SUMMARY
Human immunodeficiency virus (HIV) infection is associated with increased intestinal translocation of microbial products and enteropathy as well as alterations in gut bacterial communities. However, whether the enteric virome contributes to this infection and resulting immunodeficiency remains unknown. We characterized the enteric virome and bacterial microbiome in a cohort of Ugandan patients, including HIV-uninfected or HIV-infected subjects and those either treated with anti-retroviral therapy (ART) or untreated. Low peripheral CD4 T cell counts were associated with an expansion of enteric adenovirus sequences and this increase was independent of ART treatment. Additionally, the enteric bacterial microbiome of patients with lower CD4 T counts exhibited reduced phylogenetic diversity and richness with specific bacteria showing differential abundance, including increases in Enterobacteriaceae, which have been associated with inflammation. Thus, immunodeficiency in progressive HIV infection is associated with alterations in the enteric virome and bacterial microbiome, which may contribute to AIDS-associated enteropathy and disease progression.
Keywords: virome, microbiome, HIV, AIDS, adenovirus, AIDS enteropathy, systemic inflammation
INTRODUCTION
An estimated 35 million adults worldwide are HIV positive, with the greatest burden of disease in sub-Saharan Africa (HIV/AIDS, 2013). HIV infects and depletes CD4 T cells, leading to the development of acquired immunodeficiency syndrome (AIDS), defined by the presence of less than 200 CD4 T cells/µl circulating in the blood (denoted herein as CD4 >200 or <200) or the development of an AIDS-defining opportunistic infection or cancer (Selik et al., 2014). Antiretroviral therapy (ART) successfully controls systemic HIV replication but immune recovery is variable (Maartens et al., 2014). A hallmark of HIV disease is a rapid and profound depletion of CD4 T cells in the gut-associated lymphoid tissue (Brenchley et al., 2004; Klatt et al., 2008) and increased translocation of microbial products across this compromised epithelial barrier (Brenchley et al., 2006; Klase et al., 2015). HIV infection can also lead to enteropathy characterized by increased gastrointestinal (GI) inflammation, diarrhea and malabsorption (Brenchley, 2013). The role of enteric microbes in these disease manifestations is incompletely understood.
The human enteric microbiome contains viruses, bacteria, archaea, fungi and other eukaryotic organisms (Norman et al., 2014; Virgin, 2014). Enteric human virome and bacterial microbiome alterations have been linked to inflammatory bowel disease (IBD), obesity, and changes in host behavior (Backhed et al., 2012; Lyte, 2013; Norman et al., 2015). Enteric eukaryotic viruses can directly affect human health by instigating gastroenteritis, enteritis or colitis. Bacteriophages can perturb the bacterial community to indirectly influence gut health and may directly interact with the human immune system (Duerkop and Hooper, 2013; Virgin, 2014). In IBD diversity and richness of the bacterial microbiome is inversely correlated with that of bacteriophages, suggesting an antagonistic relationship between bacteria and bacteriophages during enteric inflammation (Norman et al., 2015). However, little else is known about the contributions of bacteriophages to other human diseases including HIV infection.
Next generation sequencing (NGS) offers a unique opportunity to examine the virome and discover potentially novel unculturable viruses (Zhao et al., 2013). Prior work using NGS demonstrated expansion of the enteric eukaryotic virome during pathogenic SIV infection of rhesus macaques, while the bacterial microbiome was not detectably changed (Handley et al., 2012). Further, adenoviruses detected by NGS during pathogenic SIV infection were associated with pathologic evidence for viral enteritis (Handley et al., 2012). These data suggest that alterations in the enteric virome may be important in the pathogenesis of AIDS and HIV enteropathy. Prior studies of the bacterial microbiome in HIV-positive individuals suggest that HIV infection may be associated with significant changes in enteric bacterial populations, including the outgrowth of potential enteropathogens (Dillon et al., 2014; Dinh et al., 2015; Li et al., 2012; Lozupone et al., 2013; Lozupone et al., 2014; McHardy et al., 2013; Mutlu et al., 2014; Perez-Santiago et al., 2013; Vujkovic-Cvijin et al., 2013). Together these studies suggest that HIV infection may alter the enteric virome and bacterial microbiome.
Here we examine the effect of HIV on the enteric virome and bacterial microbiome in a well-characterized Ugandan cohort (Siedner et al., 2015), including HIV-positive subjects stably on ART for at least 5 years and ART-naïve subjects, as well as location-matched HIV-negative subjects. Importantly, in this study we define the bacterial microbiome and virome in a region of the world that has been understudied. We show that alterations in the enteric virome and bacterial microbiome are associated with low peripheral CD4 T cell counts rather than HIV infection alone. These data reveal a relationship between alterations in enteric bacteria and viruses and HIV/AIDS-related immunocompromise that may play a role in enteropathy and the chronic systemic immune activation observed in HIV infection.
RESULTS
Cohort Characteristics
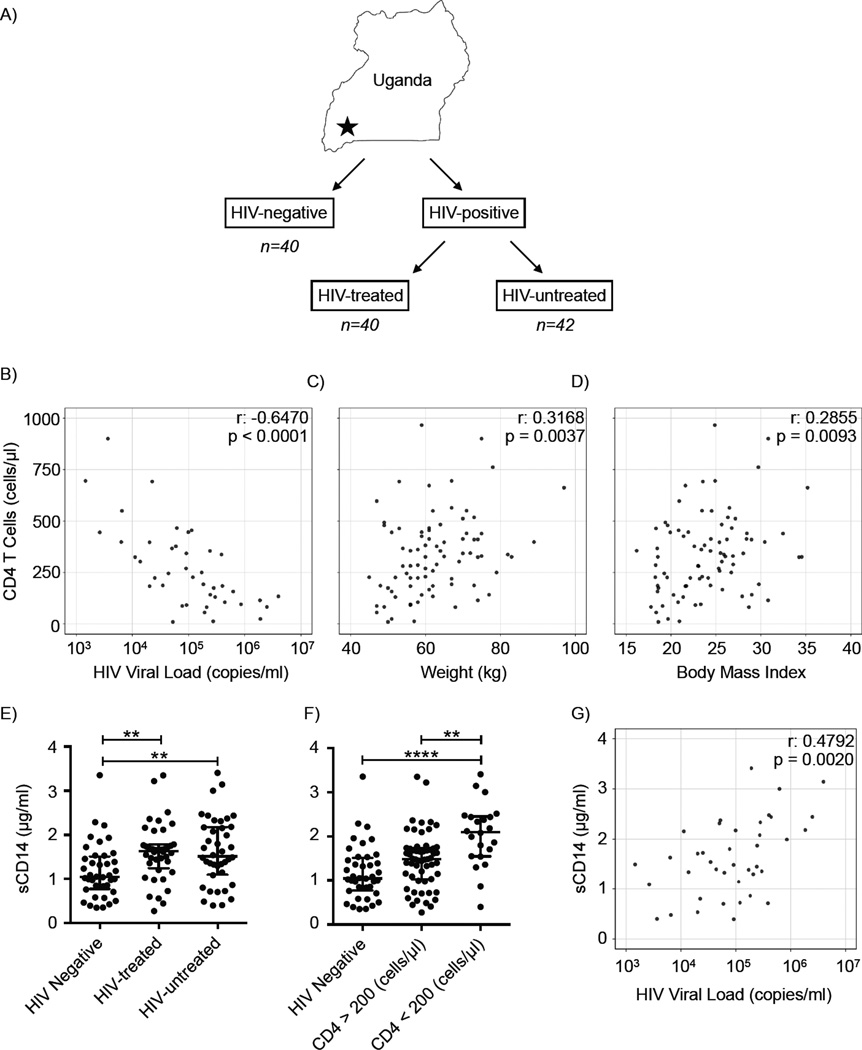
Eighty-two HIV-infected Ugandan subjects were recruited as part of the Uganda AIDS Rural Treatment Outcomes (UARTO) study at the Immune Suppression Syndrome Clinic at Mbarara Regional Referral Hospital, comprising 40 HIV-infected subjects on ART treatment (HIV-treated) and 42 HIV-infected, ART-naïve (HIV-untreated) subjects. An additional 40 HIV-negative subjects were recruited from the HIV testing clinic after a negative HIV test result (Figure 1A and Table 1). The median time on ART for treated subjects was 6.7 years (interquartile range, IQR, 6.1–7.1) and 87.5% (n=35) of ART-treated participants had undetectable viral loads (<20 copies/mL). Twenty-five participants (20.5%) had CD4 T cell counts <200 (CD4 <200) at the time of stool collection. Significant differences in CD4 T cell counts, CD4 T cell percentages, HIV viral load, prophylactic co-trimoxazole use, age, sex and other variables existed between HIV-treated and untreated groups (Table 1). Peripheral CD4 T cell counts in HIV-infected subjects were negatively correlated with circulating HIV viral load in subjects with detectable peripheral HIV RNA (Figure 1B, p < 0.0001). Additionally, weight (p = 0.0037) and body mass index (BMI, p = 0.0093) positively correlated with peripheral CD4 T cell count in HIV-infected subjects (Figure 1C, 1D). These clinical characteristics are consistent with the known effects of HIV infection (Mankal and Kotler, 2014; Palermo et al., 2011; Phillips et al., 2010).
