Abstract
Objectives:
The controversy of CpG island methylator phenotype (CIMP) in colorectal cancers (CRCs) persists, despite many studies that have been conducted on its correlation with molecular and clinicopathological features. To drive a more precise estimate of the strength of this postulated relationship, a meta-analysis was performed.
Methods:
A comprehensive search for studies reporting molecular and clinicopathological features of CRCs stratified by CIMP was performed within the PubMed, EMBASE, and Cochrane Library. CIMP was defined by either one of the three panels of gene-specific CIMP markers (Weisenberger panel, classic panel, or a mixture panel of the previous two) or the genome-wide DNA methylation profile. The associations of CIMP with outcome parameters were estimated using odds ratio (OR) or weighted mean difference (WMD) or hazard ratios (HRs) with 95% confidence interval (CI) for each study using a fixed effects or random effects model.
Results:
A total of 29 studies involving 9,393 CRC patients were included for analysis. We observed more BRAF mutations (OR 34.87; 95% CI, 22.49–54.06) and microsatellite instability (MSI) (OR 12.85 95% CI, 8.84–18.68) in CIMP-positive vs. -negative CRCs, whereas KRAS mutations were less frequent (OR 0.47; 95% CI, 0.30–0.75). Subgroup analysis showed that only the genome-wide methylation profile-defined CIMP subset encompassed all BRAF-mutated CRCs. As expected, CIMP-positive CRCs displayed significant associations with female (OR 0.64; 95% CI, 0.56–0.72), older age at diagnosis (WMD 2.77; 95% CI, 1.15–4.38), proximal location (OR 6.91; 95% CI, 5.17–9.23), mucinous histology (OR 3.81; 95% CI, 2.93–4.95), and poor differentiation (OR 4.22; 95% CI, 2.52–7.08). Although CIMP did not show a correlation with tumor stage (OR 1.10; 95% CI, 0.82–1.46), it was associated with shorter overall survival (HR 1.73; 95% CI, 1.27–2.37).
Conclusions:
The meta-analysis highlights that CIMP-positive CRCs take their own molecular feature, especially overlapping with BRAF mutations, and clinicopathological features and worse prognosis from CIMP-negative CRCs, suggesting CIMP could be used as an independent prognostic marker for CRCs.
Introduction
Aberrant DNA methylation is a hallmark of human cancer and can be summarized as global hypomethylation and regional hypermethylation. Regional hypermethylation refers to the aberrant methylation of normally unmethylated sequences, most of which are clusters of CpG sites, denoted CpG island. Specifically, regional hypermethylation of promoter-associated CpG islands of tumor-suppressor and repair genes is involved in the initiation and progression of cancer by transcription silencing.1, 2
A subgroup of human cancers is known to have frequent aberrant DNA methylation of the CpG island, referred to as the CpG island methylator phenotype (CIMP). CIMP was first identified in colorectal cancers (CRCs).3 For the determination of CIMP in CRCs, three panels of CIMP marker genes were available: a classic five-marker panel (MINT1, MINT2, MINT31, CDKN2A (p16), and hMLH1),3, 4 the Weisenberger five-marker panel (CACNA1G, IGF2, NEUROG1, RUNX3, and SOCS1),5 and a mixture panel of both. Both classic and Weisenberger CIMP-positive CRCs have been reported to be associated with proximal tumor location, microsatellite instability (MSI), and BRAF mutations.5, 6 Whereas the classic panel outperformed in predicting clinical outcomes, the Weisenberger panel was superior in detecting known clinicopathological features of CIMP but was inferior in prognostication power.7 Recently, genome-wide DNA methylation analysis was performed using the Infinium bead array to identify the CIMP subtype in human cancers, including CRCs. CRCs now can be categorized into CIMP-positive and -negative subtypes or CIMP-high, -low, and -negative subtypes.
It has been found that CIMP-positive or CIMP-high CRCs have a close association with molecular and clinicopathological features.8, 9, 10 Identifying the correlation of CIMP with molecular aberrations such as mutation of BRAF in CRCs may improve our understanding of carcinogenesis, identify strategies for subdividing patients into relevant subgroups, and highlight novel molecular target agents. Although the molecular mechanisms of CRC carcinogenesis remain unclear, both genetic and epigenetic alterations are considered to be important. Genetic alterations are responsible for the activation of oncogenes and the inactivation of tumor-suppressor genes, whereas epigenetic alterations through DNA methylation are known to play an important role in inhibiting the expression of tumor-related genes.
The presence of CIMP in CRCs has been reported to be associated with worse prognosis,8, 9 but controversial data regarding the correlation of CIMP with molecular and clinicopathological features makes it difficult to understand the internal mechanism. This is possibly because of limited sample size or confounding variables. Therefore, we initiated an international collaborative effort to evaluate the molecular features such as BRAF, KRAS mutations, MSI, and clinicopathological features and prognosis between CIMP-positive and -negative CRCs.
Methods
Search strategy
Standard guidelines for conducting and reporting a systematic review and meta-analysis were followed.11 All data available before 1 June 2015 from three electronic databases (PubMed, EMBASE, and Cochrane Library) were searched using a combination of MESH terms: colorectal cancer OR colorectal carcinoma PLUS CpG island methylator phenotype OR CIMP. All eligible studies were retrieved, and their bibliographies were hand-searched to capture for other relevant publications. Two reviewers (L.Z. and M.A.) independently screened all abstracts, following exclusion criteria for the first-round selection. Of the remaining articles, both reviewers independently evaluated the full text, following inclusion criteria for the second-round selection. Discrepancies between the two reviewers were resolved via discussion with three senior authors (J.J., W.-G.Z., and D.Y.).
Exclusion criteria
Abstracts, letters, editorials and expert opinions, reviews without original data, case reports, and studies that were not written in English were excluded. Studies or data were also excluded if: (i) they reported on non-colorectal or non-human tissues or colorectal polyps or hereditary forms of CRC; (ii) selection bias of study design existed, e.g., advanced CRCs, MSI-positive CRCs, CIMP-positive colon cancer or rectal cancer, and so on; (iii) relevant molecular or clinicopathological outcome parameters were not clearly reported; (iv) it was impossible to extract the appropriate data from the published results; and (v) there was overlap between authors or centers in the published literature and only the most recent or complete study was used.
