Abstract
Objectives:
The potential of Escherichia coli (E. coli) isolated from inflammatory bowel disease (IBD) patients to damage the integrity of the intestinal epithelium was investigated.
Methods:
E. coli strains isolated from patients with ulcerative colitis (UC) and healthy controls were tested for virulence capacity by molecular techniques and cytotoxic assays and transepithelial electric resistance (TER). E. coli isolate p19A was selected, and deletion mutants were created for alpha-hemolysin (α-hemolysin) (hly) clusters and cytotoxic necrotizing factor type 1 (cnf1). Probiotic E. coli Nissle and pathogenic E. coli LF82 were used as controls.
Results:
E. coli strains from patients with active UC completely disrupted epithelial cell tight junctions shortly after inoculation. These strains belong to phylogenetic group B2 and are all α-hemolysin positive. In contrast, probiotic E. coli Nissle, pathogenic E. coli LF82, four E. coli from patients with inactive UC and three E. coli strains from healthy controls did not disrupt tight junctions. E. coli p19A WT as well as cnf1, and single loci of hly mutants from cluster I and II were all able to damage Caco-2 (Heterogeneous human epithelial colorectal adenocarcinoma) cell tight junctions. However, this phenotype was lost in a mutant with knockout (Δ) of both hly loci (P<0.001).
Conclusions:
UC-associated E. coli producing α-hemolysin can cause rapid loss of tight junction integrity in differentiated Caco-2 cell monolayers. This effect was abolished in a mutant unable to express α-hemolysin. These results suggest that high Hly expression may be a mechanism by which specific strains of E. coli pathobionts can contribute to epithelial barrier dysfunction and pathophysiology of disease in IBD.
Introduction
Crohn's disease (CD) and ulcerative colitis (UC) are two different forms of chronic inflammatory bowel disease (IBD), the etiology of which is still unknown. CD and UC are distinguished by their clinical manifestations and inflammatory profiles.1 UC is a chronic inflammatory disorder of the colorectal mucosa, whereas CD is a chronic, segmentally localized granulomatous disease of the gastro-intestinal tract CD may even affect non-intestinal tissue such as lymph nodes and skin. Clinical practice has seen that in both diseases chronicity is interrupted by acute flares, bloody diarrhea, relapses, and remission. IBD can appear at any age, however, most often in the third decade of life.2 The highest reported prevalence values for IBD are in Europe (UC, 505 per 100,000 persons; CD, 322 per 100,000 persons).3
Genome-wide association studies in IBD have identified genetic polymorphisms contributing to susceptibility to IBD. Many of these gene polymorphisms are associated with pathways involved in intestinal homeostasis, linking host genetics to deregulated host responses to the microbiota.4 The concordance rate among monozygotic twins was 6.3% for UC and 58.3% for CD.5 This clearly indicates a role of genetic factors in CD, but also indicates an important role for environmental factors, particularly in UC. An abnormal microbiota composition and decreased complexity of the gut microbial ecosystem (commonly referred to as dysbiosis) are common features in patients with CD or UC.6 These observations have fueled efforts to identify opportunistic gut pathogens (or pathobionts) that may have a role in the pathogenesis of IBD.
Escherichia coli pathobionts exhibiting pathogen-like behaviors are more frequently cultured from IBD patients with active disease due to their selective growth advantage in inflammatory conditions.7 Moreover, adherence of the B2 phylotype E. coli to human intestinal epithelial cells is mediated through the type 1 pili interaction with mannosylated carcinoembryonic antigen-related cell adhesion molecule 6 (CEACAM6). Interestingly, CEACAM6 expression by cultured intestinal cells was shown previously to be upregulated after treatment with interferon γ and tumor necrosis factor-α.8 These findings indicate that inflammatory conditions in the gut support E. coli colonization via increased CEACAM6 expression and offer an explanation for their more frequent isolation from patients with active disease.
Among the Proteobacteria, adherent invasive variants of the B2 phylogroup E. coli (AIEC) have been proposed to have a role in the pathophysiology of IBD,9 owing to their capacity to adhere to intestinal epithelial cells, to invade intestinal epithelial cells via a macropinocytosis-like process, and to survive and replicate intracellularly in epithelial cells and macrophages.10 Others have, likewise, found increased numbers of B2 phylogroup E. coli isolated from IBD patients.11 Petersen et al.12 showed that E. coli isolates from fecal samples of primarily UC patients with active disease frequently belong to the B2 phylogenetic group and harbor genes commonly associated with extra-intestinal pathogenic E. coli (ExPEC) causing urinary tract infection and meningitis.13
Recently, a hemolysin (Hly) producing E. coli strain was shown to induce localized defects in epithelial integrity colonic cell monolayers and rat colon tissue ex vivo. Additionally, wild-type (WT) and colitis susceptible IL-10−/− mice colonized with an HlyA-expressing E. coli had elevated inflammation scores and an increased epithelial permeability compared with mice colonized with the HlyA-deficient mutant. Furthermore, qPCR analysis revealed that lesions (focal leaks) in mucosal samples from the human colon were associated with 10-fold higher levels of hlyA DNA, suggesting that Hly-expressing E. coli have a role in the pathology of intestinal inflammation in IBD.14
The aim of this study was to extend the above observations to isolates of B2 phylogroup E. coli from IBD patients by testing their effects on permeability, tight junction stability, and viability of human intestinal cell epithelial monolayers cultured in vitro. For comparison, we also tested the effect of prototype AIEC strain LF82 and the probiotic E. coli Nissle on permeability and viability of polarized human Caco-2 cells. As some strains of B2 phylogroup E. coli isolated from UC patients also possess a gene encoding cytotoxic necrotizing factor type 1 (cnf1), we investigated the role of cnf1 and hlyA in causing epithelial damage by the construction and testing of genetic mutants in cellular assays.
Methods
Study material and ethics
Permission for the study was obtained from the Regional Ethics Committee for Copenhagen County Hospitals (Permission no. KA03019), and all participants gave their informed written consents. Healthy controls were recruited among medical students. All controls had a completely normal distal colon as visualized by video sigmoidoscopy (left side colon) at study entry. None of the controls had experienced diarrhea, blood in stools, or abdominal pain or any other abdominal discomfort when the stool sample was submitted. Patients with IBD were diagnosed according to standardized criteria,15, 16 which included confirmation of inflammation by video sigmoidoscopy and a fresh set of negative stool cultures for common pathogens including Clostridium difficile.
Detailed information regarding extent of disease and current medication among the included patients has previously been described,12 neither controls nor patients had received antibiotics within the last 2 months before inclusion and all patients had an established diagnosis of IBD before inclusion in our study.