Figure 1. Study Design and Cohort Characteristics.
(A) Subject cohort and study design. Correlation between CD4 T cell count (y-axis) compared to (B) HIV viral load (p < 0.0001); (C) weight (p = 0.0037); and (D) body mass index (p= 0.0093). Circulating levels of sCD14 were graphed by (E) HIV status and ART treatment group and (F) CD4 T cell count. (G) Correlation between sCD14 levels and HIV viral load (p =0.002). p ≤ 0.01 = **, p ≤ 0.0001 = ****. Bars indicate median ± interquartile range (IQR). See also Table S1.
Table 1.
Cohort Characteristics
| Patient Characteristics | HIV Neg (n = 40) |
HIV on ART (n = 40) |
HIV no treatment (n = 42) |
P value |
|---|---|---|---|---|
| 16S rRNA sequencing (n) (total 110) | 37 | 39 | 34 | |
| VLP NGS sequencing (n) (total 65) | 21 | 21 | 23 | |
| Demographics, median (IQR) | ||||
| Age (years) | 43 (38–48) | 44 (38–49) | 29 (24 −34) | < 0.0001 |
| Male, n (%) | 20 (50) | 20 (50) | 11 (26.2) | 0.0404 |
| Height (cm) | 159 (155–163) | 159 (157–168) | 161 (156–166) | 0.3894 |
| Weight (kg) | 64 (55–74) | 61 (56–71) | 60 (53 – 68) | 0.5735 |
| BMI | 24 (22–28) | 24 (21–27) | 23 (20–27) | 0.4305 |
| Laboratory measures, median (IQR) | ||||
| CD4 T cell count (cells/µl) | NA | 396 (283–490) | 225 (113–382) | 0.0003 |
| >500, n (%) | NA | 8 (20) | 4 (9.5) | |
| 200–500, n (%) | NA | 29 (72.5) | 16 (38.1) | |
| <200, n (%) | NA | 3 (7.5) | 22 (52.4) | |
| CD4 Percent | NA | 25 (21–31) | 15 (10–24) | < 0.0001 |
| HIV Viral Load (copies/ml) | NA | 20 (20–20) | 95,571(24455–285548) | < 0.0001 |
| CD4 nadir (cells/µl) | NA | 116 (58–167) | 225 (110–382) | 0.0001 |
| Symptoms over last 30 days | ||||
| Nausea/Vomiting, n (%) | 13 (32.5) | 5 (12.5) | 15 (35.7) | 0.0389 |
| Diarrhea, n (%) | 10 (25) | 4 (10) | 9 (21) | 0.2000 |
| Constipation, n (%) | 17 (42.5) | 13 (32.5) | 10 (23.8) | 0.1969 |
| Loss of Appetite, n (%) | 21 (52.5) | 10 (25) | 20 (47.6) | 0.0286 |
| Dysgeusia, n (%) | 20 (50) | 14 (35) | 17 (40.5) | 0.3875 |
| Medications last 30d | ||||
| ART, n (%) | NA | 40 (100) | NA | |
| NRTI, n (%) | NA | 40 (100) | NA | |
| NNRTI, n (%) | NA | 35 (85) | NA | |
| PI, n (%) | NA | 6 (15) | NA | |
| Years on ART, median (IQR) | NA | 6.7 (6.1–7.1) | NA | |
| OI prophylaxis | ||||
| Bactrim last 30d, n (%) | 0 (0) | 38 (95) | 38 (90.5) | < 0.0001a |
| Other Antimicrobials 3d, n (%) | 8 (20) | 5 (12.5) | 4 (9.5) | 0.3721 |
| Other Antimicrobials 30d, n (%) | 15 (37.5) | 14 (35) | 4 (9.5) | 0.0066 |
| Living conditions | ||||
| Water source, n (%) | ||||
| Communal Tap | NA | 15 (37.5) | 22 (52.4) | 0.1913 |
| Piped In | NA | 2 (5) | 2 (4.8) | 1.0000 |
| Open Well | NA | 7 (17.5) | 8 (19) | 1.0000 |
| Protected Well | NA | 7 (17.5) | 1 (2.4) | 0.0274 |
| Other | NA | 9 (22.5) | 9 (21.4) | 1.0000 |
| Distance to water, m, median (IQR) | NA | 75 (6–675) | 15 (6–350) | 0.3071 |
| Toilet type, n (%) | ||||
| Covered pit latrine | NA | 25 (62.5) | 14 (33.3) | 0.0145 |
| Uncovered pit latrine | NA | 15 (37.5) | 25 (59.5) | 0.0512 |
| Flush toilet | NA | 0 | 3 (7.1) | 0.2412 |
| Food Security | ||||
| Grows own Produce, n (%) | NA | 29 (72.5) | 20 (47.6) | 0.0260 |
| Owns Livestock, n (%) | NA | 19 (47.5) | 12 (28.6) | 0.1107 |
| Goes Hungry, n (%) | NA | 11 (27.5) | 12 (28.6) | 1.0000 |
| Cooks in Kitchen, n (%) | NA | 25 (62.5) | 18 (42.9) | 0.0828 |
| District | ||||
| MBARARA, n (%) | 30 (75) | 26 (65) | 27 (64.2) | 0.5135 |
| ISINGIRO, n (%) | 3 (7.5) | 6 (15) | 9 (21.4) | 0.2057 |
| KIRUHURA, n (%) | 4 (10) | 2 (5) | 4 (9.5) | 0.6655 |
| Other or NA, n (%) | 3 (7.5) | 6 (15) | 2 (4.8) | 0.2485 |
ART, antiretroviral therapy; NA, not available; NS, not significant; IQR, interquartile range; For comparing continuous variables, Mann-Whitney and Kruskal-Wallis tests were used; for comparing categorical variables, Chi-Square and Fisher’s exact tests were used. OI, opportunistic infection
NS between HIV-positive groups.
Associations of sCD14, CD4 T cell counts and HIV viral load
Soluble CD14 (sCD14) binds bacterial lipopolysaccharide (LPS) and serum levels of sCD14 have been used as a reflection of microbial translocation from the gut (Marchetti et al., 2013). Circulating sCD14 levels in HIV-positive subjects are elevated and correlate with mortality (Sandler et al., 2011). We found that plasma sCD14 levels in HIV-infected subjects, either with or without ART treatment, were significantly higher than in HIV-negative subjects (Figure 1E, p = 0.0017). A hallmark of advanced HIV disease and profound immunodeficiency is peripheral CD4 <200 (Selik et al., 2014). In our cohort, subjects with CD4 <200 had significantly higher levels of circulating sCD14 than both HIV-negative subjects and HIV-infected subjects with CD4 >200 (Figure 1F, p < 0.0001). Plasma sCD14 levels positively correlated with HIV viral load (Figure 1G, p = 0.002). These data confirm that our cohort exhibits the expected positive correlation between HIV-associated immunodeficiency and a marker of disrupted intestinal function and systemic inflammation (Marchetti et al., 2013).
The enteric DNA virome in HIV-positive and HIV-negative Ugandan subjects
We next characterized the fecal DNA virome by NGS on samples enriched for viral sequences present in virus-like particles (VLPs) as described (Norman et al., 2015). Libraries were obtained from 21 HIV-negative subjects, 21 HIV-treated subjects and 23 HIV-untreated subjects (total n = 65), and averaged 1.14 +/− 0.43 million sequences per sample, of which 90 +/− 7% were of high-quality and used for all subsequent analyses (Table S1).