Inclusion criteria
The inclusion criteria were as follows: (i) CIMP status was defined by gene-specific methylation analysis with restriction to two respective gene panels of markers (classic five-marker panel and Weisenberger five-marker panel) or a mixing of the two gene panels; (ii) CIMP status was defined by genome-wide methylation analysis; (iii) the studies evaluated the relationship between CIMP and BRAF, KRAS, MSI, or clinicopathological parameters such as gender, age, tumor location, histology, differentiation, tumor stage, or overall survival; (iv) sufficient published data could be used to estimate an odds ratio (OR) or weighted mean difference (WMD) or hazard ratio (HR) with 95% confidence interval (CI).
Quality assessment
Jadad Scale and MINORS are usually used to assess the quality of randomized controlled trials and nonrandomized controlled trials, respectively.12, 13 However, they are insufficiently validated for molecular studies. Instead, we made strict criteria for included studies such as no exclusion in specimen for a single-aim study of colon cancer or rectal cancer, all stage of tumors, and no exclusion based on molecular marker. Moreover, we made a subgroup analysis to examine whether the definition or the method used for CIMP (e.g., Weisenberger panel, classic panel, mixture panel, and genome-wide DNA methylation profile) influenced the results.
Data extraction
The following data were collected from each study: first author's surname, publication date, study method, sample size, total number of patients with positive CIMP and negative CIMP, and number of patients divided by BRAF, KRAS, MSI, age, gender, tumor location, histology, differentiation status, tumor TNM stage, and overall survival in those with and without CIMP, respectively. We did not define a minimum number of patients for inclusion in our meta-analysis.
Statistical analysis
ORs with 95% CIs were used for comparisons of binary measurements (e.g., BRAF, KRAS, MSI, gender, tumor location, histology, differentiation, and tumor stage), and WMD approach was analyzed for effects on quantitative measurements (e.g., age) according to the Woolf method. A weighted average of the individual adjusted log HRs was used to summarize the association between CIMP and overall survival, with the weights inversely proportional to the variance of the log HR of each study. Heterogeneity assumption was confirmed by the χ2-based Q-test. A P value of >0.10 for the Q-test indicated a lack of heterogeneity among the studies, and therefore the OR or WMD or HR estimate for each study was calculated by the fixed effects model. Otherwise, the random effects model was used. The significance of the pooled OR or WMD or HR was determined by the Z-test and P<0.05 was considered statistically significant. Sensitivity analyses were carried out to determine whether modification of the inclusion criteria for this meta-analysis affected the final results. An estimate of potential publication bias was carried out using the funnel plot. An asymmetric plot suggested possible publication bias. Funnel plot asymmetry was assessed using Egger's linear regression test, a linear regression approach to measure funnel plot asymmetry on the natural logarithm scale of the OR, WMD, or HR. The significance of the intercept was determined by the t-test, as suggested by Egger (P<0.05 was considered representative of statistically significant publication bias). All statistical tests were performed with Review Manager Version 5.0 (The Cochrane Collaboration, Oxford, UK).
Results
Study eligibility
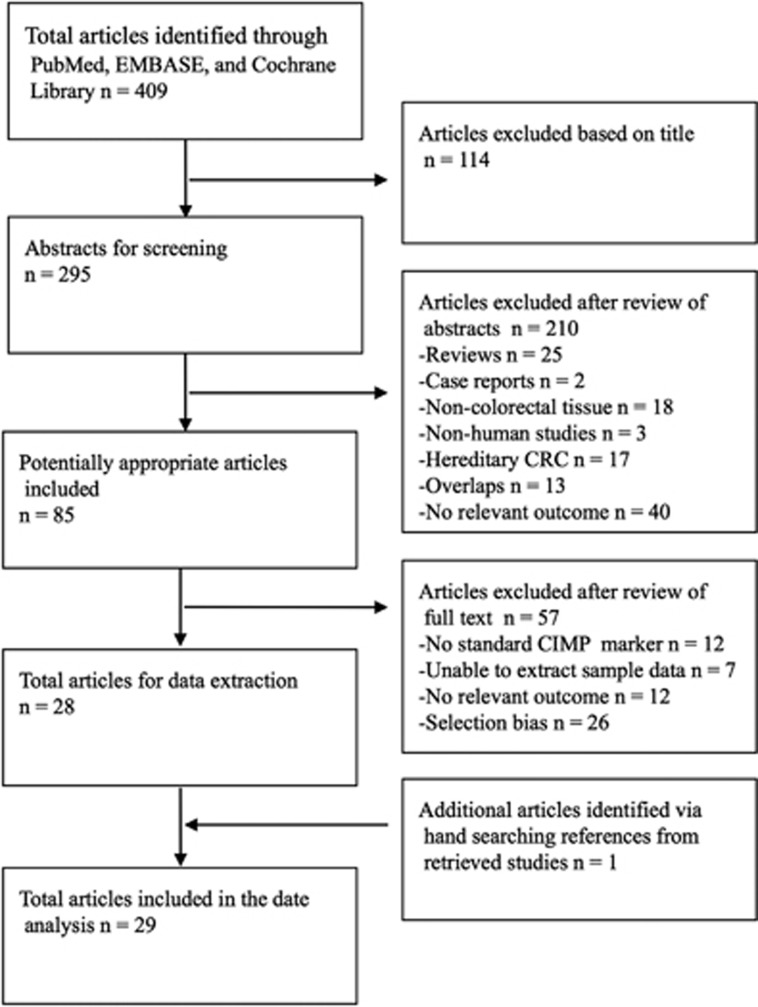
Initially, 409 articles were identified for further selection from 3 databases (Figure 1). Articles were excluded after title screening or abstract screening or full-text evaluation by the reviewers. By checking the relevant bibliography, one additional article was included.
Figure 1.
Flowchart of literature selection. CIMP, CpG island methylator phenotype; CRC, colorectal cancer.
