Abstract
Despite immense interest in employing antimalarials as autophagy inhibitors to treat cancer, it remains unclear if these agents act predominantly via autophagy inhibition or whether other pathways direct their anti-cancer properties. By comparing the treatment effects of the antimalarials chloroquine (CQ) and quinacrine (Q) on KRAS mutant lung cancer cells, we demonstrate that inhibition of the oxidative arm of the pentose phosphate pathway (oxPPP) is required for antimalarial induced apoptosis. Despite inhibiting autophagy, neither CQ treatment nor RNAi against autophagy regulators (ATGs) promote cell death. In contrast, Q triggers high levels of apoptosis, both in vitro and in vivo, and this phenotype requires both autophagy inhibition and p53-dependent inhibition of the oxPPP. Simultaneous genetic targeting of the oxPPP and autophagy is sufficient to trigger apoptosis in lung cancer cells, including cells lacking p53. Thus, in addition to reduced autophagy, oxPPP inhibition serves as an important determinant of antimalarial cytotoxicity in cancer cells.
Keywords: autophagy, apoptosis, chloroquine, cancer, pentose phosphate pathway
INTRODUCTION
Macroautophagy (autophagy) is a evolutionarily conserved lysosomal degradation process that promotes cell survival and metabolic adaptation (1). In tumors, abundant evidence supports that autophagy operates as a cell survival pathway in response to various micro-environmental stresses as well as promotes resistance to chemotherapy (2). As a result, there is tremendous clinical interest in inhibiting autophagy as a potential strategy against cancers. Importantly, antimalarials, namely chloroquine (CQ) and hydroxychloroquine (HCQ), inhibit lysosomal function and block the late stages of autophagic proteolysis. Given their long history of clinical use as antimalarials and in diseases such as rheumatoid arthritis, these lysosomotrophic compounds have gained special attention for their potential utility as pharmacological autophagy inhibitors in cancer (2, 3). Indeed, numerous clinical trials utilizing HCQ in combination with cytotoxic and targeted therapies are under evaluation in various cancers; the first of these studies indicate that HCQ can be successfully employed to therapeutically inhibit autophagy in cancer patients with minimal dose-limiting toxicities (4–10). Despite these initial positive results, much remains to learned about how antimalarials like HCQ may be best exploited therapeutically against cancer. Notably, although the current prevailing view is that the anti-cancer effects of HCQ predominantly arise from macroautophagy inhibition, multiple preclinical studies suggest that the chemosensitizing effects of this lysosotrophic agent may not be entirely autophagy-dependent (11, 12). Hence, further elucidating the pathways by which antimalarials target cancer cells remains a subject of immense therapeutic significance.
Similar to CQ and HCQ, the antimalarial quinacrine (Q) exhibits anti-cancer properties and has been demonstrated to function as a late-stage autophagy inhibitor (13–16). Remarkably, in gastrointestinal stromal tumors (GISTs), we previously found that Q was significantly more potent than CQ in inducing the apoptosis of cancer cells, both as a single agent and in combination with the tyrosine kinase inhibitor imatinib (16). Moreover, others have reported that Q is more potent than CQ in killing glioblastoma multiforme cells (17). Accordingly, by scrutinizing the phenotypic differences between these two FDA-approved agents, we sought to further mechanistically understand how these antimalarials promote cancer cell death. Here, we demonstrate that inhibition of the oxidative arm of the pentose phosphate pathway (oxPPP) is required to induce apoptosis in KRAS mutant lung cancer cells in response to antimalarial treatment.
RESULTS
Quinacrine (Q) promotes apoptosis of NSCLC cells independently of autophagy inhibition
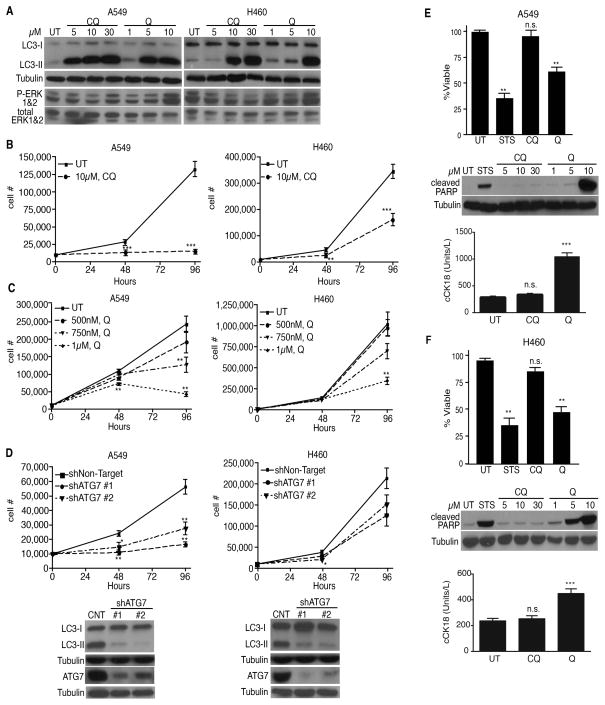
We focused on KRAS mutant non-small cell lung carcinoma (NSCLC) cells because previous studies indicate that genetic autophagy inhibition impairs the proliferation of RAS-transformed cells, while modestly impacting cell death (18–22). Treatment of KRAS mutant A549 and H460 NSCLC cells (both p53 wild type, Supplementary Fig. 1A) with either CQ or Q led to the dose-dependent accumulation of phosphotidylethanolamine lipidated LC3 (LC3-II), consistent with impaired autophagic flux. Neither of these antimalarials inhibited the levels of ERK/MAPK phosphorylation, indicating that key mitogenic signals downstream of oncogenic Ras activation remained intact (Fig. 1A). Similar to CQ, Q suppressed the proliferative outgrowth of both A549 and H460 cells (Fig. 1B, C). Stable depletion of the essential autophagy regulator ATG7 also reduced autophagosome formation (LC3-II) and proliferative expansion (Fig. 1D), thereby confirming previous studies from our lab and others that genetic autophagy inhibition suppresses KRAS mutant cancer cell proliferation (18–21).
Figure 1. Quinacrine-mediated apoptosis is independent of autophagy inhibition.
(A) LC3-II accumulation and P-ERK levels in NSCLC cells treated with chloroquine (CQ) or quinacrine (Q) for 18 h at indicated doses. (B, C) Growth curves of NSCLC cells treated with CQ (b) and Q (c) at indicated doses. (D) Growth curves following ATG7 depletion and corresponding immunoblots of LC3-II reduction and ATG7 knockdown. (E, F) Q but not CQ reduces NSCLC cell survival. Cells were treated with 30μM CQ or 10μM Q for 24h. Immunoblotting for cleaved PARP and ELISA for cleaved cytokeratin 18 (cCK18) confirms that Q promotes apoptosis. UT=untreated, STS= 1μM staurosporine (positive control). All data represent mean ± SEM from 3 independent experiments. Statistical significance was calculated using Student’s t-test for pair-wise comparisons in (B) or by ANOVA followed by Tukey’s HSD. *P≤0.05, **P≤0.01, ***P≤0.001.