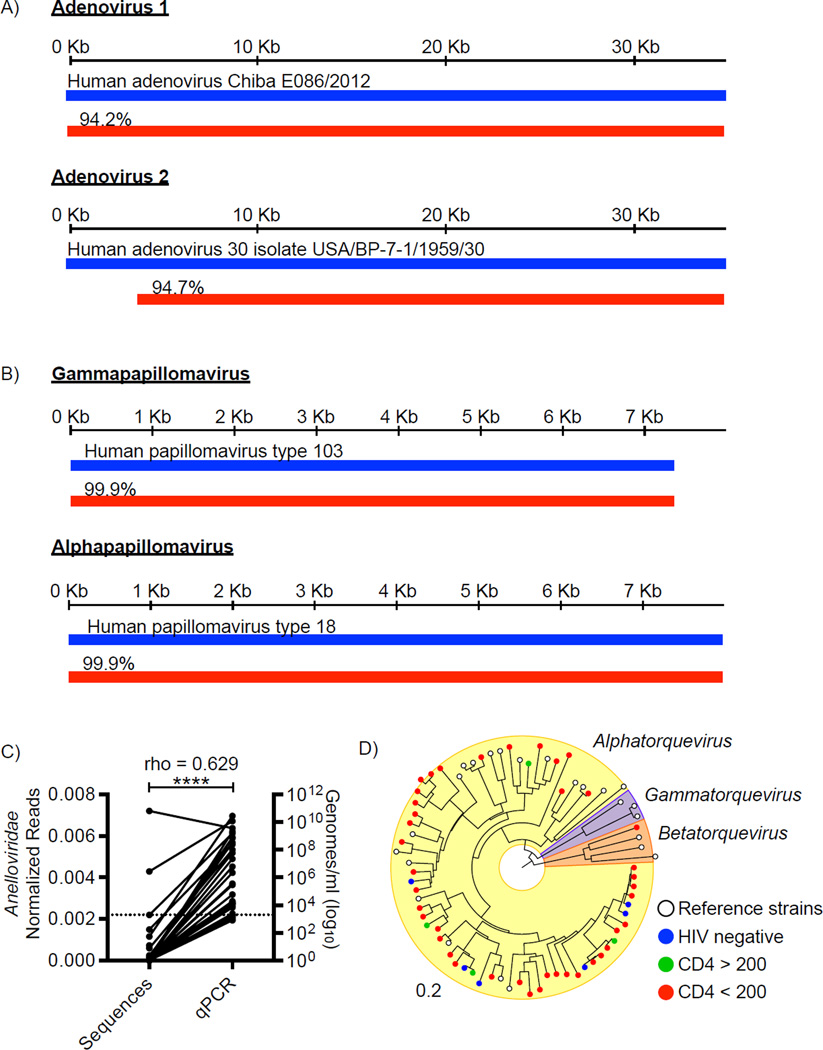
Viral sequences were classified using a two-staged approach. First, all dereplicated, high-quality sequences were queried against a virus protein database using BLASTx (Norman et al., 2015). Sequences were most frequently assigned to bacteriophages of the Caudovirales order or the Microviridae family, along with several eukaryotic virus families (e.g. Adenoviridae, Annelloviridae) and other viral families defined as “unclassified” in the NCBI Taxonomy Database (Figure S1A). All sequences assigned to eukaryotic viral taxa were binned as “potentially viral” and then queried against the NCBI non-redundant nucleic acid and protein databases using BLASTn and BLASTx encapsulated in the VirusSeeker (Vs) pipeline in order to remove sequences that aligned with higher scores (lower e-values) to non-viral reference sequences (Zhao et al., 2013)(Zhao et al., In preparation). We then compared the number of potential viral sequences from the first stage (Bx) to those found after Vs analysis. This revealed significant differences in sequence assignments to several viral families (Figure S1B). There was a statistically significant diminution in sequences assigned to Nudiviridae, Ascoviridae and other large DNA viruses using the more stringent criteria of searching against the comprehensive database; these sequences were set aside (Figure S1B). The eukaryotic viral families remaining after this stringent analysis included the Adenoviridae, Anelloviridae, Circoviridae and Papillomaviridae; dereplicated sequences assigned to these families were analyzed further after de novo assembly. Resulting contigs longer than 500bp were aligned to Adenoviridae, Anelloviridae, Circoviridae and Papillomaviridae reference genomes. Two near-full-length adenovirus genomes with high sequence identity to human adenovirus D were identified (Figure 2A). Three fecal samples contained multiple adenoviruses. Twelve unique full-length papillomavirus genomes were obtained from one HIV-positive, untreated subject (Figure 2B, Table S2).
Figure 2. Eukaryotic viruses identified in the fecal samples.
Alignment (red bars) of (A) two near full-length adenovirus contigs (34,669 and 32,313 bp) and (B) two representative Human Papillomavirus (HPV) contigs (7,362 and 7,857 bp) to the most closely related virus genome (blue bars). Percentages indicate nucleotide identity over the length of the best-aligned homologous region compared to the reference genome. C) Correlation of Alphatorquevirus copies/ml as measured by quantitative real-time qPCR with number of unique Anelloviridae-assigned sequences (p < 0.0001). (D) Phylogenetic distance of anellovirus contig sequences (colored circles) with anellovirus reference genomes (white circles). Colored circles represent viruses identified in samples from: blue, HIV-negative; green, CD4 >200; and red, CD4 <200 subjects. Bar indicates bootstrap distance. See also Figure S1, Table S2, S3.
The number of unique Anelloviridae-assigned sequences positively correlated with the genome copy numbers of anelloviruses as measured by quantitative real-time PCR analysis (qPCR) (Fig 2C, p < 0.0001). qPCR detected anelloviruses in 48% of the samples, while Illumina sequencing detected anelloviruses in 35% of the samples, suggesting Illumina sequencing of VLPs may underrepresent true viral diversity. We were able to assemble 228 unique anellovirus contigs, 51 of which contained the ORF1 gene. We performed phylogenetic analysis of these 51 contigs using a conserved region of the ORF1 gene compared to representative anellovirus ORF1 sequences (Figure 2D). One virus was most closely related to the Betatorquevirus genus, while the rest were most closely related to the Alphatorquevirus genus. Interestingly, one fecal sample harbored at least 19 different anelloviruses (Table S3).
Changes in the enteric virome associated with CD4 T cell counts
We next analyzed the relationship between the number of viral sequences in each sample and HIV infection status. There were no statistically significant differences between HIV-positive and HIV-negative subjects for Adenoviridae, Anelloviridae, Circoviridae and Papillomaviridae sequences (Figure S2, A-D). However, these subjects exhibited wide variation in CD4 T cell counts (Table 1), and lower CD4 T cell count is an indication of impaired host immune function and increased gut dysregulation. We therefore evaluated the relationship between CD4 T cell counts and the enteric virome using a clinically validated cutoff of CD4 <200 as an indication of impaired immunity.
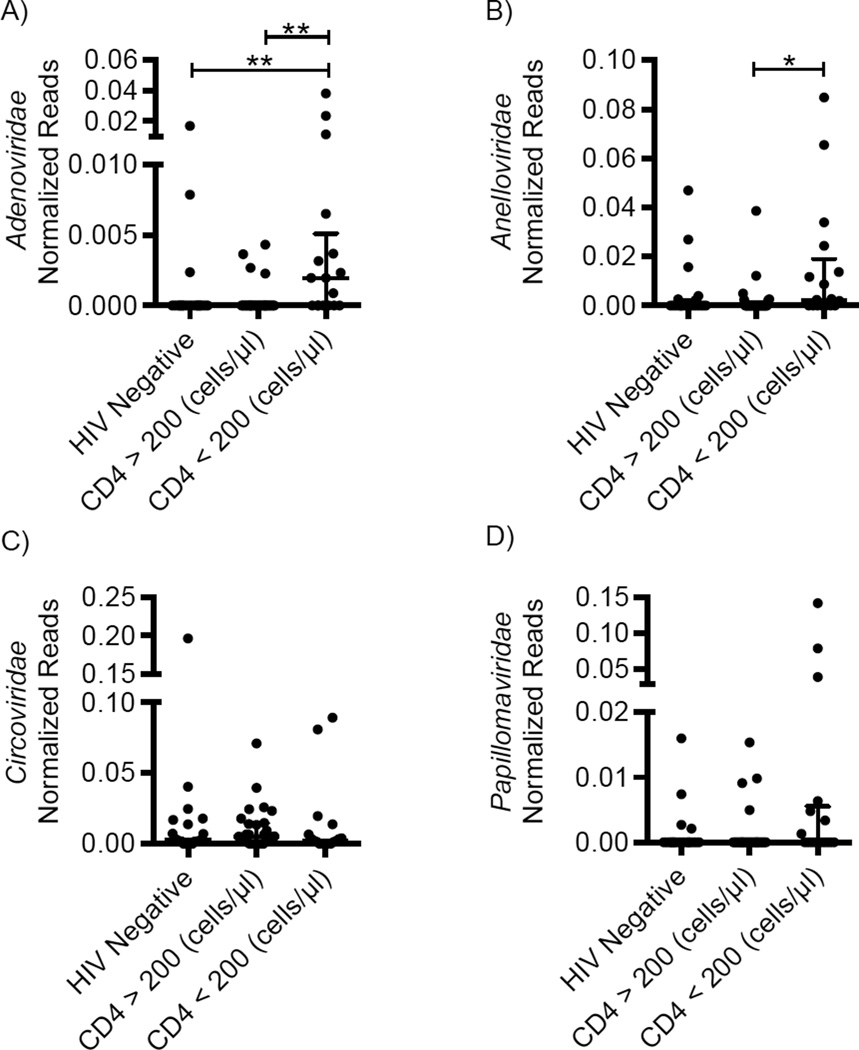
We observed significantly more Adenoviridae sequences in HIV-positive subjects with CD4 <200 when compared to both HIV-negative or HIV-positive subjects with CD4 >200 (Figure 3A, p = 0.0026). HIV-positive subjects with CD4 <200 were more likely to have detectable adenovirus sequences (Table S4, p = 0.0031), suggesting expansion of adenovirus sequence abundance and overall infection prevalence. There was no significant difference in adenovirus sequences based on ART therapy (Figure S2E, p = 0.0754). Subgroup analysis of the HIV-untreated group showed the effect was driven by low CD4 T cell counts (Figure S2I), suggesting that adenovirus expansion is associated with host immune status independent of ART therapy.