Study characteristics
A total of 29 publications including 9,393 patients met the basic inclusion criteria,5, 7, 8, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39 of which 25 investigated for BRAF mutations, 18 for KRAS mutations, 20 for MSI, 24 for gender, 13 for age, 22 for tumor location, 7 for histology, 9 for differentiation, 14 for tumor stage, and 5 for overall survival. Among these studies, sample sizes ranged from 84 to 903 (Supplementary Table S1 online). In all, 7 studies used samples from Asia,7, 19, 25, 27, 29, 35, 36 5 from Australia,5, 14, 20, 22, 36 12 from Europe,8, 15, 17, 18, 21, 23, 30, 31, 32, 33, 34, 39 and 3 from the United States.16, 26, 28 In addition, two studies used mixed samples from Hong Kong and United States,24 and the Netherland and Canada,38 respectively. Figure 2 lists the risk of bias of each included study from selection, exposure assessment, other variable assessment, outcome assessment, and confounding factors. Based on a strict exclusion and inclusion criteria, the studies with high risk in selection bias were not included. Although six studies in other variable assessment were rated as high risk, they clearly stated the methods for the assessment of CIMP, BRAF mutations, KRAS mutations, and MSI.
Figure 2.
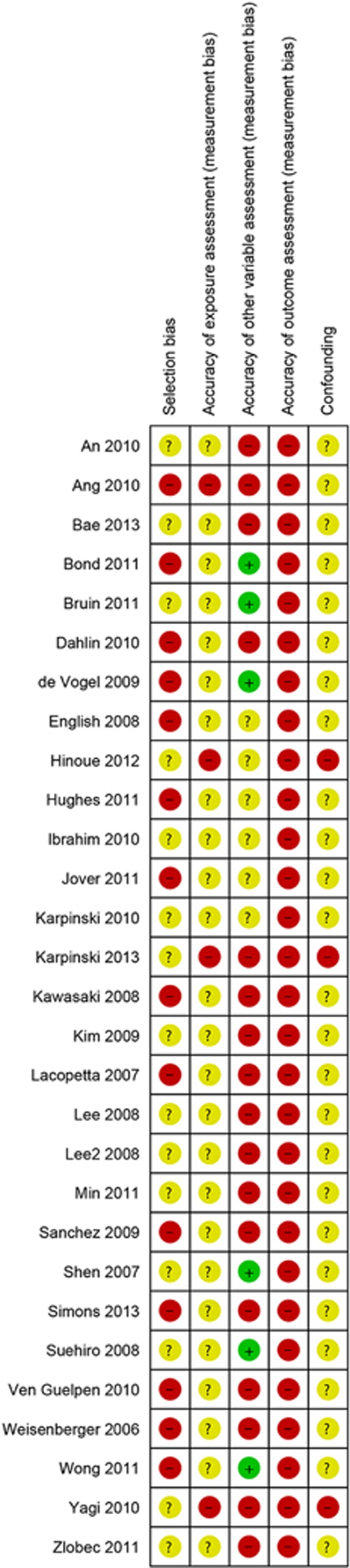
Risk of bias of each included study. Green cycle: study with high risk of bias; red cycle: study with low risk of bias; yellow cycle: study with insufficient information for assessing risk of bias.
Molecular features
BRAF mutations
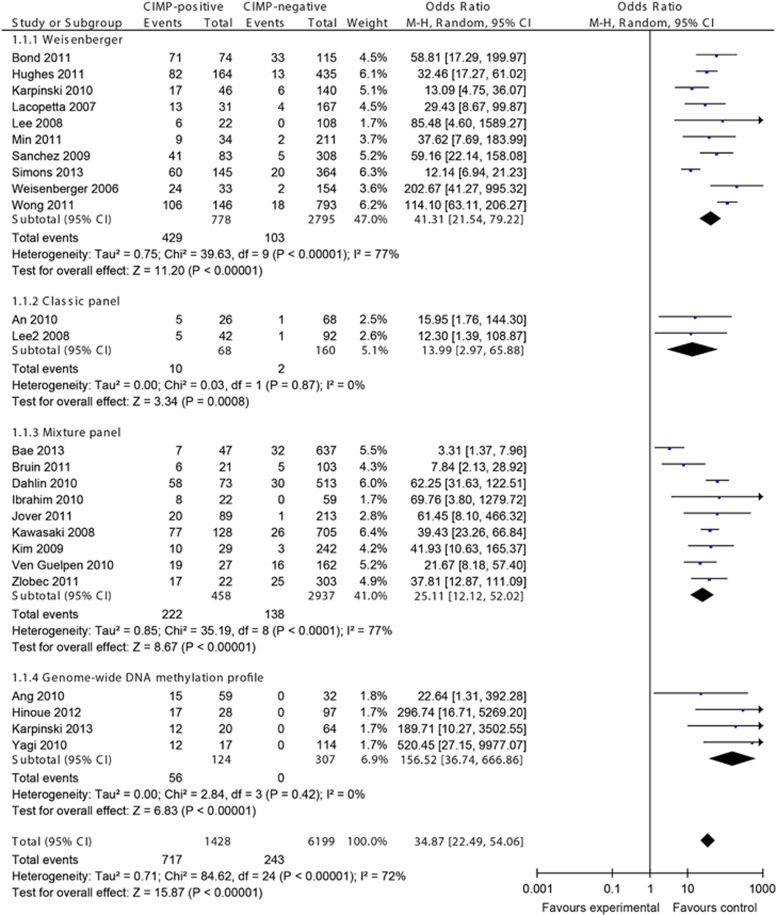
In all, 25 studies investigated the BRAF mutations in CRCs, examining a total of 7,627 CRCs: 10 studies for Weisenberger panel, 2 studies for classic panel, 9 studies for mixture panel, and 4 studies for genome-wide DNA methylation profile.
The overall OR for BRAF mutations in CIMP-positive vs. -negative CRCs was 34.87 (95% CI, 22.49–54.06; P<0.00001; Figure 3). Subgroup analyses of Weisenberger panel, classic panel, mixture panel, and genome-wide DNA methylation profile showed consistent results. Notably, we found that only the genome-wide methylation profile-defined CIMP subset of CRCs encompassed all BRAF-mutated CRCs.
