Abstract
Herpes simplex virus (HSV), a member of the Herpesviridae family, is a significant human pathogen that results in mucocutaneous lesions in the oral cavity or genital infections. Acyclovir (ACV) and related nucleoside analogues can successfully treat HSV infections, but the emergence of drug resistance to ACV has created a barrier for the treatment of HSV infections, especially in immunocompromised patients. There is an urgent need to explore new and effective tactics to circumvent drug resistance to HSV. This review summarises the current strategies in the development of new targets (the DNA helicase/primase (H/P) complex), new types of molecules (nature products) and new antiviral mechanisms (lethal mutagenesis of Janus-type nucleosides) to fight the drug resistance of HSV.
Keywords: new strategies, drug resistance, herpes simplex virus, Janus-type nucleoside analogues, lethal mutagenesis
Herpes simplex virus and drug resistance
Herpes virus detection and risk
Viral diseases are the primary cause of death among human infectious diseases worldwide.1 Herpesviridae is a large family of DNA-containing viruses that result in human infections to varying extents. This family comprises eight members, which can be grouped into three subfamilies (α, β, and γ) based on biological and genomic similarities.2 Human α-herpes viruses include herpes simplex viruses (HSV-1, HSV-2) and varicella zoster virus (VZV).3, 4, 5, 6, 7, 8 HSV infections are among the most common human diseases, and 60%–95% of the population is infected by at least one of these viruses. HSV-1 is frequently associated with oral and perioral infections, and HSV-2 generally causes genital infections. HSV leads to diseases that range from mild conditions to severe infections, such as cold sores, keratitis, corneal blindness, and encephalitis. HSV infections increase the risk for developing human immunodeficiency virus (HIV) infection and contribute to the HIV epidemic. HSV can become latent and subsequently reactivate under certain circumstances, such as emotional stress, fever, and immunosuppression. VZV is the causative agent of chickenpox and shingles. The human β-herpes viruses include cytomegalovirus, HHV-6, and HHV-7.9, 10, 11, 12, 13 The γ-herpes viruses include Epstein–Barr virus (EBV) and Kaposi's sarcoma associated with herpesvirus.14, 15, 16, 17 Acyclovir (ACV) and related drugs for the treatment of HSV infectious diseases are overwhelmingly successful,18, 19 but these anti-herpes drugs have also selected for drug-resistant strains after long-term use.20, 21, 22, 23 Therefore, there is an emergent need to explore new strategies against drug-resistant HSV.
Current drug and antiviral mechanisms
There are three classes of drugs approved for treatment of HSV infections, and all of which target viral DNA replication: acyclic guanosine analogues,24, 25, 26, 27, 28, 29, 30, 31, 32 acyclic nucleotide analogues,33, 34, 35 and pyrophosphate analogues36–37 (Table 1). Typical drugs from these three categories include valacyclovir (VCV), cidofovir, and foscarnet. ACV (9-(2-hydroxyethoxymethyl) guanine) has become a gold standard for prophylaxis and treatment of HSV infections since its introduction in the 1980s. Researchers have developed a series of anti-HSV nucleoside drugs, such as VCV, famciclovir, and ganciclovir, which are also first-line drug treatments for HSV infections.
Table 1. Three classes of drugs approved for the treatment of HSV infections.
| Class of drugs | Licensed drugs |
|---|---|
| Acyclic guanosine analogues | Acyclovir, Ganciclovir, Penciclovir, Valaciclovir, Valganciclovir, Famciclovir |
| Acyclic nucleotide analogues | Cidofovir, Adefovir dipivoxil |
| Pyrophosphate analogues | Foscarnet |
HSV, herpes simplex virus.
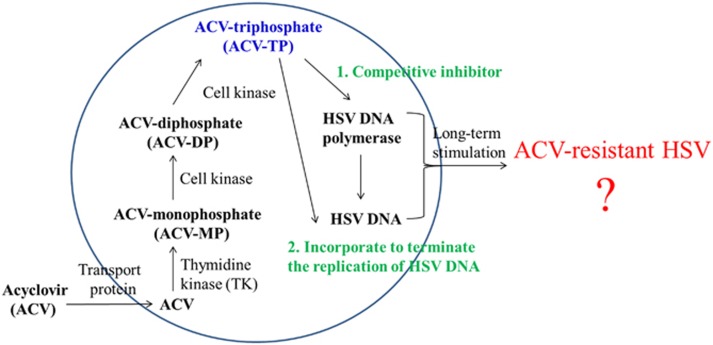
Nucleoside analogues have a similar anti-HSV mechanism. Nucleoside analogues, such as ACV, are selectively phosphorylated to a monophosphate derivative in infected cells by the virus-encoded thymidine kinase (TK). The affinity of ACV for HSV-TK is ~200 times greater than for human TK, and ACV displays remarkable safety against HSV. Cell kinases convert the monophosphate derivative of ACV to diphosphate- and triphosphate (TP)-active derivatives. The ACV-TP form is a competitive inhibitor of the viral DNA polymerase. ACV-TP also incorporates into the replicating DNA because of the absence of 3′ prime hydroxyl, which terminates the replication of viral DNA (refs. 2, 38–39) (Figure 1).
Figure 1.
The anti-HSV mechanism of ACV. ACV, acyclovir; HSV, herpes simplex virus.
