Abstract
Purpose
This study was designed to assess the protein levels of transformation/transcription domain-associated protein (TRRAP) in invasive ductal breast carcinomas, and investigated the association between TRRAP and the clinicopathological features of breast cancer.
Methods
We examined TRRAP protein expression in 470 breast cancer tissues and normal breast tissues by tissue microarray to study the correlation between TRRAP expression and clinicopathological features. This was analyzed using the chi-square test. Kaplan-Meier survival curves and log-rank tests were applied to analyze the survival status. Cox regression was applied for multivariate analysis of prognosis.
Results
The data demonstrated that expression of TRRAP was significantly lower in breast carcinomas (36.6%) than in corresponding normal breast tissues (50.8%). In addition, TRRAP protein levels negatively correlated with tumor size, and indicated poor differentiation, increased nodal involvement, and low p53-positive rates. Analysis of survival revealed that lower TRRAP expression correlated with shorter survival time. Univariate analyses identified TRRAP and progesterone receptor as independent protective factors for breast cancer prognosis. However, Ki-67, tumor size, and nodal involvement appeared to be independent risk factors.
Conclusion
The findings indicate a significant correlation between TRRAP protein levels and adverse prognosis in breast cancer. Therefore, TRRAP could be a prognostic biomarker for breast cancer. In addition, TRRAP is also a predictive biomarker of breast cancer treatment.
Keywords: Biomarkers, Breast neoplasms, Prognosis, Transformation-transcription domain-associated protein
INTRODUCTION
Breast cancer is the most common malignancy in women worldwide [1]. Although breast cancer therapy has significantly improved during the last decade, the mortality rate associated with this disease remains quite high. Recent advances in epigenetics have demonstrated that histone modifications, especially histone acetylation and deacetylation, may lead to improved treatment strategies in breast cancer. Histone acetyl-ation and deacetylation are predominantly modulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively [2]. HDACs remove acetyl groups from histone proteins, and have been suggested to exert a pro-oncogenic effect, whereas HATs have the opposite effect [3,4,5]. Therefore, cancer progression is widely perceived to be dependent on the balance between HAT and HDAC activity. In this context, the deregulation of HAT has been shown to be associated with cancer development [6]. Accordingly, exploring the underlying mechanisms or factors that can influence histone acetylation may lead to the identification of new and promising targets for breast cancer research.
Transformation/transcription domain-associated protein (TRRAP) is present in the HAT complex and assists with recruitment of the HAT complex to chromatin during gene transcription [7,8]. Studies have shown that TRRAP is not only a common component of HAT complexes in yeast, but also in mammalian cells [9]. Thus, it is reasonable to assume that TRRAP has the ability to modulate the function of the HAT complex.
TRRAP was originally identified as an interacting partner of the c-Myc protein [10]. It has been suggested that TRRAP is an adaptor protein homologous to phosphatidylinositol 3-kinase-related kinases (PIKKs) but without intrinsic kinase activity. It has been shown to contribute to various biological functions such as cell cycle progression, oncogenic transformation via c-Myc and E2F, chromatin remodeling, and embryonic development. TRRAP has also been shown to be an important component of the BRCA1 (breast cancer 1, early onset) genome surveillance repair complex and impairment of BRCA1 function predisposes women to early onset of breast and ovarian cancer [11]. Recent studies have demonstrated that overexpression of TRRAP in gliomas is associated with promotion of stem cell characteristics, and TRRAP is usually elevated in gliomas [12].
Since TRRAP is an important component of HATs, which are important in the development of cancer, and can enhance stem cell-like characteristics as well as regulate BRCA1 gene function, we hypothesized that TRRAP might play an important role in cancer. Therefore, this study investigated the association between TRRAP and breast cancer, and its clinicopathological features.