Figure 3.
Meta-analysis investigating the frequency of BRAF mutations in CpG island methylator phenotype (CIMP)-positive vs. -negative colorectal cancers (CRCs). Random effect meta-analysis showed more BRAF mutations in CIMP-positive CRCs. CI, confidence interval.
KRAS mutations
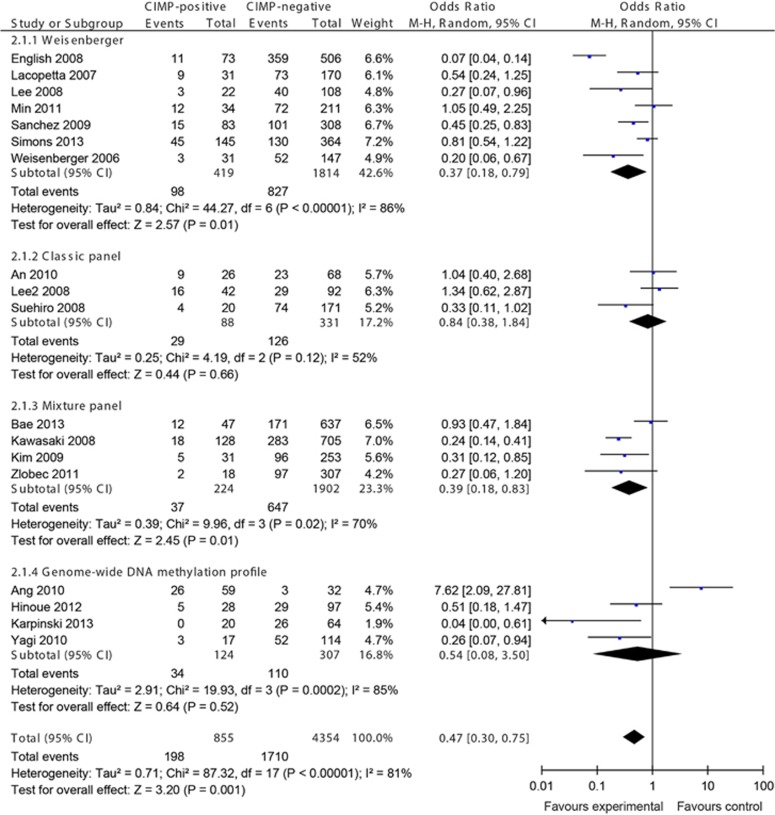
In all, 18 studies investigated the KRAS mutations in CRCs, examining a total of 5,209 CRCs: 7 studies for Weisenberger panel, 3 studies for classic panel, 4 studies for mixture panel, and 4 studies for genome-wide DNA methylation profile. The overall OR for KRAS mutations in CIMP-positive vs. -negative CRCs was 0.47 (95% CI, 0.30–0.75; P=0.001; Figure 4). Subgroup analyses with Weisenberger panel and mixture panel showed that KRAS mutations frequently occurred in CIMP-negative CRCs, whereas classic panel and genome-wide DNA methylation profile did not show any differences in comparison of CIMP-positive and -negative CRCs.
Figure 4.
Meta-analysis investigating the frequency of KRAS mutations in CpG island methylator phenotype (CIMP)-positive vs. -negative colorectal cancers (CRCs). Random effect meta-analysis showed less KRAS mutations in CIMP-positive CRCs. CI, confidence interval.
Microsatellite instability
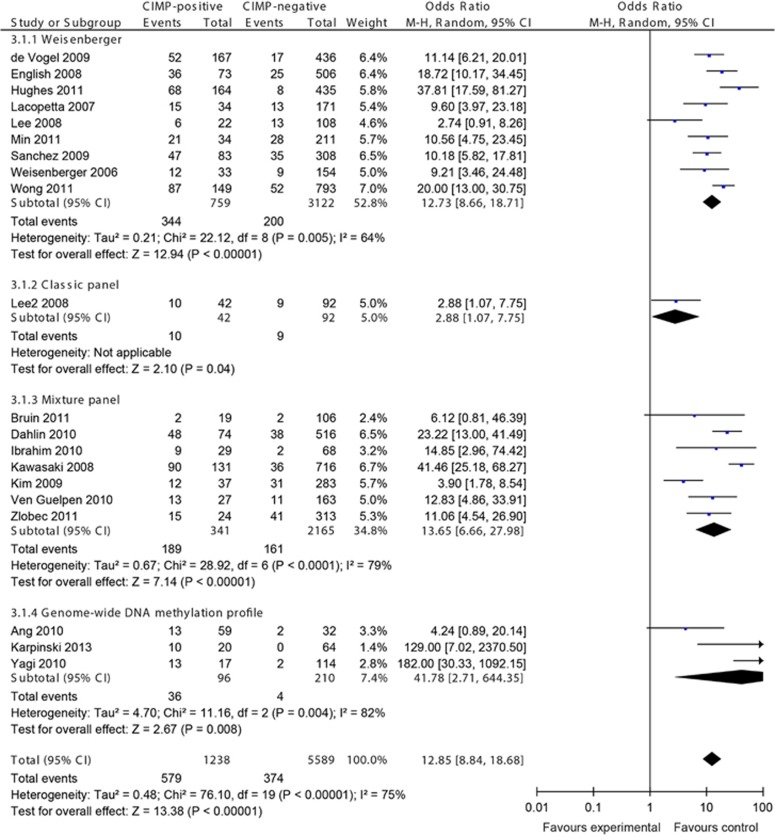
In all, 20 studies investigated the MSI status of CRCs, examining a total of 6,827 CRCs: 9 studies for Weisenberger panel, 1 study for classic panel, 7 studies for mixture panel, and 3 studies for genome-wide DNA methylation profile. A strong correlation between MSI and CIMP was achieved by OR 12.85 (95% CI, 8.84–18.68; P<0.00001; Figure 5). All of the subgroup analyses showed a similar trend, although all with large heterogeneity.