On the other hand, Q was unique in its ability to induce cell death in both A549 and H460 cells. Upon 24h of Q treatment, NSCLC cells exhibited a 35–45% reduction in cell viability (Fig. 1E–F). We confirmed that Q induced apoptosis in both lines, evidenced by increased levels of caspase-3 cleaved PARP and cleaved cytokeratin 18 fragments, both of which are specifically produced during apoptotic cell death. In contrast, CQ treatment did not significantly impact cell survival or induce apoptosis at doses robustly blocking autophagic flux (Fig. 1E–F). Similar to CQ, both ATG7 and ATG12 knockdown had minimal effects on KRAS mutant NSCLC cell survival (Supplementary Fig. 1B–F).
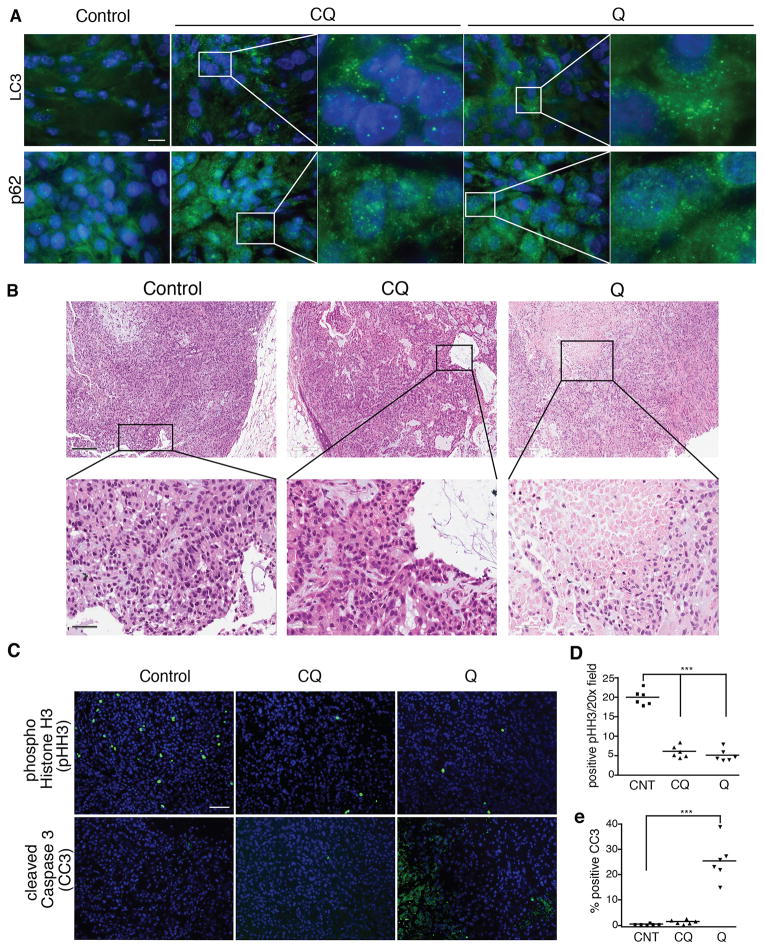
To assay whether Q triggered tumor cell death in vivo, we compared the effects of CQ and Q treatment on NSCLC xenografts. Both CQ and Q blocked the late stages of autophagy, evidenced by increased numbers of autophagosomes (punctate LC3) and p62 (SQSTM1) aggregates, both of which accumulate upon antimalarial inhibition of autophagy (Fig. 2A) (1, 23). Notably, in A549 xenografts, we also observed increased necrosis following 14 days of Q treatment compared to control or CQ treated tumors (Fig. 2B). Consistent with our in vitro studies, both CQ and Q suppressed proliferation, evidenced by a decrease in tumor cells positive for the mitosis marker, phospho-histone H3 (pHH3). However, only Q treatment promoted apoptosis in A549 xenografts, evidenced by increased cleaved caspase-3 (CC3) positive tumor cells (Fig. 2C). Furthermore, Q treated animals exhibited reduced tumor volumes and weights, compared to those treated with vehicle (CNT) and CQ, although these reductions were not statistically significant over 21 days of treatment (Supplementary Fig. 2A). Similarly, both CQ and Q reduced proliferation in H460 xenografts, whereas Q uniquely induced apoptosis (Supplementary Fig. 2B–D). Finally, in contrast to the tumor xenografts, we did not observe detectable apoptosis in several normal tissues from CQ and Q treated animals, evidenced by the lack of terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) and CC3 staining (Supplementary Fig. 2E). Overall, these results indicate that Q is selectively cytotoxic for tumors compared to normal tissues; furthermore, the death-inducing effects of Q in NSCLC cells are mediated, at least in part, by targeting pathways other than autophagy.
Figure 2. Quinacrine uniquely promotes apoptosis in lung tumors in vivo.
(A) Immunostaining for autophagosome accumulation (punctate LC3) and p62 (SQSTM1) aggregates in A549 xenograft tumors treated with CQ and Q. Bar, 50μm. (B) Increased areas of necrosis and dying cells in Q-treated tumors. Upper panel bar, 200μm; lower panel bar, 50μm. (C) Immunostaining for phospho-histone H3 (pHH3, mitosis marker) and cleaved caspase-3 (CC3, apoptosis marker) of lung tumors treated with CQ and Q. Bar, 100μm. (D, E) Quantification of pHH3 (D) and CC3 (E) positive tumor cells. Bar, 100μm. Data represent mean ± SEM from 6 independent tumors for each cohort. Statistical significance was calculated using ANOVA followed by Tukey’s HSD. ***P≤0.001.
