Abstract
The successful discovery and subsequent development of small molecule inhibitors of drug targets relies on the establishment of robust, cost-effective, quantitative, and physiologically relevant in vitro assays that can support prolonged screening and optimization campaigns. The current study illustrates the process of developing and validating an enzymatic assay for the discovery of small molecule inhibitors using alkaline phosphatase from bovine intestine as model target. The assay development workflow includes an initial phase of optimization of assay materials, reagents, and conditions, continues with a process of miniaturization and automation, and concludes with validation by quantitative measurement of assay performance and signal variability. The assay is further evaluated for dose–response and mechanism-of-action studies required to support structure–activity-relationship studies. Emphasis is placed on the most critical aspects of assay optimization and other relevant considerations, including the technology, assay materials, buffer constituents, reaction conditions, liquid handling equipment, analytical instrumentation, and quantitative assessments. Examples of bottlenecks encountered during assay development and strategies to address them are provided.
Abbreviations: AP, alkaline phosphatase; CV, coefficient of variation; DEA, diethanolamine; DiFMU, 6,8-difluoro-4-methylumbelliferone; DiFMUP, 6,8-difluoro-4-methylumbelliferyl phosphate; KM, Michaelis constant; pNP, p-nitrophenol; pNPP, p-nitrophenol phosphate; SD, standard deviation; Vmax, maximal reaction velocity; Z′, Z prime
Keywords: Assay development, Small molecule, Screening, Phosphatase, Inhibitor, Mechanism-of-action
1. Introduction
The discovery and development of small molecule modulators with desired pharmacological properties is a funneled process comprising multiple stages including: (i) identification and validation of druggable targets for specific therapeutic areas; (ii) in vitro/in silico screening, identification, and characterization/profiling of small molecules which potently and selectively engage the target of interest, enhancing or inhibiting its molecular function; (iii) toxicology, safety, and efficacy assessments of drug candidates by in vivo pre-clinical and clinical studies. In the early stages of the drug discovery process, the identification and characterization of physiologically relevant small molecule inhibitors markedly relies on the establishment and validation of robust, cost-effective, and scalable cell free and cell based assays that enable to reliably and quantitatively detect and measure variations in the activity of the target of interest or downstream signaling molecules.
The development of such an in vitro assay for screening or profiling of small molecule inhibitors is driven by scientific, technical, and budgetary considerations. Scientific considerations include the selection and optimization of materials and conditions that mimic the physiological condition of the target thus enabling the identification of relevant small molecules with desired mechanisms of action. This process may be guided in part by available literature on the target of interest and developed further by the scientific team. Technical considerations include, on one side, the type of technologies and equipment available to measure the desired enzyme activity or receptor-binding affinity, and, on the other side, the throughput, assay format, reaction scale, signal window, and level of automation that such technologies enable. Budget constraints may impose limitations to the type of materials, technologies, and amount of resources invested. Eventually, the suitability of a given assay procedure for a specific screening program must be evaluated by quantitative methods.
Failure to establish and optimize physiologically relevant assay conditions may lead to an excessive rate of false positives or negatives and identification of chemical entities that are inactive in vivo or have an undesired mechanism of action. Although some general guidelines on assay development [1] or target specific assay procedures [2], [3] can be found in literature, specific examples of assays developed following industry standards with systematic description of the procedures are limited. This study provides a comprehensive description of the development and validation of an enzymatic assay for small molecule screening, emphasizing the most critical parameters, bottlenecks, and the corrective measures to overcome them using alkaline phosphatase from bovine intestine as model target [4], [5], [6].
2. Material and methods
2.1. Material
The following reagents were purchased from Sigma-Aldrich: Trizma base (T1503), Hepes (H4034), MgCl2 hexahydrate (M2670), NaCl (S5886), KCl (P9333), ZnCl2 (208086), Tween 20 (F7949), calf intestine alkaline phosphatase (P7923), sodium orthovanadate (450243), 4-nitrophenol (241326), and 4-nitrophenyl phosphate bis(tris) salt (73737).
The following reagents were purchased from Life Technologies: 6,8-difluoro-4-methylumbelliferyl phosphate (D6567) and 6,8-difluoro-4-methylumbelliferone (6,8-difluoro-7-hydroxy-4-methylcoumarin) (D6566).
For the colorimetric assay, 96-well clear non-treated plates were purchased from Cayman Chemical (400014), and 384-well clear non-binding surface plates were purchased from Corning (3640). For the fluorometric assay, 384-well black non-binding standard plates were purchased from Greiner (781900), and 384-well black non-binding low volume plates were purchased from Corning (3676).
Polypropylene reservoirs (Socorex 330.01) and polypropylene 96-well plates (Corning 3363) were used as source container for fresh working solutions prior to their transfer to the assay plate using multichannel pipettes (Gilson and Finntip). Polypropylene 384-well plates (Corning 3657) were used as source container for automated transfers using Hummingbird Plus liquid handler (Digilab). Polypropylene 50 mL Falcon tubes (BD Biosciences 352070) were used as source container for automated transfers using Multidrop Combi dispenser (Thermo Scientific).
2.2. Reagents
The alkaline phosphatase (AP) stock was stored at 4 °C. AP intermediate dilutions were prepared in 1× assay buffer containing 50% glycerol and stored at 4 °C. Working solutions of p-nitrophenol phosphate (pNPP), 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP), Na3VO4, and AP were prepared fresh in assay buffer or H2O as described in the next section and added to reservoirs or 96-well polypropylene plates prior to transfer to the assay plate using multichannel pipettes.
2.3. Alkaline phosphatase assay
2.3.1. Colorimetric assay
Assay buffer containing TRIS was prepared at 2× final concentration and stored at room temperature. pNPP stock solution was prepared at 100 mM in dH2O and stored at −20 °C. p-Nitrophenol (pNP) stock solution was prepared at 50 mM in dH2O and stored at −20 °C.