Figure 5.
Meta-analysis investigating the frequency of microsatellite instability (MSI) in CpG island methylator phenotype (CIMP)-positive vs. -negative colorectal cancers (CRCs). Random effect meta-analysis showed more MSI in CIMP-positive CRCs. CI, confidence interval.
Clinicopathological features
Gender
In all, 24 studies investigated the correlation between CIMP and gender in CRCs, examining a total of 7,298 CRCs: 9 studies for Weisenberger panel, 3 studies for classic panel, 8 studies for mixture panel, and 4 studies for genome-wide DNA methylation profile. The overall OR for the proportions of males in CIMP-positive vs. -negative CRCs was 0.64 (95% CI, 0.56–0.72; P<0.00001; Supplementary Figure S1 online). All of the subgroup analyses showed that more females took a leading position in CIMP-positive CRCs except for the subgroup of the classic panel.
Age
A total of 3,840 CRC patients either CIMP positive or CIMP negative in 13 studies were analyzed at the age at disease diagnosis: 8 studies for Weisenberger panel, 3 studies for mixture panel, and 2 studies for genome-wide DNA methylation profile. Because of lack of data from the classic panel, only three subgroup analyses were performed. The overall WMD for the age at disease diagnosis in CIMP-positive vs. -negative CRCs was 2.77 (95% CI, 1.15–4.38; P=0.0008; Supplementary Figure S2 online). Subgroup analyses of the Weisenberger panel and the genome-wide DNA methylation profile both showed that the CIMP phenomenon was much common in elder CRCs, whereas the mixture panel showed no differences.
Tumor location
In all, 22 studies investigated the correlation between CIMP and tumor locations in CRCs, examining a total of 6,740 CRCs: 9 studies for Weisenberger panel, 3 studies for classic panel, 6 studies for mixture panel, and 4 studies for genome-wide DNA methylation profile. The overall OR for the proportions of proximal location in CIMP-positive vs. -negative CRCs was 6.91 (95% CI, 5.17–9.23; P<0.00001; Supplementary Figure S3 online). All of the subgroup analyses strongly supported that CIMP-positive CRCs more commonly occurred in the proximal location.
Histology
In all, 7 studies investigated the correlation between CIMP and histological origin in CRCs, examining a total of 2,537 CRCs: 2 studies for Weisenberger panel, 4 studies for mixture panel, and 1 study for genome-wide DNA methylation profile. The overall OR for the proportions of mucinous type in CIMP-positive vs. -negative CRCs was 3.81 (95% CI, 2.93–4.95; P<0.00001; Supplementary Figure S4 online). Consistent results among the subgroup analyses strongly supported that CIMP-positive CRCs are associated with mucinous origin.
Differentiation
Nine studies investigated the correlation between CIMP and differentiation status in CRC cells, examining a total of 3629 CRCs: three studies for Weisenberger panel, one study for classic panel, four studies for mixture panel, and one study for genome-wide DNA methylation profile. The overall OR for the proportions of poor differentiation in CIMP positive vs. negative CRCs was 4.22 (95% CI, 2.52–7.08; P<0.00001; Supplementary Figure S5 online). All of the subgroup analyses strongly supported that CIMP-positive CRCs are associated with poor differentiation.
Tumor stage
In all, 14 studies investigated the correlation between CIMP and tumor stage in CRCs, examining a total of 3,882 CRCs: 4 studies for Weisenberger panel, 2 studies for classic panel, 5 studies for mixture panel, and 3 studies for genome-wide DNA methylation profile. The overall OR for the proportions of stages III and IV in CIMP-positive vs. -negative CRCs was 1.10 (95% CI, 0.82–1.46; P=0.53; Supplementary Figure S6 online). In the subgroup analyses, CIMP-positive CRCs clearly did not associate with advanced tumor stages.
Overall survival
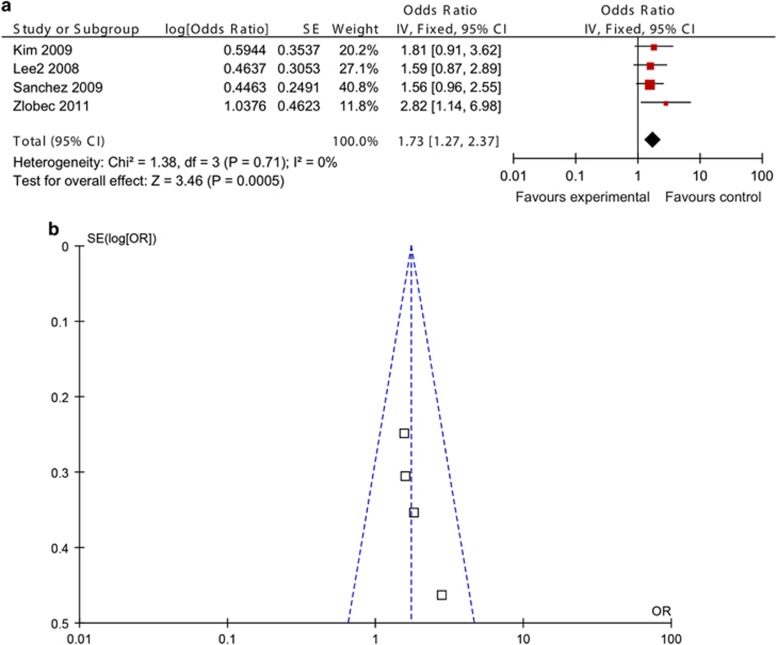
In total, four studies compared the overall survival of CIMP-positive and -negative CRCs: two studies for Weisenberger panel and two studies for mixture panel. Because of the insufficient data, we could only make a pooled analysis instead of a subgroup analysis. CIMP-positive CRCs were significantly associated with shorter overall survival (HR 1.73; 95% CI, 1.27–2.37; P=0.0005; Figure 6a). A funnel plot clearly showed that no heterogeneity existed among these four included studies (Figure 6b).
Figure 6.