Figure 3. Comparison of enteric eukaryotic virus sequences and CD4 T cell count.
Abundance of (A) Adenoviridae-, (B) Anelloviridae-, (C) Circoviridae- and (D) Papillomaviridae-assigned sequences in samples collected from HIV-negative subjects and subjects with CD4 >200 and <200. Sequences were normalized by dividing by the number of dereplicated (<95% identical), high-quality sequences. p ≤ 0.05 = *, p ≤ 0.01 = **. Bars indicate median ± interquartile range (IQR). Y-axis standardized by taking square root of the normalized value. See also Figures S2, S5, Table S4.
Anelloviridae sequences significantly differed by both CD4 T cell count (Figure 3B, p = 0.0239) and HIV treatment status (Figure S2F, p = 0.0282); however, subgroup analysis revealed this was largely driven by the HIV-untreated subjects with CD4 <200 (Figure S2J). Further, samples belonging to HIV-positive subjects with CD4 <200 were more likely to contain Anelloviridae sequences when compared to HIV-positive subjects with CD4 >200 (Table S4, p = 0.0238). However, there was no significant difference in frequency of Anelloviridae-sequences between HIV-positive subjects with CD4 <200 and HIV-negative subjects (Table S4, p =0.0990). Taken together these data suggest but do not prove that enteric Anelloviridae expansion is associated with the HIV-induced immunodeficiency.
Circoviridae were the most frequently detected type of eukaryotic virus (present in 75% of all samples). There were no statistically significant differences in the number or prevalence of Circoviridae sequences by CD4 T cell count (Figure 3C, p = 0.4530, Table S4, p = 0.8746) or HIV treatment status (Figure S2G & K, p = 0.3984). Similarly, Papillomaviridae sequences were found in 23% of all samples, but were not differentially represented based on CD4 T cell counts (Figure 3D, p = 0.1115, Table S4, p = 0.1253) or ART therapy (Figure S2H & L, p = 0.5552). This indicates that HIV-mediated effects on Adenoviridae and Anelloviridae described above were not a result of amplification bias. These data demonstrate that expansion of Adenoviridae occurs in subjects with AIDS and suggest that Anelloviridae expansion in HIV infection may also be the result of severe immunodeficiency.
Effects of HIV infection and immunodeficiency on the enteric bacterial microbiome
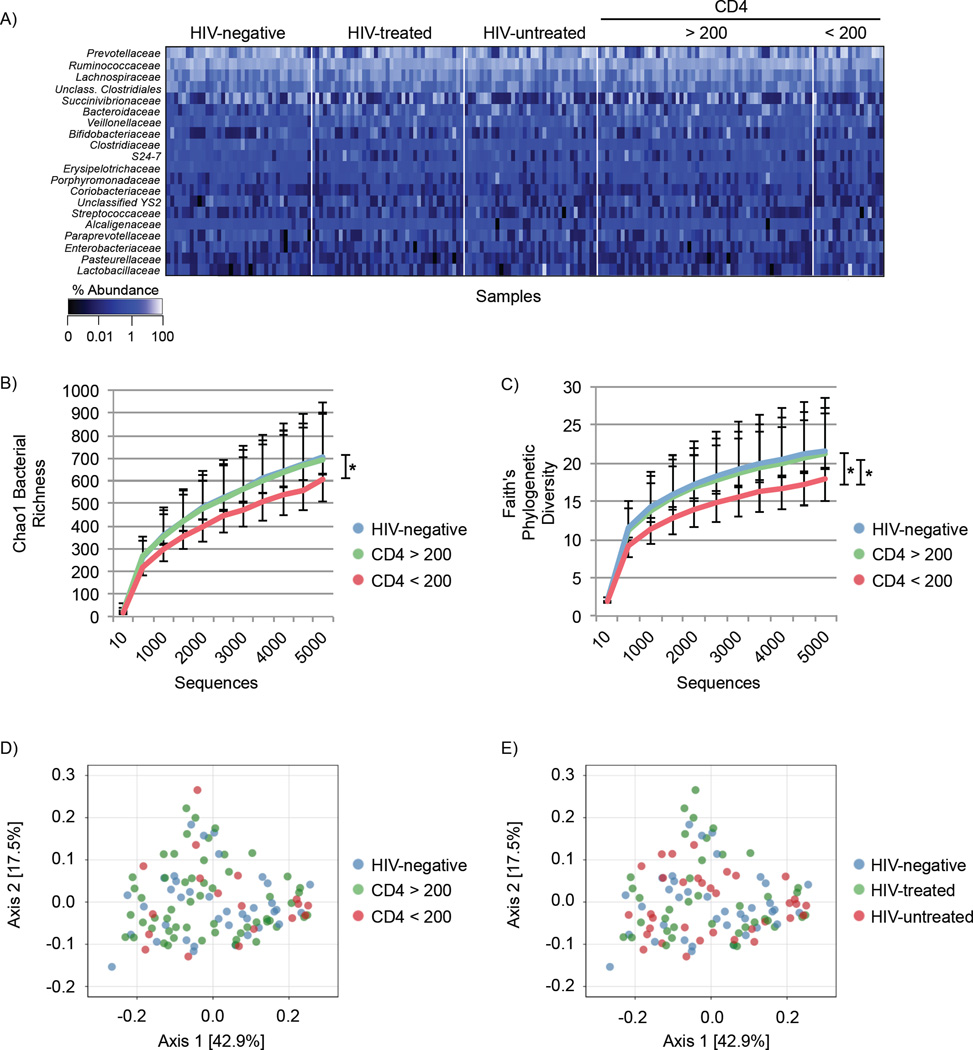
We next characterized the bacterial microbiome using 16S rRNA gene profiling for 37 HIV-negative subjects, 39 HIV-treated subjects and 34 HIV-untreated subjects (total n = 110, Figure 4A, Figure S3). Subjects with CD4 <200 had significantly decreased bacterial richness compared to subjects with CD4 >200 (Figure 4B, p = 0.009), with a near significant decrease compared to HIV negative subjects (Figure 4B, p = 0.066). Bacterial phylogenetic diversity was significantly decreased in subjects with CD4 <200 compared to both HIV-negative subjects (Figure 4C, p = 0.036) and subjects with CD4 >200 (Figure 4C, p = 0.018). However, there was no significant difference in bacterial richness or phylogenetic diversity between HIV treatment groups.
Figure 4. Bacterial community profiling.
(A) Heatmap showing relative abundance of the 20 most frequent bacterial families (y-axis) by sample (x-axis), grouped by HIV status and CD4 T cell count. Percent abundance is indicated by the gradient key. (B) Chao1 rarefied bacterial richness grouped by CD4 T cell count. (C) Comparison of bacterial Faith’s phylogenetic diversity in HIV-negative and HIV-positive subjects by CD4 T cell count. Statistical analysis was performed in QIIME using two-sample, non-parametric t-tests with Monte Carlo permutations. Error bars indicate SEM. p ≤ 0.05 = * Principle Coordinate Analysis (PCoA) plots of the weighted UniFrac distances colored by HIV status and (D) CD4 T cell count or (E) ART treatment. See also Figure S3, S4, Table S7.
We then assessed bacterial β-diversity (similarity/dissimilarity indices to measure diversity across different groups) between experimental groups by calculating weighted UniFrac distances (Lozupone and Knight, 2005). There was no significant difference in β-diversity by HIV status (p=0.588) or CD4 T cell group (p=0.231) as assessed by Permutational Multivariate Analysis of Variance Using Distance Matrices (PERMANOVA). Principle coordinates analysis (PCoA) using weighted UniFrac distances demonstrated no clustering when grouped by CD4 T cell group (Figure 4D) or HIV status (Figure 4E). In addition, bacterial community structure was not influenced by water source, recent diarrhea, antibiotic usage, gender or home geographic district (Figure S3B–F). These results demonstrate that the overall enteric bacterial community structure was similar when comparing HIV-positive to HIV-negative individuals. CD4 T cell count was the most influential factor contributing to bacterial community structure, wherein subjects with CD4 <200 had decreased phylogenetic diversity and richness.