METHODS
Patients and clinical samples
We obtained 470 patient tissue samples of confirmed histology of breast cancer, and 244 normal tissue samples, from the Department of Pathology at Affiliated Tumor Hospital of Harbin Medical University. Informed consent was obtained from all patients. The Institutional Review Board of Affiliated Tumor Hospital of Harbin Medical University approved this study (KY 2013–29). Tumor and normal breast tissues were examined diagnostically by two pathologists. All patients had invasive breast cancer, and all samples were collected before any radiotherapy or chemotherapy was applied. The tumor size at the largest diameter of the invasive carcinoma was measured in millimeters by the pathologists. Normal breast tissues were collected at least 5 cm away from the tumor tissues and were further confirmed to be cancer free. The tissue sections were prepared from formalin-fixed and paraffin-embedded blocks.
All selected patients had complete medical records from 2006 onwards. Each individual sample was analyzed by immunohistochemistry for estrogen receptor (ER), progesterone receptor (PR), human epidermal growth factor receptor 2 (HER2), Ki-67, and p53 expression. Immunohistochemical staining for ER and PR proteins was performed using a conventional detection method. Greater than 10% nuclear staining in the invasive component of tumor cells represented a positive result [13]. In addition, individual tumor samples with ≥14% Ki-67 positive staining were considered highly proliferative [14].
The intensity of anti-HER2 staining in all samples was semiquantitatively analyzed and graded as 0 to 3. Individual samples with a grade of 0 were considered negative while samples with grade 1, 2, or 3 were regarded as positive.
Examinations of all the patients were performed every 4 to 6 months for the first 5 years and every 12 months thereafter. Patients were followed regularly for a minimum of 5 years or until death or the study closing date (December 30, 2012) at the Affiliated Tumor Hospital of Harbin Medical University. Overall survival was assessed to determine prognosis.
Tissue microarray generation and immunohistochemical staining
Breast cancer and normal tissue microarrays were created by punching a hole in receptive paraffin block using a thin-walled needle with an inner diameter of 2 mm, to acquire tissue cores from the tissue block. After construction of the array block, all tissue blocks were cut with a microtome to a thickness of 4 µm.
The tissue sections were dried at 70℃ for 3 hours. This was followed by deparaffinization and hydration. Subsequently the sections were washed with phosphate-buffered saline (PBS; 3×3 minutes). The washed sections were treated with 3% H2O2 in the dark for 5 to 20 minutes and again washed with distilled water. The sections were further washed with PBS (3×5 minutes) and antigen retrieval was performed in citrate buffer (pH 6.0). This was followed by incubation of each section with 300 to 500 µL TRRAP antibody (1:250; Abnova Inc., Taipei, Taiwan) at 4℃ overnight. The next morning, after washing with PBS (3×5 minutes), each section was further incubated with 300 to 500 µL of secondary antibody (1:200; Abcam, Cambridge, England) at room temperature for 30 minutes. After washing with PBS (3×5 minutes) again, each section was incubated with 300 to 500 µL of 3,3'-diaminobenzidine (DAB) working solution at room temperature for 3 to 10 minutes, and then finally the sections were washed with distilled water.
Expression levels were assessed based on the staining intensity and distribution. The staining intensity was graded as 0, no staining; 1, weak staining, light yellow; 2, moderate staining, yellow brown; or 3, strong staining, brown. The percentage of reactivity was scored as 0 (no positive tumor cells); 1 (fewer than 10% positive tumor cells); 2 (10%–50% positive tumor cells); and 3 (more than 50% positive tumor cells) [15]. Based on these criteria, the overall expression level was scored by multiplying the intensity and reactivity values. Scores <4 reflected low expression, while all scores >4 were indicated high expression.
Inspection of all samples by pathologists suggested that >80% of the cells in each section were cancer cells. Finally, TRRAP protein expression was assessed by evaluating the proportion and intensity of staining in a series of 10 randomly selected high-power fields, which were considered representative of the average in a ×400 magnification field. Two investigators without knowledge of the clinicopathological findings scored the staining pattern of each sample independently.
Statistical analysis
The chi-square test was performed to compare the data from different groups. A two-sided p-value of <0.05 was considered statistically significant. Data were analyzed using the SPSS software version 17.0 (SPSS Inc., Chicago, USA). Overall survival was estimated using the Kaplan-Meier method.