Meta-analysis investigating overall survival in CpG island methylator phenotype (CIMP)-positive vs. -negative colorectal cancers (CRCs). (a) Meta-analysis showed shorter overall survival in CIMP-positive CRCs. (b) Funnel plot showed that no heterogeneity existed among four included studies. CI, confidence interval; OR, odds ratio.
Publication bias
Begg's funnel plot was performed to assess publication bias. The heterogeneity tests for comparing the 29 combined studies showed heterogeneity in some analyses such as BRAF, KRAS, MSI, age, tumor location, differentiation, and tumor stage. However, no single study influenced the pooled OR or WMD qualitatively as indicated by the sensitivity analyses (data not shown).
Subgroup analyses of some potential confounding factors
To analyze the potential confounding factors that might influence the data collection, we performed subgroup analyses. However, because of lack of information or insufficient data, we could only conduct the subgroup analyses of methods of tissue preservation and sources of patients for BRAF and KRAS mutations, and MSI within the Weisenberger panel (Supplementary Figures S7 online). Heterogeneous results were found to exist in analysis of KRAS mutations, but not in those of BRAF mutations or MSI. First, it was found that CIMP-negative CRCs in cryopreservation groups were frequently associated with KRAS mutations, whereas those in formalin-fixed, paraffin-embedded (FFPE) group were not (Supplementary Figure S8 online). Second, in the subgroup analysis of case–control study vs. population-based study, only case–control studies showed that there was an association between KRAS mutations and CIMP-negative CRCs (Supplementary Figure S11 online). These data suggest that methods of tissue preservation and sources of patients might increase the heterogeneity of molecular studies of CIMP.
Discussion
In this study, we aimed to review the data from the published studies in order to estimate the association between CIMP and other molecular incidents or clinicopathological features in CRCs. We found a trend toward more BRAF mutations and MSI and less KRAS mutations in CIMP-positive CRCs than CIMP-negative CRCs. We also clearly demonstrated that CIMP-positive CRCs had female preference and age dependence. Moreover, this subtype of tumors showed a significant correlation with proximal location, mucinous histology, and poor differentiation. Surprisingly, CIMP did not show a correlation with tumor stage, but was significantly associated with shorter overall survival, suggesting CIMP could be used as an independent prognostic marker.
One of the major confounding factors in the systematic review on topics relating to CIMP was the lack of a standardized definition of CIMP. Gene-specific methylation markers and genome-wide DNA methylation profile were the two major methods used to define the CIMP in CRCs. Depending on the number and set of genes used for the determination of the CIMP status, a relatively higher heterogeneity can be caused mainly by different panels of CIMP markers compared with genome-wide DNA methylation profile. Till now, two different panels of CIMP markers as well as a mixture of both were widely used in a variety of studies for CRCs. To limit most of the heterogeneity among studies, we screened and only included the studies either using the two panels of CIMP markers or a mixture of these two panels or genome-wide DNA methylation profile. In addition, subgroup analyses of different methodologies and different panels were performed.
After the concept of CIMP was initially described in CRCs, other human tumors like glioma, paraganglioma, and gastric cancer were also characterized with similar phenotype. In gliomas, Xu et al.40 revealed that IDH1 and IDH2 gene mutations led to CIMP by reducing the α-ketoglutarate and accumulating 2-hydroxyglutarate that could inhibit histone demethylases and the TET family of 5-methylcytosine hydroxylases. In paragangliomas, Letouze et al.41 reported that SDH mutation was a key factor in causing CIMP by interplaying between the Krebs cycle and 2-oxoglutarate-dependent (2-OG) histone and DNA demethylases. For gastric cancers, Kim et al.42 demonstrated an association between CIMP and oncogene mutations. In this analysis, we clearly showed that BRAF mutation was associated with CIMP in CRCs. In addition, in a recent study, Fang et al.43 provided the first demonstration that a BRAF/MEK/ERK signaling pathway, which mediates silencing of MLH1, is more generally responsible for CIMP.
In a subgroup analysis, we found that genome-wide methylation profile-defined CIMP subset of CRCs encompassed all of the BRAF-mutated CRCs, whereas other 3 panels of makers only showed 61.7–83.3% BRAF mutations in CIMP-positive CRCs. BRAF/MEK/ERK signaling pathway could explain the internal mechanism of CIMP. However, in our study, ∼49.8% of CIMP-positive CRCs lacked BRAF mutations. We speculated that the subtype of CIMP-positive CRCs without BRAF mutations might be caused by the effect of the microbiome. A recent study by Tahara et al.44 introduced the notion that fusobacterium enrichment was associated with CIMP-positive CRCs. However, they did not find an association between fusobacterium enrichment and BRAF mutations, suggesting microbiome might induce CIMP through different mechanisms.
Our results demonstrated CIMP-positive CRCs were associated with proximal location, arguing for a potentially distinct molecular pathway between proximal and distal CRCs. At least two distinct pathways, the serrated pathway and the conventional pathway, underlie most CRCs.45 The CRCs in the serrated pathway have a predilection for frequent BRAF mutations, MSI, and proximal location. On the contrary, the conventional pathway mostly occurs in the distal location with KRAS mutations and microsatellite stability. Combined with our findings, we speculated that CIMP might be a signature of the serrated CRCs.
Consistent results among different methodologies or different panels were achieved in most of the subgroup analyses, suggesting that the heterogeneity of the present four different definitions of CIMP was limited. However, there were still some exceptions in the subgroup analyses of KRAS mutations, gender, and age. For analysis of KRAS mutation rates, the overall effects of the classic panel that included three studies and the genome-wide DNA methylation profile that included four studies did not show any differences between CIMP-positive and -negative CRCs. We believe this discrepancy between the Weisenberger panel/mixture panel and the classical panel/genome-wide DNA methylation profile was caused by a relatively limited sample size in these two subgroups, because the total weights of the two were 17.2 and 16.8%. Similarly, the overall effects of the classic panel, including three studies in analysis of gender, and the mixture panel, including three studies in analysis of age, showed heterogeneous results, and their total weights were 6.0 and 21.1%.