ACV-resistant HSV
ACV and its derivatives are available for clinical application, and these agents are widely used for the treatment of HSV infections. However, long-term treatment with ACV and its derivatives may lead to drug resistance.40, 41, 42, 43, 44, 45, 46 A large difference has been observed between immunocompetent and immunocompromised patients. HSV infection in the former patients generally requires short-term anti-HSV therapy, and drug resistance does not easily occur. In contrast, the latter patients generally require long-term anti-HSV therapy, and they are likely to develop drug resistance. A low prevalence (range from 0.1% to 0.6%) of HSV resistance to ACV has been reported in normal immunocompetent patients. However, HSV resistance to ACV is more often isolated in immunocompromised patients and ranges from 3.5% to 10%.22 Some clinical surveys have reported a rate of ACV-resistant HSV isolates of up to 36%.47 For example, the prevalence of resistance to ACV among allogeneic bone marrow transplant patients has been reported to reach 30%.48 Resistant isolates result in severe, debilitating mucosal disease, and visceral dissemination. Therefore, the resistance of HSV to ACV is an important clinical problem for immunocompromised patients. The following resistance mechanisms of HSV to ACV have been reported:2 (a) decreased production of viral TK, (b) complete deficiency in viral TK activity, and (c) viral TK protein and DNA polymerase with altered substrate specify. The viral mutations conferring resistance to ACV are located in activating/phosphorylating genes (TK, UL23 kinase) and the viral DNA pol enzyme (UL30), consistently with the above mechanisms of action.42, 48, 49, 50, 51, 52 The viral mutations in the TK gene generally result in incomplete or deficient enzymes because of the addition or deletion of nucleotides in long homopolymeric runs of Gs and Cs. Approximately 95% of ACV-resistant HSV clinical isolates have a TK-deficient phenotype. The target of anti-HSV drugs is primarily the DNA pol gene of HSV. A single mutation in DNA pol enzyme may confer resistance to many anti-HSV agents.53, 54, 55 For example, most ACV-resistant HSV isolates are also resistant to penciclovir because of a mutation in viral DNA polymerase.
Strategies against drug resistance
Drug-resistant HSV mutants may result in more severe and chronic infections in immunocompromised patients, given the increasing numbers of transplant and cancer patients. Therefore, the emergence of drug-resistant HSV infections is no longer a rare event. Antiviral drugs for the treatment of HSV infections have been developed over the past 40 years. However, most drug-resistant HSV isolates have been discovered in laboratories and clinics, which may contribute to the use of a single target (such as viral DNA polymerase) in all current antiviral drugs. The identification of novel strategies for the development of new antiherpetic molecules with different mechanisms of action that are highly effective and exhibit low toxicity against drug-resistant HSV isolates is challenging. Here, we summarise some of the strategies currently in development:
New target
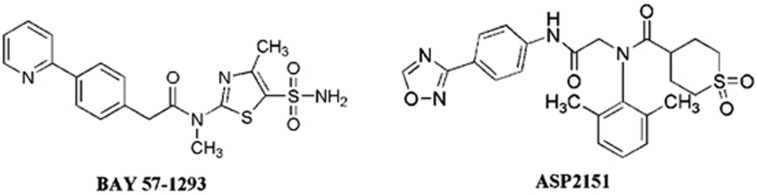
The DNA helicase/primase (H/P) complex is a target for herpes viral infection.56, 57, 58, 59, 60, 61, 62, 63, 64 The viral H/P complex is common to all members of the herpes virus family, and it may be a good target for the development of novel anti-HSV agents. The HSV-1 H/P complex includes three components (UL5, UL52, and UL8) that exhibit 5′–3′ helicase, primase, and single-stranded DNA-dependent NTPase activities, respectively. The new inhibitors of the H/P complex have diverse chemical structures, such as thiazole, thiazoleurea, and thiazolyphenyl derivatives.63 BAY 57-1293 exhibits almost 200 times greater potency against HSV than ACV in vitro65, 66, 67 (Figure 2). ASP2151 has been shown to be a safe and effective treatment for genital HSV in Phase III clinical trials59, 62, 68 (Figure 2). Some promising compounds have been identified, and several of these compounds have undergone clinical trials. However, several problems still exist. For example, the Phase I clinical trial of ASP2151 was terminated because of adverse events.69 Therefore, the development of this new type of drugs will require substantial work in the future.
Figure 2.
The chemical structures of two potent HPI active compounds against HSV. HPI, helicase-primase inhibitor; HSV, herpes simplex virus.
New types of molecules
Natural products are an important source of new molecules for use as anti-HSV agents, such as flavonoids, sugar-containing compounds, and peptides.6, 70, 71, 72 Researchers have recently found that the notoginsenoside ST-4 inhibits the entry of HSV into cells in vitro, with concentrations for 50% of maximal effect (EC50s) of 16.47 μmol·L−1 and 19.44 μmol·L−1 for HSV-1 and HSV-2, respectively.73, 74 Cheng and colleagues75, 76 have found that putranjivain A and pterocarnin A (from Euphorbia jolkini and pterocaryastenoptera, respectively) inhibit the entry and replication of viruses at concentrations of 2–8 μmol·L−1. In the 1990s, Perry et al.77 first reported that mycalamide A display antiviral activities. Mycalamide A has recently been shown to inhibit HSV-1 at 5 ng per disc.78 Traditional Chinese medicine theory (in which one compound may target several proteins, or several compounds may target one protein) has allowed identification of a large number of natural products that inhibit HSV effectively; these discoveries may hopefully solve the present problem of drug resistance. However, the development of natural antiviral drugs faces several challenges, such as the isolation and identification of the active components from complex products, large-scale production, and selective inhibition.