Oxidative Pentose Phosphate Pathway (oxPPP) inhibition is required for antimalarials to induce apoptosis in NSCLC cells
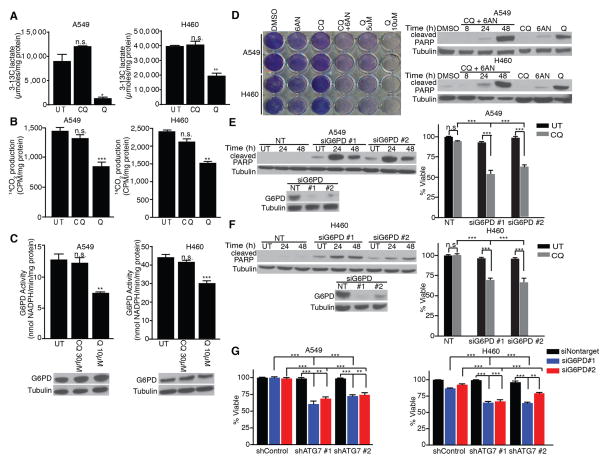
In parallel, we evaluated the effects of CQ and Q on glucose metabolism, motivated by our previous work demonstrating that genetic ATG deficiency impairs glucose uptake and glycolytic flux in RAS-transformed fibroblasts and breast cancer cells (20). Using 13C-NMR to measure [3-13C]-lactate produced from [1-13C]-glucose, we unexpectedly observed that Q treatment significantly impaired glycolytic flux in A549 and H460 cells, whereas CQ treatment did not (Fig. 3A). To extend these results, we measured how antimalarials regulated the oxidative arm of the pentose phosphate pathway (oxPPP), a side-branch of glycolysis implicated in biosynthesis, antioxidant response and cell survival (24). In contrast to CQ, Q potently inhibited the oxPPP (Fig. 3B) and attenuated the enzymatic activity of glucose-6-phosphase dehydrogenase (G6PD), the rate-limiting step of the oxPPP (Fig. 3C). We next interrogated the functional importance of oxPPP inhibition in mediating NSCLC cell death during antimalarial treatment. Treatment of A549 and H460 cells with the pharmacological oxPPP inhibitor, 6-aminonicotinamide (6-AN), induced mild cell death as a single agent. However, upon combining 6-AN with CQ, clonogenic recovery was profoundly reduced and apoptotic PARP cleavage was increased to levels comparable to that achieved with Q (Fig. 3D). Furthermore, upon RNAi-mediated G6PD depletion, CQ treatment was able to induce cell death in KRAS mutant NSCLC cells (Fig. 3E–F). Because these death-promoting effects of G6PD depletion or pharmacological inhibition were only observed upon CQ treatment, we tested whether the simultaneous inhibition of oxPPP and autophagy was sufficient to promote cell death. Indeed, the combined knockdown of ATG7 and G6PD abrogated cell survival and increased apoptotic PARP cleavage in A549 and H460 cells (Fig. 3G and Supplementary Fig. 3A–B). We propose that this unique convergence of events upon treatment with Q, but not with CQ, mediates the ability to induce cell death in KRAS mutant NSCLC cells.
Figure 3. Oxidative Pentose Phosphate Pathway (oxPPP) inhibition is required for antimalarials to induce apoptosis in NSCLC cells.
(A–C) Glycolytic activity (A, extracellular 3-13C-lactate production from 1-13C-glucose), oxidative PPP activity (B, 14CO2 production from 1-14C-glucose) and glucose 6-phosphate dehydrogenase (G6PD) activity (C) following treatment with 30μM CQ or 10μM Q. (D) Co-treatment of CQ with the pharmacological G6PD inhibitor 6AN (175μM) reduces viability and promotes apoptosis (cleaved PARP) in NSCLC cells. (E, F) G6PD knockdown combined with 30μM CQ reduces survival and promotes apoptosis (cleaved PARP immunoblot) in NSCLC cells. (G) Combined RNAi-mediated knockdown of ATG7 and G6PD is sufficient to induce cell death in A549 and H460 cells. All data represent the mean ± SEM from 3 independent experiments. Statistical significance was calculated using ANOVA followed by Tukey’s HSD. *P≤0.05, **P≤0.01, ***P≤0.001.
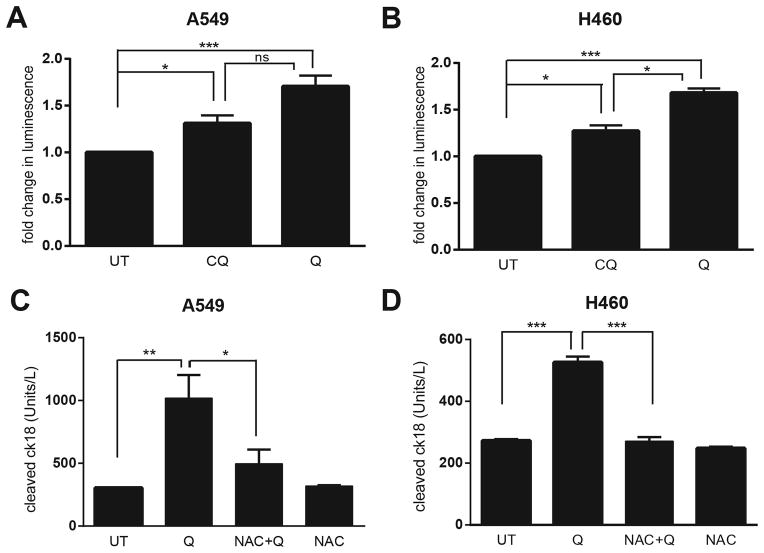
Autophagy and the oxPPP each serve distinct functions in mitigating reactive oxygen species (ROS) in cells (21, 25). Thus, we asked how the combined effects of Q on autophagy deficiency and PPP inhibition modulated ROS. Q treatment led to more pronounced levels of ROS in both A549 and H460 cells compared to control and CQ-treated cells (Fig. 4A–B). Moreover, Q-induced apoptosis was significantly attenuated upon co-treatment with the anti-oxidant N-acetyl Cysteine (NAC) (Fig. 4C–D). These results corroborate that increased oxidative stress, which we propose to be secondary to the combined inhibition of autophagy and oxPPP, is an important contributor to Q-mediated cytotoxicity in KRAS mutant NSCLC cells.
Figure 4. Quinacrine-induced cell death in NSCLC cells is associated with increased reactive oxygen species production (ROS).
(A, B) Cells were treated with 30μM CQ or 10μM Q treatment for 6h. Reactive oxygen species (ROS) production in A549 (A) or H460 (B) lung cancer cells was measured using a bioluminescence assay to detect intracellular H2O2 levels; results were normalized to those obtained with untreated controls (UT). (C, D) A549 (C) or H460 (D) cells were treated with 30μM CQ or 10μM Q for 24h and cleaved cytokeratin 18 (cCK18) levels were measured using ELISA. UT=untreated controls. NAC= 5 μM N-Acetyl Cysteine. All data represent the mean ± SEM from 3 independent experiments. Statisitical significance was calculated using ANOVA followed by Tukey’s HSD. *P≤0.05, **P≤0.01, ***P≤0.001.
