Abstract
ATP-dependent chromatin remodeling proteins are being implicated increasingly in the regulation of complex behaviors, including models of several psychiatric disorders. Here, we demonstrate that Baz1b, an accessory subunit of the ISWI family of chromatin remodeling complexes, is upregulated in the nucleus accumbens (NAc), a key brain reward region, in both chronic cocaine-treated mice and mice that are resilient to chronic social defeat stress. In contrast, no regulation is seen in mice that are susceptible to this chronic stress. Viral-mediated overexpression of Baz1b, along with its associated subunit Smarca5, in mouse NAc is sufficient to potentiate both rewarding responses to cocaine, including cocaine self-administration, and resilience to chronic social defeat stress. However, despite these similar, proreward behavioral effects, genome-wide mapping of BAZ1B in NAc revealed mostly distinct subsets of genes regulated by these chromatin remodeling proteins after chronic exposure to either cocaine or social stress. Together, these findings suggest important roles for BAZ1B and its associated chromatin remodeling complexes in NAc in the regulation of reward behaviors to distinct emotional stimuli and highlight the stimulus-specific nature of the actions of these regulatory proteins.
SIGNIFICANCE STATEMENT We show that BAZ1B, a component of chromatin remodeling complexes, in the nucleus accumbens regulates reward-related behaviors in response to chronic exposure to both rewarding and aversive stimuli by regulating largely distinct subsets of genes.
Keywords: Addiction, Baz1b, chromatin, depression, epigenetics, remodeler
Introduction
High comorbidity between depression and drug addiction suggests a convergence of molecular mechanisms and neural circuitry in the regulation of reward-related and other emotional behaviors (Ford et al., 2009; Russo and Nestler, 2013). Chronic cocaine administration and chronic social defeat stress (CSDS) each alter gene expression in the nucleus accumbens (NAc), an important brain reward region, and chromatin regulation—in particular, histone writers and erasers as well as proteins that control DNA methylation—has been shown to be important for mediating these events (Renthal et al., 2007; Hunter et al., 2009; Jiang et al., 2010; LaPlant et al., 2010; Maze et al., 2010; Covington et al., 2011; Hollis et al., 2011; Sun et al., 2013; Deng et al., 2014; Feng et al., 2015; Hodes et al., 2015). ATP-dependent chromatin remodeling complexes also play a key role in regulating nucleosome positioning to control gene expression and have recently been implicated in complex behavior such as learning and memory (Whitehouse et al., 2007; Clapier and Cairns, 2009; Yen et al., 2012; Narlikar et al., 2013; Vogel-Ciernia et al., 2013). In addition, the persistent induction of BAZ1A, an accessory subunit of the ISWI family of chromatin remodeling proteins, and the resulting upregulation of the ACF complex in the NAc has been shown to be important in mediating chronic-stress-induced depressive-like behavior (Sun et al., 2015).
The related ISWI subunit, BAZ1B (also known as WSTF, or Williams Syndrome Transcription Factor), was first discovered by mapping the deletion region of patients with the developmental disorder Williams–Beuren syndrome (Lu et al., 1998; Peoples et al., 1998). The BAZ family of proteins contain a BAZ type of bromodomain (Jones et al., 2000) and BAZ1B in particular is implicated in several chromatin remodeling/transcriptional complexes (including associating with the ISWI subunit, SMARCA5) with diverse roles in regulating DNA replication, DNA repair, and transcription (Bozhenok et al., 2002; Poot et al., 2004; Cavellán et al., 2006; Percipalle et al., 2006; Xiao et al., 2009; Vintermist et al., 2011; Culver-Cochran and Chadwick, 2013; Lundqvist et al., 2013; Sarshad et al., 2013; Takada, 2015).
Here, we characterize the role of BAZ1B in NAc after chronic exposure to two distinct emotional stimuli, cocaine and social stress. We then map the binding of BAZ1B genome wide in this brain region under both conditions to identify genomic regions implicated in its behavioral actions.
Materials and Methods
Animals.
C57BL/6J male mice (7–8 weeks old; Jackson Laboratory) were housed at constant temperature (23°C) on a 12 h light/dark cycle with ad libitum access to food and water. Experiments were initiated when the animals were 9–10 weeks old. In CSDS experiments, CD1 retired male breeder mice were used as aggressors (see below). Sprague Dawley male rats (275–300 g; Harlan Laboratories) were used for cocaine self-administration experiments. All protocols were approved by Mount Sinai's Institutional Animal Care and Use Committee (IACUC) unless specified otherwise.
Intraperitoneal cocaine administration.
Cocaine hydrochloride (Sigma-Aldrich) was dissolved in sterile 0.9% saline (w/v). Daily saline or cocaine (20 mg/kg, i.p.) injections were administered in the animals' home cages.
NAc RNA isolation and qRT-PCR.
Bilateral 14 Ga punches of NAc were obtained at varying times after the last cocaine or other treatment and frozen on dry ice. Samples were then homogenized in TRIzol and processed according to the manufacturer's instructions. RNA was purified with RNAeasy Micro columns and reverse transcribed using an iScript Kit. cDNA was quantified by qPCR using SYBR green. Each reaction was performed in duplicate and analyzed following the standard ΔΔCt method using Gapdh as a normalization control. Gapdh mRNA levels were not affected by either cocaine or CSDS treatments.
Viral-mediated gene transfer.
Herpes simplex virus (HSV) plasmids were packaged into high-titer viral particles as described previously (Maze et al., 2010). Viral titers were between 3 and 4 × 108 particles/ml. Mice were positioned in small animal stereotaxic instruments under ketamine (100 mg/kg)/xylazine (10 mg/kg) anesthesia and their cranial surfaces were exposed. Thirty-three gauge syringe needles were bilaterally lowered into NAc (anterior/posterior +1.6; medial/lateral +1.5; dorsal/ventral −4.4 mm from bregma, 10° angle) to infuse 0.5 μl of virus. Infusions occurred at a rate of 0.1 μl/min. Animals receiving HSV injections were allowed to recover for at least 24 h after surgery.
Conditioned place preference (CPP).