Differentially abundant bacterial taxa in subjects with low CD4 T cell counts
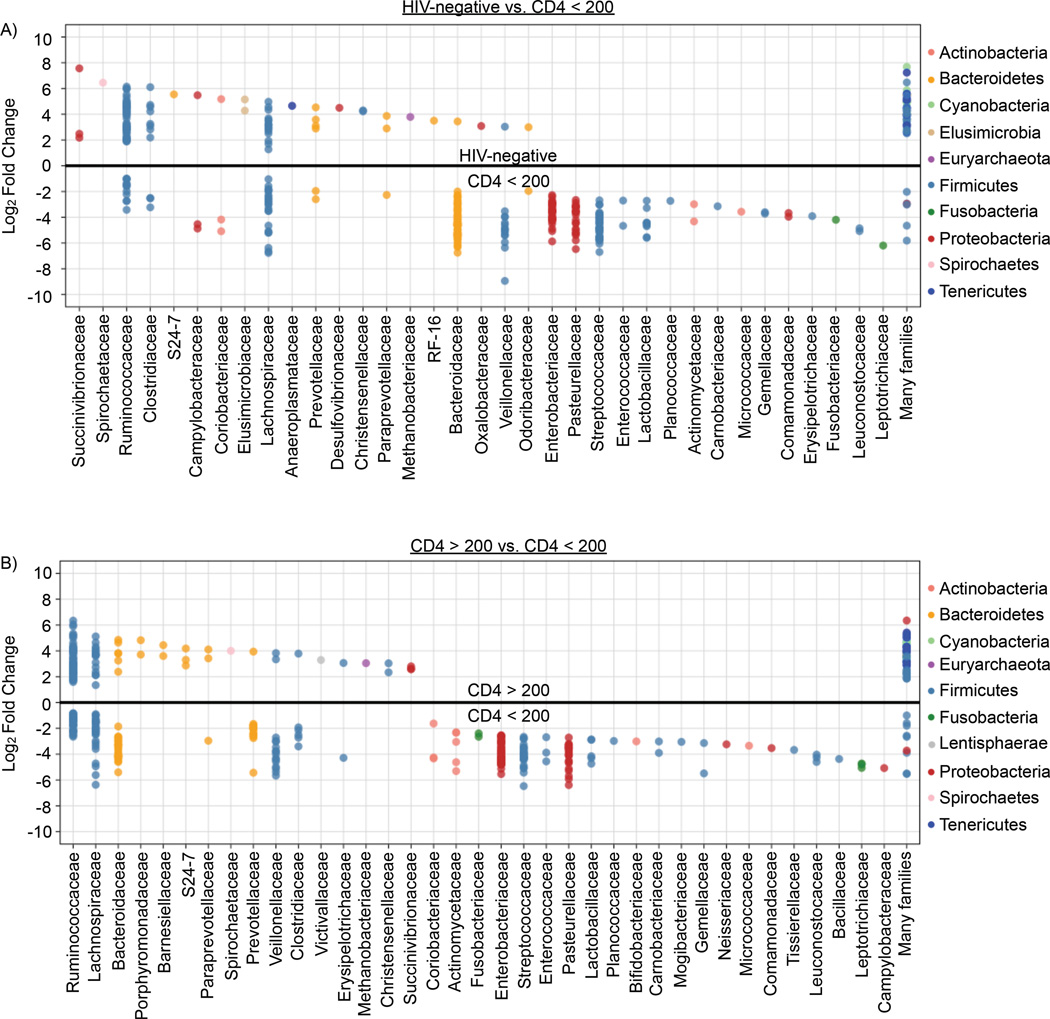
Given the decrease in bacterial phylogenetic diversity in HIV-positive subjects with CD4 <200, we next identified specific bacterial operational taxonomic units (OTUs) differentially abundant by CD4 T cell group (Tables S5, tab 1 and 2) using variance stabilization techniques based on mixture models of microbiome data (McMurdie and Holmes, 2014). These discriminant OTUs were then plotted by bacterial family association (Figure 5). OTUs belonging to 13 bacterial families (Enterobacteriaceae, Pasteurellaceae, Streptococcaceae, Enterococcaceae, Lactobacillaceae, Planococcaceae, Actinomycetalaceae, Carnobacteriaceae, Micrococcaceae, Gemellaceae, Comamonadaceae, Leuconostocaceae, and Leptotrichiaceae) discriminately associated with CD4 <200 as compared to both HIV-negative subjects (Figure 5A, Table S5 tab 1) and subjects with CD4 >200 (Figure 5B, Tables S5 tab 2). There were no differentially abundant OTUs assigned to Succinovibrionaceae, Spirochaetaceae, S24-7, and Methanobacteriaceae families in subjects with CD4 <200, but there were discriminant OTUs assigned to these families associated with both HIV-uninfected subjects as well as subjects with CD4 >200. Other differentially represented families included Ruminococcaceae, with fewer discriminant OTUs present in subjects with CD4 <200 (n = 56 OTUs in CD4 >200 vs 33 OTUs for CD4 <200 and n = 50 OTUs in HIV-Negative vs 12 OTUs for CD4 <200 OTUs) and Bacteroidaceae, with more discriminant OTUs associated with subjects with CD4 <200 (n = 1 OTUs in HIV-negative vs 67 OTUs in CD4 <200 and n = 6 OTUs in CD4 >200 vs 31 OTUs in CD4 <200). We used oligotyping (Eren et al., 2011) to further classify unassigned Ruminococcus sequences. Subjects with CD4 >200 were enriched for sequences assigned to the species R. bromii and R. callidus compared to subjects with CD4 <200. HIV-negative subjects were enriched for sequences assigned to R. bromii compared to subjects with CD4 <200. Thus severe immunodeficiency was associated with alterations in multiple bacterial OTUs.
Figure 5. Differentially abundant bacterial taxa.
Variance stabilization techniques were utilized to determine discriminant bacterial OTUs. Dots represent one OTU assigned to the indicated bacterial family (column label). The position of the dot on the y-axis indicates the abundance of that OTU (positive or negative) in relation to the opposing group. Bacterial OTUs with significant differences between (A) HIV-negative and CD4 <200 as well as (B) CD4 >200 versus <200 were graphed by log2 fold-change (y-axis) and grouped by family association (x-axis). Coloring indicates bacterial phyla to which the OTUs belong. See also Tables S5, 6.
Contribution of co-trimoxazole use
The World Health Organization and The Joint United Nations Programme on HIV and AIDS (WHO/UNAIDS) guidelines for resource-limited settings recommend co-trimoxazole (trimethoprim-sulfamethoxazole) prophylaxis in all HIV-infected patients (WHO, 2006). Therefore, the majority of HIV-positive subjects in our study were taking co-trimoxazole (Table 1). Brief antibiotic use has previously been shown to affect the enteric microbiome. To assess the contribution of long-term usage of co-trimoxazole, we performed differential abundance analysis to determine discriminant bacterial OTUs as described above. We utilized data from HIV-negative controls since only four HIV-infected subjects with 16S rRNA data were not taking co-trimoxazole. Two OTUs showed significant fold-change with co-trimoxazole use as compared to 429 discriminant OTUs between subjects with CD4 <200 and >200 (Tables S5 tab 2). There was no difference in phylogenetic diversity in subjects taking co-trimoxazole (n = 68) when compared to those that did not (n = 39, p = 0.378). These data suggest that long-term usage of co-trimoxazole has minimal contribution to the enteric microbiome alterations observed in subjects with immunodeficiency.
Multivariate analysis reveals discriminant bacterial taxa
Six metadata variables (month stool collected, co-trimoxazole use, age, MiSeq run number, antibiotic use, and BMI) were either significantly or near significantly different between CD4 T cell groups in 16S analysis. Multivariate modeling was used to determine bacterial OTUs associated with CD4 T cell count independent of these confounders using Multivariate Association with Linear Models (MaAsLin). Since cotrimoxazole use was a correlated covariate with HIV status, it was excluded from the model. Fourteen bacterial OTUs were independently associated with CD4 T cell count (Table S6) including OTUs assigned to Ruminococcaceae and Enterobacteriaceae families. Using oligotyping, we assigned the Enterobacteriaceae sequences to genus Shigella or a related Escherichia species but were unable to further assign the Ruminococcaceae. Thus, shifts in the enteric bacterial community occur in advanced HIV disease independent of other differentiating metadata factors.
Bacterial taxa are associated with plasma sCD14
Increased circulating sCD14 has been linked to increased HIV mortality (Sandler et al., 2011) and may represent a proxy for bacterial translocation from the gut (Marchetti et al., 2013). Therefore, we performed correlation analysis to identify bacterial OTUs associated with circulating sCD14 levels. We identified 144 bacterial OTUs significantly associated with sCD14 (Table S5 tab 3), including 23 assigned to the Bacteriodaceae and 23 assigned to the Ruminococcaceae. Of those, OTU 199182 assigned to Ruminococcaceae and OTU 575768 assigned to Clostridiaceae were also associated with CD4 T cell count independently of other metadata variables from the multivariate analysis above (Table S6).
Bacteriophages and HIV infection
We previously found an inverse correlation between bacterial diversity and bacteriophage richness in IBD patients (Norman et al., 2015). We therefore characterized bacteriophage community composition by measuring bacteriophage richness and alpha diversity (Figure S5). There was no significant difference in richness or Shannon diversity of bacteriophage families or genera by CD4 T cell group (Figure S5A, B, C), ART treatment (Figure S5D, E, F) or HIV infection (Figure S5G, H, I). There were also no significant correlations between bacteriophage and bacterial richness and diversity (Table S7). Thus enteric bacteriophage communities were not detectably altered by HIV infection, ART therapy, or CD4 T cell depletion in this cohort.