New antiviral mechanism
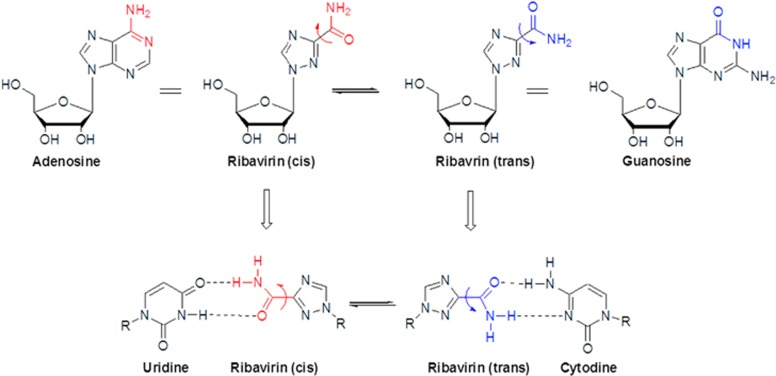
The lethal mutagenesis antiviral mechanism has been proposed as a novel chemotherapeutic strategy for drug resistance.79, 80, 81, 82, 83, 84, 85, 86, 87, 88, 89, 90, 91 Viruses survive on the basis of quasispeciestheory,92, 93 which states that viruses must maintain high levels of potentially beneficial mutations to adapt to new environments quickly via immune responses and antiviral drug therapy. However, the high frequency of mutations in the viral genome also implies a large danger of genetic phenomena. There is an intrinsic limit to the maximum number of mutations in a viral genome before the virus loses its infection activities. The viral genetic information may be lost if the virus quasispecies exceeds the limitation, or it may result in a lethal accumulation of errors (termed lethal mutagenesis). Therefore, lethal mutagenesis may be effective not only in reducing viral infection activity but also in weakening the capacity of the virus for drug resistance. Only one nucleoside analogue, ribavirin, exhibits broad spectrum of antiviral activity against DNA- and RNA-based viruses. Ribavirin is also a classic example that is mutagenic in viral cell cultures. Crotty and colleagues79, 80, 90, 94, 95 have demonstrated that ribavirin may be a template for uridine or cytidine with equal efficiency via rotation around the C3-carbonyl bond to give s-cis and s-trans conformers, which may have pushed the viral genome mutations beyond the error threshold (Figure 3).
Figure 3.
The lethal mutagenesis mechanism of ribavirin. The ribavirin cis conformer can pair with uridine by mimicking adenosine, and the trans conformer can pair with cytidine by mimicking guanosine.
However, the efficiency of ribavirin's incorporation into a viral genome is relatively low. The exploration of new mutagenic molecules to efficiently lead to the mutation of a viral genome is an excellent strategy to develop new antiviral drugs on the basis of lethal mutagenesis. Numerous researchers have focused on advancing the application of nucleoside molecules to induce viral lethal mutations88, 91 (Table 2). For example, 5-aza-5,6-dihydro-2′-deoxycytidine (KP-1212) pairs with different natural purines (guanosine and adenosine) by the diverse tautomerization of the nucleobase (amino and imino).96, 97, 98 KP-1212 inhibits HIV with an EC50 of 10 nmol·L−1, which increases the mutation frequency of proviral HIV-1 DNA by 50%–100% and does not result in resistance or genotoxicity to the host.99 The prodrug of KP-1212, KP-1461, has been used as a monotherapy for the treatment of HIV-1 infection with significant resistance in Phase IIa clinical trials, which have provided critical insight for the translation to clinical use and new avenues for drug development.91, 100
Table 2. Selected nucleoside analogue viral mutagens.
| Chemical name | Mutation |
|---|---|
| 5-aza-5,6-dihydro-2′-deoxycytidine | C/T–U transitions |
| 5-hydroxycytidine | C/T–U transitions |
| 5-aza-2′-deoxycytidine | C/G transversions |
| 2-amino-N6-hydroxyadenosine | A/G transitions |
| 8-oxiguanosine | G/U transversions |
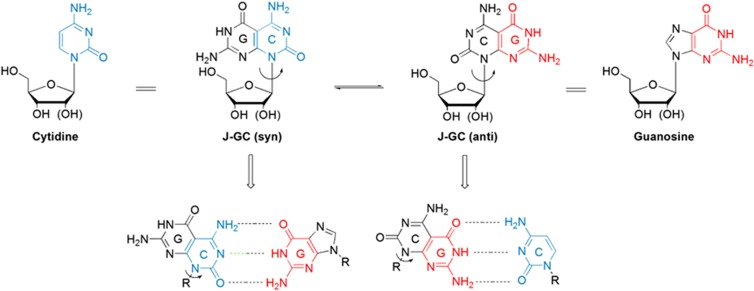
Two different faces/base-pairing systems, a novel Janus-type pyrimido[4,5-d]pyrimidine guanosine–cytosine (J-GC) ribonucleoside and 2′-deoxyribosenucleoside with a tridentate hydrogen bonding pattern, have been designed and synthesised for lethal mutagenesis.101, 102 Cristol first proposed Janus molecules (from the two-faced Roman god Janus) to describe a new symmetrical carbocyclic system. The J-GC mimics natural nucleosides and has the structure of canonical pyrimidine and pure systems in a single molecule (Figure 4). The Watson–Crick base-pairing pattern of J-GC can maintain the tridentate H-bond array. The base moiety of J-GC has one face with a Watson–Crick donor–donor–acceptor H-bond pattern of guanine and the other face with an acceptor–acceptor–donor array of cytosine. The J-GC has two different conformations (syn or anti), which allow pairing with diverse nucleosides in the viral genome via rotation around the glycosyl bond and further induces viral lethal mutations, in a similar manner to ribavirin (Figure 4).