RESULTS
TRRAP expression in breast carcinoma tissues
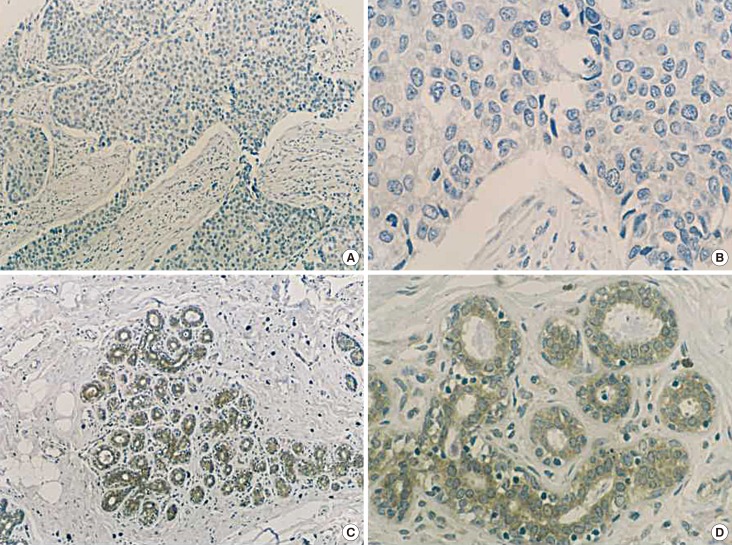
We compared the protein expression of TRRAP in breast carcinomas to that of corresponding normal breast tissues. TRRAP expression levels in invasive ductal breast carcinoma samples (Figure 1A, B) were lower than that in matched normal breast tissue samples (Figure 1C, D). A large number of samples from both categories was quantified for TRRAP expression (Table 1), and the data showed a significantly lower TRRAP expression in invasive ductal breast carcinomas than in normal tissues (p<0.001).
Figure 1. Analysis of transformation/transcription domain-associated protein (TRRAP) expression in invasive ductal carcinoma specimens by immunohistochemical (IHC) staining (IHC for TRRAP, A, ×100; B, ×400). Analysis of TRRAP expression in normal tissues by IHC staining (IHC for TRRAP, C, ×100; D, ×400).
Table 1. Summary of the TRRAP expression analysis in breast carcinomas and normal tissues.
| Histology | TRRAP, No. (%) | Total | p-value | |
|---|---|---|---|---|
| Negative | Positive | |||
| Invasive ductal carcinoma | 298 (63.4) | 172 (36.6) | 470 | <0.001 |
| Normal | 120 (49.2) | 124 (50.8) | 244 | |
TRRAP=transformation/transcription domain-associated protein.
Association between TRRAP expression and clinicopathological features
We analyzed the association between TRRAP expression and a series of clinicopathological characteristics, including patient and tumor characteristics (Table 2). Among the breast cancer patients diagnosed with invasive ductal carcinoma, 298 patients (63.4%) tested negative for TRRAP, whereas 172 (36.6%) tested positive (Table 1). There was significantly greater nodal involvement in patient samples with no TRRAP expression (42.6%) than in those positive for TRRAP (27.3%). The mean tumor size in TRRAP negative samples was 2.83±1.60, which was significantly larger than the size observed in samples positive for TRRAP expression (2.48±1.44).
Table 2. TRRAP expression in breast cancer patients with different clinicopathological features.