DISCUSSION
The enteric DNA virome in HIV/AIDS
Adenoviruses are non-enveloped DNA viruses linked to human conjunctivitis, gastroenteritis, and respiratory illnesses. Here we identified an association between expansion of enteric adenovirus sequences and advanced HIV/AIDS which strikingly parallels findings in SIV-infected macaques (Handley et al. submitted). Adenovirus antigen was previously found in GI lesions of SIV-infected macaques (Handley et al., 2012), implicating these viruses in lentivirus-associated enteropathy that contribute to translocation of microbial products from the gut. Future studies of human biopsy specimens for adenovirus-associated pathology are needed to determine whether these adenoviruses cause enteropathy.
Anelloviridae are non-enveloped small, circular ssDNA viruses often found in human serum. While not identified as causative agents of diseases, Anelloviridae are increased in the serum of medically immunosuppressed solid organ transplant recipients (De Vlaminck et al., 2013) and HIV-positive subjects with increasing immunosuppression (Christensen et al., 2000; Debiaggi et al., 2000; Li et al., 2013). However, enteric populations were not examined. Our work demonstrates enteric Anelloviridae expansion in HIV, and subgroup analysis suggests this is driven by subjects with CD4 <200. One confounding aspect of our analysis was that while we detected a relationship between anellovirus sequences and CD4 T cell count, there was no significant difference in post-hoc analysis comparing CD4 <200 and HIV-negative subjects. One possibility is that our study was underpowered to detect this difference. Alternatively, since HIV-negative subjects in this study presented to the clinic for a variety of health-related reasons, it is possible they had other conditions associated with changes in anellovirus abundance (Touinssi et al., 2001; Walton et al., 2014). This issue will need to be resolved in future studies using healthy community or household controls.
The enteric bacterial microbiome in HIV/AIDS
We found decreased richness and phylogenetic diversity of the enteric bacterial microbiome in subjects with advanced HIV/AIDS similar to our findings in SIV-infected macaques (Handley et al., 2012)(Handley et al., submitted). Additionally, OTUs assigned to Enterobacteriaceae were associated with both low CD4 T cell count in our study and low percentage of circulating CD4 T cells in rhesus macaques (Handley et al. submitted). Oligotyping identified these organisms as genus Shigella or a closely related Escherichia species. Enterobacteriaceae family members have been associated with inflammation (Lupp et al., 2007) and most Shigella are enteric pathogens (Hale and Keusch, 1996; Yang et al., 2005). These microorganisms may contribute to the gastrointestinal disease and chronic immune activation observed in HIV patients.
Interestingly, decreased Ruminococcus levels were associated with diminished peripheral CD4 T cells and increased sCD14 levels, suggesting a protective role for these bacteria. Depletion of Ruminococcus was also found in the rectal mucosa of HIV-infected, untreated subjects (McHardy et al., 2013) and with symptomatic SIV-infection of macaques (Handley et al. submitted). Ruminococcus has been associated with both protective and disruptive roles in the enteric microbiome, and this effect is highly species dependent (Crost et al., 2013; David et al., 2014; Hsiao et al., 2014; Ze et al., 2012). We identified two species, R. callidus and R. bromii, depleted in subjects with CD4 <200. Both have been shown to increase after fecal microbial transplant treatment for Clostridium difficile infection (Satokari et al., 2014) and be decreased in subjects with Crohn’s disease and ulcerative colitis (Kang et al., 2010; Rajilic-Stojanovic et al., 2013), suggesting a protective role.
Previous studies of the enteric bacterial microbiome in HIV yielded disparate conclusions regarding overall changes in the bacterial community. However, some commonalities emerged, including changes in bacterial beta diversity, enrichment of Prevotella, and depletion of Bacteroides in chronic HIV infection (Dillon et al., 2014; Dinh et al., 2015; Lozupone et al., 2013; Lozupone et al., 2014; McHardy et al., 2013; Mutlu et al., 2014; Vazquez-Castellanos et al., 2014; Vujkovic-Cvijin et al., 2013). While we observed differences in discriminant OTUs assigned to Prevotella by CD4 T cell count, there was no clear trend toward greater frequency of significant OTU changes with CD4 <200, and there was no difference in beta diversity. The fact that inclusion and exclusion criteria, duration of ART treatment, sampling method and site, sample size, sequencing technique, and statistical methods differed between studies may explain the discrepancies observed between this study and others. Additionally, prior studies were performed on subjects in the developed world with relatively well-controlled HIV (none with advanced HIV disease) whereas our cohort was from Uganda and included patients with AIDS. A Prevotella-rich and Bacteroides-poor community has been associated with agrarian cultures (De Filippo et al., 2010) and this is consistent with the average Prevotella abundance of 28% we observed in our HIV-negative subjects. It is possible that because the baseline microbiota of individuals in agrarian cultures is already Prevotella-rich and Bacteroides-poor, they would not experience Prevotella-associated changes observed with HIV infection in developed world subjects. Together these results suggest that therapies targeting the enteric bacterial microbiome that are successful in western populations may not have the same effect in the developing world.
Limitations to this study
There were several limitations to this study. We were unable to assess AIDS-defining infections and cancers since co-morbidities were unknown. Dietary information was also unavailable, and diet can have a profound effect on the enteric bacterial and viral microbiome (David et al., 2014; Minot et al., 2011). Further, HIV-negative subjects were recruited to the study when they presented to clinic for care, and may not represent healthy community controls. Finally, due to limitations related to specimen collection and preparation, we were unable to assess all components of the microbiome, including RNA viruses and intestinal parasites. Expansion of the RNA virome was observed in SIV-infected macaques that progressed to AIDS (Handley et al., 2012)(Handley et al. submitted), making this an interesting area for future investigations.
Conclusion
Here we report the largest study to date examining the enteric bacterial microbiome in HIV infection and the only one to simultaneously study the enteric DNA virome. These data also shed light on the human enteric bacterial microbiome and virome in sub-Saharan Africa, a region of the world that has been relatively understudied yet has the highest prevalence of HIV infection. We show that HIV infection, in the absence of immunodeficiency, has minimal effect on the enteric DNA virome and bacterial microbiome. Instead, AIDS and the resultant immunodeficiency is associated with expansion of enteric adenoviruses and decreased enteric bacterial diversity and richness. Bacteriophage populations were largely unchanged. These observations in HIV-infected subjects show striking parallels with findings in rhesus macaques (Handley et al., 2012)(Handley et al. submitted) suggesting a common mechanism in disease progression. Given the emergence of known or potential enteric pathogens in association with HIV-induced immunodeficiency, we speculate that immunity to or treatment of enteric pathogens might limit AIDS-associated enteropathy or decrease AIDS-related diseases by limiting GI infection, epithelial cell damage and release of inflammatory pathogen associated molecular patterns and antigens into the body. We hypothesize this might minimize systemic immune activation that is linked to AIDS progression. Together, these studies indicate that severe immunodeficiency is likely the mechanism leading to changes in the fecal microbiome, including bacteria and viruses, and that immune reconstitution, such as through early ART, may restore the healthy enteric microbiome.
EXPERIMENTAL PROCEDURES
See also Supplemental Experimental Procedures.
Study Cohort
Matched, de-identified stool and plasma samples were collected from 122 subjects enrolled from the Mbarara Regional Referral Hospital in Uganda as approved by the Institutional Review Boards at the Mbarara University of Science and Technology, Ugandan National Council of Science and Technology, and Partners Healthcare. All participants gave written informed consent. This cohort was comprised of 42 subjects with untreated HIV disease, 40 location-matched samples from subjects on long-term ART therapy (>5 years) and 40 HIV-uninfected subjects. Subjects presenting to the HIV clinic for HIV testing were recruited into either the HIV-positive untreated arm or the HIV-negative arm depending on HIV test results. Subjects with other comorbidities were not excluded. Data collected included demographics, vital signs, medication history including antibiotic use, clinical symptoms, HIV RNA, CD4 T cell counts at stool collection, CD4 at ART initiation, water source, food security, farming, and other laboratory results (Table 1). Stool samples were collected in RNAlater and frozen at −80°C. Plasma samples were collected in acid citrate dextrose tubes and frozen at −80°C. Samples were shipped on dry ice and complied with the Material Transfer Agreement between Uganda, MGH and Washington University.
Bacterial 16S rRNA Analysis
Stool (100–200mg) was pulverized and total nucleic acid extracted. Polymerase chain reaction was performed and amplicons were pooled and sequenced on the Illumina MiSeq platform (Washington University Center for Genome Sciences; 2×250 standard run). Data was analyzed using Quantitative Insights Into Microbial Ecology (QIIME, version 1.9.1). Nine samples failed DNA isolation. One sample did not achieve high enough OTUs for downstream analysis and was excluded.