Regardless of the different definition of CIMP, still many confounding factors such as methods of tissue preservation, sources of patients, smoking history, publication bias, and ethnicity might prevent us from reaching a more precise conclusion.46 We do find that whether the KRAS gene mutation rate is linked to negative CIMP depends on the tissue preservation methods and the sources of patients. However, tissue preservation and sources of patients did not affect the detection accuracy of the associations of BRAF mutations and MSI with positive CIMP. We speculate that the discrepancy might be caused by the limited sample size and the internal heterogeneity among studies, because when the negative data on the association of CIMP-negative CRCs with KRAS gene mutations were observed in formalin-fixed paraffin-embedded or population-based subgroups, a significant heterogeneity existed.
In summary, the meta-analysis highlights that CIMP-positive CRCs take their own molecular (especially overlapping with BRAF mutations) and clinicopathological features and worse prognosis from CIMP-negative CRCs, suggesting CIMP could be used as an independent prognostic marker for CRCs.
Study Highlights
Acknowledgments
We thank Yuan-Jun Yu (University of Pennsylvania) for language editing.
Guarantor of the article: Duonan Yu, MD, PhD.
Specific author contributions: Liang Zong, Duonan Yu, and Wei-Guo Zhu designed the study; Liang Zong and Masanobu Abe collected and analyzed the data; Liang Zong and Jiafu Ji interpreted the data; Liang Zong drafted the manuscript; Duonan Yu, Wei-Guo Zhu, and Jiafu Ji revised the manuscript.
Financial support: This work is supported by the National Natural Science Foundation of China (grant 81470277 to Duonan Yu), the Jiangsu Province Specially-appointed Professorship Start-up Fund (to Duonan Yu), a grant from the Ministry of Finance of China for the “Biology & Medical Science Innovation Team” program (to Duonan Yu) and the Priority Academic Program Development of Jiangsu Higher Education Institution (Veterinary Medicine, to Duonan Yu).
Potential competing interests: None.
Footnotes
Supplementary Information accompanies this paper on the Clinical and Translational Gastroenterology website (http://www.nature.com/ctg)
Supplementary Material
References
- Kim YH, Lee HC, Kim SY et al. Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann Surg Oncol 2011; 18: 2338–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H, Gabrielson E, Chen W et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet 2002; 31: 141–149. [DOI] [PubMed] [Google Scholar]
- Toyota M, Ahuja N, Ohe-Toyota M et al. CpG island methylator phenotype in colorectal cancer. Proc Natl Acad Sci USA 1999; 96: 8681–8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer 2004; 4: 988–993. [DOI] [PubMed] [Google Scholar]
- Weisenberger DJ, Siegmund KD, Campan M et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet 2006; 38: 787–793. [DOI] [PubMed] [Google Scholar]
- Curtin K, Slattery ML, Samowitz WS. CpG island methylation in colorectal cancer: past, present and future. Patholog Res Int 2011; 2011: 902674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cho NY, Yoo EJ et al. CpG island methylator phenotype in colorectal cancers: comparison of the new and classic CpG island methylator phenotype marker panels. Arch Pathol Lab Med 2008; 132: 1657–1665. [DOI] [PubMed] [Google Scholar]
- Zlobec I, Bihl M, Foerster A et al. Comprehensive analysis of CpG island methylator phenotype (CIMP)-high, -low, and -negative colorectal cancers based on protein marker expression and molecular features. J Pathol 2011; 225: 336–343. [DOI] [PubMed] [Google Scholar]
- Kim JH, Rhee YY, Bae JM et al. Loss of CDX2/CK20 expression is associated with poorly differentiated carcinoma, the CpG island methylator phenotype, and adverse prognosis in microsatellite-unstable colorectal cancer. Am J Surg Pathol 2013; 37: 1532–1541. [DOI] [PubMed] [Google Scholar]
- Arain MA, Sawhney M, Sheikh S et al. CIMP status of interval colon cancers: another piece to the puzzle. Am J Gastroenterol 2010; 105: 1189–1195. [DOI] [PubMed] [Google Scholar]
- Stroup DF, Berlin JA, Morton SC et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA 2000; 283: 2008–2012. [DOI] [PubMed] [Google Scholar]
- Jadad AR, Moore RA, Carroll D et al. Assessing the quality of reports of randomized clinical trials: is blinding necessary? Control Clin Trials 1996; 17: 1–12. [DOI] [PubMed] [Google Scholar]
- Slim K, Nini E, Forestier D et al. Methodological index for non-randomized studies (minors): development and validation of a new instrument. ANZ J Surg 2003; 73: 712–716. [DOI] [PubMed] [Google Scholar]
- Iacopetta B, Grieu F, Phillips M et al. Methylation levels of LINE-1 repeats and CpG island loci are inversely related in normal colonic mucosa. Cancer Sci 2007; 98: 1454–1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- English DR, Young JP, Simpson JA et al. Ethnicity and risk for colorectal cancers showing somatic BRAF V600E mutation or CpG island methylator phenotype. Cancer Epidemiol Biomarkers Prev 2008; 17: 1774–1780. [DOI] [PubMed] [Google Scholar]
- Sanchez JA, Krumroy L, Plummer S et al. Genetic and epigenetic classifications define clinical phenotypes and determine patient outcomes in colorectal cancer. Br J Surg 2009; 96: 1196–1204. [DOI] [PubMed] [Google Scholar]
- de Vogel S, Wouters KA, Gottschalk RW et al. Genetic variants of methyl metabolizing enzymes and epigenetic regulators: associations with promoter CpG island hypermethylation in colorectal cancer. Cancer Epidemiol Biomarkers Prev 2009; 18: 3086–3096. [DOI] [PubMed] [Google Scholar]
- Karpinski P, Myszka A, Ramsey D et al. Polymorphisms in methyl-group metabolism genes and risk of sporadic colorectal cancer with relation to the CpG island methylator phenotype. Cancer Epidemiol 2010; 34: 338–344. [DOI] [PubMed] [Google Scholar]