p53 induction is required for NSCLC cell death in response to antimalarial treatment
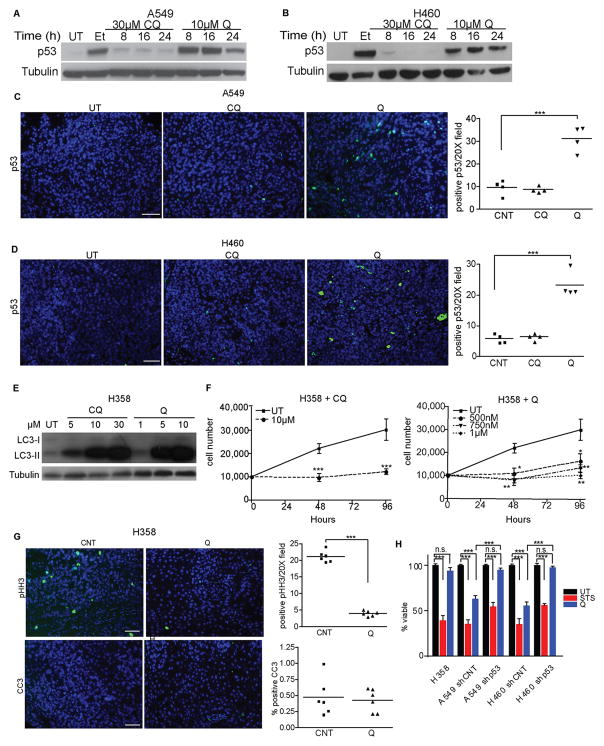
Previous studies indicate CQ and Q can activate the p53 tumor suppressor pathway via diverse mechanisms (13–15, 26, 27). Indeed, Q treatment led to robust p53 induction in A549 and H460 cells in vitro (Fig. 5A–B) and increased numbers of p53 positive cells were evident in Q-treated A549 and H460 xenografts in vivo (Fig. 5C–D). In contrast, CQ failed to significantly induce p53 in either cell type in vitro or in vivo (Fig 5A–D). To ascertain the functional significance of p53 induction downstream of Q treatment, we interrogated the effects of antimalarials on the p53 null, KRAS mutant NSCLC cell line, H358 (Supplementary Fig. 1A). Importantly, CQ and Q blocked autophagic flux in H358 similarly to p53 wild type NSCLC cells (Fig. 5E and Supplementary Fig. 4A) and suppressed proliferative outgrowth in vitro (Fig. 5F). Moreover, Q treatment attenuated mitotic activity (pHH3) in H358 xenografts (Fig. 5G). Thus, the autophagy-inhibitory and anti-proliferative effects of CQ and Q were not p53 dependent. On the other hand, H358 cells were highly resistant to Q-induced cell death both in vivo (Fig. 5G) as well as in vitro (Fig. 4H). Furthermore, the stable shRNA-mediated depletion of p53 in A549 and H460 cells (Supplementary Fig. 4B–C) significantly reversed Q-induced cytotoxicity (Fig. 5H).
Figure 5. p53 is required for cell death in response to antimalarial treatment.
(A, B) p53 induction in A549 (A) or H460 (B) lung cancer cells upon CQ or Q treatment for the indicated times and doses. UT=untreated controls, Et= 50μM etoposide (positive control) (C, D) Immunostaining for p53 in A549 (C) and H460 (D) xenograft tumors treated for 3d with CQ and Q. Bar, 100μm. Right graphs: Quantification of p53 positive cells from tumors. Data are the mean ± SEM from 4 independent tumors for each condition. (E) LC3-II accumulation in p53 null H358 cells upon 18 h treatment with CQ or Q at indicated doses. (F) Growth curves of H358 cells treated with CQ or Q at indicated doses. Data represent the mean ± SEM from 3 independent experiments. (G) Immunostaining for pHH3 (proliferation) and CC3 (apoptosis) in H358 xenograft tumors treated with Q. Bar, 100μm. Right graphs: Quantification of pHH3 and CC3 positive cells. Data are the mean ± SEM from 6 independent tumors for each condition. (H) Reduced Q-mediated cell death in p53 null H358 cells and upon shRNA-mediated p53 knockdown in A549 and H460 cells. Q=10 μM quinacrine; STS=1μM staurosporine (positive control). Data represent the mean ± SEM from 3 independent experiments. Statistical significance was calculated using Student’s t-test for pair-wise comparisons in (F) or by ANOVA followed by Tukey’s HSD. *P≤0.05, **P≤0.01, ***P≤0.001.
Combined inhibition of oxPPP and autophagy is sufficient to elicit lung cancer cell death irrespective of p53 status
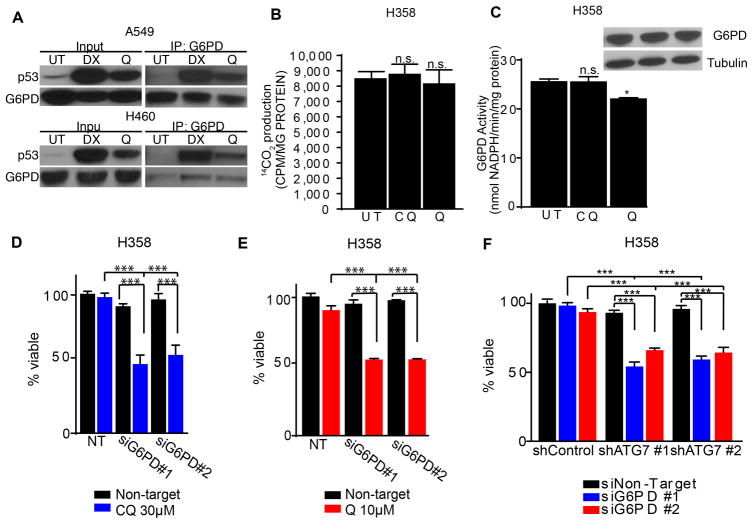
The tumor suppressor p53 has emerged as an important regulator of glucose metabolism (28). Intriguingly, recent studies demonstrate that p53 inhibits oxPPP flux by directly binding and inhibiting G6PD (29). Indeed, co-immunoprecipitation studies confirmed a physical interaction between G6PD and p53 upon Q treatment of NSCLC cells (Fig. 6A). Furthermore, similar to CQ, Q failed to significantly inhibit the oxPPP in H358 cells (Fig. 6B). Remarkably, we did note that Q treatment slightly reduced G6PD activity in these cells (Fig. 6C), suggesting factors other than p53 status may influence Q inhibition of G6PD enzymatic activity. Nonetheless, our results indicated that Q-induced cell death and overall oxPPP inhibition in NSCLC cells were both p53 dependent. Since p53 can mediate multiple pro-apoptotic pathways (28), we evaluated the specific requirement of oxPPP inhibition downstream of p53 by testing whether G6PD knockdown was sufficient to promote antimalarial cytotoxicity in p53-deficient cells. Indeed, G6PD knockdown in H358 cells diminished cell survival and increased apoptotic PARP cleavage following CQ or Q treatment (Fig. 6D–E and Supplementary Fig. 5A). Finally, we tested the effects of combined knockdown of G6PD and ATG7 on the viability of H358 cells. Indeed, despite the absence of p53, the concomitant genetic inhibition of oxPPP and autophagy was sufficient to promote the death of H358 cells (Fig. 6F and Supplementary Fig. 5B–C). Thus, the simultaneous genetic targeting of autophagy and the oxPPP was sufficient to trigger apoptosis in lung cancer cells, irrespective of p53 status.