VLP Preparation, Sequencing, and Sequence Assignment
VLPs were enriched from pulverized human stool and DNA was amplified using Phi29 polymerase (GenomiPhi V2 kit, GE Healthcare) and fragmented. Six samples failed amplification. NEBNext Ultra DNA kit was used for library construction (New England Biolabs). Equimolar pools were sequenced on Illumina MiSeq platform (Washington University Center for Genome Sciences; 2 × 250bp run). Viral sequences were identified using a custom bioinformatics pipeline VirusSeeker.
sCD14 Measurements in Plasma
sCD14 concentration in plasma was measured by ELISA (R&D Systems Human sCD14 Quantikine ELISA kit #DC140) per manufacturer’s instructions. Plasma samples from two subjects were unavailable for measurement.
Statistical Analysis
Descriptive measures were used to summarize the data. Continuous variables were summarized using median and IQR; categorical variables were summarized using frequency and percent (%). Spearman’s rank correlations were used to examine bivariate associations between study variables. Fisher’s exact and Chi-square tests were used to compare categorical variables between the study groups. Mann-Whitney test and Kruskal Wallis test (indicated by p-value in text) with Dunn’s post hoc analyses (p-values in figures) were used for comparing continuous variables. No correction for multiple comparisons was performed unless otherwise stated. Statistical analyses and graphing were performed in R and Prism version 6.05 for Windows (GraphPad Software, La Jolla California USA, www.graphpad.com). All p-values were two-sided and p < 0.05 was considered significant.
Supplementary Material
Acknowledgments
UARTO was funded by R01 MH54907 and R21 HL124712-01 (Bangsberg). The following were supported by NIH training grants: CLM (5T32AI007172-34), MTB (5T32CA009547), JMN (5T32AI007163-35) and BCK (T32 HL007317-36). DBG is supported by a medical student research supplement to training grant T32DK007191. HWV, CLM, SAH, and JMN were supported by R24 ODO19793, R01 OD011170, R01 AI111918, and R01 DK101354. MJS was supported by K23 MH099916 and the Harvard Center for AIDS Research. JNM was supported by NIH grants P30 AI027763, R01 MH054907, and UM1 CA181255. We would like to thank the Genome Technology Access Center (GTAC) in the Department of Genetics and Jessica Hoisington-Lopez from the Center for Genome Sciences at Washington University School of Medicine for use of their instruments and sequencing. GTAC is partially supported by NCI Cancer Center Support Grant #P30 CA91842 to the Siteman Cancer Center and by ICTS/CTSA Grant# UL1TR000448 from the National Center for Research Resources (NCRR). This publication is solely the responsibility of the authors and does not necessarily represent the official view of NCRR or NIH. We would like to thank study participants and research nurses for sample collection.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
The EMBL-EBI accession number for the 16S rRNA and VLP sequences is PRJEB9524.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures, seven tables, and Supplemental Experimental Procedures and can be found with this article online.
AUTHOR CONTRIBUTIONS
Conceptualization and Methodology, CLM, DSK, HWV, AL, SAH, MS, DRB; Software: CLM, GZ; Formal Analysis – CLM, MSG; Investigation: CLM, DBG, JML, MTB, MF, JMN, BCK, CBW; Resources: YB, PWH, JNM, DRB, MS, DSK; Data curation: MS, CLM; Writing – original draft: CLM; Writing – reviewing and editing: all authors; Visualization: CLM, GZ, ESL, SAH; Supervision: HWV; Funding acquisition: DRB, DSK, HWV.
REFERENCES
- Backhed F, Fraser CM, Ringel Y, Sanders ME, Sartor RB, Sherman PM, Versalovic J, Young V, Finlay BB. Defining a healthy human gut microbiome: current concepts, future directions, and clinical applications. Cell host & microbe. 2012;12:611–622. doi: 10.1016/j.chom.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Brenchley JM. Mucosal immunity in human and simian immunodeficiency lentivirus infections. Mucosal immunology. 2013;6:657–665. doi: 10.1038/mi.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Price DA, Schacker TW, Asher TE, Silvestri G, Rao S, Kazzaz Z, Bornstein E, Lambotte O, Altmann D, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. NatMed. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- Brenchley JM, Schacker TW, Ruff LE, Price DA, Taylor JH, Beilman GJ, Nguyen PL, Khoruts A, Larson M, Haase AT, et al. CD4+ T cell depletion during all stages of HIV disease occurs predominantly in the gastrointestinal tract. JExpMed. 2004;200:749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen JK, Eugen-Olsen J, M SL, Ullum H, Gjedde SB, Pedersen BK, Nielsen JO, Krogsgaard K. Prevalence and prognostic significance of infection with TT virus in patients infected with human immunodeficiency virus. The Journal of infectious diseases. 2000;181:1796–1799. doi: 10.1086/315440. [DOI] [PubMed] [Google Scholar]
- Crost EH, Tailford LE, Le Gall G, Fons M, Henrissat B, Juge N. Utilisation of mucin glycans by the human gut symbiont Ruminococcus gnavus is strain-dependent. PloS one. 2013;8:e76341. doi: 10.1371/journal.pone.0076341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vlaminck I, Khush KK, Strehl C, Kohli B, Luikart H, Neff NF, Okamoto J, Snyder TM, Cornfield DN, Nicolls MR, et al. Temporal response of the human virome to immunosuppression and antiviral therapy. Cell. 2013;155:1178–1187. doi: 10.1016/j.cell.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debiaggi M, Zara F, Sacchi P, Bruno R, Mazzucco M, Poma R, Raffaldi F, Gerace L, Perini M, Pistorio A, et al. Transfusion-transmitted virus infection in HIV-1-seropositive patients. Clin Microbiol Infect. 2000;6:246–250. doi: 10.1046/j.1469-0691.2000.00069.x. [DOI] [PubMed] [Google Scholar]
- Dillon SM, Lee EJ, Kotter CV, Austin GL, Dong Z, Hecht DK, Gianella S, Siewe B, Smith DM, Landay AL, et al. An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal immunology. 2014;7:983–994. doi: 10.1038/mi.2013.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh DM, Volpe GE, Duffalo C, Bhalchandra S, Tai AK, Kane AV, Wanke CA, Ward HD. Intestinal microbiota, microbial translocation, and systemic inflammation in chronic HIV infection. J Infect Dis. 2015;211:19–27. doi: 10.1093/infdis/jiu409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duerkop BA, Hooper LV. Resident viruses and their interactions with the immune system. Nature immunology. 2013;14:654–659. doi: 10.1038/ni.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eren AM, Zozaya M, Taylor CM, Dowd SE, Martin DH, Ferris MJ. Exploring the diversity of Gardnerella vaginalis in the genitourinary tract microbiota of monogamous couples through subtle nucleotide variation. PloS one. 2011;6:e26732. doi: 10.1371/journal.pone.0026732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale TL, Keusch GT. Shigella. 1996 [PubMed] [Google Scholar]
- Handley SA, Thackray LB, Zhao G, Presti R, Miller AD, Droit L, Abbink P, Maxfield LF, Kambal A, Duan E, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151:253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIV/AIDS, J.U.N.P.o. Global report: UNAIDS report on the global AIDS epidemic 2013. (WHO Library Cataloguing-in-Publication Data) 2013 [Google Scholar]
- Hsiao A, Ahmed AM, Subramanian S, Griffin NW, Drewry LL, Petri WA, Jr, Haque R, Ahmed T, Gordon JI. Members of the human gut microbiota involved in recovery from Vibrio cholerae infection. Nature. 2014;515:423–426. doi: 10.1038/nature13738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S, Denman SE, Morrison M, Yu Z, Dore J, Leclerc M, McSweeney CS. Dysbiosis of fecal microbiota in Crohn’s disease patients as revealed by a custom phylogenetic microarray. Inflammatory bowel diseases. 2010;16:2034–2042. doi: 10.1002/ibd.21319. [DOI] [PubMed] [Google Scholar]
- Klase Z, Ortiz A, Deleage C, Mudd JC, Quinones M, Schwartzman E, Klatt NR, Canary L, Estes JD, Brenchley JM. Dysbiotic bacteria translocate in progressive SIV infection. Mucosal immunology. 2015;8:1009–1020. doi: 10.1038/mi.2014.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klatt NR, Villinger F, Bostik P, Gordon SN, Pereira L, Engram JC, Mayne A, Dunham RM, Lawson B, Ratcliffe SJ, et al. Availability of activated CD4+ T cells dictates the level of viremia in naturally SIV-infected sooty mangabeys. The Journal of clinical investigation. 2008;118:2039–2049. doi: 10.1172/JCI33814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Deng X, Linsuwanon P, Bangsberg D, Bwana MB, Hunt P, Martin JN, Deeks SG, Delwart E. AIDS alters the commensal plasma virome. Journal of virology. 2013;87:10912–10915. doi: 10.1128/JVI.01839-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SK, Leung RK, Guo HX, Wei JF, Wang JH, Kwong KT, Lee SS, Zhang C, Tsui SK. Detection and identification of plasma bacterial and viral elements in HIV/AIDS patients in comparison to healthy adults. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. 2012;18:1126–1133. doi: 10.1111/j.1469-0691.2011.03690.x. [DOI] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Applied and environmental microbiology. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Li M, Campbell TB, Flores SC, Linderman D, Gebert MJ, Knight R, Fontenot AP, Palmer BE. Alterations in the gut microbiota associated with HIV-1 infection. Cell host & microbe. 2013;14:329–339. doi: 10.1016/j.chom.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Rhodes ME, Neff CP, Fontenot AP, Campbell TB, Palmer BE. HIV-induced alteration in gut microbiota: driving factors, consequences, and effects of antiretroviral therapy. Gut Microbes. 2014;5:562–570. doi: 10.4161/gmic.32132. [DOI] [PubMed] [Google Scholar]