Place conditioning was performed as described previously (Maze et al., 2010) with slight modifications. Several visual and nonvisual (tactile) cues enabled the animals to distinguish two side chambers of a three-chamber apparatus. All conditioning and test sessions were performed under dim illumination. Baseline preference was monitored and balanced 1 d before surgery. After surgery and recovery, mice were conditioned for 30 min over 2 d to the saline-paired side in the morning and the cocaine-paired side (7.5 mg/kg, i.p.) in the afternoon. On the final day, mice were placed again in the central compartment at noon and allowed to move freely between the two side chambers for 20 min. CPP scores were calculated as time spent in the cocaine-paired chamber minus the time spent in the saline-paired chamber.
Locomotor activity.
After recovery from surgery, mice were habituated in a rectangular-shaped arena equipped with motion sensors for 1 d for 30 min and then monitored for locomotor activity after a saline injection (LaPlant et al., 2010; Scobie et al., 2014). Cocaine (10 mg/kg. i.p.) was then administered daily in the locomotor box for 5 d, with locomotor activity monitored for 30 min thereafter.
Cocaine self-administration.
This study was conducted in accordance with the guidelines set up by the IACUC of the State University of New York at Buffalo. The details of this procedure have been described previously (Scobie et al., 2014; Gancarz et al., 2015). Briefly, naive male Sprague Dawley rats were implanted with jugular catheters, allowed 7 d of recovery, after which they were randomly assigned to self-administer either saline or cocaine (1 mg/kg/infusion). Rats were tested for self-administration for 10 daily 2 h sessions, during which responses at the active snout-poke hole resulted in intravenous injections of cocaine (or saline) according to a fixed ratio 1 schedule. Infusions were accompanied by a 5 s illumination of the stimulus light above the active snout-poke hole. Responses at the inactive snout-poke hole resulted in no programmed consequences. After the last day of self-administration, the rats were returned to their colony rooms and left undisturbed for 7 d. They were subsequently retested for a single-self-administration session in an identical manner as in previous self-administration sessions and killed 24 h after the reexposure test for tissue collection.
For the cocaine dose–response experiment, a separate cohort of rats were first exposed to 5 d of cocaine self-administration, during which animals underwent self-administration training as described above. On day 6, animals were subsequently trained on a within-session dose–response procedure for 5 additional days (Scobie et al., 2014; Gancarz et al., 2015). Briefly, the self-administration period was divided into 5 30 min components, each preceded by a 2 min timeout period. This arrangement allowed the assessment of a range of cocaine doses in a single session. The cocaine dose per injection was regulated by adjusting infusion volumes and pump durations. Rats were exposed to 5 doses of cocaine (0.03, 0.06, 0.10, 0.30, and 1.00 mg/kg/infusion) for 30 min. The order of doses tested was pseudorandomized such that the same doses were never tested in the same order during training. After training, animals were counterbalanced according to self-administration performance and infused with either HSV-GFP or HSV-BAZ1B + SMARCA5 in the NAc. After recovery from stereotaxic surgeries, animals were placed in the operant chambers and retested on the within-session dose–response procedure described above. In addition, animals were tested in a paradigm that is thought to be an indirect measure of drug seeking (Li et al., 2015; Werner et al., 2015). Briefly, animals were placed back in the operant chambers with all of the same cues, but without drug infusions, and responses in the previously active snout-poke hole were measured for 1 h.
Western blotting.
Frozen NAc tissue was homogenized in 30 μl of buffer containing 10 mm Tris, 150 mm NaCl, 1 mm EDTA, 0.1% SDS, 1% Triton X-100, 1% sodium deoxycholate, and protease inhibitors (Roche) using an ultrasonic processor (Cole Parmer). Protein concentrations were determined using a DC protein assay (Bio-Rad) and 50 μg of protein was loaded onto 4–15% gradient Tris-HCl polyacrylamide gels for electrophoresis fractionation (Bio-Rad). Proteins were transferred to nitrocellulose membranes, blocked with Odyssey blocking buffer (Li-Cor) and incubated overnight at 4°C with primary antibodies (BAZ1B: Abcam ab51256, 1/500; SMARCA5: Abcam ab3749, 1/500) in Odyssey blocking buffer. After thorough washing with 1× Tris-buffered saline plus 0.1% Tween 20, membranes were incubated with IRDye secondary antibodies (1/5000 to 1/10000; Li-Cor) dissolved in Odyssey blocking buffer for 1 h at room temperature. For analysis, the blots were imaged with the Odyssey Infrared Imaging system (Li-Cor) and quantified by densitometry using ImageJ. The amount of protein blotted onto each lane was normalized to levels of GAPDH (Cell Signaling Technology 2118, 1/30000) or β-actin (Cell Signaling Technology 3700, 1/10000), the levels of which were not affected by cocaine or CSDS treatments.
CSDS.
CSDS was performed exactly as described previously (Berton et al., 2006; Krishnan et al., 2007). Briefly, an experimental C57BL/6J mouse was placed into the home cage of a CD1 mouse for 10 min, during which time it was physically defeated by the CD1 mouse. After the physical interaction, the CD1 and experimental mouse were maintained in sensory contact for 24 h using a perforated Plexiglas partition dividing the resident home cage in two. The experimental mice were exposed to a new CD1 mouse for 10 consecutive days. Other than not being exposed to physical defeat by a CD1 mouse, control mice were housed under similar conditions as CSDS mice with a perforated Plexiglas separating two control mice. The control mice were also handled daily like the CSDS mice so that they were exposed to a novel control mouse for 10 consecutive days. A social interaction (SI) test was performed 24 h after the last defeat or control session on both CSDS and control mice (Berton et al., 2006). For the SI test, mice were placed in an open field that included an interaction zone and two opposing corner zones. A social target (novel CD1 mouse) was placed in a metal meshplastic box in the interaction zone, which allows sensory but not physical interaction. Ethovision XT (Noldus) tracking software was used to measure the time that the test mouse spent in the interaction zone with and without the target CD1 present (2.5 min). The SI ratio was calculated as follows: (time in interaction zone with target CD1)/(time in interaction zone without CD1) × 100. CSDS mice with an SI ratio of <100 were categorized as susceptible and those with SI ratios of 100 and above were termed resilient. This determination has been shown to be a highly reliable measure of two distinct phenotypic responses to CSDS and has been validated in numerous studies (Krishnan et al., 2007; LaPlant et al., 2010; Sun et al., 2015).
ChIP, library preparation, and sequencing.
