing region is critical for Hfq-mediated regulation. An A-rich sequence immediately 3′ of the complementary sequence of a molecular beacon, which fluoresces upon base pairing with a target, significantly enhanced Hfq-mediated annealing in vitro [22]. This enhancement was lost as the distance between the (A-A-N)n site and the complementary sequence increased unless the sequences were brought into proximity with a secondary structure. In vivo, potential mRNA targets of the Spot 42 sRNA were not regulated if the A-rich sequence overlapped the sRNA binding site, but did show regulation if the sequence was repositioned upstream of the base pairing region [23]. Similarly, repositioning the (A-A-N)n at various positions in the rpoS 5′UTR sequence, either upstream or downstream of the natural site, reduced the ability of Hfq to facilitate annealing with the DsrA sRNA in vitro and sRNA activation of rpoS translation in vivo [24]. Although the distances between the Hfq- and sRNA-binding sites are greater than 20 nucleotides in a number of regulated mRNAs, it is likely that, as for rpoS, the RNA secondary and tertiary structures place the Hfq binding site adjacent to the sRNA binding site.
While the first A-rich sequences to be characterized were synthetic or derived from mRNAs, multiple studies have now shown that some sRNAs also have A-rich sequences that allow them to bind the distal face [16,19,25-29]. In general, Hfq-binding sRNAs in E. coli can be classified into two groups, though a few have intermediate properties [25] (Figure 1e). Class I sRNAs, which constitute the majority, bind to the proximal and rim domains of Hfq and base pair with mRNAs that have distal face binding sites. Class II sRNAs, which bind the proximal and distal sites of Hfq, base pair with mRNAs with rim binding sites. Binding on both proximal and distal sites appears to have a stabilizing effect on Class II sRNAs, and Class I sRNAs can be stabilized by the addition of A-R-N motifs.
Rim binding to UA-rich sequences in sRNAs and mRNAs
The rim of E. coli Hfq contains a patch of positively charged surface residues (R16, R17 and R19) on the outside of a shallow groove leading out from the proximal sRNA binding site on each of the six monomers, and recent work has shown that the rim is a secondary binding site for UA-rich sequences in sRNAs [12,25,26,30-33]. In vitro, mutations of the positively charged patch impaired RybB sRNA binding, and wild type Hfq could not bind RybB and SgrS sRNAs with mutations in the internal UA-rich regions [12,30]. In addition, a recent structure of full-length E. coli Hfq with the Salmonella enterica RydC sRNA shows direct phosphate-backbone contacts between R16 and R17 of Hfq and U23 and U24 within the body of RydC [32]. In vivo, mutations of the charged residues reduced the levels of Class I sRNAs, indicating reduced sRNA binding and stability [25]. As for the A-rich sequences that contact the distal face, the distance between the UA-rich sequence on the sRNA and the region of base pairing impacts regulation [12]. Together these studies indicate that many sRNAs are anchored to the proximal side of Hfq by the single stranded polyU tail of the Rho-independent terminator, with additional contacts between the sRNA and the charged rim residues. Recently, some mRNAs, particularly targets of Class II sRNAs that bind the proximal and distal faces, have also been found to require the charged rim residues for binding [25,34]. The arginine patch, while conserved in most Gram-negative bacteria, is almost completely absent in Hfq from a number of Gram-positive species such as B. anthracis and S. aureus (reviewed in [3,6]), but it is possible that RNAs contact other rim residues in these species.