Fecal samples were cultured at Statens Serum Institut (SSI): bacteriological analysis, E. coli phenotypic characterization, determination of phylogenetic group, and ExPEC virulence gene detection were performed as described previously in Petersen et al.12 E. coli clinical isolates p7, p10, p13, p19A, p22, p25, p26, p27, and p32; healthy control E. coli isolates C2, C4, and C6 were characterized by PCR for virulence genes (data not shown) in this study. The probiotic E. coli Nissle 1917 and the pathogenic E. coli LF82 (ref. 17) were used as a negative and positive control, respectively.
Patient characteristics and diseases association and location are described in Table 1.
Table 1. Characteristics of patients and controls included in the study.
| ID number | Disease association | Medication | Disease localizationa | Gender | Age |
|---|---|---|---|---|---|
| p7 | Active UC | 5-ASA | Proctosigmoid colon | Male | 71 |
| p13 | Active UC | 5-ASA Azathioprine, Prednisolone, | Pancolitis | Male | 39 |
| p19A | Active UC | 5-ASA, Azathioprine | Proctosigmoid colon | Male | 71 |
| p22 | Active UC | 5-ASA | Proctosigmoid colon | Female | 40 |
| p25 | Active UC | 5-ASA | Pancolitis | Male | 34 |
| p10 | Inactive UC | 5-ASA | Proctosigmoid colon | Female | 40 |
| p26 | Inactive UC | 5-ASA | Proctosigmoid colon | Male | 53 |
| p27 | Inactive UC | Azathioprine | Rectum | Male | 37 |
| p32 | Inactive UC | None | Pancolitis | Female | 40 |
| C2 | Healthy | None | None | Female | 25 |
| C4 | Healthy | None | None | Male | 24 |
| C6 | Healthy | None | None | Female | 29 |
| LF82 | Crohn's disease | None | Ileum | Not known | Not known |
| E. coli Nissle | Healthy | None | None | Male | Not known |
UC, ulcerative colitis; 5-ASA, 5-aminosalicylic acid.
Present when active disease, previous when inactive disease.
Cell infection assay and measurement of transepithelial electric resistance
Heterogeneous human epithelial colorectal adenocarcinoma cells (Caco-2 BBE cell line, ATCC CRL 2102) were maintained at 37 °C in a humidified 5% CO2 atmosphere in Dulbecco's modified Eagle medium (DMEM; Invitrogen, Paisley, UK) containing Glutamax, 10% heat-inactivated fetal bovine serum (PAA Laboratories, Colbe, DE), 100 U/ml penicillin, 100 μg/ml streptomycin (PenStrep, Sigma, St. Louis, MO), 1% non-essential amino acids (Lonza, Basel, Switzerland), and 1% l-glutamine.
Caco-2 cells (between passages 55 and 60) were seeded at a density of 2.6 × 105 cells/cm2 in a 24-transwell system containing Tissue Culture-treated filter (0.4 μm pore size, BD Biosciences Falcon type # 353494, Erembodegem, Belgium) and grown for 16 days until they differentiated into polarized monolayers. After 14 days, the transepithelial electric resistance (TER) reached 600–800 Ohms/cm2 (Volt/Ohm meter; World Precision Instruments, Sarasota, FL).
For bacterial co-incubation experiments, the medium was removed by aspiration from the Transwell filters, and the filters were inserted in the cellZscope apparatus (Nanoanalytics, Münster, DE). Cell-culture medium without antibiotics was then added to the upper chambers (450 μl) and lower chambers (800 μl), and the apparatus was placed in a humidified incubator at 37°C containing 5% CO2/95% O2 atmosphere for 2 h before the addition of bacteria. Bacteria were grown overnight in LB (Luria broth) media at 37 °C, centrifuged, resuspended in DMEM, and added to the upper chambers (filter) in the cellZscope at a multiplicity of infection (MOI) of 10. TER measurements were recorded continuously for up to 24 h, and TER values were normalized to the initial TER value (100%) and absolute TER is mean of four independent measurements. As a control, TER was measured for uninfected Caco-2 cell monolayers (controls in figures). Three independent experiments were performed for p19A WT and its mutant strains.
Detection of occludin by immunofluorescence and western blotting
To visualize the effect of bacteria on Caco-2 cells, occludin was detected by immunofluorescence microscopy18 and western blotting. Caco-2 cell monolayers were grown as described above and infected with bacteria for up to 15 h at a MOI of max. 100. Caco-2 cell monolayers were either fixed for immunofluorescence or lysed in 100 μl of lysis buffer (Promega, Madison, WI) on ice for 5–10 min. The cell lysate was centrifuged at 13,000 g for 12 min to pellet debris, and the supernatant was used for western blotting. Fifty micrograms of Caco-2 cell proteins were resolved by 10% SDS-PAGE and transferred onto 0.2 μm polyvinylidene fluoride (PVDF) membranes. Membranes were blocked for 1 h with 3% non-fat milk powder diluted in 0.05% Tween-20 (TBST), then incubated with primary antibody in 3% non-fat milk powder diluted in TBST overnight at 4 °C. Hereafter, membranes were visualized with secondary (Sigma-Aldrich, GmbH, Germany, DE) antibody for 1 h at room temperature. Rabbit polyclonal anti-actin antibody (A2066; Sigma-Aldrich) and rabbit anti-Occludin antibody (ABT146 Merck KGaA, Darmstadt, Germany, DE) were used in this study.
Hemolysin assay
The presence of α-hemolysin was demonstrated on 5% sheep blood agar plates (SSI no. 31349, Statens Serum Institut, Diagnostica, Hillerød, Denmark) after 3–4 h of incubation at 37 °C as opposed to enterohemolysin, which was detectable only after overnight incubation at 37 °C.
Hemolysis determination by titration assay
Defibrinated horse blood (SSI no. 23699 Statens Serum Institut, Diagnostica) was washed twice in hemolysis buffer (0.0077 m Tris-HCl, 0.137 m NaCl and 0.02 m CaCl2 pH 7.4) and centrifuged at 300 g for 5 min. Washed red blood cells were resuspended in hemolysis buffer to a final concentration of 2% red blood cells. Overnight bacterial culture (approx. CFU (colony forming units) 2 × 108), 5 ml LB, 37 °C, was centrifuged; and both the bacterial pellet and the bacterial growth supernatant were tested for hemolytic activity. The bacterial pellet was resuspended in 5 ml hemolysis buffer. Two-fold serial dilutions (1:2 to 1:1024) in microtiter plates of either 150 μl of bacterial suspension or 150 μl of bacterial supernatant were performed in phosphate-buffered saline (pH 7.4; Sigma-Aldrich, St. Louis, MO), and finally 150 μl 2% red blood cells suspension was added and incubated for 2 h at 37 °C. After incubation, the plate was centrifuged for 10 min at 700 g, 150 μl of supernatant was transferred to a new microtiter plate, and the OD (optical density) measured at 562 nm. Hemolytic titration assays were performed at least twice with essentially the same results. Hemolysis buffer and phosphate-buffered saline were used as negative controls.