The final reaction conditions were 50 mM Tris–HCl pH 7.5, 135 mM NaCl, 7.5 mM KCl, 5 mM MgCl2, 0.1 mM ZnCl2, 0.3 mM Tween 20 or as specified in the text. pNPP and AP concentrations varied as specified in the text. AP was prepared at 2× final concentration in 2× assay buffer, whereas pNPP was prepared at 2× final concentration in dH2O. Reactions were initiated by adding equal volumes of AP and pNPP to the assay plate (50 μL each to 96-well non-treated plates or 25 μL each to 384-well non-binding plates) using a manual multichannel pipette. Plates were spun down and A425 was monitored continuously at room temperature with an Analyst GT microplate reader (Molecular Devices).
2.3.2. Fluorometric assay
Assay buffer containing HEPES was prepared at 1× final concentration and stored at 4 °C. DiFMUP and 6,8-difluoro-4-methylumbelliferone (DiFMU) stock solutions were prepared at 10 mM in DMSO and stored at −20 °C. Na3VO4 stock solution was prepared at 50 mM in H2O and stored at −20 °C.
The final reaction conditions were 50 mM HEPES pH 6.5, 135 mM NaCl, 7.5 mM KCl, 5 mM MgCl2, 0.1 mM ZnCl2, 0.3 mM Tween 20 or as specified in the text. DiFMUP and AP concentrations varied as specified in the text. For reactions without inhibitor in standard volume plates, AP was prepared at 2× final concentration in 1× assay buffer, and DiFMUP was prepared at 2× final concentration in 1× assay buffer. Reactions were initiated by adding 25 μL of 2× AP and 25 μL of 2× DiFMUP to the assay plate using a manual multichannel pipette. For reactions with Na3VO4 in standard volume plates, AP was prepared at 2.5× final concentration in 1× assay buffer, DiFMUP was prepared at 2× final concentration in 1× assay buffer, and Na3VO4 was prepared at 10× final concentration in H2O. Reactions were initiated by adding 20 μL of 2.5× AP, 5 μL of 10× Na3VO4, and 25 μL of 2× DiFMUP to the assay plate using a manual multichannel pipette. For reactions with Na3VO4 in low volume plates, AP or a mixture of AP and Na3VO4 were prepared at 3× final concentration in 1× assay buffer, and DiFMUP was prepared at 1.5× final concentration in 1× assay buffer. Reactions were initiated by adding 5 μL of 3× AP or AP plus Na3VO4, and 10 μL of 1.5× DiFMUP to the assay plate using a Multidrop Combi dispenser (Thermo Scientific). Plates were spun down and incubated at 37 °C. Fluorescence intensity (ex: 358 nm, em: 455 nm) was monitored continuously at 37 °C or at a single end-point as indicated in the text and figure legends with a PHERAstar microplate reader (BMG Labtech).
The concentration of enzyme in each reaction was calculated according to the nominal concentration of the original stock provided by the manufacturer (2000 DEA Units in 15 μL) and expressed as DEA μUnits μL−1. For the validation tests, 0.1 μL of DMSO was transferred to the low volume plates prior to dispensing of the other reagents using Hummingbird Plus liquid handler (Digilab).
For both colorimetric and fluorometric assays, blank reactions contained the same constituents as the test reactions except AP.
2.4. Data analysis
Initial reaction velocities were estimated by converting blank subtracted Absorbance or Fluorescence units from the reaction progress curves into product concentration units using pNP or DiFMU calibration curves, respectively, and calculating the slope of the normalized curves in the initial linearity phase following the equation:
| (1) |
where v0 is the initial reaction velocity (nmols min−1), ΔP is the increment in amount of product produced in the linear phase (nmols), and Δt is the time window of the linear phase (min).
Enzyme kinetic parameters were calculated by plotting initial reaction velocities against substrate concentration and fitting the data points by non-linear regression to the classical Michaelis Menten steady state model (2) or a variant of the model that accounts for positive and negative cooperativity to the substrate binding site (3):
| (2) |
| (3) |
where v0 is the initial reaction velocity (nmols min−1), S is the concentration of substrate (μM), Vmax is the maximal reaction velocity (nmols min−1), KM is the Michaelis constant (μM), and n is the Hill coefficient that quantifies the level of cooperativity.
The Vmax units retrieved from the regression analysis (nmols min−1) were converted into standard Vmax units (μmols min−1 mg−1) by normalizing to the mass of protein (mg) in the reaction. The mass of protein per reaction was estimated from the nominal specific activity (≥4000 DEA Units mg−1) and concentration (2000 DEA Units in 15 μL) provided by the manufacturer and the corresponding dilution factor.
The catalytic constant or turnover number (kcat) was calculated assuming a molecular mass of 70 kDa per subunit of enzyme [5].
Percent enzyme inhibition was calculated by normalizing raw fluorescence intensity signal at 60 min in the presence of inhibitor to the maximum signal in the absence of inhibitor following the equation:
| (4) |
where % inhibition is the percent of enzyme inhibition, RFUI is the signal intensity at 60 min in the presence of inhibitor, and RFUC is the signal intensity at 60 min in the absence of inhibitor.
Half maximal inhibitory concentrations (IC50s) were estimated by plotting percent inhibition as a function of inhibitor concentration and fitting the corresponding concentration–response curves to the following four parameter logistic model:
| (5) |
where % inhibition is the percent of enzyme inhibition, A and B are the minimum and maximum projected percent inhibition of the curve (bottom and top asymptotes), respectively, C is the relative IC50, and D is the Hill coefficient.
The Z prime (Z′) was calculated according to the following equation:
| (6) |
where σmax and σmin are the standard deviations of the maximum and minimum signals, respectively, and μmax and μmin are the means of the maximum and minimum signals, respectively.