C-terminal tail contacts with some sRNAs
The C-terminal tail, which varies significantly in sequence and length, was absent in the first Hfq structures as it is intrinsically disordered and flexible. Early studies to dissect the cellular function of the C-terminus of E. coli Hfq also yielded conflicting conclusions (reviewed in [3]). However, new evidence is accumulating that the C-terminal tail is important for the interaction with at least some sRNAs. The structure of Hfq with the RydC sRNA showed the C-terminal tail makes distributive contacts over the surface of RydC [32]. This observation, together with the finding that the in vivo half-life of RydC was reduced in the absence of the C-terminus, is consistent with binding. Deletion of the C-terminal tail in E. coli Hfq also reduced in vitro binding affinity to the V. cholera Qrr sRNAs [35], and tryptophan quenching experiments showed the C-terminus can bind the hfq mRNA [18]. Interestingly, Hfq from Clostridium difficile, which has an extremely asparagine and glutamine-rich tail, can complement many of the functions of E. coli Hfq [36]. In this heterologous context, deletion of the tail impacted some of the regulation by C. difficile Hfq. These in vitro and in vivo findings, along with the highly variable sequence, suggest that, while the C-terminal tail binds some but not all RNAs, binding relies less on specific binding pockets and residues and more on the intrinsically disordered nature of this domain.
Promoting sRNA-mRNA interactions
The fact that sRNAs and mRNAs make several, varied contacts allows for rapid exchange of RNAs on Hfq (reviewed in [37]) and is consistent with multiple models for Hfq-facilitated pairing. Innovative experimental approaches are now providing direct evidence that Hfq impacts multiple steps; changing the structures of RNAs (Figure 1b), bringing RNAs into proximity (Figure 1c), neutralizing the negative charge of the two pairing RNAs, stimulating the nucleation of the first base pairs (Figure 1d) as well as facilitating the further annealing of the two RNA strands (Figure 1e).
Early structure probing studies indicated that Hfq could change the secondary structures of bound RNAs (reviewed in [3]). This role has been further documented with a range of experiments. SAXS (small-angle X-ray scattering) and SANS (small-angle neutron scattering) analysis combined with circular dichroism of Hfq bound to the RprA and OxyS sRNAs revealed E. coli Hfq binds the sRNAs in a 1:1 stoichiometry and changes the structures of the sRNAs but not Hfq [28]. Similarly, SAXS and SHAPE (selective 2′-hydroxyl acylation and primer extension) analysis showed that E. coli Hfq makes rim contacts with a U5 motif in conjunction with distal contacts with the (A-A-N)n motif of rpoS, changing the tertiary conformation of the mRNA into a more compact structure and positioning the region involved in base pairing near an sRNA bound to the proximal face [34]. The observations that mutations of rim residues or deletion of the C-terminal tail of E. coli Hfq impair formation of an Hfq-RydC-cfa ternary complex further support the model that Hfq plays an active role in positioning RNAs for optimal base pairing [32].
Other studies have shown that the rim goes beyond positioning the RNAs and carries out a more catalytic role in promoting base pairing [31,38]. Assays of sRNA-mRNA annealing using stopped flow spectroscopy and an RNA beacon revealed that wild type E. coli Hfq, but not a derivative lacking the rim arginine residues, increased the annealing of complementary RNAs up to 100-fold by reducing both entropic and electrostatic barriers [31]. Experiments combining the molecular beacon with a target RNA carrying a photo-caged guanosine derivative within the complementary region allowed assessment of base pairing before and after uncaging [38]. These experiments showed that Hfq directly overcomes the initial energetic barrier for nucleation of the RNA helix. Moreover, given that annealing was reduced if Hfq was proteolytically removed after nucleation, the chaperone must also promote extension of the duplex.
Finding RNAs in the cell
One of the most intriguing and unresolved questions surrounding Hfq function is how this protein is able to facilitate the base pairing of cognate RNA pairs in the context of thousands of cellular RNAs within the minute time frames of the environmental stress responses in which the sRNAs act. While estimates of the number of Hfq hexamers vary from ≈400 to 10,000, it is clear that Hfq is limiting under most conditions (reviewed in [37]). Recent studies, which have examined Hfq and RNA subcellular localization and levels in E. coli, are giving clues into how Hfq effectively carries out its matchmaking function within the cell.