Construction of genetic deletion mutants
Isogenic mutants of the E. coli clinical isolate p19A were constructed by allelic exchange with antibiotic resistance encoding cassettes using the λ-Red recombinase method as previously described.19 All primers used are shown in Table 2. For deletion of the hly cluster, 289- bp and 422- bp regions flanking the hly gene cluster were amplified by PCR using the primer pairs UphlyC-F/UphlyC-R and DwhlyD/DwhlyD-R and added to a kanamycin cassette. To construct the double hly mutant (ΔhlyI, ΔhlyII), the λ-Red procedure was repeated on the single hly mutant (ΔhlyI) using a tetracycline resistance encoding cassette PCR amplified by primers 379 and 380 containing 50-bp overhangs homologous to up- and downstream regions, respectively, of the hly gene cluster. The cnf1 cluster was deleted using a tetracycline resistance encoding cassette PCR amplified by the use of primers Upcnf-F and Dwcnf-R containing 50-bp overhangs homologous to up- and downstream regions the cnf1 gene, respectively. Allelic replacement was mediated via the thermo-sensitive helper plasmid pKOBEGApra, encoding λ-Red recombinase functions. Allelic replacements were verified by PCR.
Table 2. Primers used for construction of mutants.
| Primer name | Sequence 5′ to 3′ |
|---|---|
| UphlyC-F | CGGGCTAACCAATATGCT |
| UphlyC-R | GAAGCAGCTCCAGCCTACACCCTCCGTGAAATTCTGATACT |
| DwhlyD-F | GGACCATGGCTAATTCCCATAAGAAAAGAGCAGAGCGA |
| DwhlyD-R | GTAACAACCCCACCTTCA |
| 379 | CACCACGAGTTAATAACTGAAGTAAAAAACAAGACAGATTT- |
| CAATTTTTCATTAACAGGCAAGAATTGCCGGCGGAT | |
| 380 | CTGTTAGTCTGACTGTAACTGATATAAGTAACTGTATAAACTT- |
| TCTGGTTCGGTATTTCACACCGCATAGC | |
| Upcnf-F | GATTAGGTATTCTGATAAGGTGTAGTAAAATATTAATCTTCACA- |
| GAGGAGCAAGAATTGCCGGCGGAT | |
| Upcnf-R | GCGCTAACAAAACAGCACAAGGGTAACTTATAACAATGGCCAAT- |
| AAATAATTTCCCGGGTATTTCACACCGCATAGCAG | |
| hlyA-F | ACCTTGTCAGGACGGCAGAT |
| hlyA-R | CCGTGCCATTCTTTTCATCA |
| RrpoA1 | TTGATATCGAGCAAGTGAGTTCG |
| RrpoA2 | GCATCGATGAGAGCAGAATACG |
Quantification of hemolysin expression
Total RNA was phenol/chloroform extracted from LB growing cultures at OD600 0.8 (approx. 5 × 106 CFU/ml) followed by DNase I digestion (# EN0525; Thermoscientific, Herlev, Denmark). The RNA was then purified using Qiagen column (cat. no. 74104) treated with a dsDNase (# EN0771; Thermoscientific) and directly used for cDNA preparation using a First Strand cDNA synthesis kit (# K0702; Thermoscientific). For the amplification of hlyA, primers hlyA forward and hlyA reverse were used (Table 2).20 The gene RpoA was used as a housekeeping/reference gene and amplified by primer pair RrpoA1 RrpoA2 (Table 2).21 The quantitative-PCR assay was performed using Takara SYBR Premix Ex Taq II (RR820A, Mountain View, CA) in a BioRAD CFX96 (Hercules, CA). The PCR was performed using the manufacturer's recommendations: preheating at 95 °C for 30 s followed by 40 cycles of 95 °C for 5 s and 60 °C for 30 s for elongation.
Cytotoxicity by neutral red assay
Caco-2 BBE cell line (between passages 60 and 68) was maintained at 37 °C in a humidified 5% CO2/95% O2 atmosphere, in DMEM containing Glutamax, 10% heat-inactivated fetal bovine serum, 1% non-essential amino acids, and 1% l-glutamine for 7 days. After 7 days, the media were removed from the confluent cell layer by aspiration, and the monolayer was washed twice with phosphate-buffered saline. Trypsinated cell suspension was seeded in 24-transwell plates (seeding density of 0.05 × 106/cm2) and incubated overnight before co-incubation with bacteria. Caco-2 cells were infected with an overnight culture of E. coli grown in DMEM at an MOI of 10 and maintained at 37 °C in a humidified 5% CO2/95% O2 atmosphere for 4 h. The monolayer was then washed once with DMEM, and then DMEM containing 50 μg/ml neutral red (N4638, Sigma-Aldrich, Brøndby, Denmark) was added and incubated at 37 °C for 30 min. Hereafter, the cells were washed rapidly with a suspension containing 40% formaldehyde and 10% CaCl2. Neutral red was extracted with 1% acetic acid-50% ethanol and quantified in a spectrometer (OD 450 nm). The amount of extracted neutral red is expressed as a percentage of the amount recovered from uninfected cells.
Statistics
The software “GraphPad Prism 5” was used for statistical analysis. TER and hemolytic titration results were analyzed using the two-way ANOVA test when compared with blank or negative control. Neutral Red test results were analyzed using the one-way ANOVA test.