Figure 6. Simultaneous genetic targeting of autophagy and the oxPPP is sufficient to trigger apoptosis in p53 null lung cancer cells.
(A) p53 is induced upon 10 μM Q treatment in A549 and H460 cells and co-immunoprecipitates with G6PD. DX=1μM doxorubicin (positive control). (B, C) Oxidative PPP activity (B, 14CO2 production from 1-14C-glucose) and G6PD activity (C) following treatment with CQ or Q. (D) CQ promotes cell death upon G6PD knockdown in H358 cells. (E) Q promotes cell death upon G6PD knockdown in H358 cells. (F) Combined RNAi-mediated knockdown of ATG7 and G6PD is sufficient to induce cell death in H358 cells. All data represent the mean ± SEM from 3 independent experiments. Statistical significance was calculated using ANOVA followed by Tukey’s HSD. *P≤0.05, **P≤0.01, ***P≤0.001.
DISCUSSION
Currently, there is immense interest in repurposing antimalarials as autophagy inhibitors to treat cancer (2, 3). Here, by evaluating the treatment effects of chloroquine (CQ) and quinacrine (Q), we provide insight into the mechanisms underlying the anti-tumorigenic properties of these two FDA-approved antimalarial lysosomotrophic agents. First, our results suggest that the ability of antimalarials to suppress proliferation in KRAS mutant lung cancer cells is at least partly due to autophagy inhibition, because genetic autophagy deficiency achieved via ATG knockdown elicits analogous anti-proliferative effects. On the other hand, autophagy inhibition in itself does not promote significant apoptosis in KRAS mutant NSCLC cells. Rather, inhibition of the oxidative arm of the pentose phosphate pathway (oxPPP) cooperates with autophagy inhibition to promote antimalarial cytotoxicity in these cells. In addition, using genetic loss-of-function approaches, we demonstrate that the simultaneous targeting of autophagy and the oxPPP is needed to efficiently trigger the apoptosis of these lung cancer cells.
In contrast to our studies, others have found that autophagy inhibition by itself is sufficient to elicit cancer cell death. For example, the basal breast carcinoma cell line, MDA-MB-468 undergoes robust cell death in response to CQ treatment or ATG knockdown and murine KRAS mutant lung cancers exhibit apoptosis and regression in response to acute ATG7 deletion in vivo (30, 31). These studies indicate that the cooperation that we have observed between autophagy and oxPPP inhibition during Q treatment may be context-specific. However, since the status of the oxPPP was not specifically examined in these studies, it remains uncertain whether alterations in glucose metabolism or other metabolic susceptibilities predispose these tumors to the death-promoting effects of autophagy inhibition. Nonetheless, our results indicate that co-targeting the oxPPP may represent a valuable strategy to promote the apoptosis of cancer cells that do not robustly respond to autophagy inhibition alone.
To most effectively scrutinize mechanisms underlying cell death, we have employed antimalarials as single agents in this study. In reality, these agents are most commonly being repurposed as chemosensitizers in combination with other therapies; in fact, abundant work in preclinical models supports the rationale for targeting autophagy as a combination strategy in multiple tumor types and in response to diverse chemotherapeutic agents (2, 3). Moreover, the first early phase clinical trials utilizing HCQ in combination with anticancer therapies have recently been reported in humans; although clinical efficacy was not the primary endpoint of these initial phase I or I/II studies, it is noteworthy that remarkable clinical responses were observed in certain patients with refractory tumors(4–10). Moreover, a 100% clinical response rate and over 70% complete remission rate was observed in dogs with naturally occurring lymphoma following treatment with doxorubicin combined with HCQ (4). Given the initial optimism from these clinical trials, a more complete understanding of the mechanisms by which antimalarials induce tumor cell death is crucial for the development of future therapeutic strategies employing these agents. Based on our findings, we hypothesize that CQ or Q will most effectively induce cancer cell apoptosis when combined with a second treatment modality targeting the oxPPP. Although dose-limiting limiting toxicities may hamper direct targeting of the oxPPP in cancer patients, several currently utilized clinical agents are known to suppress glucose metabolism. Accordingly, one can predict that such agents, due to the resulting decline in oxPPP activity, will similarly cooperate with autophagy inhibitors in promoting cell death. Indeed, support for this concept comes from our previous studies in GIST; because the tyrosine kinase inhibitor imatinib potently inhibits glucose uptake and glycolytic metabolism in GIST cells, inhibiting autophagy via ATG knockdown or antimalarials potently synergizes with imatinib to trigger apoptosis and prevent the development of acquired resistance in GIST (16).
We also demonstrate that the dual blockade of autophagy and oxPPP promotes cytotoxicity regardless of p53 status. These findings may be of particular importance in the treatment of NSCLC and pancreatic ductal carcinomas (PDAC), two highly lethal cancers that commonly harbor oncogenic KRAS mutations in combination with the loss or mutation of p53. Notably, studies in genetically engineered mouse models of KRAS mutant pancreatic cancer have revealed complex interconnections between autophagy and p53 in PDAC progression and tumor metabolism(32, 33). When p53 inactivation occurs by somatic loss of heterozygosity (LOH), autophagy inhibition impairs KRAS driven PDAC progression; moreover, CQ leads to mitochondrial respiration defects(33). On the other hand, upon embryonic deletion of p53 in the pancreas, autophagy inhibition via genetic ATG7 deletion or HCQ treatment unexpectedly accelerates the onset of PDAC; remarkably, cells isolated from these rapidly growing tumors exhibit increased levels of both glycolytic and oxPPP intermediates(32). These results broach the possibility that oxPPP inhibition may be exploited to promote tumor cell death and prevent carcinoma progression in certain PDACs lacking both autophagy and p53.
Finally, as clinical trials go forward, an important challenge is identifying useful biomarkers and surrogates to predict antimalarial response against tumors, especially in the advent of new divalent CQ derivatives, such as Ly-05 (34). Our results suggest that the oxPPP activity may serve as one useful monitor of response in therapeutic regimens employing antimalarials. Overall, they highlight the importance of identifying and scrutinizing other biological parameters, not just autophagy inhibition, to more effectively evaluate the utility of antimalarials in the clinical oncology setting.