| Characteristic | TRRAP, No. (%) | p-value | |
|---|---|---|---|
| Negative | Positive | ||
| Age at diagnosis (yr)* | 49.6±9.7 | 48.7±9.2 | NS |
| Age at menarche (yr)* | 15.4±1.7 | 15.2±1.8 | NS |
| Age at menopause (yr)* | 48.9±4.4 | 49.4±4.0 | NS |
| Primiparity (yr)* | 25.3±3.4 | 25.4±3.1 | NS |
| Breastfeeding (mo)* | 17.0±13.9 | 15.4±11.1 | NS |
| No. of parity* | 1.7±1.1 | 1.5±1.0 | NS |
| No. of abortions* | 0.8±1.0 | 0.7±1.0 | NS |
| Size (cm)* | 2.8±1.6 | 2.5±1.4 | 0.018 |
| Grade | 0.001 | ||
| Well differentiated | 49 (16.4) | 39 (22.7) | |
| Moderately | 133 (44.6) | 95 (55.2) | |
| Poorly | 116 (38.9) | 38 (22.1) | |
| Nodal involvement | 0.020 | ||
| 0 | 127 (43.0) | 85 (49.4) | |
| 1–3 | 45 (14.4) | 40 (23.3) | |
| >3 | 126 (42.6) | 47 (27.3) | |
| ER | NS | ||
| Negative | 167 (56.0) | 84 (48.8) | |
| Positive | 131 (44.0) | 88 (51.2) | |
| PR | NS | ||
| Negative | 129 (43.3) | 63 (36.6) | |
| Positive | 169 (56.7) | 109 (63.4) | |
| HER2 | NS | ||
| Negative | 94 (31.5) | 37 (21.5) | |
| Positive | 204 (68.5) | 135 (79.0) | |
| p53 | 0.001 | ||
| Negative | 66 (22.2) | 19 (11.1) | |
| Positive | 232 (77.9) | 153 (89.0) | |
| Ki-67 (%) | NS | ||
| <14 | 241 (80.9) | 129 (75.0) | |
| ≥14 | 57 (19.1) | 43 (25.0) | |
TRRAP=transformation/transcription domain-associated protein; NS=not significant; ER=estrogen receptor; PR=progesterone receptor; HER2= human epidermal growth factor receptor 2.
*Mean±SD.
An investigation of the association between TRRAP expression and differentiation grades also revealed some significant differences. Tumors that were negative for TRRAP expression were more likely to be less differentiated than those positive for TRRAP. Specifically in tumors with no TRRAP expression, 16.4%, 44.6%, and 38.9% of the tumors were well, moderately, and poorly differentiated, respectively. However, in tumors positive for TRAAP, the corresponding percentages were 22.7%, 55.2%, and 22.1%, respectively.
The rate of positive p53 expression was significantly lower in tumors with negative TRRAP expression (77.9%) than in those with positive TRRAP expression (89.0%) (Table 2). However, levels of ER, PR, HER2, and Ki-67 were similar between tumors regardless of TRRAP expression level.
The correlation between TRRAP expression and disease prognosis
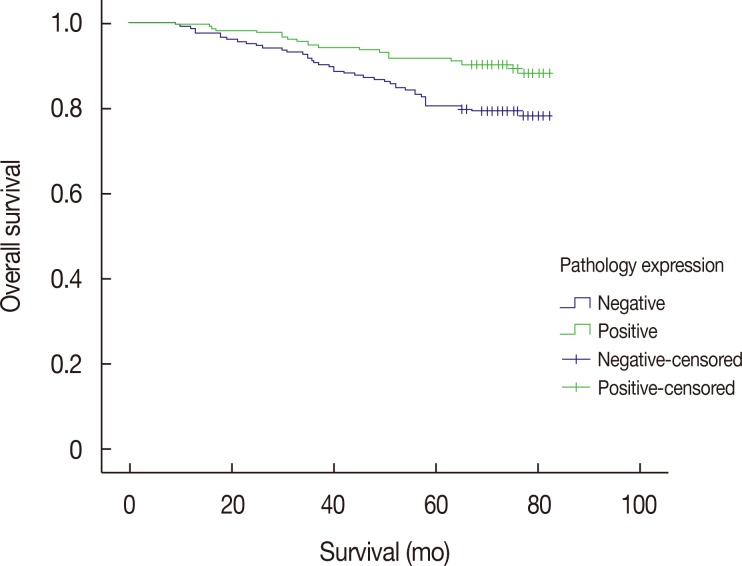
We assessed the potential clinical significance of TRRAP expression in breast cancer prognosis by performing univariate and multivariate analyses (Table 3). Univariate analysis demonstrated that TRRAP expression and PR were independent protective factors for breast cancer prognosis, while Ki-67, tumor size, and nodal involvement were independent risk factors. Similarly, the multivariate Cox regression model also revealed that TRRAP expression, Ki-67, and nodal involvement were independent prognostic markers. Moreover, the Kaplan-Meier survival curve analysis suggested that lack of TRRAP expression was associated with shorter survival times (Figure 2).