Results
Hemolytic strains of E. coli isolated from IBD patients with active disease disrupt the epithelial cell barrier integrity tested by TER
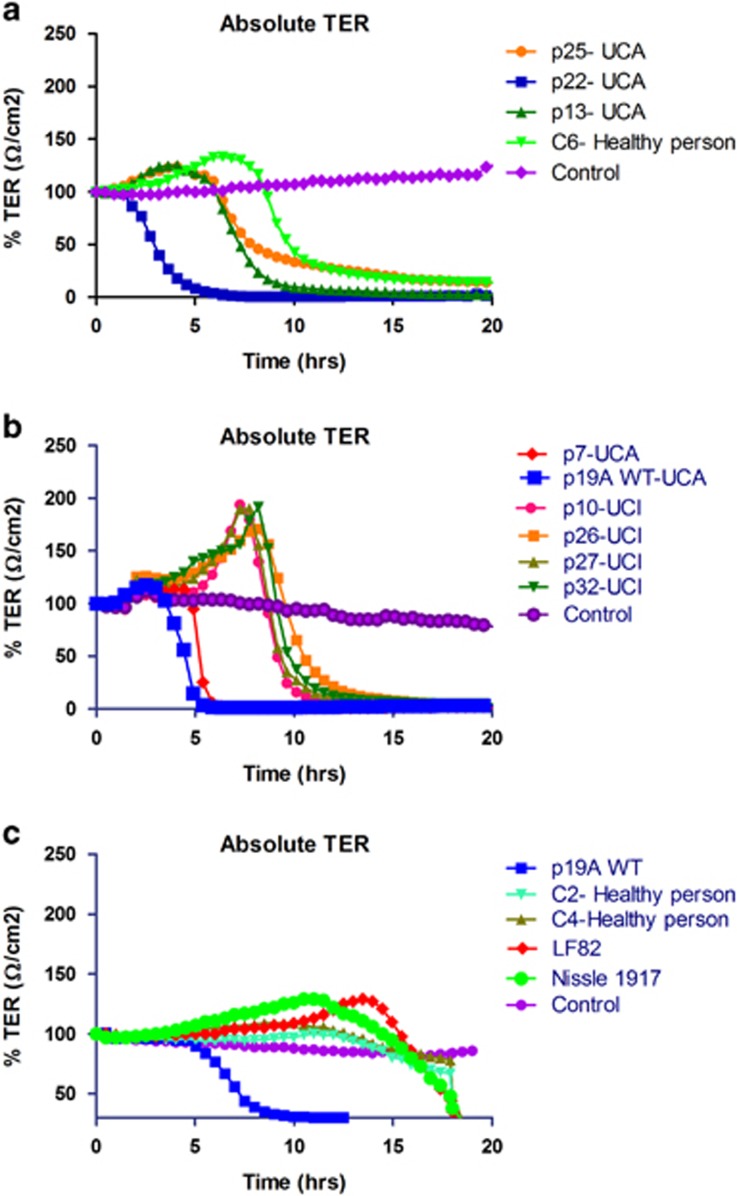
In this study, we investigated the effect of twelve E. coli strains isolated from nine patients with IBD and three control subjects on intestinal epithelial integrity, using TER measurements of Caco-2 cell monolayers grown in Transwells (Table 3). Three of the five phylotype B2 E. coli strains, p7, p19A, and p22, isolated from UC patients with active disease induced a rapid decrease in TER at an MOI of 10, starting after about 2 h and resulting in a complete loss of TER by 6 h (Figures 1a and b). All four E. coli strains isolated from patients with inactive UC or healthy controls decreased the TER after 10–12 h, which was similar for the probiotic E. coli Nissle and the adhesive and invasive E. coli (AIEC) strain LF82 (isolated from an ileal biopsy of a patient with CD11) (Figures 1a–c). The loss of TER after about 10–15 h is due to the growth of E. coli and acidification of the medium. As expected, the TER of untreated Caco-2 cell monolayers was not significantly changed over the 20 h of incubation. Isolates p7, p19A, and p22 were identified as the only α-hemolytic strains among those tested, therefore implicating α-hemolysin in the disruption of TER (Table 3).
Table 3. Origin, molecular, and physiological characteristics of E. coli strains used in this study.
| E. coli strain | Phylogenetic group | Disease association | Hemolytic activity | TER reduction (h) | Two-way ANOVA (TER: 5-12 h) |
|---|---|---|---|---|---|
| p7 | B2 | Active UC | Alfa (<4 h) | 6 | P<0.05*** |
| p13 | B2 | Active UC | None | >10 | ns |
| p19A | B2 | Active UC | Alfa (<4 h) | 6 | P<0.05*** |
| p22 | B2 | Active UC | Alfa (<4 h) | 6 | P<0.05*** |
| p25 | B2 | Active UC | Ent (24 h) | >10 | ns |
| p10 | A | Inactive UC | None | >10 | ns |
| p26 | A | Inactive UC | Ent (24 h) | >10 | ns |
| p27 | A | Inactive UC | Ent (24 h) | >10 | ns |
| p32 | B2 | Inactive UC | None | >10 | ns |
| C2 | A | Healthy | None | >10 | ns |
| C4 | B1 | Healthy | None | >10 | ns |
| C6 | D | Healthy | None | >10 | ns |
| LF82 | B2 | Crohn's disease | None | >10 | ns |
| E. coli Nissle | B2 | Healthy | None | >10 | ns |
ANOVA, analysis of variance; TER, transepithelial electric resistance; UC, ulcerative colitis.
***Statistical significant.
Figure 1.
Effect of E. coli clinical isolates from inflammatory bowel disease (IBD) patients and controls on a monolayer of Caco-2 cells measured by transepithelial electric resistance (TER). (a) E. coli strains p13, p22, and p25 from patients with active ulcerative colitis (UCA) revealed that p22 disrupted the epithelial cell barrier in less than 6 h after co-incubation with Caco-2 cells, while strains p13 and p25, and E. coli C6 from a healthy control did not disrupt the epithelial barrier until after 10 h of co-incubation. (b) E. coli strains p7 and p19A wild type (WT) from patients with UCA disrupted the epithelial cell barrier in less than 6 h after co-incubation with Caco-2 cells. E. coli p10, p26, p27, and p32 from patients with inactive ulcerative colitis (UCI) did not disrupt the epithelial barrier until after 10 h of co-incubation. (c) E. coli strain p19A WT from UCA patient was compared with the adherent invasive Crohn's disease (CD) associated E. coli LF82, the probiotic E. coli Nissle and two E. coli strains, C2 and C4, isolated from healthy controls, it is seen that the probiotic and control isolates did not disrupt epithelial barrier until after 15 h of co-incubation. Simultaneously with all experiments presented (a–c) control TER of the media was performed on Caco-2 cells without addition of bacteria. TER values were normalized to the initial TER value (100%). Absolute TER values are mean of four measurements.
IBD-associated strain p19A contains cnf1 and two hly gene clusters
Among the α-hemolysin-positive strains from patients with active UC, p19A was chosen for further investigation. In a previous study, we have shown that E. coli strain p19A belongs to the phylogenetic group B2, and harbors cnf1 and hly genes.22 To determine the possible role of E. coli hly and cnf1 in barrier disruption, deletion mutants of the individual toxin-encoding genes were constructed. The first hly mutant constructed (ΔhlyI) was still hemolytic, indicating that p19A contained two hly clusters. The presence of two hly clusters has been reported for some ExPEC isolates, and in these strains one of the hly clusters is often located upstream of the cnf1 gene.23 Indeed PCR analysis of strain p19A revealed that the intact hly cluster remaining in the ΔhlyI mutant was located upstream of the cnf1 gene. Thus, mutants of p19A lacking the second hly cluster (ΔhlyII) and also both hly clusters were constructed (ΔhlyI, II).