MATERIALS AND METHODS
Cell culture
The NSCLC lines A549, H460 and H358 were acquired from the American Type Culture Collection and passaged for less than 6 months following resuscitation; cell lines were not authenticated or tested for Mycoplasma contamination. A549 cells were grown in DMEM containing 25 mm glucose (UCSF Cell Culture Facility) supplemented with 10% fetal bovine serum (FBS), penicillin, and streptomycin. H460 and H358 were cultured in RPMI-1640 supplemented with 10% FBS, penicillin, and streptomycin. Chloroquine (CQ) and quinacrine (Q) were dissolved in water and cells were treated at the indicated doses. Because Q treatment of cells elicits high levels of autofluorescence, the analysis of cultured cells via fluorescence-based activity assays was not technically feasible. N-acetyl Cysteine (NAC) was dissolved directly into tissue culture media at a final concentration of 5 μM and pH was readjusted to 7.4 prior to use.
Antibodies and chemicals
Commercial antibodies include: phospho-Histone H3 (Cell Signaling Technology (CST) 9701), cleaved caspase-3 Alexa Fluor 488 (CST 9669S), phospho-p44/42 MAPK (P-ERK) (Invitrogen 44–680G), p44/42 MAPK (ERK1/2) (Invitrogen 13–6200), ATG12 (CST 2010), ATG7 (Santa Cruz Biotechnology sc8668), p62 (Progen Biotechnik GP62C), tubulin (Sigma T6199), p53 (DO1, Calbiochem OP43), cleaved PARP (CST 9451), G6PD (Abcam ab993) LC3 5F10 for IF (Axxora NT0231-00). For immunobloting, we utilized an LC3 antibody which has been described previously and is now commercially available (EMD Millipore ABC232) (35). Chemicals include: chloroquine (CQ), quinacrine (Q), 6-aminonicotinamide (6-AN), N-acetyl Cysteine (NAC), etoposide (Et) and doxorubicin (DX), all from Sigma-Aldrich, staurosporine (STS, EMD Chemicals), 1-13C glucose (Cambridge Isotopes), 1-14C glucose and 6-14C glucose (Perkin Elmer), and Hoechst (Invitrogen).
shRNA and siRNA mediated RNA interference
pLKO.1 lentiviral expression plasmids containing shRNAs against ATG7 and ATG12 were purchased from Sigma-Aldrich (Mission shRNA) and shRNA against p53 was purchased from Addgene (plasmid 19119). Viral particles were produced using a third-generation lentiviral packaging system in HEK293T cells. Following infection and drug selection, early passage stable pools of ATG knockdown cells (maximum of 5 passages) were utilized because extended propagation results in the loss of RNAi-mediated silencing and autophagy inhibition.
The target sequences for hairpins directed against ATG7 (NM_006395) are shATG7 #1 (TRCN0000007584): GCCTGCTGAGGAGCTCTCCA and shATG7 #2 (TRCN0000007587): CCCAGCTATTGGAACACTGTA; the target sequence directed against ATG12 (NM_004707) is shATG12 #1 (TRCN0000007393): TGTTGCAGCTTCCTACTTCAA; and the target sequence directed against p53 (Addgene plasmid 19119) is: CCGACTCCAGTGGTAATCTACTTCAAGAGAGTAGATTACCACTGGAGTCTTTTT. For siRNA-mediated knockdown, Stealth RNAi oligos against human G6PD (HSS103891 and HSS103892) were purchased from Invitrogen and cells were transfected with 100nM of each oligonucleotide using Oligofectamine (Invitrogen) following manufacturer’s instructions.
Cell proliferation, cell death and reactive oxygen species (ROS) assays
For growth curves, 1 × 104 cells were grown in triplicate in the presence of the indicated treatments. Cells were harvested, stained with trypan blue and live cells were enumerated using a hemocytometer at the indicated timepoints. For studies of cell death, trypan blue exclusion was utilized to quantify cell viability because Q treatment elicits high auto-fluorescence in cells, which precludes the use of fluorescent-based cell death assays. The caspase cleaved cytokeratin 18 (cCK18) M30 fragment was measured using the M30 Cytodeath ELISA (Diapharma P10900), according to the manufacturer’s protocol; this assay quantitatively measures a stable apoptosis-generated neoepitope in CK-18 (Asp396). Levels of reactive oxygen species (ROS) were detected using the ROS-Glo H2O2 luminescence kit (Promega G8820) according to the manufacturer’s protocol.
Immunoblotting
Cells were lysed in RIPA lysis buffer plus 10 mM NaF, 10 mM β-glycerophosphate, 1 mM Na3VO3, 10 nM calyculin A, and protease inhibitors. Lysates were clarified by centrifugation for 15 min at 4°C, and protein concentrations were assessed using a bicinchoninic acid (BCA) protein assay (Thermo, Waltham, MA). Samples containing equal amounts of protein were boiled in SDS sample buffer, resolved using SDS–PAGE, and transferred to poly-vinylidene diflouride membrane. Membranes were blocked in phosphate-buffered saline (PBS) + 0.1% Tween 20 with 5% nonfat dry milk, incubated with the indicated primary antibodies overnight at 4°C, washed, incubated with horseradish peroxidase-conjugated secondary antibodies, and analyzed by enhanced chemiluminescence.
Immunofluorescence and Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) analysis
Tumors and normal tissues (lung, liver, and small intestine) were harvested, fixed in 4% paraformaldehyde and sent for paraffin embedding and sectioning (4μm) and hematoxylin and eosin (H&E) staining at the UCSF Helen Diller Family Comprehensive Cancer Center Mouse Pathology Core. For immunofluorescence staining, slides were subject to heat-induced antigen retrieval (Antigen Unmasking solution, Citrate Based, Vector Labs) for 20 minutes at 95C. Q autofluorescence was not evident in formalin-fixed paraffin embedded sections. Slides were cooled, blocked in 10% goat serum, followed by overnight primary antibody incubation at 4C in a humidified chamber. Slides were washed in PBS and incubated with Alexa 488 conjugated secondary antibodies (Invitrogen) for 1h at 20C. After washing with PBS, slides were counterstained with Hoechst (2 μg/mL) for 10 min, washed and mounted with Prolong Gold (Invitrogen). TUNEL assays were performed using ApopTag® kits (EMD Millipore) according to manufacturer’s protocol.
Phase and immunofluorescence image acquisition and analysis
Phase images of H&E sections were acquired on Aperio’s ScanScope XT affixed with a 20X and 40X objective using Aperio ImageScope software (v11.0.1.725). IF images were acquired on an Axiovert 200 microscope (Carl Zeiss) equipped with a Spot RT camera (Diagnostic Instruments) and a mercury lamp. Images were acquired using MetaMorph software (v6.0) and arranged using Adobe Illustrator CS5 (v15.0.1).