The rate of Hfq diffusion in the cell was estimated by monitoring single-molecule trajectories [39]. These measurements showed three distinct states with different diffusion constants. The interpretation of these observations were that Hfq has three binding states: free unbound Hfq (fastest diffusion constant), Hfq bound to RNA and/or other proteins (intermediate diffusion constant), and Hfq bound to RNA during transcription and thus tethered to transcription complexes (slowest diffusion constant, which disappears after transcription is blocked with rifampicin). Overall, these findings suggest that Hfq binds at least some RNAs as they are synthesized [39]. A recent study combined smFISH (single-molecule fluorescence in situ hybridization) with super-resolution microscopy, which enables subcellular localization of RNA molecules, to quantify single sRNA, mRNA and sRNA-mRNA complexes and thus determine the in vivo kinetics of sRNA target search and sRNA-mRNA co-degradation [40]. These experiments showed that the association rate for duplex formation is far slower than the dissociation rate.
A recent modeling study examining the effects of different RNA levels reported that, given random order binding of sRNAs and mRNAs observed previously (reviewed in [37]), maximum sRNA-dependent regulation occurs at specific Hfq concentrations, which varies for sRNA-mRNA pairs [41]. Too little or too much Hfq leads to suboptimal sRNA activity, with the latter resulting from sequestration of sRNAs and mRNAs in singly bound Hfq complexes. Furthermore, deep sequencing experiments have revealed a broad spectrum of additional RNAs that bind Hfq, further increasing competition for the RNA chaperone. Several of the abundant RNAs, including fragments of unprocessed tRNAs and bacteriophage-encoded transcripts, base pair with and antagonize the functions of sRNAs and have been denoted “sponge RNAs” [42,43]. Together these studies emphasize how the relative levels of a specific sRNA, its mRNA target as well as other sRNAs, mRNAs, sponge RNAs and Hfq all impact the extent of regulation.
Conclusions
The recent in vitro and in vivo studies have shown that Hfq binds sRNAs and mRNAs in multiple configurations on four different surfaces to affect stability of the sRNAs as well promote sRNA-mRNA base pairing by enhancing multiple steps. As a consequence, Hfq-mediated regulation by sRNAs can be controlled intricately by the levels and the sequences of each of the binding partners. However, a note of caution is warranted. Most of the studies were carried out on a limited set of sRNAs and mRNAs in E. coli, and findings regarding the importance of specific residues or steps in the base pairing for one sRNA-mRNA pair may not necessarily apply to another pair. In addition, as mentioned at multiple points, the RNA binding surfaces on Hfq, particularly the rim and C-terminal domains, vary substantially from one organism to another, and Hfq molecules from one organism frequently fail to complement the full function of Hfq in another organism [36,44,45]. Nevertheless, we anticipate that the application of mutational and structural analysis combined with fluorescence-based in vitro and in vivo approaches described here to other sRNA-mRNA pairs and other Hfq homologs undoubtedly will further elucidate how the chaperone functions as such an effective, yet flexible RNA matchmaker.
Highlights.
Hfq contacts RNA via proximal, distal, rim and C-terminal surfaces
RNAs interact with Hfq in multiple ways
Mechanisms of Hfq-facilitated RNA pairing are being elucidated by new technologies
Acknowledgements
We thank S. Gottesman and S. Woodson and members of their groups for comments. Work in the Storz laboratory is supported by the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development (Project ZIA HD001608).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and recommended reading
Papers of particular interest published within the period of review have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Franze de Fernandez MT, Eoyang L, August JT. Factor fraction required for the synthesis of bacteriophage Qβ-RNA. Nature. 1968;219:588–590. doi: 10.1038/219588a0. [DOI] [PubMed] [Google Scholar]