- Lupp C, Robertson ML, Wickham ME, Sekirov I, Champion OL, Gaynor EC, Finlay BB. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell host & microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- Lyte M. Microbial endocrinology in the microbiome-gut-brain axis: how bacterial production and utilization of neurochemicals influence behavior. PLoS pathogens. 2013;9:e1003726. doi: 10.1371/journal.ppat.1003726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maartens G, Celum C, Lewin SR. HIV infection: epidemiology, pathogenesis, treatment, and prevention. Lancet. 2014;384:258–271. doi: 10.1016/S0140-6736(14)60164-1. [DOI] [PubMed] [Google Scholar]
- Mankal PK, Kotler DP. From wasting to obesity, changes in nutritional concerns in HIV/AIDS. Endocrinology and metabolism clinics of North America. 2014;43:647–663. doi: 10.1016/j.ecl.2014.05.004. [DOI] [PubMed] [Google Scholar]
- Marchetti G, Tincati C, Silvestri G. Microbial translocation in the pathogenesis of HIV infection and AIDS. Clin Microbiol Rev. 2013;26:2–18. doi: 10.1128/CMR.00050-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McHardy IH, Li X, Tong M, Ruegger P, Jacobs J, Borneman J, Anton P, Braun J. HIV Infection is associated with compositional and functional shifts in the rectal mucosal microbiota. Microbiome. 2013;1:26. doi: 10.1186/2049-2618-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS computational biology. 2014;10:e1003531. doi: 10.1371/journal.pcbi.1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minot S, Sinha R, Chen J, Li H, Keilbaugh SA, Wu GD, Lewis JD, Bushman FD. The human gut virome: inter-individual variation and dynamic response to diet. Genome research. 2011;21:1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutlu EA, Keshavarzian A, Losurdo J, Swanson G, Siewe B, Forsyth C, French A, Demarais P, Sun Y, Koenig L, et al. A compositional look at the human gastrointestinal microbiome and immune activation parameters in HIV infected subjects. PLoS pathogens. 2014;10:e1003829. doi: 10.1371/journal.ppat.1003829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, Keller BC, Kambal A, Monaco CL, Zhao G, Fleshner P, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell. 2015;160:447–460. doi: 10.1016/j.cell.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman JM, Handley SA, Virgin HW. Kingdom-agnostic metagenomics and the importance of complete characterization of enteric microbial communities. Gastroenterology. 2014;146:1459–1469. doi: 10.1053/j.gastro.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palermo B, Bosch RJ, Bennett K, Jacobson JM. Body mass index and CD4+ T-lymphocyte recovery in HIV-infected men with viral suppression on antiretroviral therapy. HIV clinical trials. 2011;12:222–227. doi: 10.1310/HCT1204-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Santiago J, Gianella S, Massanella M, Spina CA, Karris MY, Var SR, Patel D, Jordan PS, Young JA, Little SJ, et al. Gut Lactobacillales are associated with higher CD4 and less microbial translocation during HIV infection. AIDS. 2013;27:1921–1931. doi: 10.1097/qad.0b013e3283611816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips AN, Lampe FC, Smith CJ, Geretti AM, Rodger A, Lodwick RK, Cambiano V, Tsintas R, Johnson MA. Ongoing changes in HIV RNA levels during untreated HIV infection: implications for CD4 cell count depletion. AIDS. 2010;24:1561–1567. doi: 10.1097/QAD.0b013e32833a6056. [DOI] [PubMed] [Google Scholar]
- Rajilic-Stojanovic M, Shanahan F, Guarner F, de Vos WM. Phylogenetic analysis of dysbiosis in ulcerative colitis during remission. Inflammatory bowel diseases. 2013;19:481–488. doi: 10.1097/MIB.0b013e31827fec6d. [DOI] [PubMed] [Google Scholar]
- Sandler NG, Wand H, Roque A, Law M, Nason MC, Nixon DE, Pedersen C, Ruxrungtham K, Lewin SR, Emery S, et al. Plasma levels of soluble CD14 independently predict mortality in HIV infection. The Journal of infectious diseases. 2011;203:780–790. doi: 10.1093/infdis/jiq118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satokari R, Fuentes S, Mattila E, Jalanka J, de Vos WM, Arkkila P. Fecal transplantation treatment of antibiotic-induced, noninfectious colitis and long-term microbiota follow-up. Case Rep Med. 2014;2014:913867. doi: 10.1155/2014/913867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selik RM, Mokotoff ED, Branson B, Owen SM, Whitmore S, Hall HI. Revised surveillance case definition for HIV infection—United States, 2014. MMWR. 2014;63:1–10. [PubMed] [Google Scholar]
- Siedner MJ, Kim JH, Nakku RS, Bibangambah P, Hemphill L, Triant VA, Haberer JE, Martin JN, Mocello AR, Boum Y, et al. Persistent immune activation and carotid atherosclerosis in HIV-infected Ugandans receiving antiretroviral therapy. The Journal of infectious diseases. 2015 doi: 10.1093/infdis/jiv450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touinssi M, Gallian P, Biagini P, Attoui H, Vialettes B, Berland Y, Tamalet C, Dhiver C, Ravaux I, De Micco P, et al. TT virus infection: prevalence of elevated viraemia and arguments for the immune control of viral load. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2001;21:135–141. doi: 10.1016/s1386-6532(01)00157-3. [DOI] [PubMed] [Google Scholar]
- Vazquez-Castellanos JF, Serrano-Villar S, Latorre A, Artacho A, Ferrus ML, Madrid N, Vallejo A, Sainz T, Martinez-Botas J, Ferrando-Martinez S, et al. Altered metabolism of gut microbiota contributes to chronic immune activation in HIV-infected individuals. Mucosal immunology. 2014 doi: 10.1038/mi.2014.107. [DOI] [PubMed] [Google Scholar]
- Virgin HW. The virome in mammalian physiology and disease. Cell. 2014;157:142–150. doi: 10.1016/j.cell.2014.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vujkovic-Cvijin I, Dunham RM, Iwai S, Maher MC, Albright RG, Broadhurst MJ, Hernandez RD, Lederman MM, Huang Y, Somsouk M, et al. Dysbiosis of the gut microbiota is associated with HIV disease progression and tryptophan catabolism. Science translational medicine. 2013;5 doi: 10.1126/scitranslmed.3006438. 193ra191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walton AH, Muenzer JT, Rasche D, Boomer JS, Sato B, Brownstein BH, Pachot A, Brooks TL, Deych E, Shannon WD, et al. Reactivation of multiple viruses in patients with sepsis. PloS one. 2014;9:e98819. doi: 10.1371/journal.pone.0098819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Guidelines on co-trimoxazole prophylaxis for HIV-related infections among children, adolescents and adults: Recommendations for a public health approach. Geneva, Switzerland: World Health Organization Press; 2006. [Google Scholar]
- Yang F, Yang J, Zhang X, Chen L, Jiang Y, Yan Y, Tang X, Wang J, Xiong Z, Dong J, et al. Genome dynamics and diversity of Shigella species, the etiologic agents of bacillary dysentery. Nucleic acids research. 2005;33:6445–6458. doi: 10.1093/nar/gki954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze X, Duncan SH, Louis P, Flint HJ. Ruminococcus bromii is a keystone species for the degradation of resistant starch in the human colon. The ISME journal. 2012;6:1535–1543. doi: 10.1038/ismej.2012.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Krishnamurthy S, Cai Z, Popov VL, da Rosa APT, Guzman H, Cao S, Virgin HW, Tesh RB, Wang D. Identification of Novel Viruses Using VirusHunter--an Automated Data Analysis Pipeline. PloS one. 2013;8:e78470. doi: 10.1371/journal.pone.0078470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley SA, Desai C, Zhao G, Droit L, Monaco CL, Schroeder A, Nkolola JP, Norman ME, Miller AD, Wang D, et al. SIV Infection-mediated Changes in the Gastrointestinal Bacterial Microbiome and Virome are Associated With Immunodeficiency and Prevented by Vaccination. doi: 10.1016/j.chom.2016.02.010. (submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Wu G, Cao S, Virgin HW, Félix MA, Wang D. VirusSeeker and MicrobeSeeker: computational pipelines for microbial discovery and classification from metagenomic datasets. Nature Biotechnology, In preparation. 2014 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.