Abstract
Pathway choice is a critical event in the repair of DNA double-strand breaks. In a recent paper published in Nature, Orthwein et al. define a mechanism by which homologous recombination is controlled in G1 cells to favor non-homologous end joining.
Homologous recombination (HR) is an essential process that produces genetic variation during meiosis and protects the genome during mitotic cell division1. Inherited mutations in various HR factors, including the BRCA1, BRCA2 and PALB2 tumor suppressors, predispose to the development of cancer. Although HR is generally beneficial for maintaining genome integrity, HR events between homologous chromosomes can also be deleterious and lead to loss of genetic information. HR is therefore suppressed during G1 phase and in non-dividing cells, yet, the exact mechanism behind this phenomenon has remained elusive. New work from the laboratory of Daniel Durocher describes a mechanism that is both necessary and sufficient for the suppression of HR in G1 cells2.
DNA double-strand breaks (DSBs) are one of the most dangerous types of DNA lesion and need to be eliminated to prevent the accumulation of mutations. DSB repair is carried out by two main pathways, HR and non-homologous end joining (NHEJ)1. Whereas NHEJ is an error-prone process that simply fuses the two broken ends together, HR is essentially error-free as it uses the genetically identical sister chromatid as a template for repair. Due to the cell cycle-dependent availability of sister chromatids, HR is restricted to the S and G2 phases of the cell cycle.
In the HR repair pathway, the DSB ends are first resected to produce extended single-stranded DNA (ssDNA) tails by the coordinated actions of a series of helicase and nuclease activities (e.g., MRN, CtIP and EXO1)1. CtIP plays a particularly important role in regulating resection, which is mediated through its interaction with BRCA13. In the following cascade of events, BRCA1 interacts directly with the BRCA2-PALB2 complex, which in turn is recruited to the ssDNA where it acts as a chaperone that stimulates the formation of RAD51 nucleoprotein filaments that drive homology-directed HR repair to restore the integrity of the DNA4,5.
Whereas most HR events take place between the newly replicated sister chromatids, recombination between homologous chromosomes can result in loss of heterozygosity, a potentially mutagenic event that can lead to the inactivation of tumor suppressors or activation of oncogenes. HR must therefore be tightly regulated and effectively suppressed in G1 phase, at the time when only homologous chromosomes are available for repair. At such times, NHEJ is the favored mechanism for DSB repair.
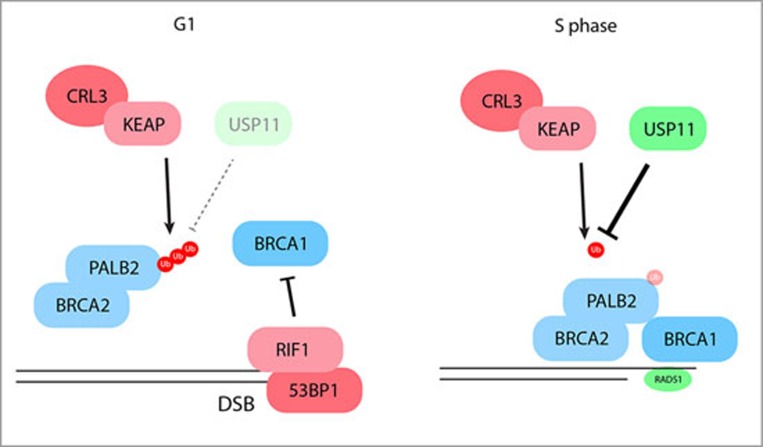
A number of mechanisms regulate HR to a specific phase of the cell cycle. For example, CtIP is activated for interaction with BRCA1 by CDK-dependent phosphorylation, which occurs in the S and G2 phases of the cell cycle. Conversely, HR is suppressed in G1 phase by the pro-NHEJ factors 53BP16, RIF17 and REV78, which impair the recruitment of BRCA1 and thereby inhibit DNA end resection. Consequently, disruption of 53BP1 leads to the recruitment of BRCA1 to DSBs in G1 phase. In the recent Nature paper from Durocher's laboratory, Orthwein et al.2 discovered that although BRCA1 is localized to DSBs during G1 phase in 53BP1-deficient cells, it fails to recruit the BRCA2-PALB2 complex, which is consistent with the lack of HR activity in these cells.