- 2•.Vogel J, Luisi BF. Hfq and its constellation of RNA. Nat. Rev. Microbiol. 2011;9:578–589. doi: 10.1038/nrmicro2615. [This broad review covers almost all aspects of Hfq-mediated regulation including descriptions of structural and functional features of Hfq.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Sobrero P, Valverde C. The bacterial protein Hfq: much more than a mere RNA-binding factor. Crit. Rev. Microbiol. 2012;38:276–299. doi: 10.3109/1040841X.2012.664540. [This very comprehensive review summarizes information about the surface charge of Hfq from different bacteria, known hfq deletion phenotypes, and the controversy surrounding the role of the C-terminal tail.] [DOI] [PubMed] [Google Scholar]
- 4.Valentin-Hansen P, Eriksen M, Udesen C. The bacterial Sm-like protein Hfq: a key player in RNA transactions. Mol. Microbiol. 2004;51:1525–1533. doi: 10.1111/j.1365-2958.2003.03935.x. [DOI] [PubMed] [Google Scholar]
- 5.Sauer E. Structure and RNA-binding properties of the bacterial LSm protein Hfq. RNA Biol. 2013;10:610–618. doi: 10.4161/rna.24201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 7.Wilusz CJ, Wilusz J. Lsm proteins and Hfq: Life at the 3′ end. RNA Biol. 2013;10:592–601. doi: 10.4161/rna.23695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mura C, Randolph PS, Patterson J, Cozen AE. Archaeal and eukaryotic homologs of Hfq: A structural and evolutionary perspective on Sm function. RNA Biol. 2013;10:636–651. doi: 10.4161/rna.24538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schumacher MA, Pearson RF, Møller T, Valentin-Hansen P, Brennan RG. Structures of the pleiotropic translational regulator Hfq and an Hfq-RNA complex: a bacterial Sm-like protein. EMBO J. 2002;21:3546–3556. doi: 10.1093/emboj/cdf322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl. Acad. Sci. USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otaka H, Ishikawa H, Morita T, Aiba H. PolyU tail of rho-independent terminator of bacterial small RNAs is essential for Hfq action. Proc. Natl. Acad. Sci. USA. 2011;108:13059–13064. doi: 10.1073/pnas.1107050108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishikawa H, Otaka H, Maki K, Morita T, Aiba H. The functional Hfq-binding module of bacterial sRNAs consists of a double or single hairpin preceded by a U-rich sequence and followed by a 3′ poly(U) tail. RNA. 2012;18:1062–1074. doi: 10.1261/rna.031575.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang W, Wang L, Wu J, Gong Q, Shi Y. Hfq-bridged ternary complex is important for translation activation of rpoS by DsrA. Nucleic Acids Res. 2013;41:5938–5948. doi: 10.1093/nar/gkt276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sauer E, Weichenrieder O. Structural basis for RNA 3′-end recognition by Hfq. Proc. Natl. Acad. Sci. USA. 2011;108:13065–13070. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kovach AR, Hoff KE, Canty JT, Orans J, Brennan RG. Recognition of U-rich RNA by Hfq from the Gram-positive pathogen Listeria monocytogenes. RNA. 2014;20:1548–1559. doi: 10.1261/rna.044032.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang A, Schu DJ, Tjaden BC, Storz G, Gottesman S. Mutations in interaction surfaces differentially impact E. coli Hfq association with small RNAs and their mRNA targets. J. Mol. Biol. 2013;425:3678–3697. doi: 10.1016/j.jmb.2013.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Panja S, Santiago-Frangos A, Schu DJ, Gottesman S, Woodson SA. Acidic residues in the Hfq chaperone increase the selectivity of sRNA binding and annealing. J. Mol. Biol. 2015;427:3491–3500. doi: 10.1016/j.jmb.2015.