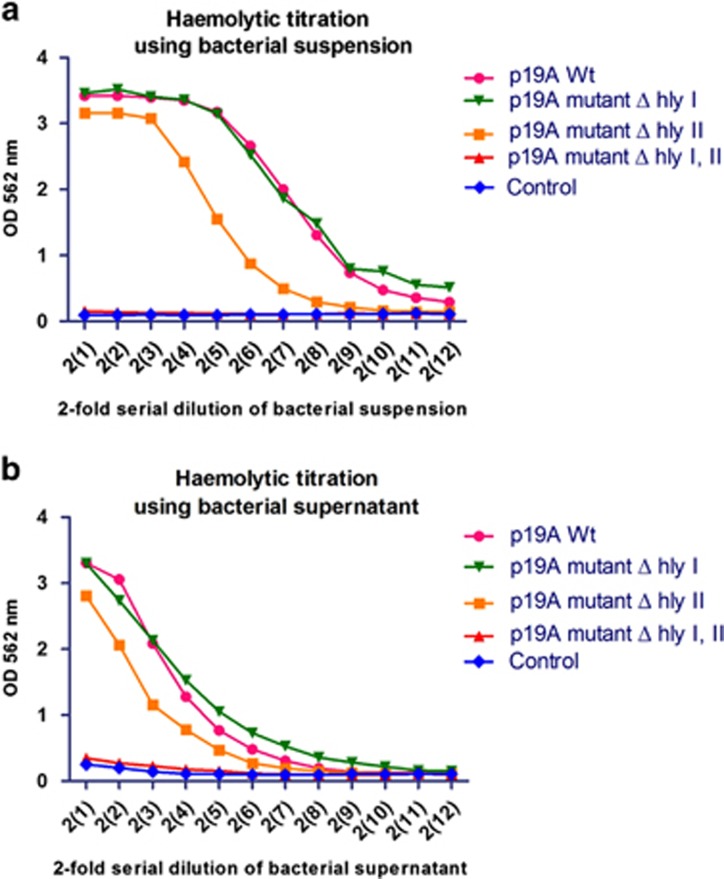
Hemolytic titration assays were performed with bacteria and bacterial culture supernatants in order to investigate the hemolytic activity of clinical isolate E. coli p19A WT and the hly and cnf1 mutants. The hemolytic activity was only completely abolished in the double mutant lacking hly clusters I and II (P<0.05) (Figure 2). Deletion of hly cluster II only partially decreased hemolytic activity, compared with the WT, suggesting that hly cluster I does contribute to the overall hemolytic activity of the WT strain, despite the fact that no reduction in hemolysis was observed in the hly cluster I mutant.
Figure 2.
Hemolysin activity of clinical isolate p19A wild type (WT) from active ulcerative colitis (UCA) patient and its isogenic mutants. (a) Bacterial cell suspension of clinical isolate p19A WT and the two single hemolysin mutants (p19A ΔhlyI and p19A ΔhlyII) showed almost strong hemolytic activity, while in p19A hemolysin double mutant (p19A Δhly I and II) the activity was completely hampered. We used buffer as a control. (b) Bacterial growth supernatant of the above cultures revealed the same hemolytic activity while the double mutant had abolished activity. We used growth medium as a control. OD, optical density.
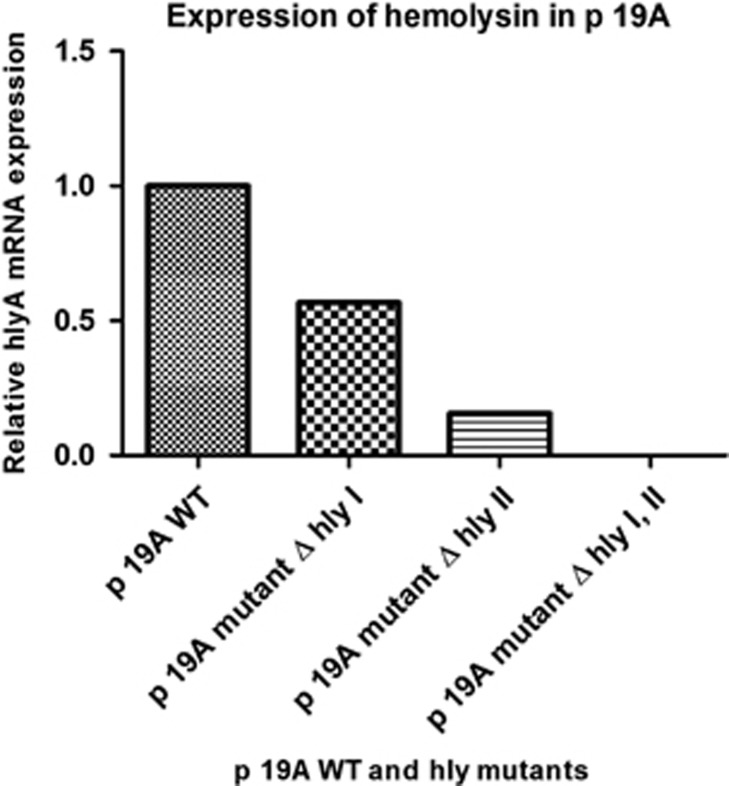
RT-PCR was used to quantify the relative amounts of the hly transcript in E. coli p19A WT and the different hly deletion mutants, using rpoA transcripts as an internal control. The relative expression of hly was twofold higher in the WT p19A than in the ΔhlyI mutant and fourfold higher than in the ΔhlyII mutant. As expected, only the double ΔhlyI, II mutant of p19A lacked hly gene expression (Figure 3).
Figure 3.
Quantification of hemolysin expression in clinical isolate p19A. It is clearly seen that p19A wild type (WT) expresses more hly compared with the single mutants. Expression of hemolysin is completely abolished in the double mutant p19A Δhly I and II. Relative hlyA mRNA was measured by quantitative RT-PCR. The rpoA mRNA level was used as an internal quantitative control.
hly expression in IBD-associated strain p19A causes rapid loss of epithelial integrity
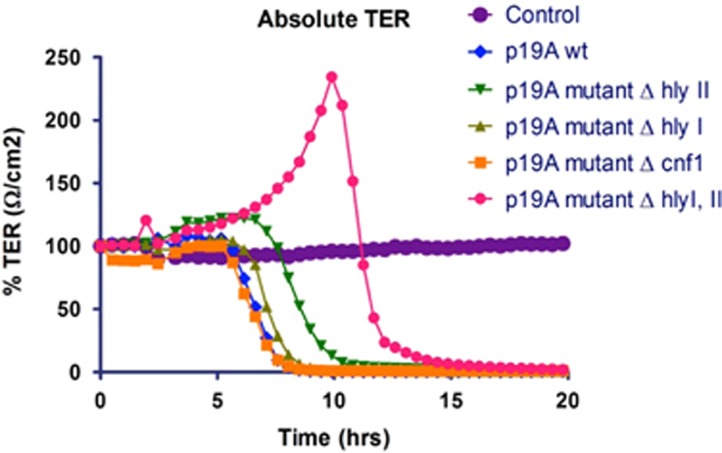
To study the effect of hly and cnf1 expression on the intestinal epithelial barrier integrity, TER measurements were performed with p19A WT and its deletion mutants. At an MOI of 10, the WT, single mutants ΔhlyI, ΔhlyII, and Δcnf1 strains caused loss of TER in Caco-2 cell monolayers in less than 6 h (Figure 4). Deletion of both hly clusters (ΔhlyI, II) in p19A WT abrogated the rapid loss of epithelial integrity, and the effects on TER were comparable to the probiotic Nissle and other E. coli strains not expressing hly (12 h; P<0.0001***).