Through immunoprecipitation experiments Orthwein et al. showed that while BRCA2 and PALB2 interact throughout the cell cycle, BRCA1 and PALB2 only interact efficiently in S phase, suggesting that there might be a mechanism that restricts their interaction to S and G2 phases, while also blocking it in G1 phase. The region of PALB2 that is responsible for its cell cycle-regulated interaction with BRCA1 was localized to its N-terminal domain, which corresponds to a known interaction site for KEAP1, a substrate adaptor for the CUL3-RING (CRL3) ubiquitin ligase. Remarkably, they found that deletion of the KEAP1 gene using CRISPR-Cas9 technology restored the BRCA1-PALB2 interaction in G1 cells, and led to the recruitment of BRCA2-PALB2 to sites of DNA damage in 53BP1-deficient G1 cells.
Since KEAP1 is involved in protein ubiquitylation, Orthwein et al. hypothesized that ubiquitylation of PALB2 in the BRCA1-interacting region might block their interaction. Indeed, mutation of lysines in the interacting region of PALB2 restored its interaction with BRCA1 in G1 cells. Furthermore, pull-down experiments showed that ubiquitylation of PALB2 on Lysine-20 by KEAP1-CRL3 prevented its interaction with BRCA1. However, as neither the activity of the KEAP1-CRL3 ubiquitin ligase nor its interaction with BRCA1 is cell cycle regulated, Orthwein et al. reasoned that a deubiquitylation step could be the rate-limiting regulator of the BRCA1-PALB2 interaction. They highlighted the deubiquitylating enzyme USP11 as a potential candidate for this activity due to its interaction with BRCA1, BRCA2 and PALB2, and indeed found that USP11 disruption impaired the interaction between BRCA1 and PALB2. Moreover, they found that USP11 was unstable and interacted poorly with PALB2 in G1 cells, and that USP11 was rapidly lost by proteasomal degradation in G1 phase after DNA damage. By contrast, expression of USP11 in S-phase was high and insensitive to DNA damage. Taken together, these data led the authors to propose that the opposing activities of USP11 and KEAP1-CRL3 regulate cell cycle-dependent interactions between BRCA1 and PALB2 (Figure 1).
Figure 1.
Schematic representation indicating how the opposing activities of USP11 and KEAP1-CRL3 regulate cell cycle-dependent interactions between BRCA1 and PALB2, and thereby mediate pathway choice in DSB repair.
To extend these remarkable observations, Orthwein et al. disrupted this regulatory network to allow HR in G1 cells. They expected that depletion of KEAP1 in 53BP1-deficient cells might be sufficient for RAD51 foci formation following ionizing radiation (IR), but this was not the case because end resection remained a limiting factor. To counteract this, the authors expressed a constitutively active form of CtIP (T847E)9, which augmented resection and led to the efficient formation of IR-induced RAD51 foci in 53BP1- and KEAP1-deficient G1 cells. To address whether these RAD51 foci in G1 cells corresponded to productive HR events, they used a fluorescent-based gene-targeting assay. Whereas CtIP (T847E)expressed in 53BP1-deficient cells alone was insufficient to induce productive HR, depletion of KEAP1 or expression of a non-ubiquitylable version of PALB2 led to a robust increase in gene-targeting events. Collectively, this study therefore demonstrates that activation of DNA end resection, combined with the recruitment of BRCA2 to DSBs, are both necessary and sufficient to produce HR in G1 cells.
Gene targeting has great potential for therapeutic purposes, but the fact that most cells in the body are non-dividing has so far limited its use10. We suspect that the new knowledge highlighted in this work will further improve gene-targeting therapies to help fight human diseases.
References
- Symington LS, Gautier J. Annu Rev Genet 2011; 45:247–271. [DOI] [PubMed]
- Orthwein A, Noordermeer SM, Wilson MD, et al. Nature 2015; 528:422–426. [DOI] [PMC free article] [PubMed]
- Cruz-Garcia A, Lopez-Saavedra A, Huertas P, et al. Cell Rep 2014; 9:451–459. [DOI] [PubMed]
- Shahid T, Soroka J, Kong EH, et al. Nat Struct Mol Biol 2014; 21:962–968. [DOI] [PMC free article] [PubMed]
- Jensen RB, Carreira A, Kowalczykowski SC. Nature 2010; 467:678–683. [DOI] [PMC free article] [PubMed]
- Bunting SF, Callén E, Wong N, et al. Cell 2010; 141:243–254. [DOI] [PMC free article] [PubMed]
- Escribano-Díaz C, Orthwein A, Fradet-Turcotte A, et al. Mol Cell 2013; 49:872–883. [DOI] [PubMed]
- Xu G, Chapman JR, Brandsma I, et al. Nature 2015; 521:541–544. [DOI] [PMC free article] [PubMed]
- Huertas P, Jackson SP. J Biol Chem 2009; 284:9558–9565. [DOI] [PMC free article] [PubMed]
- Cox DB, Platt RJ, Zhang F. Nat Med 2015; 21:121–131. [DOI] [PMC free article] [PubMed]