13C-NMR acquisition and analysis
Cells were grown in DMEM medium containing 25 mmol/L of [1-13C] glucose to 80% confluency, upon which medium was collected for NMR analysis and cells were harvested for protein quantification using the BCA assay. Briefly, medium was lyophilized and reconstituted in 1mL of deuterium oxide. To acquire 13C spectra, the reconstituted sample was acquired on a 600-MHz INOVA spectrometer (Varian; Palo Alto, CA) using a 30° flip angle, 3.5-s repetition time, and broadband proton decoupling. Analysis of the NMR spectra was performed using Bruker Topspin software version 3.0 (Bruker Biospin GmbH, Germany). The concentrations of metabolites were determined with respect to an external reference, 2,2,3,3-tetradeutero-3-(trimethylsilyl)propionic acid (TSP), with a known concentration.
Analysis of Oxidative Pentose Phosphate Pathway
One million cells were plated onto 10 cm dishes in triplicate, grown overnight and treated with vehicle control (water) or with CQ or Q at the indicated doses for 3h at 37C. Cells were washed twice with PBS and the tissue culture media was replaced with media containing 0.8μCi of 1-14C-glucose (Perkin Elmer) or 6-14C-glucose containing vehicle (water) or the indicated antimalarial; cells were incubated for an additional 7h prior to analysis. To capture gaseous 14CO2, Whatman filter paper was taped to the inside of culture dish lid and the plate was then sealed with Parafilm; the experiment was terminated by adding 500μL of perchloric acid to the cells; the plate was resealed and left in the incubator overnight. Filter papers were then carefully removed and placed into vials containing Ready-Safe Liquid Scintillation Fluid (Beckman Coulter), and 14CO2 activity was measured on a Beckman LS6000 Liquid Scintillation Analyzer. The incubation of cells with 6-14C-glucose, used to correct for 14CO2 production from the citric acid cycle, yielded minimal 14CO2 activity.
Glucose-6-phosphate dehydrogenase (G6PD) activity assay
G6PD activity was determined using the G6PD activity assay kit from Biovision (Milpitas, CA). 1×106 cells were grown overnight in triplicate on 10cm dishes and either treated with CQ, Q at the indicated doses or left untreated for 16 h. Cells were washed in cold PBS and cells were harvested in 0.5mL of lysis buffer described above. Protein concentration was quantified using the Pierce BCA assay; samples were normalized to protein concentration before analysis with the G6PD activity kit according to manufacturer’s instructions.
Antimalarial treatment of lung cancer xenografts
Animal studies were conducted in accordance with approved UCSF Institutional Animal Care and Use Committee Protocols. Xenografts were established in 4-week old nu/nu male mice (Simonsen Laboratories) by rear flank SC injection of 1 × 106 (H460), 4 × 106 (A549) or 10 × 106 (H358) cells in a 1:1 dilution of Matrigel™ (BD Biosciences). Tumors were grown to 0.5 cm (maximal dimension) and animals were randomized prior to starting antimalarial treatment. Mice were randomized to receive CQ, Q or vehicle (water) daily. CQ was administered at 60 mg/kg via intraperitoneal (IP) injection, Q at 50 mg/kg by oral gavage and vehicle (water) by oral gavage as previously described (16). Tumors were evaluated for autophagosome (punctate LC3) and p62/SQSTM accumulation, apoptosis and proliferation following 14 days of treatment. For quantification of imaging data from tissue sections, 10 fields were randomly chosen prior to quantification. Analysis of tumor growth in tumor-bearing mice treated with CQ, Q or vehicle (water) was performed in parallel; the dimensions of tumors were measured using calipers, and tumor volume was calculated using the formula: (length × width2)/2. Following 21 days of treatment, mice were euthanized and individual tumors were weighed. No statistical method was used to predetermine sample size, and no data were excluded.
Statistical analyses
Each experiment was repeated at least three independent times and no data were excluded for analysis. GraphPad Prism software (v5.0b) was used for generation of graphs and statistical analysis. P values were determined by Student’s t-test for pair-wise comparisons or by ANOVA followed by Tukey’s HSD for multiple comparisons as indicated. For statistical comparisons, all data exhibited a normal distribution and the variance between groups was similar.
Supplementary Material
Acknowledgments
Grant support to JD includes the NIH (R01CA126792 and R01CA188404), California Tobacco Related Diseases Research Program (18XT-0106), and Samuel Waxman Cancer Research Foundation.
Abbreviations
- ATG
Autophagy-related gene
- CQ
chloroquine
- NSCLC
non-small cell lung carcinoma
- oxPPP
oxidative pentose phosphate pathway
- Q
quinacrine
Footnotes
CONFLICTS OF INTEREST:
The authors have no conflicts of interest to disclose.
AUTHOR CONTRIBUTIONS:
J.D. and B.R. conceived the study. E.S., S.R., T.M. and J.D. designed the experiments. E.S., S.R. and T.M. performed the experiments. E.S., S.R., T.M. and J.D. analyzed the data. J.D. supervised the study and wrote the paper with input from the other authors.
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- 1.Murrow L, Debnath J. Autophagy as a stress-response and quality-control mechanism: implications for cell injury and human disease. Annual Review of Pathology: Mechanisms of Disease. 2013;8:105–37. doi: 10.1146/annurev-pathol-020712-163918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kimmelman AC. The dynamic nature of autophagy in cancer. Genes Dev. 2011;25(19):1999–2010. doi: 10.1101/gad.17558811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amaravadi RK, Lippincott-Schwartz J, Yin XM, Weiss WA, Takebe N, Timmer W, et al. Principles and current strategies for targeting autophagy for cancer treatment. Clin Cancer Res. 2011;17(4):654–66. doi: 10.1158/1078-0432.CCR-10-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barnard RA, Wittenburg LA, Amaravadi RK, Gustafson DL, Thorburn A, Thamm DH. Phase I clinical trial and pharmacodynamic evaluation of combination hydroxychloroquine and doxorubicin treatment in pet dogs treated for spontaneously occurring lymphoma. Autophagy. 2014;10(8):1415–25. doi: 10.4161/auto.29165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahalingam D, Mita M, Sarantopoulos J, Wood L, Amaravadi RK, Davis LE, et al. Combined autophagy and HDAC inhibition: a phase I safety, tolerability, pharmacokinetic, and pharmacodynamic analysis of hydroxychloroquine in combination with the HDAC inhibitor vorinostat in patients with advanced solid tumors. Autophagy. 2014;10(8):1403–14. doi: 10.4161/auto.29231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rangwala R, Chang YC, Hu J, Algazy KM, Evans TL, Fecher LA, et al. Combined MTOR and autophagy inhibition: phase I trial of hydroxychloroquine and temsirolimus in patients with advanced solid tumors and melanoma. Autophagy. 2014;10(8):1391–402. doi: 10.4161/auto.