Figure 4.
Effect of clinical isolate E.coli p19A and its hemolysin deletion mutants on a monolayer of Caco-2 cells measured by transepithelial electric resistance (TER). Wild type (WT) p19A and its single hemolysin- and cnf1-deletion mutants all disrupted the epithelial barrier in less than 6 h, whereas the p19A ΔhlyI, II double deletion mutant did not have any effect on TER. As a control, no bacteria were added to the Caco-2 cells.
hly expression is linked to rapid dissolution of occludin from the tight junctions of epithelial cell monolayers
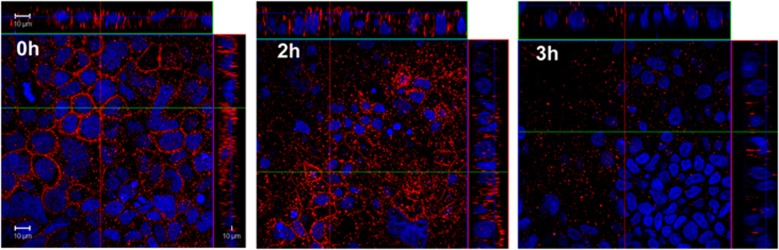
To investigate the effect of hemolytic strains on tight junctions, Caco-2 cell monolayers were incubated with E. coli p19A for <1, 2, or 3 h, and then fixed and stained for occludin and nuclear DNA (Figure 5). No dissolution of occludin from the tight junction was observed with E. coli Nissle or the Hly-negative AIEC strain LF82. A significant reduction in the immunofluorescent staining of occludin was evident, from 2 to 3 h of co-culture with WT p19A (P<0.001) as confirmed by western blotting with antibodies to occludin (Figure 5). Similar results were obtained with all three Hly-producing E. coli strains p7, p19A, or p22, but not with the p19A ΔhlyI, ΔhlyI II double mutant demonstrating a link between Hly expression and loss of tight junction occludin (data not shown).
Figure 5.
Disruption of occluding on Caco-2 cells after incubation with clinical isolate p19A. Confocal images of Caco-2 cell monolayers stained for occludin (red) and nuclei (blue) after apical incubation with E. coli p19A (MOI: 50) after 2 and 3 h (P <0.01 and P<0.001, respectively). MOI, multiplicity of infection.
Effect of p19A WT on epithelial tight junctions disruption and loss of TER is not due to cytotoxicity
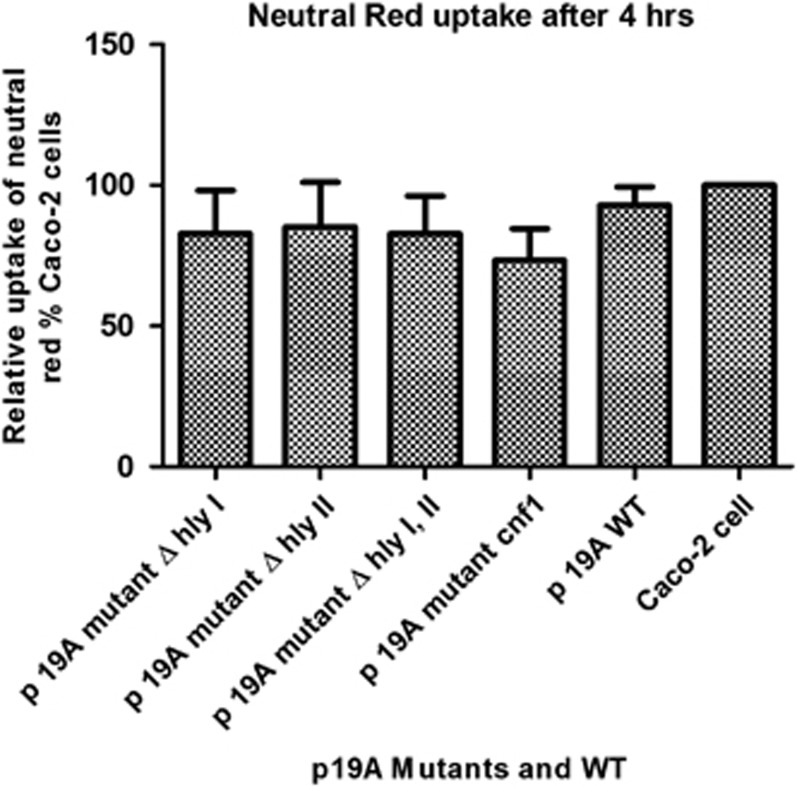
To investigate a possible cytotoxic effect of hly and cnf1 genes on epithelial cells in the above assays, Neutral Red uptake by viable Caco-2 cells was measured after 4 h of co-culture with p19A WT and its mutants at the same MOI. Neutral Red assays were performed six times on 3 different days. No significant differences were found between p19A WT and mutants, which rules out the possibility that cytotoxicity of Hly or Cnf1 causes the rapid loss of occludin and a decrease in TER (Figure 6).
Figure 6.
Cytotoxicity of clinical isolate p19A and its hemolysin mutants on Caco-2 cells. No significant differences in Caco-2 cell viability were found between the p19A wild type (WT) and mutants. Neutral Red uptake in Caco-2 cells was measured after co-incubation with E. coli p19A WT and mutants.
Discussion
The epithelial cell layer is an essential constituent of the gut and a highly specialized interface between the host and its environment. Desmosomes, adherence junctions, and tight junctions hold the cells of the intestinal epithelial layer together. Tight junctions are important in controlling paracellular permeability to ions and small molecules and preventing translocation of luminal antigens and bacteria into the lamina propria.24 In this paper, we demonstrated that IBD-associated E. coli strains from UC patients who produce α-hemolysin cause disruption of epithelial tight junctions of intestinal cell monolayers, leading to the loss of TER. Three of five UC-associated E. coli strains (p7, p19A, and p22) isolated from patients with active UC induced a rapid loss of TER at low MOI without any loss of cell viability.
The IBD-associated strains causing loss of epithelial integrity were all of the phylotype B2 and consistent with previous reports showing an increased abundance of the phylotype B2 E. coli in UC and CD patients with active disease.11, 12, 25, 26 The role of E. coli pathobionts in the pathophysiology in IBD was attributed to their capability to adhere and invade epithelial cells and replicate in macrophages, and the most well-studied prototype strain is LF82. In contrast to p19A, strain LF82 does not cause rapid dissolution of epithelial tight junctions, clearly indicating that the phylotype B2 of UC-associated strains differs markedly in pathogenic mechanisms. The type 1 fimbriae of AIEC were shown to bind to CEACAM6, which is expressed at higher levels in inflamed intestinal epithelial cells of IBD patients.27 Our UC-associated E. coli p19A strain has the same capacity as LF82 to adhere to epithelial cells (data not shown).