Figure 4.
The potential mutagenic molecule. Janus nucleoside analogues (for example, J-GC) can pair with guanosine and cytidine by rotating around the glycosyl bond. J-GC, Janus-type pyrimido[4,5-d]pyrimidine guanosine–cytosine.
Janus-type pyrimido[4,5-d]pyrimidine adenosine–thymidine (J-AT) nucleosides have been synthesised to expand this tridentate J-GC nucleoside system to a bidentate J-AT nucleoside system and obtain a combination of all four chemical letters of the genetic nucleoside alphabet.103 The base moiety of J-AT has one face with a Watson–Crick H-bond acceptor–donor pattern of thymidine and the other face with a donor–acceptor pattern of adenine. J-AT may be able to pair with diverse nucleosides in the viral genome by rotation around the glycosyl bond. Different mono-substituted nucleosides have been synthesised by replacing one N–H on the thymine ring or the adenine ring with corresponding sugar residues attaching to N1, N3, or N8 of a Janus-type adenosine–thymidine system through divergent synthetic routes, such as Vorbruggen or transglycosylation reactions.104, 105, 106, 107, 108 The preliminary antiviral activity testing has demonstrated that the J-GC ribonucleoside is active against the hepatitis B virus, which supports the application of Janus-type nucleosides in the field of drug-resistant HSV and the great potential for antiviral drug development. These researchers have also found that the Janus-type nucleosides form different morphogenesis nanostructures (flower-like superstructures, nanobundles, and nanoparticles) by self-assembling in solutions and have demonstrated that the novel self-assembled nucleoside nanoparticle can efficiently act as a drug delivery system in the treatment of oral cancer.107, 108 These molecules for the development of this theory for antiviral use are just beginning. However, this subject will likely yield the best advances in strategies against drug resistance.
Conclusion
ACV and related nucleoside analogues have been gold standard molecules for the treatment of HSV infections during the past decades. However, the emergence of ACV drug-resistant HSV is rising rapidly with the increasing numbers of transplant and cancer patients. Therefore, new antiviral drugs with different antiviral actions, including new antiviral targets, new antiviral mechanisms, and new antiviral molecules, are required. Janus-type nucleosides have two different faces (mimicking the natural purine and pyrimidine systems) in one molecule, and these drugs may pair with diverse natural bases via rotation around the glycosyl bond, which further induces viral lethal mutation. Therefore, unique Janus-type nucleoside analogues possess great potential in the exploitation of new lethal mutagenesis drugs as novel strategies for antiviral chemotherapy.
Acknowledgments
We thank the National Natural Science Foundations of China (document no.: 81321002, 81500860, 81300888) and a grant from 111 Project of Ministry of Education, China, for financial support.
References
- Howard CR, Fletcher NF. Emerging virus diseases: can we ever expect the unexpected? Emerg Microbes Infect 2012; 1 (12): e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Bestman-Smith J, Boivin G. Resistance of herpesviruses to antiviral drugs: clinical impacts and molecular mechanisms. Drug Resist Updat 2002; 5 (2): 88–114. [DOI] [PubMed] [Google Scholar]
- Wilson SS, Fakioglu E, Herold BC. Novel approaches in fighting herpes simplex virus infections. Expert Rev Anti Infect Ther 2009; 7 (5): 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markus A, Grigoryan S, Sloutskin A et al. Varicella-zoster virus (VZV) infection of neurons derived from human embryonic stem cells: direct demonstration of axonal infection, transport of VZV, and productive neuronal infection. J Virol 2011; 85 (13): 6220–6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrel S, Ait-Arkoub Z, Voujon D et al. Molecular characterization of herpes simplex virus 2 strains by analysis of microsatellite polymorphism. J Clin Microbiol 2013; 51 (11): 3616–3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galdiero S, Falanga A, Tarallo R et al. Peptide inhibitors against herpes simplex virus infections. J Pept Sci 2013; 19 (3): 148–158. [DOI] [PubMed] [Google Scholar]
- Jones M, Dry IR, Frampton D et al. RNA-seq analysis of host and viral gene expression highlights interaction between varicella zoster virus and keratinocyte differentiation. PLoS Pathog 2014; 10 (1): e1003896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suazo PA, Ibañez FJ, Retamal-Díaz AR et al. Evasion of early antiviral responses by herpes simplex viruses. Mediators Inflamm 2015; 2015: 593757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura M, Takemoto M, Yamanishi K et al. Human herpesvirus 6 major immediate early promoter has strong activity in T cells and is useful for heterologous gene expression. Virol J 2011; 8: 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashimoto Y, Ohta A, Nishiyama Y et al. Development of a human herpesvirus 6 species-specific immunoblotting assay. J Clin Microbiol 2012; 50 (4): 1245–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnepf N, Scieux C, Resche-Riggon M et al. Fully automated quantification of cytomegalovirus (CMV) in whole blood with the new sensitive Abbott RealTime CMV assay in the era of the CMV international standard. J Clin Microbiol 2013; 51 (7): 2096–2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill JA, Boeckh MJ, Sedlak RH et al. Human herpesvirus 6 can be detected in cerebrospinal fluid without associated symptoms after allogeneic hematopoietic cell transplantation. J Clin Virol 2014; 61 (2): 289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aichelburg MC, Weseslindtner L, Mandorfer M et al. Association of CMV-specific T cell-mediated immunity with CMV DNAemia and development of CMV disease in HIV-1-infected individuals. PLoS One 2015; 10 (8): e0137096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza JL, Takami A, Trung LQ et al. Resveratrol prevents EBV transformation and inhibits the outgrowth of EBV-immortalized human B cells. PLoS One 2012; 7 (12): e51306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antsiferova O, Müller A, Rämer PC et al. Adoptive transfer of EBV specific CD8+ T cell clones can transiently control EBV infection in humanized mice. PLoS Pathog 2014; 10 (8): e1004333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HB, Zhang H, Zhang JP et al. Neuropilin 1 is an entry factor that promotes EBV infection of nasopharyngeal epithelial cells. Nat Commun 2015; 6: 6240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Son M, Ryu E et al. Quercetin-induced apoptosis prevents EBV infection. Oncotarget 2015; 6 (14): 12603–12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. Antivirals: past, present and future. Biochem Pharmacol 2013; 85 (6): 727–744. [DOI] [PubMed] [Google Scholar]
- De Clercq E. A 40-year journey in search of selective antiviral chemotherapy. Annu Rev Pharmacol Toxicol 2011; 51: 1–24. [DOI] [PubMed] [Google Scholar]
- Czartoski T, Liu C, Koelle DM et al. Fulminant, acyclovir-resistant, herpes simplex virus type 2 hepatitis in an immunocompetent woman. J Clin Microbiol 2006; 44 (4): 1584–1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnitzler P, Koch C, Reichling J. Susceptibility of drug-resistant clinical herpes simplex virus type 1 strains to essential oils of ginger, thyme, hyssop, and sandalwood. Antimicrob Agents Chemother 2007; 51 (5): 1859–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piret J, Boivin G. Resistance of herpes simplex viruses to nucleoside analogues: mechanisms, prevalence, and management. Antimicrob Agents Chemother 2011; 55 (2): 459–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toriyama K, Inoue T, Suzuki T et al. Necrotizing keratitis caused by acyclovir-resistant herpes simplex virus. Case Rep Ophthalmol 2014; 5 (3): 325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer HJ, Beauchamp L, de Miranda P et al. 9-(2-hydroxyethoxymethyl) guanine activity against viruses of the herpes group. Nature 1978; 272 (5654): 583–585. [DOI] [PubMed] [Google Scholar]
- Sáez-Llorens X, Yogev R, Arguedas A et al. Pharmacokinetics and safety of famciclovir in children with herpes simplex or varicella-zoster virus infection. Antimicrob Agents Chemother 2009; 53 (5): 1912–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogungbenro K, Matthews I, Looby M et al. Population pharmacokinetics and optimal design of paediatric studies for famciclovir. Br J Clin Pharmacol 2009; 68 (4): 546–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattamanchi A, Saracino M, Selke S et al. Treatment with valacyclovir, famciclovir, or antiretrovirals reduces human herpesvirus-8 replication in HIV-1 seropositive men. J Med Virol 2011; 83 (10): 1696–1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janoly-Dumenil A, Rouvet I, Bleyzac N et al. A pharmacodynamic model of ganciclovir antiviral effect and toxicity for lymphoblastoid cells suggests a new dosing regimen to treat cytomegalovirus infection. Antimicrob Agents Chemother 2012; 56 (7): 3732–3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedel AN, Hemmelgarn TS, Kohli R. Retrospective review of the incidence of cytomegalovirus infection and disease after liver transplantation in pediatric patients: comparison of prophylactic oral ganciclovir and oral valganciclovir. Liver Transpl 2012; 18 (3): 347–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake AL, Roxby AC, Ongecha-Owuor F et al. Valacyclovir suppressive therapy reduces plasma and breast milk HIV-1 RNA levels during pregnancy and postpartum: a randomized trial. J Infect Dis 2012; 205 (3): 366–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopal MG, Shannoma, Kumar BCS et al. A comparative study to evaluate the efficacy and safety of acyclovir and famciclovir in the management of herpes zoster. J Clin Diagn Res 2013; 7 (12): 2904–2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perti T, Saracino M, Baeten JM et al. High-dose valacyclovir decreases plasma HIV-1 RNA more than standard-dose acyclovir in persons coinfected with HIV-1 and HSV-2: a randomized crossover trial. J Acquir Immune Defic Syndr 2013; 63 (2): 201–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gokulgandhi MR, Barot M, Bagui M et al. Transporter-targeted lipid prodrugs of cyclic cidofovir: a potential approach for the treatment of cytomegalovirus retinitis. J Pharm Sci 2012; 101 (9): 3249–3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momper JD, Zhao Y, Shapiro R et al. Pharmacokinetics of low-dose cidofovir in kidney transplant recipients with BK virus infection. Transpl Infect Dis 2013; 15 (1): 34–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vossen MG, Gattringer