ChIP, library preparation, and sequencing were performed as described previously (Feng et al., 2014; Sun et al., 2015). For each ChIP-sequencing (ChIP-seq) replicate, bilateral 14 Ga NAc punches were pooled from five to 10 mice. Tissue was lightly fixed to cross-link DNA with associated proteins and the material was further sheared using a Bioruptor to obtain mostly 100–300 bp fragments and immunoprecipitated using sheep anti-rabbit magnetic beads conjugated to an antibody that specifically recognizes BAZ1B. Immunoprecipitated DNA and total (input) genomic DNA were prepared for ChIP-seq using an Illumina kit according to the manufacturer's instructions. Each experimental condition was analyzed with independent biological triplicates. Amplification and size selection were confirmed with a BioAnalyzer. The resulting libraries were sequenced on an Illumina HiSeq 2500 with 100 bp read length.
ChIP-seq data analysis.
ChIP-seq data were aligned to the mouse genome (mm9) by CASAVA 1.8, and only unique reads were retained for analysis. FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was applied for quality control and then SAMTools (http://samtools.sourceforge.net) was used to remove potential PCR duplicates (Li et al., 2009). PhantomPeak (https://code.google.com/p/phantompeakqualtools/) was applied to estimate the quality and enrichment of the ChIP-seq dataset (Landt et al., 2012). Additional ENCODE quality metrics, such as the normalized strand coefficient (NSC) and the relative strand correlation (RSC), were calculated. For all samples in our research, NSC was ≥1.05 and RSC was ≥0.8. Basic filtering and quality control confirmed that these samples were of strong quality and exceeded ENCODE standards (Landt et al., 2012). Differential analysis between treatment groups was performed with an in-house program called diffReps. For more details, please refer to http://code.google.com/p/diffreps/ (Shen et al., 2013). The Corrgram package in R software was used to calculate and visualize the correlation between different chromatin remodelers and histone marks. TDF files (all duplicative/redundant reads >2 removed) were applied in IGV for genome browser views of ChIP-seq tracks.
Statistical analysis.
Student's t tests were used whenever two groups were compared. One-way and two-way ANOVAs were performed wherever necessary to determine significance for all other data. Significant main effects (p < 0.05) were further analyzed using post hoc tests.
Results
BAZ1B and SMARCA5 in cocaine action
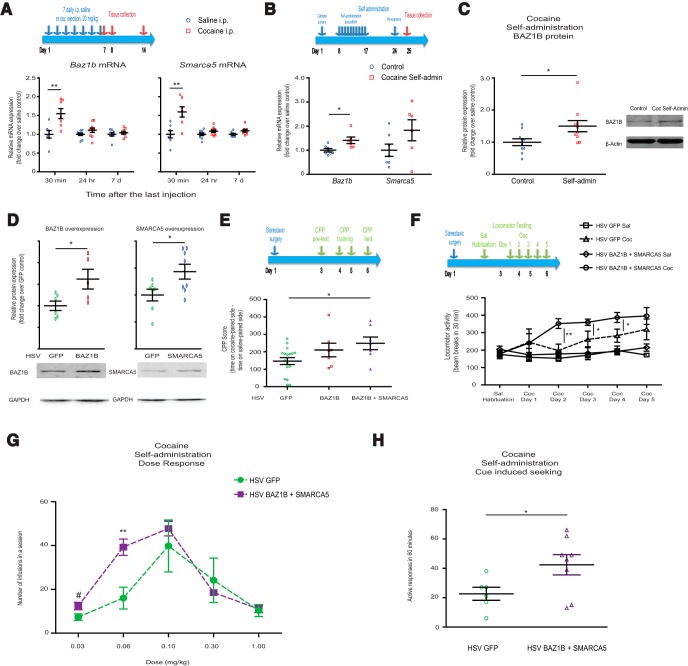
We first assessed the effect of chronic administration of cocaine on expression levels of Baz1b (an accessory ISWI subunit) and Smarca5 (a core ISWI ATPase subunit) in the NAc. We found that mRNA levels of both Baz1b (t(12) = 3.309, p = 0.0062) and Smarca5 (t(12) = 3.537, p = 0.0041) were transiently but significantly increased 30 min after 7 daily cocaine injections (20 mg/kg, i.p.) compared with saline controls (Fig. 1A). This increase returned to control levels 24 h and 7 d after the last injection (Fig. 1A). A similar induction of NAc BAZ1B mRNA (t(11) = 2.798, p = 0.017) and protein levels (t(18) = 2.456, p = 0.024) was observed after chronic cocaine self-administration (Fig. 1B,C), whereas a nonstatistically significant trend for an increase in Smarca5 mRNA levels was observed after cocaine self-administration (t(11) = 1.694, p = 0.118).
Figure 1.
BAZ1B in NAc in cocaine action. A, Baz1b and Smarca5 mRNA levels in NAc 30 min, 24 h, and 7 d after 7 daily intraperitoneal injections of 20 mg/kg cocaine or saline (0.9% w/v). B, Baz1b and Smarca5 mRNA levels in NAc 24 h after cocaine self-administration. C, BAZ1B protein levels in NAc 24 h after cocaine self-administration. D, Validation of HSV viral overexpression of BAZ1B and SMARCA5 in mouse NAc. E, BAZ1B + SMARCA5 overexpression in NAc enhances cocaine reward as measured by CPP. F, BAZ1B + SMARCA5 overexpression in NAc enhances cocaine-induced locomotor activity. G, BAZ1B + SMARCA5 overexpression in NAc enhances cocaine self-administration at lower doses. H, BAZ1B + SMARCA5 overexpression in NAc enhances cue-induced drug seeking. For A, B, E, F, experimental paradigms are shown above each panel. #0.05 < p < 0.10, *p < 0.05, **p < 0.01 in respective comparisons.
To examine directly whether increased Baz1b levels in NAc are causally important in regulating cocaine reward behavior, we used HSV-mediated gene transfer to rapidly overexpress BAZ1B (t(9) = 2.584, p = 0.030) and SMARCA5 (t(13) = 2.413, p = 0.031) in this brain region (Fig. 1D). In an unbiased cocaine CPP paradigm, which provides an indirect measure of drug reward, animals with BAZ1B overexpression alone in NAc did not differ in cocaine preference compared with HSV-GFP controls (Fig. 1E; one-way ANOVA: F(2,28) = 3.578, p = 0.0413; GFP vs BAZ1B: Dunnett's post hoc, q = 1.580, p > 0.05). In contrast, animals with cooverexpression in NAc of BAZ1B plus SMARCA5 showed a significant increase in cocaine preference (Fig. 1E; Dunnett's post hoc, q = 2.526, p < 0.05). Overexpression of SMARCA5 alone had no effect (data not shown). These observations are consistent with the knowledge, derived from cultured cells, that BAZ1B and SMARCA5 together form a functional complex that regulates nucleosome positioning (see Discussion). Similarly, BAZ1B + SMARCA5 overexpression in NAc increased locomotor responses to repeated cocaine exposures compared with HSV-GFP controls, whereas it did not affect locomotor behavior in response to saline injections (Fig. 1F, two-way ANOVA: group F(3,84) = 19.13, p < 0.0001; day F(5,84) = 3.623, p = 0.0051; interaction F(15,84) = 1.465, NS).