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson KE, Orans J, Kovach AR, Link TM, Brennan RG. Mapping Hfq-RNA interaction surfaces using tryptophan fluorescence quenching. Nucleic Acids Res. 2014;42:2736–2749. doi: 10.1093/nar/gkt1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L, Wang W, Li F, Zhang J, Wu J, Gong Q, Shi Y. Structural insights into the recognition of the internal A-rich linker from OxyS sRNA by Escherichia coli Hfq. Nucleic Acids Res. 2015;43:2400–2411. doi: 10.1093/nar/gkv072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horstmann N, Orans J, Valentin-Hansen P, Shelburne SAI, Brennan RG. Structural mechanism of Staphylococcus aureus Hfq binding to an RNA A-tract. Nucleic Acids Res. 2012;40:11023–11035. doi: 10.1093/nar/gks809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Someya T, Baba S, Fujimoto M, Kawai G, Kumasaka T, Nakamura K. Crystal structure of Hfq from Bacillus subtilis in complex with SELEX-derived RNA aptamer: insight into RNA-binding properties of bacterial Hfq. Nucleic Acids Res. 2012;40:1856–1867. doi: 10.1093/nar/gkr892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panja S, Woodson SA. Hfq proximity and orientation controls RNA annealing. Nucleic Acids Res. 2012;40:8690–8697. doi: 10.1093/nar/gks618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beisel CL, Updegrove TB, Janson BJ, Storz G. Multiple factors dictate target selection by Hfq-binding small RNAs. EMBO J. 2012;31:1961–1974. doi: 10.1038/emboj.2012.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peng Y, Soper TJ, Woodson SA. Positional effects of AAN motifs in rpoS regulation by sRNAs and Hfq. J. Mol. Biol. 2014;426:275–285. doi: 10.1016/j.jmb.2013.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25••.Schu DJ, Zhang A, Gottesman S, Storz G. Alternative Hfq-sRNA interaction modes dictate alternative mRNA recognition. EMBO J. 2015;34:2557–2573. doi: 10.15252/embj.201591569. [Schu and Zhang et al. showed that sRNAs can be classified into two predominant groups based on their dependence on different surface regions of Hfq for stability and turnover. Class I sRNAs depend on proximal and rim Hfq sites, and Class II depend on proximal and distal Hfq sites] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Małecka EM, Stróżecka J, Sobańska D, Olejniczak M. Structure of bacterial regulatory RNAs determines their performance in competition for the chaperone protein Hfq. Biochemistry. 2015;54:1157–1170. doi: 10.1021/bi500741d. [DOI] [PubMed] [Google Scholar]
- 27.Ellis MJ, Trussler RS, Haniford DB. Hfq binds directly to the ribosome binding site of IS10 transposase mRNA to inhibit translation. Mol. Microbiol. 2015;96:633–650. doi: 10.1111/mmi.12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henderson CA, Vincent HA, Casamento A, Stone CM, Phillips JO, Cary PD, Sobott F, Gowers DM, Taylor JE, Callaghan AJ. Hfq binding changes the structure of Escherichia coli small noncoding RNAs OxyS and RprA, which are involved in the riboregulation of rpoS. RNA. 2013;19:1089–1104. doi: 10.1261/rna.034595.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Updegrove T, Wilf N, Sun X, Wartell RM. Effect of Hfq on RprA-rpoS mRNA pairing: Hfq-RNA binding and the influence of the 5′ rpoS mRNA leader region. Biochemistry. 2008;47:11184–11195. doi: 10.1021/bi800479p. [DOI] [PubMed] [Google Scholar]
- 30.Sauer E, Schmidt S, Weichenrieder O. Small RNA binding to the lateral surface of Hfq hexamers and structural rearrangements upon mRNA target recognition. Proc. Natl. Acad. Sci. USA. 2012;109:9396–9401. doi: 10.1073/pnas.1202521109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Panja S, Schu DJ, Woodson SA. Conserved arginines on the rim of Hfq catalyze base pair formation and exchange. Nucleic Acids Res. 2013;41:7536–7546. doi: 10.1093/nar/gkt521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32••.Dimastrogiovanni D, Fröhlich KS, Bandyra KJ, Bruce HA, Hohensee S, Vogel J, Luisi BF. Recognition of the small regulatory RNA RydC by the bacterial Hfq protein. eLife. 2014;3:e05375. doi: 10.7554/eLife.05375. [Dimastrogiovanni et al. present the first X-ray crystallographic structure of full-length Hfq bound to a full-length sRNA, showing RydC sRNA contacting the rim and C-terminal tail of Hfq.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murina V, Lekontseva N, Nikulin A. Hfq binds ribonucleotides in three different RNA-binding sites. Acta Cryst. 2013;D69:1504–1513. doi: 10.1107/S090744491301010X. [DOI] [PubMed] [Google Scholar]