KB, Jäger W et al. Single-dose pharmacokinetics of cidofovir in continuous venovenous hemofiltration. Antimicrob Agents Chemother 2014; 58 (4): 1952–1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Azzi A, Goyette N et al. Recombinant phenotyping of cytomegalovirus UL54 mutations that emerged during cell passages in the presence of either ganciclovir or foscarnet. Antimicrob Agents Chemother 2011; 55 (9): 4019–4027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minces LR, Nguyen MH, Mitsani D et al. Ganciclovir-resistant cytomegalovirus infections among lung transplant recipients are associated with poor outcomes despite treatment with foscarnet-containing regimens. Antimicrob Agents Chemother 2014; 58 (1): 128–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E, Holý A. Acyclic nucleoside phosphonates: a key class of antiviral drugs. Nat Rev Drug Discov 2005; 4 (11): 928–940. [DOI] [PubMed] [Google Scholar]
- De Clercq E. The discovery of antiviral agents: ten different compounds, ten different stories. Med Res Rev 2008; 28 (6): 929–953. [DOI] [PubMed] [Google Scholar]
- Castelo-Soccio L, Bernardin R, Stern J et al. Successful treatment of acyclovir-resistant herpes simplex virus with intralesional cidofovir. Arch Dermatol 2010; 146 (2): 124–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand M, Islam K, Edlund K et al. 2-[4,5-Difluoro-2-(2-fluorobenzoylamino)-benzoylamino]benzoic acid, an antiviral compound with activity against acyclovir-resistant isolates of herpes simplex virus types 1 and 2. Antimicrob Agents Chemother 2012; 56 (11): 5735–5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Coen DM. Quantification and analysis of thymidine kinase expression from acyclovir-resistant G-string insertion and deletion mutants in herpes simplex virus-infected cells. J Virol 2012; 86 (8): 4518–4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner LD, Beckingsale P. Acyclovir-resistant herpetic keratitis in a solid-organ transplant recipient on systemic immunosuppression. Clin Ophthalmol 2013; 7: 229–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakiuchi S, Nonoyama S, Wakamatsu H et al. Neonatal herpes encephalitis caused by a virologically confirmed acyclovir-resistant herpes simplex virus 1 strain. J Clin Microbiol 2013; 51 (1): 356–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D, Kaye SB, Hopkins M et al. Common and new acyclovir resistant herpes simplex virus-1 mutants causing bilateral recurrent herpetic keratitis in an immunocompetent patient. J Infect Dis 2014; 209 (3): 345–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latief MA, Chikama T, Ko JA et al. Inactivation of acyclovir-sensitive and -resistant strains of herpes simplex virus type 1 in vitro by photodynamic antimicrobial chemotherapy. Mol Vis 2015; 21: 532–537. [PMC free article] [PubMed] [Google Scholar]
- Langston AA, Redei I, Caliendo AM et al. Development of drug-resistant herpes simplex virus infection after haploidentical hematopoietic progenitor cell transplantation. Blood 2002; 99 (3): 1085–1088. [DOI] [PubMed] [Google Scholar]
- Morfin F, Thouvenot D. Herpes simplex virus resistance to antiviral drugs. J Clin Virol 2003; 26 (1): 29–37. [DOI] [PubMed] [Google Scholar]
- Watson-Jones D, Wald A, Celum C et al. Use of acyclovir for suppression of human immunodeficiency virus infection is not associated with genotypic evidence of herpes simplex virus type 2 resistance to acyclovir: analysis of specimens from three phase III trials. J Clin Microbiol 2010; 48 (10): 3496–3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei G, Topalis D, Fiten P et al. In vitro-selected drug-resistant varicella-zoster virus mutants in the thymidine kinase and DNA polymerase genes yield novel phenotype-genotype associations and highlight differences between antiherpesvirus drugs. J Virol 2012; 86 (5): 2641–2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaba AH, Cohen LK, Glaubach T et al. Longitudinal characterization of herpes simplex virus (HSV) isolates acquired from different sites in an immune-compromised child: a new HSV thymidine kinase mutation associated with resistance. J Pediatric Infect Dis Soc 2012; 1 (2): 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrel S, Deback C, Agut H et al. Genotypic characterization of UL23 thymidine kinase and UL30 DNA polymerase of clinical isolates of herpes simplex virus: natural polymorphism and mutations associated with resistance to antivirals. Antimicrob Agents Chemother 2010; 54 (11): 4833–4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon TH, Levin MJ, Leary JJ et al. Herpes simplex virus resistance to acyclovir and penciclovir after two decades of antiviral therapy. Clin Microbiol Rev 2003; 16 (1): 114–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarisky RT, Bartus HR, Dennis SA et al. Absence of rapid selection for acyclovir or penciclovir resistance following suboptimal oral prodrug therapy of HSV-infected mice. BMC Infect Dis 2001; 1: 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarisky RT, Quail MR, Clark PE et al. Characterization of herpes simplex viruses selected in culture for resistance to penciclovir or acyclovir. J Virol 2001; 75 (4): 1761–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz UA, Fischer R, Kleymann G et al. Potent in vivo antiviral activity of the herpes simplex virus primase-helicase inhibitor BAY 57-1293. Antimicrob Agents