Next, we investigated whether BAZ1B and SMARCA5 overexpression in NAc affected cocaine self-administration. Using a within-session dose–response self-administration procedure, we found that overexpression of BAZ1B and SMARCA5 in NAc increased cocaine self-administration (Fig. 1G, 2-way ANOVA: dose F(4,48) = 17.80, p < 0.0001; interaction F(4,48) = 2.658, p = 0.044), particularly at the lower doses (0.03 mg/kg: t(12) = 2.127, p = 0.0549; 0.06 mg/kg: t(12) = 3.826, p = 0.0024). In addition, after withdrawal, animals with BAZ1B + SMARCA5 overexpression in NAc exhibited increased drug-seeking behavior compared with GFP control animals when placed back in the operant chambers with all of the same cues but no drug infusions (Fig. 1H, t(12) = 2.216, p = 0.0467). Together, these lines of evidence suggest that the BAZ1B-SMARCA5 complex increases sensitivity to cocaine reward.
BAZ1B and SMARCA5 in CSDS
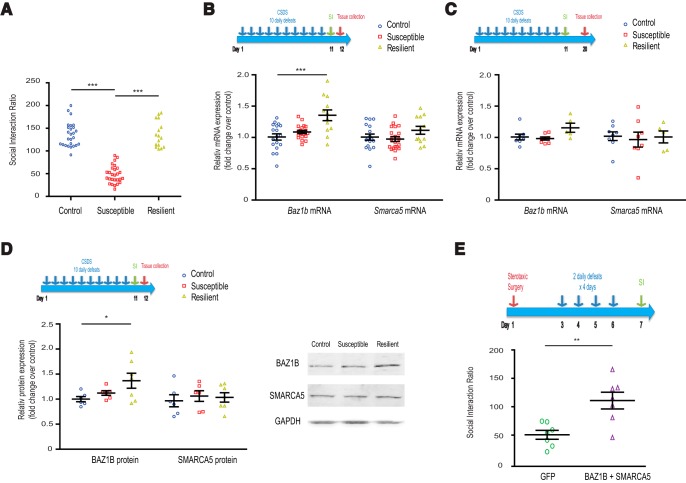
We next examined the regulation of Baz1b in NAc by CSDS, an ethologically validated mouse model of depression (Berton et al., 2006; Krishnan et al., 2007; Hollis et al., 2011). Over 10 consecutive days, C57BL/6J male mice were subjected to daily 10 min aggressive encounters with CD1 mice, followed by sensory but not physical contact for the remainder of the day. After CSDS, ∼65% of test mice exhibited depression-related behavioral abnormalities including social avoidance and were termed susceptible, whereas the remaining ∼35% did not exhibit these behavioral abnormalities and were termed resilient (Krishnan et al., 2007; Fig. 2A). Baz1b mRNA levels in NAc of resilient animals were elevated 48 h after the last CSDS session compared with nonstressed controls (Fig. 2B; one-way ANOVA: F(2,47) = 10.80, p = 0.0001; control vs resilient: Dunnett's post hoc q = 4.540, p < 0.001), with no differences observed 10 d after CSDS (Fig. 2C; One-way ANOVA: F(2,20) = 3.441, p = 0.0543; control vs resilient: Dunnett's post hoc q = 2.153, p > 0.05). BAZ1B protein levels were similarly increased in resilient animals 48 h after CSDS (Fig. 2D; one-way ANOVA: F(2,18) = 3.394, p = 0.0591; control vs resilient: Dunnett's post hoc q = 2.536, p < 0.05). No differences were observed for NAc BAZ1B mRNA and protein levels in susceptible animals nor for NAc SMARCA5 mRNA and protein levels in susceptible or resilient animals compared with controls (Fig. 2B–D).
Figure 2.
BAZ1B in NAc in CSDS. A, Representative SI ratios for control, susceptible, and resilient animals 24 h after 10 d of CSDS. B, Baz1b and Smarca5 mRNA levels in NAc 48 h after CSDS. C, Baz1b and Smarca5 mRNA levels in NAc 10 d after CSDS. D, BAZ1B and SMARCA5 protein levels in NAc 48 h after CSDS. E, BAZ1B + SMARCA5 overexpression in NAc increases SI ratios after a 4 d accelerated defeat paradigm, indicating a pro-resilience effect. For B–E, experimental paradigms are shown above each panel. *p < 0.05, **p < 0.01, ***p < 0.001 in respective comparisons.
To test directly the hypothesis that elevated BAZ1B levels play a causal role in regulating resilience to social stress, we overexpressed BAZ1B plus SMARCA5 selectively in NAc of adult animals and then examined stress susceptibility versus resilience using an accelerated repeated social defeat protocol that induces behavioral deficits in HSV-GFP control mice within the time frame of maximal HSV-mediated transgene expression (Krishnan et al., 2007; Sun et al., 2015). Animals overexpressing BAZ1B + SMARCA5 in NAc showed increased social interaction compared with HSV-GFP controls (Fig. 2E; t(12) = 3.614, p = 0.0036.), indicating that induction of NAc BAZ1B increases resilience to social defeat stress.