The coefficient of variation (CV) was calculated according to the following equation:
| (7) |
where σ and μ are the standard deviation and the mean of a population of values, respectively.
Regression analyses were done using either XLfit (iDBS) or GraphPad Prism (GraphPad Software).
3. Results
3.1. Assay development
3.1.1. Assay development workflow and selection of initial assay conditions
Supplemental Fig. 1 summarizes the assay development and optimization workflow followed in this study. AP from calf intestine was selected as model target to illustrate the assay development, validation and optimization process. The initial substrate chosen for determination of AP activity was p-nitrophenol phosphate (pNPP) which dephosphorylates into p-nitrophenol (pNP), a soluble chromophore with a maximum molar extinction coefficient at 405 nm under alkaline conditions [7]. pNPP is a low cost substrate, commonly available from most vendors. Furthermore, this homogeneous technology enables continuous determination of phosphatase activity in real time. During the initial stages of assay development, buffer composition and assay conditions were determined from the literature (50 mM Tris–HCl pH 8.6, 5 mM MgCl2, 2 mM pNPP) and subsequently optimized. Reactions were initiated by mixing equal volumes (25 μL in 384-well plates or 50 μL in 96-well plates, respectively) of AP in 2× assay buffer and 4 mM pNPP in dH2O. Blank reactions contained the same constituents as the test reactions except AP. The readout of the colorimetric reaction was conducted at room temperature using the Analyst GT microplate reader (Molecular Devices) equipped with a 425/35 nm bandpass filter.
3.1.2. Selection of assay plates
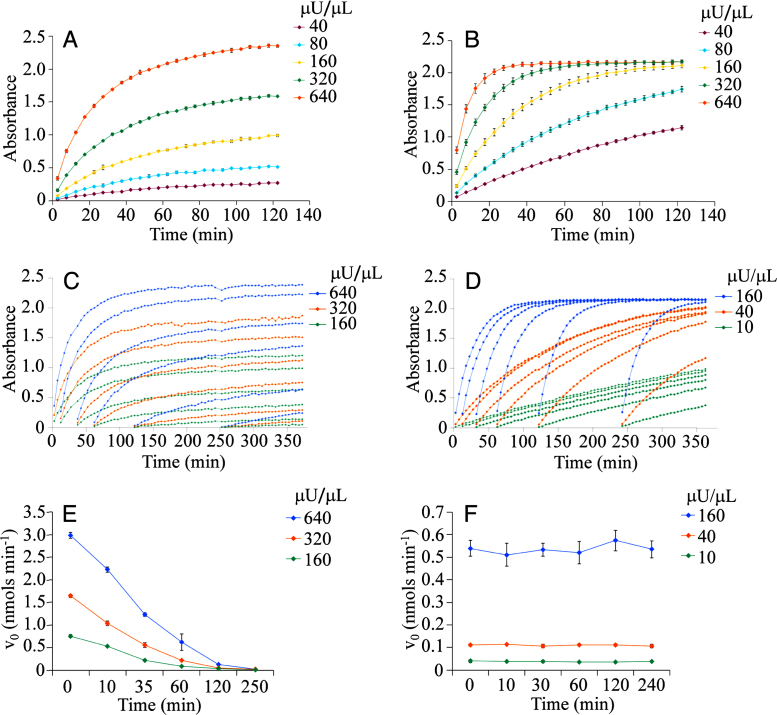
One critical aspect to consider at the initial stages of assay development is the selection of a plate surface with the adequate protein binding capacity. Fig. 1A and B illustrates the effect of the surface binding capacity on AP activity. When a range of enzyme concentrations were assayed in non-binding surface (NBS) plates, the reaction progress curves converged to a common equilibrium value. In contrast, in non-treated (NT) plates the curves diverged, an evidence of enzyme inactivation over the reaction course. The effect of the binding capacity on enzyme stability was verified by pre-incubating the enzyme in NT and NBS plates and initiating the reactions by substrate addition at different pre-incubation times (Fig. 1C–F). As anticipated, initial reaction velocities decayed upon pre-incubation of the enzyme in NT but not in NBS plates, confirming the instability of AP in NT plates. Therefore NBS plates were selected for further optimization of this assay.
Fig. 1.
Effect of protein binding capacity of assay plate on AP stability. Reaction progress curves of the AP colorimetric assay at the indicated concentrations of enzyme (DEA μU/μL) in non-treated (A and C) and non-binding surface (B and D) plates. Reactions were initiated by addition of substrate before (A and B) or after (C and D) pre-incubation of the enzyme in the assay plate. Initial reaction velocities as a function of pre-incubation time in non-treated (E) and non-binding surface (F) plates estimated from panels C and D.
3.1.3. Optimization of assay buffer
A second important aspect that must guide the development of an in vitro assay is the cellular and physiological conditions of the target in vivo (cofactors, salt concentration, pH, and association with lipids or membranes). Despite the animal origin of the target under investigation, the assay buffer composition was altered to mimic more closely the native conditions of the human intestine for illustrative purposes. Sodium and potassium concentrations in the small intestine range from 130 to 140 mM and from 5 to 10 mM, respectively, whereas the pH range varies from 6.6 to 7.5 [8]. Besides, AP is a homodimeric enzyme with each subunit containing two Zn2+ and one Mg2+ ions in the catalytic site [6]. Based on these considerations, the buffer composition was modified to 50 mM Tris–HCl, 135 mM NaCl, 7.5 mM KCl, 5 mM MgCl2, 0.1 mM ZnCl2. The pH was preliminary set to 7.5 and subsequently optimized.