Chemother 2002; 46 (6): 1766–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Liuzzi M, Paris W et al. Oral bioavailability and in vivo efficacy of the helicase-primase inhibitor BILS 45 BS against acyclovir-resistant herpes simplex virus type 1. Antimicrob Agents Chemother 2003; 47 (6): 1798–1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufman HE, Varnell ED, Gebhardt BM et al. Efficacy of a helicase-primase inhibitor in animal models of ocular herpes simplex virus type 1 infection. J Ocul Pharmacol Ther 2008; 24 (1): 34–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata K, Weinberg A, Chono K et al. Susceptibility of herpes simplex virus isolated from genital herpes lesions to ASP2151, a novel helicase-primase inhibitor. Antimicrob Agents Chemother 2012; 56 (7): 3587–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadrick WR, Ndjomou J, Kolli R et al. Discovering new medicines targeting helicases: challenges and recent progress. J Biomol Screen 2013; 18 (7): 761–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohni KN, Smith S, Dee AR et al. Herpes simplex virus type 1 single strand DNA binding protein and helicase/primase complex disable cellular ATR signaling. PLoS Pathog 2013; 9 (10): e1003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsumata K, Chono K, Kato K et al. Pharmacokinetics and pharmacodynamics of ASP2151, a helicase-primase inhibitor, in a murine model of herpes simplex virus infection. Antimicrob Agents Chemother 2013; 57 (3): 1339–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller SK, Kuchta RD. The DNA helicase-primase complex as a target for herpes viral infection. Expert Opin Ther Targets 2013; 17 (10): 1119–1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Sukla S, Goldner T et al. Pharmacokinetics-pharmacodynamics of the helicase-primase inhibitor pritelivir following treatment of wild-type or pritelivir-resistant virus infection in a murine herpes simplex virus 1 infection model. Antimicrob Agents Chemother 2014; 58 (7): 3843–3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleymann G, Fischer R, Betz UA et al. New helicase-primase inhibitors as drug candidates for the treatment of herpes simplex disease. Nat Med 2002; 8 (4): 392–398. [DOI] [PubMed] [Google Scholar]
- Baumeister J, Fischer R, Eckenberg P et al. Superior efficacy of helicase-primase inhibitor BAY 57-1293 for herpes infection and latency in the guinea pig model of human genital herpes disease. Antivir Chem Chemother 2007; 18 (1): 35–48. [DOI] [PubMed] [Google Scholar]
- Biswas S, Jennens L, Field HJ. The helicase primase inhibitor, BAY 57-1293 shows potent therapeutic antiviral activity superior to famciclovir in BALB/c mice infected with herpes simplex virus type 1. Antiviral Res 2007; 75 (1): 30–35. [DOI] [PubMed] [Google Scholar]
- Tyring S, Wald A, Zadeikis N et al. ASP2151 for the treatment of genital herpes: a randomized, double-blind, placebo- and valacyclovir-controlled, dose-finding study. J Infect Dis 2012; 205 (7): 1100–1110. [DOI] [PubMed] [Google Scholar]
- Astellas A. Phase 1, randomized, double-blind, multiple dose, multi-center study to compare the safety of ASP2151 to valacylcovir and placebo in healthy male and female subjects. Available at http://ichgcp.net/clinical-trials-registry/NCT00870441 accessed 21 January 2013.
- Vo TS, Ngo DH, Ta QV et al. Marine organisms as a therapeutic source against herpes simplex virus infection. Eur J Pharm Sci 2011; 44 (1/2): 11–20. [DOI] [PubMed] [Google Scholar]
- Namazi R, Zabihollahi R, Behbahani M et al. Inhibitory activity of avicennia marina, a medicinal plant in persian folk medicine, against HIV and HSV. Iran J Pharm Res 2013; 12 (2): 435–443. [PMC free article] [PubMed] [Google Scholar]
- Zhong MG, Xiang YF, Qiu XX et al. Natural products as a source of anti-herpes simplex virus agents. RSC Adv 2013; 3(2): 313–328. [Google Scholar]
- Vadlapudi AD, Vadlapatla RK, Mitra AK. Update on emerging antivirals for the management of herpes simplex virus infections: a patenting perspective. Recent Pat Antiinfect Drug Discov 2013; 8 (1): 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pei Y, Du Q, Liao PY et al. Notoginsenoside ST-4 inhibits virus penetration of herpes simplex virus in vitro. J Asian Nat Prod Res 2011; 13 (6): 498–504. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Lin TC, Yang CM et al. Mechanism of action of the suppression of herpes simplex virus type 2 replication by pterocarnin A. Microbes Infect 2004; 6 (8): 738–744. [DOI] [PubMed] [Google Scholar]
- Cheng HY, Lin TC, Yang CM et al. Putranjivain A from Euphorbia jolkini inhibits both virus entry and late stage replication of herpes simplex virus type 2 in vitro. J Antimicrob Chemother 2004; 53 (4): 577–583. [DOI] [PubMed] [Google Scholar]
- Perry NB, Blunt JW, Munro MHG et al. Mycalamide A, an antiviral compound from a New Zealand sponge of the genus Mycale. J Am ChemSoc 1988; 110 (14): 4850–4851. [Google Scholar]
- Perry NB, Blunt JW, Munro MHG et al. Antiviral and antitumor agents from a New Zealand sponge, Mycale sp. 2. Structures and solution conformations of mycalamides A and B. J Org Chem 1990; 55 (1): 223–227. [Google Scholar]
- Crotty S, Maag D, Arnold JJ et al. The broad-spectrum antiviral ribonucleoside ribavirin is an RNA virus mutagen. Nat Med 2000; 6 (12): 1375–1379. [DOI] [PubMed] [Google Scholar]