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rangwala R, Leone R, Chang YC, Fecher LA, Schuchter LM, Kramer A, et al. Phase I trial of hydroxychloroquine with dose-intense temozolomide in patients with advanced solid tumors and melanoma. Autophagy. 2014;10(8):1369–79. doi: 10.4161/auto.29118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rosenfeld MR, Ye X, Supko JG, Desideri S, Grossman SA, Brem S, et al. A phase I/II trial of hydroxychloroquine in conjunction with radiation therapy and concurrent and adjuvant temozolomide in patients with newly diagnosed glioblastoma multiforme. Autophagy. 2014;10(8):1359–68. doi: 10.4161/auto.28984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vogl DT, Stadtmauer EA, Tan KS, Heitjan DF, Davis LE, Pontiggia L, et al. Combined autophagy and proteasome inhibition: a phase 1 trial of hydroxychloroquine and bortezomib in patients with relapsed/refractory myeloma. Autophagy. 2014;10(8):1380–90. doi: 10.4161/auto.29264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolpin BM, Rubinson DA, Wang X, Chan JA, Cleary JM, Enzinger PC, et al. Phase II and pharmacodynamic study of autophagy inhibition using hydroxychloroquine in patients with metastatic pancreatic adenocarcinoma. The oncologist. 2014;19(6):637–8. doi: 10.1634/theoncologist.2014-0086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy. 2012;8(2) doi: 10.4161/auto.8.2.18554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maes H, Kuchnio A, Peric A, Moens S, Nys K, De Bock K, et al. Tumor vessel normalization by chloroquine independent of autophagy. Cancer cell. 2014;26(2):190–206. doi: 10.1016/j.ccr.2014.06.025. [DOI] [PubMed] [Google Scholar]
- 13.Gallant JN, Allen JE, Smith CD, Dicker DT, Wang W, Dolloff NG, et al. Quinacrine synergizes with 5-fluorouracil and other therapies in colorectal cancer. Cancer Biol Ther. 2011;12(3):239–51. doi: 10.4161/cbt.12.3.17034. [DOI] [PubMed] [Google Scholar]
- 14.Gurova KV, Hill JE, Guo C, Prokvolit A, Burdelya LG, Samoylova E, et al. Small molecules that reactivate p53 in renal cell carcinoma reveal a NF-kappaB-dependent mechanism of p53 suppression in tumors. Proc Natl Acad Sci U S A. 2005;102(48):17448–53. doi: 10.1073/pnas.0508888102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ehsanian R, Van Waes C, Feller SM. Beyond DNA binding - a review of the potential mechanisms mediating quinacrine’s therapeutic activities in parasitic infections, inflammation, and cancers. Cell Commun Signal. 2011;9:13. doi: 10.1186/1478-811X-9-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gupta A, Roy S, Lazar AJ, Wang WL, McAuliffe JC, Reynoso D, et al. Autophagy inhibition and antimalarials promote cell death in gastrointestinal stromal tumor (GIST) Proc Natl Acad Sci U S A. 2010;107(32):14333–8. doi: 10.1073/pnas.1000248107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Geng Y, Kohli L, Klocke BJ, Roth KA. Chloroquine-induced autophagic vacuole accumulation and cell death in glioma cells is p53 independent. Neuro Oncol. 2010;12(5):473–81. doi: 10.1093/neuonc/nop048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo JY, Chen HY, Mathew R, Fan J, Strohecker AM, Karsli-Uzunbas G, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25(5):460–70. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guo JY, Karsli-Uzunbas G, Mathew R, Aisner SC, Kamphorst JJ, Strohecker AM, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27(13):1447–61. doi: 10.1101/gad.219642.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lock R, Roy S, Kenific CM, Su JS, Salas E, Ronen SM, et al. Autophagy facilitates glycolysis during Ras-mediated oncogenic transformation. Mol Biol Cell. 2011;22(2):165–78. doi: 10.1091/mbc.E10-06-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang S, Wang X, Contino G, Liesa M, Sahin E, Ying H, et al. Pancreatic cancers require autophagy for tumor growth. Genes Dev. 2011;25(7):717–29. doi: 10.1101/gad.2016111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lock R, Kenific CM, Leidal AM, Salas E, Debnath J. Autophagy-dependent production of secreted factors facilitates oncogenic RAS-driven invasion. Cancer discovery. 2014;4(4):466–79. doi: 10.1158/2159-8290.CD-13-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buchakjian MR, Kornbluth S. The engine driving the ship: metabolic steering of cell proliferation and death. Nat Rev Mol Cell Biol. 2010;11(10):715–27. doi: 10.1038/nrm2972. [DOI] [PubMed] [Google Scholar]
- 25.Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, et al. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461(7260):109–13. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bakkenist CJ, Kastan MB. DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 27.Maclean KH, Dorsey FC, Cleveland JL, Kastan MB. Targeting lysosomal degradation induces p53-dependent cell death and prevents cancer in mouse models of lymphomagenesis. J Clin Invest. 2008;118(1):79–88. doi: 10.1172/JCI33700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vousden KH, Ryan KM. p53 and metabolism. Nat Rev Cancer. 2009;9(10):691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 29.Jiang P, Du W, Wang X, Mancuso A, Gao X, Wu M, et al. p53 regulates biosynthesis through direct inactivation of glucose-6-phosphate dehydrogenase. Nat Cell Biol. 2011;13(3):310–6. doi: 10.1038/ncb2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maycotte P, Gearheart CM, Barnard R, Aryal S, Mulcahy Levy JM, Fosmire SP, et al. STAT3-mediated autophagy dependence identifies subtypes of breast cancer where autophagy inhibition can be efficacious. Cancer research. 2014;74(9):2579–90. doi: 10.1158/0008-5472.CAN-13-3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karsli-Uzunbas G, Guo JY, Price S, Teng X, Laddha SV, Khor S, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer discovery. 2014;4(8):914–27. doi: 10.1158/2159-8290.CD-14-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rosenfeldt MT, O’Prey J, Morton JP, Nixon C, MacKay G, Mrowinska A, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504(7479):296–300. doi: 10.1038/nature12865. [DOI] [PubMed] [Google Scholar]
- 33.Yang A, Rajeshkumar NV, Wang X, Yabuuchi S, Alexander BM, Chu GC, et al. Autophagy is critical for pancreatic tumor growth and progression in tumors with p53 alterations. Cancer discovery. 2014;4(8):905–13. doi: 10.1158/2159-8290.CD-14-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McAfee Q, Zhang Z, Samanta A, Levi SM, Ma XH, Piao S, et al. Autophagy inhibitor Lys05 has single-agent antitumor activity and reproduces the phenotype of a genetic autophagy deficiency. Proc Natl Acad Sci U S A. 2012;109(21):8253–8. doi: 10.1073/pnas.1118193109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fung C, Lock R, Gao S, Salas E, Debnath J. Induction of Autophagy during Extracellular Matrix Detachment Promotes Cell Survival. Mol Biol Cell. 2008;19(3):797–806. doi: 10.1091/mbc.E07-10-1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.