Regarding the cellular environment, intestinal AP is anchored to the outer leaflet of the plasma membrane by a glycosyl-phosphatidylinositol moiety [6]. Therefore, the effect of membrane-mimicking agents on AP activity was tested. Supplemental Fig. 2 illustrates the effect of one zwitterionic (CHAPS) and two non-ionic (Triton X-100 and Tween 20) detergents at two concentrations above their critical micelle concentration (cmc) on the activity of AP. The three detergents increased AP activity by up to 40% irrespective of the concentration tested. Therefore, the assay buffer was supplemented with 0.3 mM Tween 20.
3.1.4. Iterative optimization of substrate type and concentration, pH, and measurement technology
The third critical constituent of the assay reaction mix that requires fine tuning is the concentration of substrate. Typically, the enzymatic screening assays to identify small molecule inhibitors are conducted at a concentration of substrate near the KM to ensure an even representation of small molecule hit compounds with different inhibition modalities. The pH optima for some enzymes, including AP, in turn varies with substrate concentration [9]. Therefore, if information on the physiological pH is not available, both pH and substrate concentration must be optimized simultaneously.
A precise determination of KM requires the calculation of initial reaction velocities at a range of substrate concentrations spanning the KM. Before performing this experiment, the lowest testable concentration of substrate enabled by the available technology needs to be established. Calibration curves of the substrate or product provide an indication of the system detection limit. For the AP colorimetric assay, 16 μM pNPP was estimated as the lowest testable concentration of substrate (data not shown). Subsequently, enzyme concentration was adjusted to the lowest substrate concentration in a titration experiment (Supplemental Fig. 3). An appropriate enzyme concentration must generate sufficient data points in the linear range to enable accurate estimation of initial reaction velocities. Based on these considerations, AP concentration was set to 5 DEA μU/μL for pNPP concentrations equal to or higher than 16 μM.
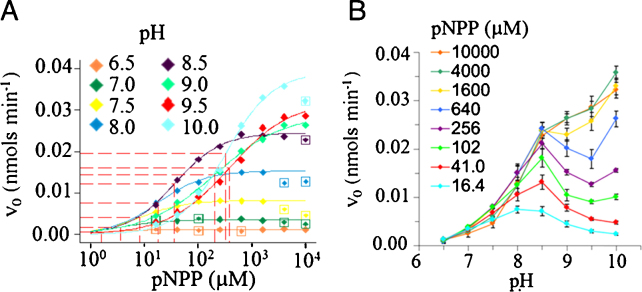
To determine the kinetic parameters of AP at different pH values, reaction progress curves were obtained at a range of substrate concentrations for each pH (Supplemental Fig. 4A–D). The estimated initial velocities were plot against substrate concentration and fit by non-linear regression analysis to the Michaelis Menten model (Fig. 2A). The corresponding Vmax, kcat, and KM estimated from the analysis are shown in Table 1. Moreover, the estimated catalytic efficiency (kcat/KM) of AP for pNPP was maximal at pH 7.5 (Table 1). Likewise, a re-plot of initial velocity versus pH at different substrate concentrations illustrates the shift in pH optima from alkaline at 10 mM pNPP to neutral at 16 μM pNPP as previously reported [9] (Fig. 2B). Therefore, these studies indicated that the optimal substrate concentration for small molecule inhibitor screen at pH 7.5 would be 8 μM pNPP. However, 8 μM pNP fell below the detection limits of the available equipment due to the low extinction coefficient of pNP at neutral pH, precluding further advancement of the development process using the current conditions.
Fig. 2.
Determination of optimal pH and substrate concentration for the hydrolysis of pNPP by AP at room temperature. (A) Variation of initial reaction velocity as a function of pNPP concentration at pH 6.5–10.0. Plots were fit by non-linear regression analysis to the Michaelis-Menten model as described in the text. (B) Dependence of AP pH optima on pNPP concentration. Initial reaction velocities from panel A were re-plot as a function of pH for each substrate concentration tested to show the shift in optimal pH toward neutrality at low pNPP concentrations.
Table 1.
Kinetic parameters for the hydrolysis of pNPP by AP at room temperature and different pHs.
| pH | Vmax (μmol min−1 mg−1)* | kcat (s−1) | KM (μM)* | kcat/KM (M−1 s−1) | Hill coefficient |
|---|---|---|---|---|---|
| 6.5 | 20 ± 2 | 23 | 2 ± 8 | 1.44 × 107 | 1 |
| 7.0 | 58 ± 7 | 68 | 4 ± 12 | 1.84 × 107 | 1 |
| 7.5 | 132 ± 5 | 154 | 8 ± 3 | 1.86 × 107 | 1 |
| 8.0 | 246 ± 12 | 287 | 18 ± 5 | 1.60 × 107 | 1 |
| 8.5 | 392 ± 14 | 458 | 37 ± 7 | 1.24 × 107 | 1 |
| 9.0 | 463 ± 42 | 540 | 206 ± 82 | 0.26 × 107 | 0.68 |
| 9.5 | 515 ± 86 | 601 | 385 ± 250 | 0.16 × 107 | 0.76 |
| 10.0 | 626 ± 40 | 731 | 330 ± 74 | 0.22 × 107 | 1 |
95% confidence intervals are indicated for Vmax and KM.
Two alternative strategies were considered to increase assay sensitivity using the available equipment: (i) alkalinization of the reaction with NaOH at the assay end-point and prior to the readout; (ii) or use of an alternative technology which provided increased signal at neutral pH values. The first option required minor changes to the assay protocol but was less likely to produce a signal increment of the magnitude required to meet subsequent validation tests. In contrast, the second option required major changes to assay materials, analytical instrumentation, and conditions but was more likely to produce the desired response. Consequently, pNPP was replaced by 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP), a fluorogenic substrate that, upon hydrolysis, generates 6,8-difluoro-4-methylumbelliferone (DiFMU), a fluorophore with excitation and emission maxima at 358 and 450 nm, respectively, that enables continuous determination of phosphatase activity at acidic, neutral, and alkaline pH values [10].