All the UC-associated E. coli strains that caused loss of tight junctions in epithelial cell monolayers were hemolytic. Four types of hemolysin have been demonstrated in E. coli: alpha-hemolysin (HlyA), plasmid- and phage-carried enterohemolysin (EhxA and HlyA) and silent hemolysin (SheA); EhxA and HlyA belong to the RTX (repeats in toxin) related family, which lyse erythrocytes from different mammalian species.28, 29, 30 It is known that a number of E. coli pathotypes, i.e., urinary tract pathogenic E. coli, enteropathogenic E. coli, and enterotoxigenic E. coli are all able to produce α-hemolysin.20 The E. coli α-hemolysin is known to be able to lyse erythrocytes through binding to the surface protein glycoporin,31, 32, 33 but also other cell types including leukocytic cells, bladder, and renal tubular cells in a dose-dependent manner.25, 26, 27, 28, 29 Lysis of immune cells is greatly influenced by the presence of cell receptors CD11a and CD18, which are expressed on B and T cells, as well as neutrophils monocytes and dendritic cells.30, 31
The role of HlyA in tight junction disruption was further investigated in E. coli strain p19A, which possessed two hlyA clusters as previously reported for some isolates of uropathogenic E. coli belonging to phylotype B2.34 We showed that both hlyA gene clusters in p19A contributed to the damaging effects on the epithelial integrity, suggesting that intestinal E. coli strains possessing more than one hlyA locus may have increased pathological consequences in intestinal inflammation. Although our UC-associated strains did not induce epithelial cell apoptosis, an hly-expressing uropathogenic E. coli was previously shown to cause localized regions of apoptosis in HT29/B6 cell monolayers. The difference between these findings and our results may be due to the use of a higher MOI than in our study, the use of different strains, or the amount of hlyA expressed.35
Our study showed that around 50% of phylotype B2 E. coli isolated from UC patients can adhere to epithelial cells and disrupt epithelial tight junctions via an HlyA-dependent mechanism, provides strong evidence that this is an important novel pathogenic mechanism in UC; and distinct of AIEC LF82 in CD. Lesions in tight junctions of intestinal epithelium from IBD patients with active disease have been associated with a reduction in several tight junction proteins including claudin 1 and 4, occludin and tricellulin,36 and the synthetic octapeptide (AT1001), which prevents the opening of tight junctions, improves colitis in susceptible IL-10−/− mice.37 Further evidence for the importance of HlyA in the epidemiology of IBD comes from a previous study showing that an HlyA-producing strain of E. coli but not an HlyA-deficient mutant was a potentiator of inflammatory activity in the colon of susceptible IL-10−/− mice and monocolonized germ-free mice due to its effects on the epithelial barrier function.14 During active UC and high inflammation and increased CEACAM6 expression, binding of specific E. coli is facilitated.
This study is a mandate for further investigation of epithelial barrier disruption in other UC cohorts and geographic locations. Preliminary evidence from genomic sequencing suggests that some E. coli strains carry large conjugative plasmids, suggesting that lateral gene transfer of hly loci could contribute to the spread of pathogenic traits.
A recent meta-study including 10 randomized trials from CD patients and 9 randomized trials from UC patients yielded an odds ratio of 2.17 (95% confidence interval, 1.54–3.05) in favor of antibiotic therapy.38 These results suggest that antibiotics improve clinical outcomes in patients with IBD. Another meta-study published in 2011 by Khan et al.39 concluded that antibiotic therapy may induce remission in active CD and UC, although the diverse number of antibiotics tested means the data are difficult to interpret. This systematic review proposed further trials of antibiotic therapy in IBD.
Approaches for combating bacteria that adversely affect the barrier function (e.g., HlyA-expressing E. coli) might provide new treatment options for IBD. This might include antibiotic therapy, vaccination or competition by probiotic bacteria lacking HlyA and other virulence factors that can cause harm to the host.
Study Highlights
Acknowledgments
We thank the Met-Vet-Net Association for a travel grant to H.C.M.-L. We also thank post doc Mette Elena Skindersø, SSI, Nico and Anja Taverne-Thiele, University of Wageningen, for their help with cell assays and for helpful discussions. G.C., Copenhagen University, was supported by the Lundbeck foundation. We also thank the laboratory staff at Zodiac (J. Wells' laboratory) for their help and support during H.C.M.-L. stay in their laboratory. Marian Jørgensen is thanked for proof reading the manuscript.
Guarantor of the article: Karen Angeliki Krogfelt, PhD.
Specific author contributions: Participated in the design of the study: Karen Angeliki Krogfelt, Andreas Munk Petersen, Hengameh Chloé Mirsepasi-Lauridsen, Zhengyu Du, Carsten Struve, Jurgen Karczewski, and Jerry M. Wells; drafted the manuscript: Hengameh Chloé Mirsepasi-Lauridsen, Andreas Munk Petersen, Jerry M. Wells, Carsten Struve, and Karen Angeliki Krogfelt; responsible for the experimental setting: Hengameh Chloé Mirsepasi-Lauridsen, Zhengyu Du, Carsten Struve, Godefroid Charbon, and Jurgen Karczewski. All authors have read and approved the final manuscript.
Financial support: None.
Potential competing interests: None.