- Crotty S, Cameron CE, Andino R. RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci USA 2001; 98 (12): 6895–6900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P, Shakhnovich EI. Lethal mutagenesis in viruses and bacteria. Genetics 2009; 183 (2): 639–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin G, Gandon S. Lethal mutagenesis and evolutionary epidemiology. Philos Trans R Soc Lond B Biol Sci 2010; 365 (1548): 1953–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox EJ, Loeb LA. Lethal mutagenesis: targeting the mutator phenotype in cancer. Semin Cancer Biol 2010; 20 (5): 353–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno H, Grande-Pérez A, Domingo E et al. Arenaviruses and lethal mutagenesis. Prospects for new ribavirin-based interventions. Viruses 2012; 4 (11): 2786–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dapp MJ, Patterson SE, Mansky LM. Back to the future: revisiting HIV-1 lethal mutagenesis. Trends Microbiol 2013; 21 (2): 56–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull JJ, Joyce P, Gladstone E et al. Empirical complexities in the genetic foundations of lethal mutagenesis. Genetics 2013; 195 (2): 541–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega-Prieto AM, Sheldon J, Grande-Pérez A et al. Extinction of hepatitis C virus by ribavirin in hepatoma cells involves lethal mutagenesis. PLoS One 2013; 8 (8): e71039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnac LF, Mansky LM, Patterson SE. Structure-activity relationships and design of viral mutagens and application to lethal mutagenesis. J Med Chem 2013; 56 (23): 9403–9414. [DOI] [PubMed] [Google Scholar]
- Pauly MD, Lauring AS. Effective lethal mutagenesis of influenza virus by three nucleoside analogs. J Virol 2015; 89 (7): 3584–3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S, Cameron C, Andino R. Ribavirin's antiviral mechanism of action: lethal mutagenesis? J Mol Med 2002; 80 (2): 86–95. [DOI] [PubMed] [Google Scholar]
- Graci JD, Cameron CE. Therapeutically targeting RNA viruses via lethal mutagenesis. Future Virol 2008; 3 (6): 553–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauring AS, Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog 2010; 6 (7): e1001005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E, Sheldon J, Perales C. Viral quasispecies evolution. Microbiol Mol Biol Rev 2012; 76 (2): 159–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas JM, González-Candelas F, Moya A et al. Effect of ribavirin on the mutation rate and spectrum of hepatitis C virus in vivo. J Virol 2009; 83 (11): 5760–5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidwell RW, Huffman JH, Khare GP et al. Broad-spectrum antiviral activity of Virazole: 1-beta-D-ribofuranosyl-1,2,4-triazole-3-carboxamide. Science 1972; 177 (4050): 705–706. [DOI] [PubMed] [Google Scholar]
- Rawson JM, Heineman RH, Beach LB et al. 5,6-Dihydro-5-aza-2'-deoxycytidine potentiates the anti-HIV-1 activity of ribonucleotide reductase inhibitors. Bioorg Med Chem 2013; 21 (22): 7222–7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Fedeles BI, Singh V et al. Tautomerism provides a molecular explanation for the mutagenic properties of the anti-HIV nucleoside 5-aza-5,6-dihydro-2'-deoxycytidine. Proc Natl Acad Sci USA 2014; 111 (32): E3252–E3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng CS, Fedeles BI, Singh V et al. Two-dimensional IR spectroscopy of the anti-HIV agent KP1212 reveals protonated and neutral tautomers that influence pH-dependent mutagenicity. Proc Natl Acad Sci USA 2015; 112 (11): 3229–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KS, Brabant W, Styrchak S et al. KP-1212/1461, a nucleoside designed for the treatment of HIV by viral mutagenesis. Antiviral Res 2005; 67 (1): 1–9. [DOI] [PubMed] [Google Scholar]
- Anon. Nove anti-HIV agent enters Phase IIa clinical trial. Expert Rev Anti Infect Ther 2007; 5 (4): 540–541. [DOI] [PubMed] [Google Scholar]
- Yang HZ, Pan MY, Jiang DW et al. Synthesis of Janus type nucleoside analogues and their preliminary bioactivity. Org Biomol Chem 2011; 9 (5): 1516–1522. [DOI] [PubMed] [Google Scholar]
- Zhao H, Huang W, Wu X et al. Synthesis of a complete Janus-type guanosine-cytosine base and its 2'-deoxyribonucleoside. Chem Lett 2011; 40 (7): 684–686. [Google Scholar]
- Pan MY, Hang W, Zhao XJ et al. Janus-type AT nucleosides: synthesis, solid and solution state structures. Org Biomol Chem 2011; 9 (16): 5692–5702. [DOI] [PubMed] [Google Scholar]
- Zhao H, Huang W, Wu X et al. Different superstructures formed by Janus-type nucleosides. Chem Commun (Camb) 2012; 48 (49): 6097–6099. [DOI] [PubMed] [Google Scholar]
- Zhao H, He S, Yang M et al. Micro-flowers changing to nano-bundle aggregates by translocation of the sugar moiety in Janus TA nucleosides. Chem Commun (Camb) 2013; 49 (36): 3742–3744. [DOI] [PubMed] [Google Scholar]
- Zhao H, Guo X, He S et al. Complex self-assembly of pyrimido[4,5-d]pyrimidine nucleoside supramolecular structures. Nat Commun 2014; 5: 3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Zhao H, Guo X et al. The readout of base-pair information in adenine-thymine α-D-Arabinonucleosides. Chemistry 2014; 20 (47): 15473–15481. [DOI] [PubMed] [Google Scholar]
- Zhao H, Feng H, Liu D et al. Self-assembling monomeric nucleoside molecular nanoparticles loaded with 5-FU enhancing therapeutic efficacy against oral cancer. ACS Nano 2015; 9 (10): 9638–9651. [DOI] [PubMed] [Google Scholar]