The other factor that can have a significant impact on assay performance and validation tests is the instrument used for particular assay readout. Therefore, if multiple microplate readers equipped with appropriate optics to support the technology of interest are available, the selection must be driven by instrument performance. For the fluorometric assay, PHERAstar (BMG Labtech) and EnVision (Perkin Elmer) were evaluated to quantify DiFMU. To choose between them, the settings (light intensity, gain, and focal height) of both readers were optimized with DiFMU and DiFMUP standard solutions and their signal-to-background ratio at the optimized settings were compared (Supplemental Fig. 5). PHERAstar was selected for further assay development due to its higher sensitivity.
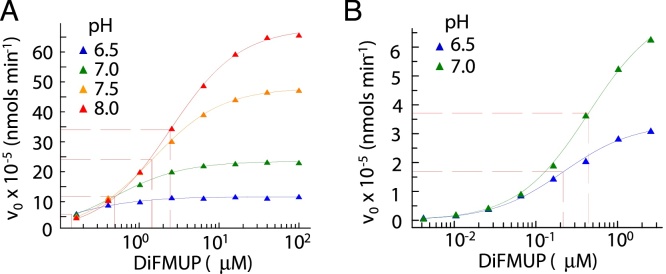
The kinetic parameters and optimal pH of an enzyme vary with the substrate class. Therefore, optimal pH and substrate concentration for DiFMUP were determined as described for pNPP (Fig. 3). As the optimal pH was expected to fall within 6.5–7.5, TRIS was replaced by HEPES, which has a more effective buffering capacity at physiological pH [6.8–8.2]. All other assay buffer components were kept unchanged. Further, the assay temperature was shifted to 37 °C to mimic more closely the physiological conditions of the target. Table 2 displays the estimated kinetic parameters for the hydrolysis of DiFMUP by AP at 37 °C from pH 6.5–8. At 95% confidence interval, the estimated Vmax and KM in the fluorometric assay was tighter than in the colorimetric assay, indicating higher robustness and sensitivity of the fluorescence-based technology. As with pNPP, the catalytic constant (kcat) of AP against DiFMUP increases whereas the substrate affinity (reciprocal of KM) decreases with increasing pH, resulting in a maximum catalytic efficiency (kcat/KM) at pH 6.5. Based on these results, DiFMUP concentration was set to 0.2 μM and pH to 6.5 for screening of inhibitors.
Fig. 3.
Determination of optimal pH and substrate concentration for the hydrolysis of DiFMUP by AP at 37 °C. (A and B) Variation of initial reaction velocity as a function of DiFMUP concentration at pH 6.5–8.0 (A) and pH 6.5–7.0 (B). Plots were fit by non-linear regression analysis to the Michaelis-Menten model as described in the text. Due to the decrease in KM at lower pH values, curves at pH 6.5 and 7.0 were re-evaluated using lower DiFMUP concentrations to accurately calculate the KM at those pH values (B).
Table 2.
Kinetic parameters for the hydrolysis of DiFMUP by AP at 37 °C and different pHs.
| pH | Vmax (μmol min−1 mg−1)* | kcat (s−1) | KM (μM)* | kcat/KM (M−1 s−1) | Hill coefficient |
|---|---|---|---|---|---|
| 6.5 | 32 ± 2 | 37 | 0.20 ± 0.04 | 1.96 × 108 | 1 |
| 7.0 | 71 ± 3 | 82 | 0.50 ± 0.04 | 1.65 × 108 | 1 |
| 7.5 | 126 ± 3 | 147 | 1.4 ± 0.2 | 1.03 × 108 | 1 |
| 8.0 | 181 ± 1 | 211 | 2.8 ± 0.1 | 0.77 × 108 | 1 |
95% confidence intervals are indicated for Vmax and KM.
3.1.5. Optimization of enzyme concentration
Enzyme concentration is the last component of the assay reaction mix that requires fine adjustment upon establishing substrate concentration. Ideally, the amount of enzyme in the assay reaction has to be sufficiently low to ensure: (i) reaction linearity within a time interval adequately long to accommodate the intended plate throughput; (ii) less than 10–15% substrate depletion at the assay end-point to ensure steady state conditions throughout the assay course; (iii) and enzyme concentration significantly lower than the expected Ki or Ki′ of the inhibitors. However, the final enzyme concentration in the reaction has to be sufficiently high to provide acceptable signal window.
AP concentration was optimized for the pre-set assay conditions in a titration experiment. Supplemental Fig. 6 displays the reaction progress curves for the hydrolysis of DiFMUP by AP obtained at a range of enzyme concentrations. As the reading time per plate in the preset PHERAstar settings was 2.5 min, a hypothetical screen with an intended throughput of 20 plates per run would require sustained reaction linearity for at least 50 min after the readout of the first assay plate and no more than 10–15% of substrate depletion upon reading of the last plate. Based on these considerations, final AP concentration was set to 0.0781 DEA μU/μL since it is the highest enzyme concentration that satisfied these criteria with at least 80 min of reaction time. Higher AP concentrations would shorten the linearity interval whereas lower concentrations would compromise signal window.
3.1.6. Determination of DMSO tolerance
Since small molecule library compounds are typically dissolved in DMSO, it is crucial to determine the DMSO tolerance of the in vitro assay, that is, the minimum concentration of DMSO that reduces enzyme activity significantly (>10%). Supplemental Fig. 7 shows that DMSO concentrations up to 1.5% caused less than 10% decrease in reaction velocity. This observation implies that library compound stocks should be prepared at concentrations that permit screening at or below 1.5% DMSO in the final reaction volume.