Genome-wide mapping of BAZ1B binding in NAc under cocaine and CSDS conditions
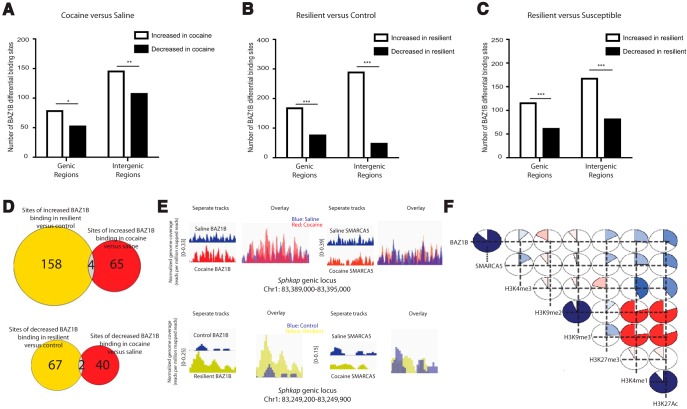
We used ChIP-seq to map BAZ1B binding genome wide in NAc under control, cocaine, and CSDS conditions. Using diffreps (Shen et al., 2013), we determined sites of differential BAZ1B binding between saline- and cocaine-treated animals, as well as between control, susceptible, and resilient animals. There were 223 increased BAZ1B binding events (at 78 genic and 145 intergenic sites) after chronic cocaine compared with saline and 159 decreased BAZ1B-binding events (at 52 genic and 107 intergenic sites) (Fig. 3A; genic: χ2(df = 1) = 5.200, p = 0.0113; intergenic: χ2(df = 1) = 5.730, p = 0.0084). This is consistent with our observation of BAZ1B induction in NAc after cocaine exposure (Fig. 1A–C). Likewise, we observed increased genome-wide binding of BAZ1B in NAc of resilient animals compared with control mice (167 increased and 75 decreased) and susceptible mice (115 increased and 61 decreased) (Fig. 3B,C). The increased genome-wide BAZ1B binding in resilient animals was observed in both genic and intergenic regions (resilient vs control: genic: χ2(df = 1) = 34.975, p < 0.0001; intergenic: χ2(df = 1) = 173.376, p < 0.0001; resilient vs susceptible: genic: χ2(df = 1) = 16.568, p < 0.0001; intergenic: χ2(df = 1) = 29.823, p < 0.0001).
Figure 3.
Genome-wide mapping of BAZ1B in NAc after cocaine or CSDS. A, Differential BAZ1B binding sites in NAc between chronic cocaine and saline treatment. B, Differential BAZ1B binding sites in NAc between resilient and control animals. C, Differential BAZ1B binding sites in NAc between resilient and susceptible animals. D, Overlap between upregulated (top) and downregulated (bottom) BAZ1B genic binding sites in NAc in resilient versus control and cocaine versus saline conditions. E, IGV (integrative genomics viewer) visualization of BAZ1B and SMARCA5 ChIP-seq tracks at Sphkap genomic loci in either separate (left) or overlaid (right) tracks in NAc of cocaine and saline groups (top), as well as control and resilient groups (bottom). F, Correlation between the binding of BAZ1B, SMARCA5, and several histone marks at promoter regions genome-wide in NAc. Corrgram package analysis was used to calculate correlation/r-values. To help with visualization, horizontal and vertical dotted lines were extended from each factor/chromatin mark. The piechart where the two lines intersect represents the correlation value between the two factors/marks. The size of the shaded region within the pie reflects correlation/r-values (bigger shaded region represents higher correlation or r-value), whereas the color of the shaded region represents the direction of correlation (blue reflects positive correlation and red reflects negative correlation). p < 2.2 × 10−16 for all correlations. For A–C, χ2 test, *p < 0.05, **p < 0.01, ***p < 0.001 in respective comparisons.
We next investigated whether the upregulated BAZ1B binding in NAc of cocaine-exposed mice and of resilient mice after CSDS occur at similar or distinct subsets of genes. Of the 162 and 69 genes that showed increased NAc BAZ1B binding in cocaine-treated and resilient animals, respectively, there was only overlap of 4 genes (221040812Rik, Anks1b, Col19a1, and Sphkap), suggesting that BAZ1B promotes cocaine reward and stress resilience mainly by regulating different subsets of genes (Fig. 3D, top). Similarly, decreased BAZ1B binding in cocaine-treated and resilient animals occurred in a distinct subset of genes with minimal overlap (Fig. 3D, bottom). We examined one of the four genes that showed increased BAZ1B binding in NAc under both cocaine and resilient conditions, Sphkap (sphingosine kinase type 1-interacting protein), and confirmed increased BAZ1B binding in both conditions relative to their respective controls (Fig. 3E). Interestingly, at these sites that showed increased BAZ1B binding, there was no obvious increase in SMARCA5 binding, but rather subtle shifting of SMARCA5 location within these regions (Fig. 3E).
To examine BAZ1B's potential transcriptional role, we correlated BAZ1B binding in promoter regions genome-wide with published ChIP-seq data for SMARCA5 and for several well studied histone marks (Feng et al., 2014; Feng et al., 2015; Sun et al., 2015; Fig. 3F). There was a strong positive correlation between BAZ1B and SMARCA5 binding (r = 0.873), suggesting that BAZ1B is at least partly working with SMARCA5 to exert its regulation. Agreeing with the previously reported transcriptionally activating role of BAZ1B in other systems (Vintermist et al., 2011; Culver-Cochran and Chadwick, 2013; Lundqvist et al., 2013; Sarshad et al., 2013; Takada, 2015), we found positive correlations between BAZ1B binding and three activating histone marks, H3K4me3 (r = 0.117), H3K4me1 (r = 0.279), and H3K27Ac (r = 0.332), and a negative correlation between BAZ1B and two repressive marks, H3K9me2 (r = −0.165) and H3K9me3 (r = −0.101). Surprisingly, BAZ1B had a significant positive correlation with the repressive mark H3K27me3 (r = 0.168).
Discussion
Results from the present study demonstrate that BAZ1B is upregulated in NAc by two types of chronic emotional stimuli, cocaine and social defeat stress. Our functional experiments indicate that induction of BAZ1B in NAc enhanced behavioral responses to cocaine both for noncontingent (CPP and locomotor activity) and contingent (self-administration and drug-seeking/relapse) paradigms, whereas such induction enhanced resilience to chronic stress. Together, these data establish that BAZ1B in NAc regulates behavioral responses to emotional stimuli: elevated levels of BAZ1B in this brain region heighten responses to rewarding stimuli and promote adaptive responses to aversive stimuli. Interestingly, the upregulation of BAZ1B by either stimulus does not persist for the duration of behavioral effects elicited by these two stimuli: Baz1b levels return to control values within hours or days of cocaine or stress exposure despite the fact that the behavioral sequelae of these treatments persist for weeks or months. In addition, Baz1b upregulation caused by chronic cocaine is more transient (minutes to hours after intraperitoneal cocaine) compared with that caused by CSDS (days), an observation that was also reported for another chromatin regulator, Dmnt3a (LaPlant et al., 2010).