Table 3. Univariate and multivariate Cox regression analysis showing hazard ratio to assess the clinical significance of TRRAP and other proteins in breast cancer.
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| HR | 95% CI | p-value | HR | 95% CI | p-value | |
| TRRAP | 0.484 | 0.281–0.833 | 0.009 | 0.509 | 0.291–0.892 | 0.018 |
| ER | 0.777 | 0.485–1.244 | 0.293 | - | - | - |
| PR | 0.597 | 0.376–0.948 | 0.029 | - | - | - |
| HER2 | 0.810 | 0.493–1.329 | 0.404 | - | - | - |
| p53 | 0.682 | 0.396–1.174 | 0.167 | - | - | - |
| Ki-67 | 2.062 | 1.280–3.324 | 0.003 | 1.988 | 1.215–3.253 | 0.006 |
| Tumor size | 2.038 | 1.013–4.102 | 0.046 | - | - | - |
| Lymph node | 4.162 | 2.278–7.603 | < 0.001 | 3.685 | 2.009–6.758 | < 0.001 |
TRRAP=transformation/transcription domain-associated protein; HR=hazard ratio; CI=confidence interval; ER=estrogen receptor; PR=progesterone receptor; HER2=human epidermal growth factor receptor 2.
Figure 2. Kaplan-Meier curve analysis for overall survival based on the transformation/transcription domain-associated protein (TRRAP) expression status in breast cancer patients (p=0.007).
DISCUSSION
Despite improvements in cancer therapeutics, a large number of patients diagnosed with invasive breast carcinomas will eventually die from this disease. Therefore, further identification of effective molecular targets and biomarkers for tumor classification is urgently required. Therefore, in this study, we investigated the role of the TRRAP protein in invasive ductal breast carcinomas, and analyzed its association with different clinicopathological features. Our data revealed that TRRAP expression levels in breast carcinoma tissue were lower than in normal patient-matched tissues. Moreover, in these breast cancer patients, the expression levels of TRRAP were also associated with tumor size, grade, nodal involvement, and p53 expression. There was no significant association between TRRAP expression and specific breast cancer subtypes. In add-ition, we also found that negative TRRAP expression was associated with shorter survival times. Thus, our analysis suggests that TRRAP expression may be negatively involved in breast tumorigenesis.
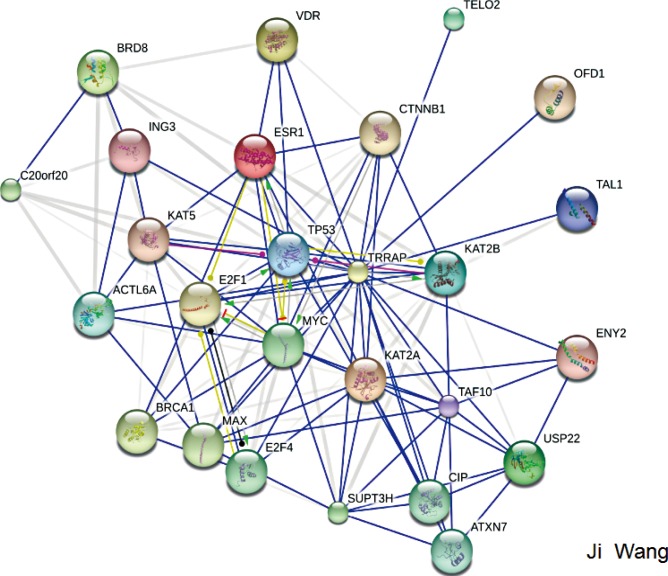
The molecular mechanisms regarding how TRRAP influences the development of breast cancer has been unclear. As shown in Figure 3, we explored the different interaction partners (either direct or indirect) of TRRAP using the Ingenuity Pathway Analysis (IPA) software (QIAGEN, Duesseldorf, Germany). The analysis speculated that TRRAP can interact and may regulate the function of many genes, such as MYC, BRCA1, CTNNB1, KAT2A (K[lysine] acetyl transferase 2A), KAT2B, E2F4, and p53.