3.2. Assay validation using known small molecule inhibitors of AP
3.2.1. Reversibility studies
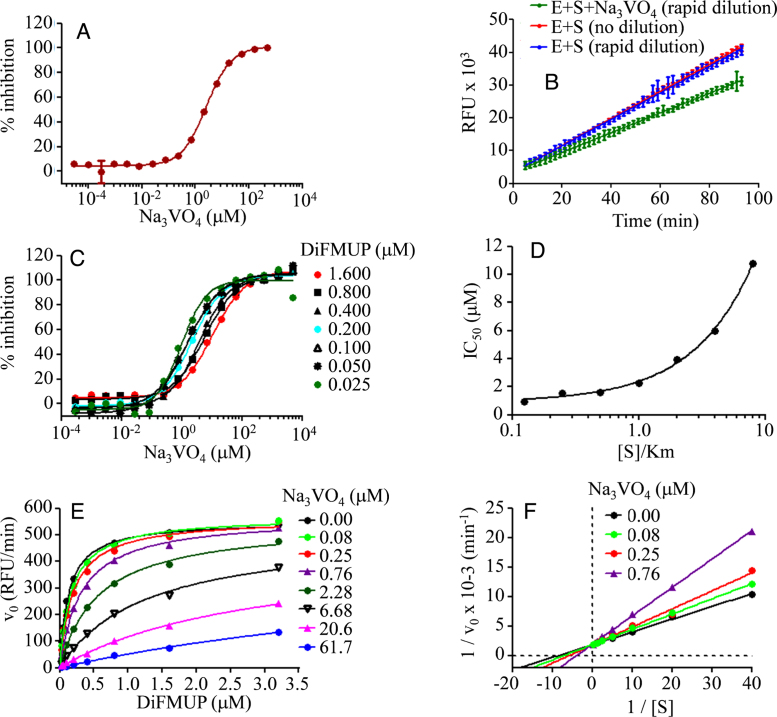
To verify the suitability of the optimized assay conditions for compound screening and mechanism of action studies, a standard competitive inhibitor of AP, sodium orthovanadate (Na3VO4), was used. Initially, concentration–response assays were run to determine the half maximal inhibitory concentration (IC50) of Na3VO4 at 0.2 μM DiFMUP (Fig. 4A). Subsequently, the reversibility of the enzyme-inhibitor complex was evaluated by pre-incubating AP at 100-fold the final assay concentration with Na3VO4 at 10-fold the IC50 for 30 min and subsequently diluting the mixture 100-fold with substrate to a final enzyme concentration of 1× and inhibitor concentration of 0.1× IC50. The expected reaction progress curves should be curvilinear if the inhibition is slowly reversible, linear with 91% recovery of enzymatic activity if the inhibition is rapidly reversible or linear with 9% recovery if the inhibition is irreversible [11]. The reaction progress curve obtained for AP after rapid dilution of Na3VO4 was linear with 73% fractional activity (Fig. 4B), indicating that Na3VO4 is a rapidly reversible inhibitor of AP.
Fig. 4.
Mechanism of action studies using Na3VO4. (A) Concentration-dependent inhibition of AP by Na3VO4 at pH 6.5, 37 °C, and 0.2 μM DiFMUP. Fluorescence intensity was recorded at 60 min of reaction and percent inhibition was calculated and plot as a function of Na3VO4 concentration. Plots were fit by non-linear regression analysis to a four parameter logistic model as described in the text to determine the IC50. (B) Reversibility test. Enzyme was co-incubated in the presence or absence of Na3VO4 for 30 min and rapidly diluted with substrate as described in the text. The corresponding reaction progress curves were recorded. (C) Effect of DiFMUP concentration on Na3VO4 IC50. Concentration–response curves at a range of DiFMUP concentrations [0.025–1.6 μM] were obtained and plot as in A. (D) Variation of Na3VO4 IC50 as a function of DiFMUP concentration. (E) Variation of initial reaction velocity as a function of DiFMUP concentration at a range of Na3VO4 concentrations [0.084–61.7 μM]. Plots were fit by non-linear regression analysis to the Michaelis-Menten model as described in the text. (F) Lineweaver–Burk or double reciprocal plot of the data displayed in panel E shows the intersection of the lines in the Y axis, and indication of a competitive mechanism of inhibition.
3.2.2. Mechanism of action studies
To elucidate the inhibition modality of reversible inhibitors, initial reaction velocities must be determined in the presence of a range of substrate concentrations spanning the KM and inhibitor concentrations spanning the estimated IC50. This type of study enables to inspect the variation of IC50 with substrate concentration, and the variation of the enzyme kinetic parameters with inhibitor concentration, the two diagnostic tests for mechanism-of-action assessment. Fig. 4C and D, and Table 3 show the increase in the estimated IC50 of Na3VO4 with increasing DiFMUP concentrations, a first indication of a competitive mode of action. Consistently, the apparent KM but not the Vmax of the reaction increases with increasing substrate concentrations (Fig. 4E and F, and Table 4), confirming the competitive nature of the AP inhibition by Na3VO4. These validation tests verified the suitability of the selected assay conditions for subsequent inhibitory screening, IC50 determination, and mechanism-of-action studies, and concluded the assay optimization phase.
Table 3.
Effect of DiFMUP concentration on Na3VO4 IC50.
| DiFMUP (μM) | 0.025 | 0.050 | 0.100 | 0.200 | 0.400 | 0.800 | 1.600 |
|---|---|---|---|---|---|---|---|
| IC50 (μM)* | 1.0 ± 0.4 | 1.5 ± 0.4 | 1.6 ± 0.3 | 2.3 ± 0.4 | 4.0 ± 0.4 | 6.1 ± 0.9 | 11 ± 3 |
95% confidence intervals are indicated for IC50.
Table 4.