- 34••.Peng Y, Curtis JE, Fang X, Woodson SA. Structural model of an mRNA in complex with the bacterial chaperone Hfq. Proc. Natl. Acad. Sci. USA. 2014;111:17134–17139. doi: 10.1073/pnas.1410114111. [Using a combination of chemical footprinting, small-angle X-ray scattering and molecular modeling to analyze Hfq bound to rpoS mRNA, Peng et al. showed rpoS interacts with the rim and distal side of Hfq. Such interactions caused rpoS to wrap around Hfq and position the sRNA binding site adjacent to the sRNA.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vincent HA, Henderson CA, Ragan TJ, Garza-Garcia A, Cary PD, Gowers DM, Malfois M, Driscoll PC, Sobott F, Callaghan AJ. Characterization of Vibrio cholerae Hfq provides novel insights into the role of the Hfq C-terminal region. J. Mol. Biol. 2012;420:56–69. doi: 10.1016/j.jmb.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caillet J, Gracia C, Fontaine F, Hajnsdorf E. Clostridium difficile Hfq can replace Escherichia coli Hfq for most of its function. RNA. 2014;20:1567–1578. doi: 10.1261/rna.043372.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wagner EGH. Cycling of RNAs on Hfq. RNA. 2013;10:619–626. doi: 10.4161/rna.24044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38•.Panja S, Paul R, Greenberg MM, Woodson SA. Light-Triggered RNA Annealing by an RNA Chaperone. Angew. Chem. Int. Ed. Engl. 2015;54:7281–7284. doi: 10.1002/anie.201501658. [These authors developed a novel approach to temporally control annealing activity by engineering a ‘photo-caged’ guanosine within the complementary region of a synthetic target RNA. Using this approach, Panja et al. demonstrated that Hfq initiates RNA-RNA nucleation and facilitates additional base pairing.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39•.Persson F, Lindén M, Unoson C, Elf J. Extracting intracellular diffusive states and transition rates from single-molecule tracking data. Nat. Methods. 2013;10:265–269. doi: 10.1038/nmeth.2367. [Persson et al. developed a method to estimate Hfq diffusion states in the cell, which provided evidence that Hfq associates with transcriptional complexes.] [DOI] [PubMed] [Google Scholar]
- 40•.Fei J, Singh D, Zhang Q, Park S, Balasubramanian D, Golding I, Vanderpool CK, Ha T. RNA biochemistry. Determination of in vivo target search kinetics of regulatory noncoding RNA. Science. 2015;347:1371–1374. doi: 10.1126/science.1258849. [The use of super-resolution imaging of single sRNA and mRNA molecules in this study allowed the determination of kinetic parameters governing sRNA-mRNA binding and target regulation in vivo.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sagawa S, Shin JE, Hussein R, Lim HN. Paradoxical suppression of small RNA activity at high Hfq concentrations due to random-order binding. Nucleic Acids Res. 2015;43:8502–8515. doi: 10.1093/nar/gkv777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tree JJ, Granneman S, McAteer SP, Tollervey D, Gally DL. Identification of bacteriophage-encoded anti-sRNAs in pathogenic Escherichia coli. Mol. Cell. 2014;55:199–213. doi: 10.1016/j.molcel.2014.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lalaouna D, Carrier MC, Semsey S, Brouard JS, Wang J, Wade JT, Massé E. A 3′ external transcribed spacer in a tRNA transcript acts as a sponge for small RNAs to prevent transcriptional noise. Mol. Cell. 2015;58:393–405. doi: 10.1016/j.molcel.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 44.Vrentas C, Ghirlando R, Keefer A, Hu Z, Tomczak A, Gittis AG, Murthi A, Garboczi DN, Gottesman S, Leppla SH. Hfqs in Bacillus anthracis: Role of protein sequence variation in the structure and function of proteins in the Hfq family. Protein Sci. 2015;24:1808–1819. doi: 10.1002/pro.2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rochat T, Bouloc P, Yang Q, Bossi L, Figueroa-Bossi N. Lack of interchangeability of Hfq-like proteins. Biochimie. 2012;94:1554–1559. doi: 10.1016/j.biochi.2012.01.016. [DOI] [PubMed] [Google Scholar]