Figure 3. Identification of molecular interactions of transformation/transcription domain-associated protein (TRRAP) from Ingenuity Pathway Analysis.
Among these genes, as a confirmation of the IPA analysis, we observed that in our study, p53 expression was significantly lower in the tumors of patients that tested negative for TRRAP expression and vice-versa. The p53 protein has been shown to be an important tumor suppressor gene and acts as a key transcription factor in cellular stress response pathways [16,17]. An earlier study showed that p53 can be acetylated by HATs and deacetylated by HDACs [18]. Enhancement of p53 acetylation leads to protein stabilization and activation [19]. The balance between p53 acetylation and deacetylation is often disrupted in diseases such as cancer [20]. Another study revealed that pharmacologic activation of HATs promotes cancer cell apoptosis through direct hyperacetylation of p53 [21]. In breast cancer cells, acetylation of p53 has been shown to induce cell death [22]. Based on all these published findings, and the present observations, it would be reasonable to deduce that loss of TRRAP expression can promote deacetyl-ation of p53 by inactivating HATs, eventually leading to the development of breast cancer.
However, other TRRAP interacting partners such as KAT2A and KAT2B are histone acetyl transferases and primarily function as transcriptional activators [23]. E2F2 and E2F4 belong to the E2F family and are involved in control of the cell cycle and act as tumor suppressor proteins [24]. BRCA1 impairment predisposes the onset of ovarian and breast cancer, and has been shown to interact with TRRAP, which might be involved in BRCA1 gene regulation [11]. Thus, these interacting partners further support the hypothesis that TRRAP is a tumor suppressor in cancer. However, additional studies are warranted to explore the specific function of TRRAP in the development of breast cancer.
The breast cancer field currently analyzes everything through a lens of specific molecular subtypes, especially in the clinic. The different molecular subtypes are based on the expression levels of ER, PR, and HER2 protein markers. Breast cancer that is negative for all three markers (i.e., triple-negative breast cancer) has attracted considerable attention due to its aggressive biological behavior and poor clinical outcome [25]. In this context, we explored the possibility of an association between TRRAP protein expression and the different breast cancer subtypes and found no significant association.
Finally, we also explored the association between TRRAP and various other clinical outcomes. The Kaplan-Meier survival curve data showed that positive TRRAP expression was associated with improved survival. This observation suggests that TRRAP may suppress or inhibit the proliferation of breast cancer cells. After adjusting for the influence of other confounders, multivariate Cox proportional hazard model analysis showed that positive TRRAP expression could reduce the risk of death by 49.1% as opposed to negative TRRAP expression. This information further confirmed that lack of TRRAP expression was associated with poor breast cancer prognosis.
In conclusion, our results indicate that the presence of TRRAP protein negatively correlates with breast carcinoma progression and thus seems to act as a tumor suppressor. To the best of our knowledge, this is the first detailed study that simultaneously analyzed TRRAP expression in a large number of normal and breast cancer tissues, and investigated a correlation to clinical outcomes, such as overall survival in breast cancer patients. Further understanding of the functional and molecular properties of TRRAP will aid in providing greater insight into its role in breast tumorigenesis.