Effect of Na3VO4 concentration on apparent AP kinetic parameters.
| Na3VO4 (μM) | 0.000 | 0.084 | 0.250 | 0.760 | 2.28 | 6.68 |
|---|---|---|---|---|---|---|
| Vmax (RFU min−1)* | 550 ± 18 | 567 ± 18 | 561 ± 35 | 566 ± 24 | 546 ± 32 | 513 ± 53 |
| KM (μM)* | 0.13 ± 0.02 | 0.17 ± 0.02 | 0.19 ± 0.04 | 0.31 ± 0.04 | 0.57 ± 0.09 | 1.3 ± 0.3 |
95% confidence intervals are indicated for Vmax and KM.
3.3. Assay miniaturization and automation
The purpose of assay miniaturization is to minimize the consumption of reagents (and therefore reduce cost) and increase the assay throughput without compromising the signal window or assay performance. This process may involve changes in assay format, reaction scale, and microplate reader settings without modifications to assay conditions. The purpose of automation is to standardize operations for higher throughput and day-to-day consistency. This process also involves changes in the handling of the assay reagents. Eventually, the impact of implementing changes in the assay format and operations on performance and signal stability must be evaluated.
The AP assay was transferred from standard to low volume plates and scaled down from 50 μL to 20, 15, and 10 μL reaction volume. Reader settings (gain and focal height) were re-optimized for the new assay format using a standard solution of DiFMU. Automation was implemented with a Hummingbird Plus liquid handler (Digilab) for the addition of DMSO or compounds and a Multidrop Combi dispenser (Thermo Scientific) equipped with standard and small tubing cassettes for the addition of assay buffer, enzyme, and substrate. The impact of these changes on assay performance and signal variability was evaluated by testing the different conditions in a full plate format, each one including 192 wells for maximum (enzyme and substrate) and minimum (assay buffer and substrate) signal controls. Fluorescence was recorded at 30, 45, 60, and 75 min to track assay performance throughout the assay course, and Z prime (Z′) and coefficient of variation (CV) of the maximum signal at each combination of miniaturized volume, tubing cassette, and assay end-point were computed (Supplemental Fig. 8). The Z′ is a statistical parameter that measures the suitability of an assay for compound screening based on the signal window and the dispersion of the maximum and minimum signals. The CV is a statistical parameter that measures the dispersion or variability of a population of values based on the ratio of the standard deviation to the mean. Among all conditions tested, the maximum assay performance was obtained in 15 μL reactions using the small tubing cassette for dispensing of reagents (Supplemental Fig. 8).
3.4. Final assessment of assay performance for small molecule screening
The suitability of the optimized assay conditions, format, and operations for small molecule screening and profiling needs to be evaluated by quantitative methods. Guidelines for formal assay validation procedures, analyses, and acceptance criteria are available in the Assay Guidance Manual by Eli Lilly and Company and the National Center for Advancing Translational Sciences: http://assay.nih.gov/assay/index.php/Section2:Plate_Uniformity_and_Signal_Variability_Assessment. Typically, it is recommended to conduct an assessment of spatial uniformity and signal variability within plates and between plates and days by running a limited number of assay plates in independent days. The plates must include a significant number of replicate wells for the theoretical maximum (MAX; enzyme and substrate), medium (MID; enzyme, inhibitor, and substrate) and minimum (MIN; assay buffer and substrate) signals. This test provides a quantitative estimation of the assay robustness and stability.
In general, an assay is considered acceptable for small molecule screening if the assay parameters meet the following criteria:
-
1.
Spatial uniformity. Signal variability due to material drift or edge effects must not exceed 20%.
-
2.
Intra-plate variability. For all plates and days, the CV of each signal must be lower than 20%, the SD of the normalized medium signal (in percent inhibition) lower than 20%, and the Z′ higher than 0.4.
-
3.
Inter-plate and inter-day variability. The difference in the normalized medium signal (in percent inhibition) between plates and days must be lower than 15%.
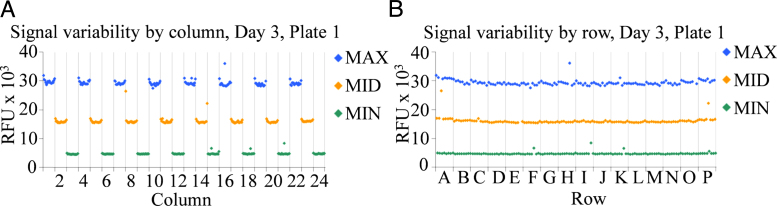
The assessment of assay performance of AP assay comprised three runs of three full assay plates, each one including 128 wells for MAX, MID, and MIN signals spread across the plate in an interleaved format. Fluorescence was read at 30, 45, 60, and 75 min as end-points. Scatter plots of the maximum, medium, and minimum signals against well coordinates by rows and by columns for each plate, day, and time point revealed no significant positional effects (Fig. 5).
Fig. 5.
Assessment of spatial uniformity for the AP fluorometric assay. Spatial variability of the maximum (MAX), medium (MID), and minimum (MIN) signals by column (A) and row (B) for the plate uniformity study described in the text. The scatter plots correspond to one representative plate out of the total of nine plates tested and one time point (60 min) out of the total of four time points recorded for each plate and day.
Likewise, intra-plate, inter-plate, and inter-day signal variability parameters for each plate, day, and time point met assay acceptance criteria excluding no more than three data points per plate (Table 5, Table 6). This final assessment of assay performance confirmed the suitability of the developed assay for screening of small molecule inhibitors.
Table 5.