References
- Baumgart DC, Sandborn WJ. Inflammatory bowel disease: clinical aspects and established and evolving therapies. Lancet 2007; 369: 1641–1657. [DOI] [PubMed] [Google Scholar]
- Loftus CG, Loftus EV Jr., Harmsen WS et al. Update on the incidence and prevalence of Crohn's disease and ulcerative colitis in Olmsted County, Minnesota, 1940-2000. Inflamm Bowel Dis 2007; 13: 254–261. [DOI] [PubMed] [Google Scholar]
- Molodecky NA, Soon IS, Rabi DM et al. Increasing incidence and prevalence of the inflammatory bowel diseases with time, based on systematic review. Gastroenterology 2012; 142: 46–54.e42; quiz e30. [DOI] [PubMed] [Google Scholar]
- Annese V, Latiano A, Andriulli A. Genetics of inflammatory bowel disease: the beginning of the end or the end of the beginning? Dig Liver Dis 2003; 35: 442–449. [DOI] [PubMed] [Google Scholar]
- Tysk C, Lindberg E, Jarnerot G et al. Ulcerative colitis and Crohn's disease in an unselected population of monozygotic and dizygotic twins. A study of heritability and the influence of smoking. Gut 1988; 29: 990–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manichanh C, Borruel N, Casellas F et al. The gut microbiota in IBD. Nat Rev Gastroenterol Hepatol 2012; 9: 599–608. [DOI] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN et al. Host-drived nitrate boosts growth of E. coli in the inflamed gut. Science 2013; 339: 708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnich N, Carvalho FA, Glasser A-L et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest 2007; 117: 1566–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudeau J, Glasser AL, Masseret E et al. Invasive ability of an Escherichia coli strain isolated from the ileal mucosa of a patient with Crohn's disease. Infect Immun 1999; 67: 4499–4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darfeuille-Michaud A. Adherent-invasive Escherichia coli: a putative new E. coli pathotype associated with Crohn's disease. Int J Med Microbiol 2002; 292: 185–193. [DOI] [PubMed] [Google Scholar]
- Kotlowski R, Bernstein CN, Sepehri S et al. High prevalence of Escherichia coli belonging to the B2+D phylogenetic group in inflammatory bowel disease. Gut 2007; 56: 669–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen AM, Nielsen EM, Litrup E et al. A phylogenetic group of Escherichia coli associated with active left-sided inflammatory bowel disease. BMC Microbiol 2009; 9: 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JL, Fratamico PM, Gunther NW. Extraintestinal pathogenic Escherichia coli. Foodborne Pathog Dis 2007; 4: 134–163. [DOI] [PubMed] [Google Scholar]
- Bücker R, Schulz E, Günzel D et al. α-Haemolysin of Escherichia coli in IBD: a potentiator of inflammatory activity in the colon. Gut 2014; 63: 1893–1901. [DOI] [PubMed] [Google Scholar]
- Langholz E, Munkholm P, Davidsen M et al. Course of ulcerative colitis: analysis of changes in disease activity over years. Gastroenterology 1994; 107: 3–11. [DOI] [PubMed] [Google Scholar]
- Munkholm P, Langholz E, Nielsen OH et al. Incidence and prevalence of Crohn's disease in the county of Copenhagen, 1962-87: a sixfold increase in incidence. Scand J Gastroenterol 1992; 27: 609–614. [DOI] [PubMed] [Google Scholar]
- Miquel S, Peyretaillade E, Claret L et al. Complete genome sequence of Crohn's disease-associated adherent-invasive E. coli strain LF82. PLoS One 2010; 5: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi O, Karczewski J, Stolte EH et al. Vectorial secretion of interleukin-8 mediates autocrine signalling in intestinal epithelial cells via apically located CXCR1. BMC Res Notes 2013; 6: 431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struve C, Bojer M, Krogfelt KA. Identification of a conserved chromosomal region encoding Klebsiella pneumoniae type 1 and type 3 fimbriae and assessment of the role of fimbriae in pathogenicity. Infect Immun 2009; 77: 5016–5024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos Y, Beutin L. Common origin of plasmid encoded alpha-hemolysin genes in Escherichia coli. BMC Microbiol 2010; 10: 110–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimitsu K, Su'etsugu M, Yamaguchi Y et al. Modes of overinitiation, dnaA gene expression, and inhibition of cell division in a novel cold-sensitive hda mutant of Escherichia coli. J Bacteriol 2008; 190: 5368–5381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vejborg RM, Hancock V, Petersen AM et al. Comparative genomics of Escherichia coli isolated from patients with inflammatory bowel disease. BMC Genomics 2011; 12: 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landraud L, Gibert M, Popoff MR et al. Expression of cnf1 by Escherichia coli J96 involves a large upstream DNA region including the hlyCABD operon, and is regulated by the RfaH protein. Mol Microbiol 2003; 47: 1653–1667. [DOI] [PubMed] [Google Scholar]
- Yu Q-H, Yang Q. Diversity of tight junctions (TJs) between gastrointestinal epithelial cells and their function in maintaining the mucosal barrier. Cell Biol Int 2009; 33: 78–82. [DOI] [PubMed] [Google Scholar]
- Martinez-Medina M, Aldeguer X, Lopez-Siles M et al. Molecular diversity of Escherichia coli in the human gut: new ecological evidence supporting the role of adherent-invasive E. coli (AIEC) in Crohn's disease. Inflamm Bowel Dis 2009; 15: 872–882. [DOI] [PubMed] [Google Scholar]
- Conte MP, Schippa S, Zamboni I et al. Gut-associated bacterial microbiota in paediatric patients with inflammatory bowel disease. Gut 2006; 55: 1760–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnich N, Carvalho FA, Glasser A-L et al. CEACAM6 acts as a receptor for adherent-invasive E. coli, supporting ileal mucosa colonization in Crohn disease. J Clin Invest 2007; 117: 1566–1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch RA. Pore-forming cytolysins of gram-negative bacteria. Mol Microbiol 1991; 5: 521–528. [DOI] [PubMed] [Google Scholar]
- Beutin L, Prada J, Zimmermann S et al. Enterohemolysin, a new type of hemolysin produced by some strains of enteropathogenic E. coli (EPEC). Zentralbl Bakteriol Mikrobiol Hyg A 1988; 267: 576–588. [DOI] [PubMed] [Google Scholar]
- Rennie RP, Arbuthnott JP. Partial characterisation of Escherichia coli haemolysin. J Med Microbiol 1974; 7: 179–188. [DOI] [PubMed] [Google Scholar]
- Cortajarena AL, Goñi FM, Ostolaza H. Glycophorin as a receptor for Escherichia coli alpha-hemolysin in erythrocytes. J Biol Chem 2001; 276: 12513–12519. [DOI] [PubMed] [Google Scholar]
- Cavalieri SJ, Bohach GA, Snyder IS. Escherichia coli alpha-hemolysin: characteristics and probable role in pathogenicity. Microbiol Rev 1984; 48: 326–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougayrede JP, Homburg S, Taieb F et al. Escherichia coli induces DNA double-strand breaks in eukaryotic cells. Science 2006; 313: 848–851. [DOI] [PubMed] [Google Scholar]
- Cooke EM, Ewins SP. Properties of strains of Escherichia coli isolated from a variety of sources. J Med Microbiol 1975; 8: 107–111. [DOI] [PubMed] [Google Scholar]
- Troeger H, Richter JF, Beutin L et al. Escherichia coli alpha-haemolysin induces focal leaks in colonic epithelium: a novel mechanism of bacterial translocation. Cell Microbiol 2007; 9: 2530–2540. [DOI] [PubMed] [Google Scholar]
- Hering NA, Fromm M, Schulzke J-D. Determinants of colonic barrier function in inflammatory bowel disease and potential therapeutics. J Physiol 2012; 590: 1035–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrieta MC, Madsen K, Doyle J et al. Reducing small intestinal permeability attenuates colitis in the IL10 gene-deficient mouse. Gut 2009; 58: 41–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SL, Wang ZR, Yang CQ. Meta-analysis of broad-spectrum antibiotic therapy in patients with active inflammatory bowel disease. Exp Ther Med 2012; 4: 1051–1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan KJ, Ullman TA, Ford AC et al. Corrigendum: antibiotic therapy in inflammatory bowel disease: a systematic review and meta-analysis. Am J Gastroenterol 2011; 106: 661–673. [DOI] [PubMed] [Google Scholar]