Footnotes
CONFLICT OF INTEREST: The authors declare that they have no competing interests.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Hebbes TR, Thorne AW, Crane-Robinson C. A direct link between core histone acetylation and transcriptionally active chromatin. EMBO J. 1988;7:1395–1402. doi: 10.1002/j.1460-2075.1988.tb02956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bannister AJ, Kouzarides T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–395. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lane AA, Chabner BA. Histone deacetylase inhibitors in cancer therapy. J Clin Oncol. 2009;27:5459–5468. doi: 10.1200/JCO.2009.22.1291. [DOI] [PubMed] [Google Scholar]
- 5.Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26:5420–5432. doi: 10.1038/sj.onc.1210610. [DOI] [PubMed] [Google Scholar]
- 6.Emanuele S, Lauricella M, Tesoriere G. Histone deacetylase inhibitors: apoptotic effects and clinical implications (Review) Int J Oncol. 2008;33:637–646. [PubMed] [Google Scholar]
- 7.Li H, Cuenin C, Murr R, Wang ZQ, Herceg Z. HAT cofactor Trrap regulates the mitotic checkpoint by modulation of Mad1 and Mad2 expression. EMBO J. 2004;23:4824–4834. doi: 10.1038/sj.emboj.7600479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, et al. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 9.Murr R, Vaissière T, Sawan C, Shukla V, Herceg Z. Orchestration of chromatin-based processes: mind the TRRAP. Oncogene. 2007;26:5358–5372. doi: 10.1038/sj.onc.1210605. [DOI] [PubMed] [Google Scholar]
- 10.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD. The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell. 1998;94:363–374. doi: 10.1016/s0092-8674(00)81479-8. [DOI] [PubMed] [Google Scholar]
- 11.Oishi H, Kitagawa H, Wada O, Takezawa S, Tora L, Kouzu-Fujita M, et al. An hGCN5/TRRAP histone acetyltransferase complex co-activates BRCA1 transactivation function through histone modification. J Biol Chem. 2006;281:20–26. doi: 10.1074/jbc.M510157200. [DOI] [PubMed] [Google Scholar]
- 12.Charles NA, Holland EC. TRRAP and the maintenance of stemness in gliomas. Cell Stem Cell. 2010;6:6–7. doi: 10.1016/j.stem.2009.12.005. [DOI] [PubMed] [Google Scholar]
- 13.Matkovic B, Juretic A, Separovic V, Novosel I, Separovic R, Gamulin M, et al. Immunohistochemical analysis of ER, PR, HER-2, CK 5/6, p63 and EGFR antigen expression in medullary breast cancer. Tumori. 2008;94:838–844. doi: 10.1177/030089160809400611. [DOI] [PubMed] [Google Scholar]
- 14.Cheang MC, Chia SK, Voduc D, Gao D, Leung S, Snider J, et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J Natl Cancer Inst. 2009;101:736–750. doi: 10.1093/jnci/djp082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remmele W, Stegner HE. Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe. 1987;8:138–140. [PubMed] [Google Scholar]
- 16.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609–622. doi: 10.1016/j.cell.2009.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749–758. doi: 10.1038/nrc2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brooks CL, Gu W. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell. 2011;2:456–462. doi: 10.1007/s13238-011-1063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li AG, Piluso LG, Cai X, Gadd BJ, Ladurner AG, Liu X. An acetylation switch in p53 mediates holo-TFIID recruitment. Mol Cell. 2007;28:408–421. doi: 10.1016/j.molcel.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Zhang H, Luo M, Jin Z, Wang D, Sun M, Zhao X, et al. Expression and clinicopathological significance of FSIP1 in breast cancer. Oncotarget. 2015;6:10658–10666. doi: 10.18632/oncotarget.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mantelingu K, Kishore AH, Balasubramanyam K, Kumar GV, Altaf M, Swamy SN, et al. Activation of p300 histone acetyltransferase by small molecules altering enzyme structure: probed by surface-enhanced Raman spectroscopy. J Phys Chem B. 2007;111:4527–4534. doi: 10.1021/jp067655s. [DOI] [PubMed] [Google Scholar]
- 22.Dastjerdi MN, Salahshoor MR, Mardani M, Hashemibeni B, Roshankhah S. The effect of CTB on P53 protein acetylation and consequence apoptosis on MCF-7 and MRC-5 cell lines. Adv Biomed Res. 2013;2:24. doi: 10.4103/2277-9175.108005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sterner DE, Berger SL. Acetylation of histones and transcription-related factors. Microbiol Mol Biol Rev. 2000;64:435–459. doi: 10.1128/mmbr.64.2.435-459.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khaleel SS, Andrews EH, Ung M, DiRenzo J, Cheng C. E2F4 regulatory program predicts patient survival prognosis in breast cancer. Breast Cancer Res. 2014;16:486. doi: 10.1186/s13058-014-0486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reis-Filho JS, Tutt AN. Triple negative tumours: a critical review. Histopathology. 2008;52:108–118. doi: 10.1111/j.1365-2559.2007.02889.x. [DOI] [PubMed] [Google Scholar]