The transient nature of these regulation events suggests that BAZ1B may control other genes to exert longer-term behavioral effects. Our ChIP-seq data demonstrate altered binding of BAZ1B at numerous target genes after either stimulus. However, these genome-wide data show that these distinct emotional stimuli regulate virtually nonoverlapping subsets of genes to exert their functional effects. This is a surprising finding because, in both conditions (cocaine and CSDS), BAZ1B induction in NAc can be seen as enhancing reward. Our data thus underscore the important principle that an individual chromatin regulatory protein acting in the same tissue can exert very different molecular actions based on the larger context of the stimulus. The very small number of genes (e.g., Sphkap) that exhibited similar BAZ1B regulation after both chronic cocaine and CSDS may be molecular candidates in NAc that control global emotional states in response to rewarding or aversive stimuli, whereas the largely distinct subsets may represent those necessary in response to specific stimuli. Sphkap encodes an interesting protein that is thought to link cAMP and sphingosine signaling pathways (Kovanich et al., 2010). The ChIP-seq data reported here therefore provide new avenues for understanding the biological mechanisms in NAc that control cocaine reward and stress resilience. An important question to address in the future is how the same complex (BAZ1B-SMARCA5) can regulate distinct subsets of genes in response to different stimuli (e.g., whether distinct cofactors are recruited, etc.).
We recently implicated a related gene, Baz1a, in NAc in mediating susceptibility to CSDS (Sun et al., 2015), whereas here, we demonstrate an opposite role for Baz1b in promoting stress resilience. This suggests that closely related ISWI subunits can form different chromatin remodeling complexes with very distinct functional consequences. Although controversy remains concerning the exact complexes that Baz1b forms in different systems (Yoshimura et al., 2014), our overexpression studies as well as the high level of coincidence of BAZ1B binding with SMARCA5 binding genome-wide in NAc suggest that BAZ1B exerts its effect at least partly through association with SMARCA5. Likewise, BAZ1A is also thought to work in large part by complexing with SMARCA5 (see Sun et al., 2015). An interesting parallel between Baz1a and Baz1b is that whereas only the accessory subunit (i.e., Baz1a or Baz1b) is regulated by CSDS without coregulation of the active ATPase (i.e., Smarca5), viral overexpression typically requires both components to exert a behavioral effect. However, whereas increased BAZ1A binding at genomic regions is typically associated with increased SMARCA5 binding (Sun et al., 2015), this does not seem to be true for the BAZ1B-SMARCA5 complex, at least at the Sphkap locus, for example (Fig. 3E). Rather, increased BAZ1B binding after chronic cocaine or CSDS seems to redistribute subtly the binding of SMARCA5 around the affected locus rather than increasing its binding levels. Whether these differences contribute to different downstream effects remain interesting biochemical and mechanistic questions to address about these complexes in the future.
Consistent with previously reported transcriptional activation roles of BAZ1B in cell culture systems, we show here that BAZ1B binding at gene promoters in NAc is positively correlated with several histone marks associated with transcriptional activation (H3K4me3, H3K4me1, and H3K27Ac) and negatively correlated with histone marks associated with transcriptional repression (H3K9me3 and H3K9me2). However, it is curious that BAZ1B binding is also positively correlated with the repressive H3K27me3 mark. This novel finding suggests that BAZ1B may contribute to the regulation of genes with bivalent chromatin domains that have been implicated in poising certain genes for especially dynamic regulation (Watson et al., 2012). Whether BAZ1B contributes to the regulation of such bivalent chromatin domains in NAc or other brain regions is a worthy subject for future investigation.
Together, results of the present study establish that induction of Baz1b in NAc both promotes cocaine reward behavior and promotes resilience to chronic social stress, indicating that it is important for the regulation of reward and other emotional behaviors. These findings will guide future investigations aimed at better understanding the biological basis of cocaine- and stress-related disorders, a prerequisite for devising more effective treatments.
Footnotes
This work was supported by the National Institutes of Health–National Institute of Mental Health (Grant P50 MH096890), the National Institutes of Health–National Institute on Drug Abuse (Grants P01 DA008227 and R01 DA037257), and the Hope for Depression Research Foundation. We thank Patrick Varga-Weisz for contribution of the BAZ1B and SMARCA5 plasmids.
The authors declare no competing financial interests.
References
- Berton O, McClung CA, Dileone RJ, Krishnan V, Renthal W, Russo SJ, Graham D, Tsankova NM, Bolanos CA, Rios M, Monteggia LM, Self DW, Nestler EJ. Essential role of BDNF in the mesolimbic dopamine pathway in social defeat stress. Science. 2006;311:864–868. doi: 10.1126/science.1120972. [DOI] [PubMed] [Google Scholar]
- Bozhenok L, Wade PA, Varga-Weisz P. WSTF-ISWI chromatin remodeling complex targets heterochromatic replication foci. EMBO J. 2002;21:2231–2241. doi: 10.1093/emboj/21.9.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavellán E, Asp P, Percipalle P, Farrants AK. The WSTF-SNF2h chromatin remodeling complex interacts with several nuclear proteins in transcription. J Biol Chem. 2006;281:16264–16271. doi: 10.1074/jbc.M600233200. [DOI] [PubMed] [Google Scholar]
- Clapier CR, Cairns BR. The biology of chromatin remodeling complexes. Annu Rev Biochem. 2009;78:273–304. doi: 10.1146/annurev.biochem.77.062706.153223. [DOI] [PubMed] [Google Scholar]