- Min BH, Bae JM, Lee EJ et al. The CpG island methylator phenotype may confer a survival benefit in patients with stage II or III colorectal carcinomas receiving fluoropyrimidine-based adjuvant chemotherapy. BMC Cancer 2011; 11: 344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JJ, Hawkins NJ, Ward RL et al. Methylation of the 3p22 region encompassing MLH1 is representative of the CpG island methylator phenotype in colorectal cancer. Mod Pathol 2011; 24: 396–411. [DOI] [PubMed] [Google Scholar]
- Hughes LA, Simons CC, van den Brandt PA et al. Body size, physical activity and risk of colorectal cancer with or without the CpG island methylator phenotype (CIMP). PLoS One 2011; 6: e18571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond CE, Umapathy A, Ramsnes I et al. p53 mutation is common in microsatellite stable, BRAF mutant colorectal cancers. Int J Cancer 2012; 130: 1567–1576. [DOI] [PubMed] [Google Scholar]
- Simons CC, Hughes LA, Smits KM et al. A novel classification of colorectal tumors based on microsatellite instability, the CpG island methylator phenotype and chromosomal instability: implications for prognosis. Ann Oncol 2013; 24: 2048–2056. [DOI] [PubMed] [Google Scholar]
- Suehiro Y, Wong CW, Chirieac LR et al. Epigenetic-genetic interactions in the APC/WNT, RAS/RAF, and P53 pathways in colorectal carcinoma. Clin Cancer Res 2008; 14: 2560–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An B, Kondo Y, Okamoto Y et al. Characteristic methylation profile in CpG island methylator phenotype-negative distal colorectal cancers. Int J Cancer 2010; 127: 2095–2105. [DOI] [PubMed] [Google Scholar]
- Shen L, Catalano PJ, Benson AB 3rd et al. Association between DNA methylation and shortened survival in patients with advanced colorectal cancer treated with 5-fluorouracil based chemotherapy. Clin Cancer Res 2007; 13: 6093–6098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cho NY, Choi M et al. Clinicopathological features of CpG island methylator phenotype-positive colorectal cancer and its adverse prognosis in relation to KRAS/BRAF mutation. Pathol Int 2008; 58: 104–113. [DOI] [PubMed] [Google Scholar]
- Kawasaki T, Ohnishi M, Suemoto Y et al. WRN promoter methylation possibly connects mucinous differentiation, microsatellite instability and CpG island methylator phenotype in colorectal cancer. Mod Pathol 2008; 21: 150–158. [DOI] [PubMed] [Google Scholar]
- Kim JH, Shin SH, Kwon HJ et al. Prognostic implications of CpG island hypermethylator phenotype in colorectal cancers. Virchows Arch 2009; 455: 485–494. [DOI] [PubMed] [Google Scholar]
- Dahlin AM, Palmqvist R, Henriksson ML et al. The role of the CpG island methylator phenotype in colorectal cancer prognosis depends on microsatellite instability screening status. Clin Cancer Res 2010; 16: 1845–1855. [DOI] [PubMed] [Google Scholar]
- Van Guelpen B, Dahlin AM, Hultdin J et al. One-carbon metabolism and CpG island methylator phenotype status in incident colorectal cancer: a nested case-referent study. Cancer Causes Control 2010; 21: 557–566. [DOI] [PubMed] [Google Scholar]
- Ibrahim AE, Arends MJ, Silva AL et al. Sequential DNA methylation changes are associated with DNMT3B overexpression in colorectal neoplastic progression. Gut 2011; 60: 499–508. [DOI] [PubMed] [Google Scholar]
- Jover R, Nguyen TP, Perez-Carbonell L et al. 5-Fluorouracil adjuvant chemotherapy does not increase survival in patients with CpG island methylator phenotype colorectal cancer. Gastroenterology 2011; 140: 1174–1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruin SC, He Y, Mikolajewska-Hanclich I et al. Molecular alterations associated with liver metastases development in colorectal cancer patients. Br J Cancer 2011; 105: 281–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae JM, Kim JH, Cho NY et al. Prognostic implication of the CpG island methylator phenotype in colorectal cancers depends on tumour location. Br J Cancer 2013; 109: 1004–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi K, Akagi K, Hayashi H et al. Three DNA methylation epigenotypes in human colorectal cancer. Clin Cancer Res 2010; 16: 21–33. [DOI] [PubMed] [Google Scholar]
- Ang PW, Loh M, Liem N et al. Comprehensive profiling of DNA methylation in colorectal cancer reveals subgroups with distinct clinicopathological and molecular features. BMC Cancer 2010; 10: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinoue T, Weisenberger DJ, Lange CP et al. Genome-scale analysis of aberrant DNA methylation in colorectal cancer. Genome Res 2012; 22: 271–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpinski P, Walter M, Szmida E et al. Intermediate- and low-methylation epigenotypes do not correspond to CpG island methylator phenotype (low and -zero) in colorectal cancer. Cancer Epidemiol Biomarkers Prev 2013; 22: 201–208. [DOI] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell 2011; 19: 17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letouze E, Martinelli C, Loriot C et al. SDH mutations establish a hypermethylator phenotype in paraganglioma. Cancer Cell 2013; 23: 739–752. [DOI] [PubMed] [Google Scholar]
- Kim JG, Takeshima H, Niwa T et al. Comprehensive DNA methylation and extensive mutation analyses reveal an association between the CpG island methylator phenotype and oncogenic mutations in gastric cancers. Cancer Lett 2013; 330: 33–40. [DOI] [PubMed] [Google Scholar]
- Fang M, Ou J, Hutchinson L et al. The BRAF oncoprotein functions through the transcriptional repressor MAFG to mediate the CpG island methylator phenotype. Mol Cell 2014; 55: 904–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahara T, Yamamoto E, Suzuki H et al. Fusobacterium in colonic flora and molecular features of colorectal carcinoma. Cancer Res 2014; 74: 1311–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettington M, Walker N, Clouston A et al. The serrated pathway to colorectal carcinoma: current concepts and challenges. Histopathology 2013; 62: 367–386. [DOI] [PubMed] [Google Scholar]
- Anderson JC, Rangasamy P, Rustagi T et al. Risk factors for sessile serrated adenomas. J Clin Gastroenterol 2011; 45: 694–699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.