- Covington HE, 3rd, Maze I, Sun H, Bomze HM, DeMaio KD, Wu EY, Dietz DM, Lobo MK, Ghose S, Mouzon E, Neve RL, Tamminga CA, Nestler EJ. A role for repressive histone methylation in cocaine-induced vulnerability to stress. Neuron. 2011;71:656–670. doi: 10.1016/j.neuron.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver-Cochran AE, Chadwick BP. Loss of WSTF results in spontaneous fluctuations of heterochromatin formation and resolution, combined with substantial changes to gene expression. BMC Genomics. 2013;14:740. doi: 10.1186/1471-2164-14-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng JV, Wan Y, Wang X, Cohen S, Wetsel WC, Greenberg ME, Kenny PJ, Calakos N, West AE. MeCP2 phosphorylation limits psychostimulant-induced behavioral and neuronal plasticity. J Neurosci. 2014;34:4519–4527. doi: 10.1523/JNEUROSCI.2821-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Wilkinson M, Liu X, Purushothaman I, Ferguson D, Vialou V, Maze I, Shao N, Kennedy P, Koo J, Dias C, Laitman B, Stockman V, LaPlant Q, Cahill ME, Nestler EJ, Shen L. Chronic cocaine-regulated epigenomic changes in mouse nucleus accumbens. Genome Biol. 2014;15:R65. doi: 10.1186/gb-2014-15-4-r65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Shao N, Szulwach KE, Vialou V, Huynh J, Zhong C, Le T, Ferguson D, Cahill ME, Li Y, Koo JW, Ribeiro E, Labonte B, Laitman BM, Estey D, Stockman V, Kennedy P, Courousse T, Mensah I, Turecki G, et al. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat Neurosci. 2015;18:536–544. doi: 10.1038/nn.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford JD, Gelernter J, DeVoe JS, Zhang W, Weiss RD, Brady K, Farrer L, Kranzler HR. Association of psychiatric and substance use disorder comorbidity with cocaine dependence severity and treatment utilization in cocaine-dependent individuals. Drug Alcohol Depend. 2009;99:193–203. doi: 10.1016/j.drugalcdep.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gancarz AM, Wang ZJ, Schroeder GL, Damez-Werno D, Braunscheidel KM, Mueller LE, Humby MS, Caccamise A, Martin JA, Dietz KC, Neve RL, Dietz DM. Activin receptor signaling regulates cocaine-primed behavioral and morphological plasticity. Nat Neurosci. 2015;18:959–961. doi: 10.1038/nn.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodes GE, Pfau ML, Purushothaman I, Ahn HF, Golden SA, Christoffel DJ, Magida J, Brancato A, Takahashi A, Flanigan ME, Menard C, Aleyasin H, Koo JW, Lorsch ZS, Feng J, Heshmati M, Wang M, Turecki G, Neve R, Zhang B, et al. Sex differences in nucleus accumbens transcriptome profiles associated with susceptibility versus resilience to subchronic variable stress. J Neurosci. 2015;35:16362–16376. doi: 10.1523/JNEUROSCI.1392-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis F, Duclot F, Gunjan A, Kabbaj M. Individual differences in the effect of social defeat on anhedonia and histone acetylation in the rat hippocampus. Horm Behav. 2011;59:331–337. doi: 10.1016/j.yhbeh.2010.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci U S A. 2009;106:20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Jakovcevski M, Bharadwaj R, Connor C, Schroeder FA, Lin CL, Straubhaar J, Martin G, Akbarian S. Setdb1 histone methyltransferase regulates mood-related behaviors and expression of the NMDA receptor subunit NR2B. J Neurosci. 2010;30:7152–7167. doi: 10.1523/JNEUROSCI.1314-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones MH, Hamana N, Nezu Ji, Shimane M. A novel family of bromodomain genes. Genomics. 2000;63:40–45. doi: 10.1006/geno.1999.6071. [DOI] [PubMed] [Google Scholar]
- Kovanich D, van der Heyden MA, Aye TT, van Veen TA, Heck AJ, Scholten A. Sphingosine kinase interacting protein is an A-kinase anchoring protein specific for type I cAMP-dependent protein kinase. Chembiochem. 2010;11:963–971. doi: 10.1002/cbic.201000058. [DOI] [PubMed] [Google Scholar]
- Krishnan V, Han MH, Graham DL, Berton O, Renthal W, Russo SJ, Laplant Q, Graham A, Lutter M, Lagace DC, Ghose S, Reister R, Tannous P, Green TA, Neve RL, Chakravarty S, Kumar A, Eisch AJ, Self DW, Lee FS, et al. Molecular adaptations underlying susceptibility and resistance to social defeat in brain reward regions. Cell. 2007;131:391–404. doi: 10.1016/j.cell.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Landt SG, Marinov GK, Kundaje A, Kheradpour P, Pauli F, Batzoglou S, Bernstein BE, Bickel P, Brown JB, Cayting P, Chen Y, DeSalvo G, Epstein C, Fisher-Aylor KI, Euskirchen G, Gerstein M, Gertz J, Hartemink AJ, Hoffman MM, Iyer VR, et al. ChIP-seq guidelines and practices of the ENCODE and modENCODE consortia. Genome Res. 2012;22:1813–1831. doi: 10.1101/gr.136184.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPlant Q, Vialou V, Covington HE, 3rd, Dumitriu D, Feng J, Warren BL, Maze I, Dietz DM, Watts EL, Iniguez SD, Koo JW, Mouzon E, Renthal W, Hollis F, Wang H, Noonan MA, Ren Y, Eisch AJ, Bolanos CA, Kabbaj M, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Rubio FJ, Zeric T, Bossert JM, Kambhampati S, Cates HM, Kennedy PJ, Liu QR, Cimbro R, Hope BT, Nestler EJ, Shaham Y. Incubation of methamphetamine craving is associated with selective increases in expression of Bdnf and trkb, glutamate receptors, and epigenetic enzymes in cue-activated fos-expressing dorsal striatal neurons. J Neurosci. 2015;35:8232–8244. doi: 10.1523/JNEUROSCI.1022-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu X, Meng X, Morris CA, Keating MT. A novel human gene, WSTF, is deleted in Williams syndrome. Genomics. 1998;54:241–249. doi: 10.1006/geno.1998.5578. [DOI] [PubMed] [Google Scholar]
- Lundqvist J, Hansen SK, Lykkesfeldt AE. Vitamin D analog EB1089 inhibits aromatase expression by dissociation of comodulator WSTF from the CYP19A1 promoter-a new regulatory pathway for aromatase. Biochim Biophys Acta. 2013;1833:40–47. doi: 10.1016/j.bbamcr.2012.10.012. [DOI] [PubMed] [Google Scholar]
- Maze I, Covington HE, 3rd, Dietz DM, LaPlant Q, Renthal W, Russo SJ, Mechanic M, Mouzon E, Neve RL, Haggarty SJ, Ren Y, Sampath SC, Hurd YL, Greengard P, Tarakhovsky A, Schaefer A, Nestler EJ. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narlikar GJ, Sundaramoorthy R, Owen-Hughes T. Mechanisms and functions of ATP-dependent chromatin-remodeling enzymes. Cell. 2013;154:490–503. doi: 10.1016/j.cell.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peoples RJ, Cisco MJ, Kaplan P, Francke U. Identification of the WBSCR9 gene, encoding a novel transcriptional regulator, in the Williams-Beuren syndrome deletion at 7q11.23. Cytogenet Cell Genet. 1998;82:238–246. doi: 10.1159/000015110. [DOI] [PubMed] [Google Scholar]
- Percipalle P, Fomproix N, Cavellán E, Voit R, Reimer G, Krüger T, Thyberg J, Scheer U, Grummt I, Farrants AK. The chromatin remodelling complex WSTF-SNF2h interacts with nuclear myosin 1 and has a role in RNA polymerase I transcription. EMBO Rep. 2006;7:525–530. doi: 10.1038/sj.embor.7400657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot RA, Bozhenok L, van den Berg DL, Steffensen S, Ferreira F, Grimaldi M, Gilbert N, Ferreira J, Varga-Weisz PD. The Williams syndrome transcription factor interacts with PCNA to target chromatin remodelling by ISWI to replication foci. Nat Cell Biol. 2004;6:1236–1244. doi: 10.1038/ncb1196. [DOI] [PubMed] [Google Scholar]
- Renthal W, Maze I, Krishnan V, Covington HE, 3rd, Xiao G, Kumar A, Russo SJ, Graham A, Tsankova N, Kippin TE, Kerstetter KA, Neve RL, Haggarty SJ, McKinsey TA, Bassel-Duby R, Olson EN, Nestler EJ. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Russo SJ, Nestler EJ. The brain reward circuitry in mood disorders. Nat Rev Neurosci. 2013;14:609–625. doi: 10.1038/nrn3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarshad A, Sadeghifar F, Louvet E, Mori R, Böhm S, Al-Muzzaini B, Vintermist A, Fomproix N, Östlund AK, Percipalle P. Nuclear myosin 1c facilitates the chromatin modifications required to activate rRNA gene transcription and cell cycle progression. PLoS Genet. 2013;9:e1003397. doi: 10.1371/journal.pgen.1003397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scobie KN, Damez-Werno D, Sun H, Shao N, Gancarz A, Panganiban CH, Dias C, Koo J, Caiafa P, Kaufman L, Neve RL, Dietz DM, Shen L, Nestler EJ. Essential role of poly(ADP-ribosyl)ation in cocaine action. Proc Natl Acad Sci U S A. 2014;111:2005–2010. doi: 10.1073/pnas.1319703111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Shao NY, Liu X, Maze I, Feng J, Nestler EJ. diffReps: detecting differential chromatin modification sites from ChIP-seq data with biological replicates. PLoS One. 2013;8:e65598. doi: 10.1371/journal.pone.0065598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Kennedy PJ, Nestler EJ. Epigenetics of the depressed brain: role of histone acetylation and methylation. Neuropsychopharmacology. 2013;38:124–137. doi: 10.1038/npp.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Damez-Werno DM, Scobie KN, Shao NY, Dias C, Rabkin J, Koo JW, Korb E, Bagot RC, Ahn FH, Cahill ME, Labonte B, Mouzon E, Heller EA, Cates H, Golden SA, Gleason K, Russo SJ, Andrews S, Neve R, et al. ACF chromatin-remodeling complex mediates stress-induced depressive-like behavior. Nat Med. 2015;21:1146–1153. doi: 10.1038/nm.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takada I. DGCR14 induces Il17a gene expression through the RORgamma/BAZ1B/RSKS2 complex. Mol Cell Biol. 2015;35:344–355. doi: 10.1128/MCB.00926-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vintermist A, Böhm S, Sadeghifar F, Louvet E, Mansén A, Percipalle P, Ostlund Farrants AK. The chromatin remodelling complex B-WICH changes the chromatin structure and recruits histone acetyl-transferases to active rRNA genes. PLoS One. 2011;6:e19184. doi: 10.1371/journal.pone.0019184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel-Ciernia A, Matheos DP, Barrett RM, Kramar EA, Azzawi S, Chen Y, Magnan CN, Zeller M, Sylvain A, Haettig J, Jia Y, Tran A, Dang R, Post RJ, Chabrier M, Babayan AH, Wu JI, Crabtree GR, Baldi P, Baram TZ, et al. The neuron-specific chromatin regulatory subunit BAF53b is necessary for synaptic plasticity and memory. Nat Neurosci. 2013;16:552–561. doi: 10.1038/nn.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson CT, Disanto G, Sandve GK, Breden F, Giovannoni G, Ramagopalan SV. Age-associated hyper-methylated regions in the human brain overlap with bivalent chromatin domains. PLoS One. 2012;7:e43840. doi: 10.1371/journal.pone.0043840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner CT, Milovanovic M, Christian DT, Loweth JA, Wolf ME. Response of the ubiquitin-proteasome system to memory retrieval after extended-access cocaine or saline self-administration. Neuropsychopharmacology. 2015;40:3006–3014. doi: 10.1038/npp.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehouse I, Rando OJ, Delrow J, Tsukiyama T. Chromatin remodelling at promoters suppresses antisense transcription. Nature. 2007;450:1031–1035. doi: 10.1038/nature06391. [DOI] [PubMed] [Google Scholar]
- Xiao A, Li H, Shechter D, Ahn SH, Fabrizio LA, Erdjument-Bromage H, Ishibe-Murakami S, Wang B, Tempst P, Hofmann K, Patel DJ, Elledge SJ, Allis CD. WSTF regulates the H2A.X DNA damage response via a novel tyrosine kinase activity. Nature. 2009;457:57–62. doi: 10.1038/nature07668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen K, Vinayachandran V, Batta K, Koerber RT, Pugh BF. Genome-wide nucleosome specificity and directionality of chromatin remodelers. Cell. 2012;149:1461–1473. doi: 10.1016/j.cell.2012.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura K, Kitagawa H, Fujiki R, Tanabe M, Takezawa S, Takada I, Yamaoka I, Yonezawa M, Kondo T, Furutani Y, Yagi H, Yoshinaga S, Masuda T, Fukuda T, Yamamoto Y, Ebihara K, Li DY, Matsuoka R, Takeuchi JK, Matsumoto T, et al. Retraction for Yoshimura et al. Distinct function of 2 chromatin remodeling complexes that share a common subunit, Williams syndrome transcription factor (WSTF) Proc Natl Acad Sci U S A. 2014;111:2398. doi: 10.1073/pnas.1323397111. [DOI] [PMC free article] [PubMed] [Google Scholar]