Intra-plate signal variability of the AP fluorometric assay.
| Parameter | Z’ |
CV (MAX) |
CV (MIN) |
CV (MID) |
SD of MID (%) |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Time (min) | 30 | 45 | 60 | 75 | 30 | 45 | 60 | 75 | 30 | 45 | 60 | 75 | 30 | 45 | 60 | 75 | 30 | 45 | 60 | 75 |
| Plate 1. Day 1 | 0.63 | 0.74 | 0.79 | 0.67 | 6.5 | 5.4 | 4.6 | 8.5 | 9.4 | 8.5 | 8.2 | 7.7 | 4.1 | 3.6 | 3.4 | 3.3 | 3.3 | 2.5 | 2.2 | 2.0 |
| Plate 1. Day 2 | 0.79 | 0.83 | 0.84 | 0.74 | 2.1 | 2.2 | 2.3 | 5.9 | 10.8 | 11.1 | 12.3 | 11.6 | 2.7 | 2.7 | 3.1 | 3.1 | 2.2 | 1.9 | 2.0 | 1.9 |
| Plate 1. Day 3 | 0.63 | 0.74 | 0.81 | 0.84 | 8.3 | 5.9 | 4.8 | 4.3 | 2.7 | 5.3 | 2.7 | 2.9 | 2.8 | 2.6 | 2.6 | 2.6 | 2.2 | 1.8 | 1.7 | 1.6 |
| Plate 2. Day 1 | 0.80 | 0.84 | 0.86 | 0.87 | 2.5 | 2.6 | 2.7 | 2.7 | 9.1 | 8.0 | 7.8 | 8.3 | 2.2 | 2.3 | 2.5 | 2.6 | 1.7 | 1.6 | 1.6 | 1.6 |
| Plate 2. Day 2 | 0.84 | 0.87 | 0.89 | 0.90 | 2.6 | 2.4 | 2.3 | 2.4 | 5.3 | 5.2 | 4.7 | 4.5 | 2.2 | 2.4 | 2.4 | 2.4 | 1.8 | 1.7 | 1.6 | 1.5 |
| Plate 2. Day 3 | 0.88 | 0.90 | 0.90 | 0.90 | 2.1 | 2.2 | 2.4 | 2.5 | 3.4 | 2.8 | 2.8 | 3.0 | 12.2 | 9.6 | 8.0 | 7.1 | 10.0 | 6.8 | 5.2 | 4.4 |
| Plate 3. Day 1 | 0.77 | 0.83 | 0.83 | 0.85 | 3.2 | 2.8 | 3.2 | 3.1 | 9.1 | 9.0 | 9.0 | 8.7 | 11.0 | 8.8 | 7.2 | 6.3 | 9.0 | 6.2 | 4.7 | 3.9 |
| Plate 3. Day 2 | 0.84 | 0.88 | 0.81 | 0.83 | 2.4 | 2.1 | 2.1 | 2.2 | 5.9 | 6.2 | 19.7 | 18.5 | 5.8 | 9.7 | 7.6 | 6.2 | 4.8 | 6.9 | 5.0 | 3.9 |
| Plate 3. Day 3 | 0.76 | 0.81 | 0.84 | 0.85 | 2.5 | 2.5 | 2.6 | 2.8 | 12.8 | 12.2 | 12.1 | 11.5 | 2.1 | 2.1 | 2.4 | 2.5 | 1.7 | 1.5 | 1.6 | 1.6 |
| Acceptance criteria | >0.5 | ≤20% | ||||||||||||||||||
Table 6.
Inter-plate and inter-day signal variability of the AP fluorometric assay.
| aDifference in MID signal between plates (%) | Day 1 |
Day 2 |
Day 3 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Plate 1 | Plate 2 | Plate 3 | Plate 1 | Plate 2 | Plate 3 | Plate 1 | Plate 2 | Plate 3 | ||||
| 30 min | Day 1 | Plate 1 | – | 1.8 | 0.4 | 2.3 | 0.3 | 3.7 | 4.3 | 4.4 | 3.1 | 45 min |
| Plate 2 | 1.3 | – | 1.4 | 0.4 | 2.1 | 1.8 | 2.4 | 2.5 | 1.3 | |||
| Plate 3 | 0.5 | 1.8 | – | 1.8 | 0.7 | 3.3 | 3.9 | 4.0 | 2.7 | |||
| Day 2 | Plate 1 | 2.0 | 0.7 | 2.5 | – | 2.5 | 1.4 | 2.0 | 2.1 | 0.8 | ||
| Plate 2 | 3.0 | 4.3 | 2.5 | 5.0 | – | 3.9 | 4.5 | 4.6 | 3.3 | |||
| Plate 3 | 3.9 | 2.6 | 4.4 | 1.9 | 6.9 | – | 0.6 | 0.7 | 0.6 | |||
| Day 3 | Plate 1 | 5.1 | 3.8 | 5.6 | 3.1 | 8.0 | 1.2 | – | 0.1 | 1.2 | ||
| Plate 2 | 3.3 | 2.0 | 3.8 | 1.3 | 6.3 | 0.6 | 1.8 | – | 1.3 | |||
| Plate 3 | 2.9 | 1.6 | 3.4 | 0.9 | 5.9 | 1.0 | 2.1 | 0.3 | – | |||
| 60 min | Day 1 | Plate 1 | – | 1.0 | 0.3 | 1.1 | 0.1 | 6.0 | 1.7 | 2.6 | 2.0 | 75 min |
| Plate 2 | 2.7 | – | 0.7 | 0.1 | 0.9 | 5.0 | 0.7 | 1.6 | 1.0 | |||
| Plate 3 | 0.5 | 2.3 | – | 0.8 | 0.1 | 5.7 | 1.4 | 2.3 | 1.7 | |||
| Day 2 | Plate 1 | 1.4 | 1.4 | 0.9 | – | 0.9 | 4.9 | 0.6 | 1.5 | 0.9 | ||
| Plate 2 | 0.1 | 2.6 | 0.4 | 1.3 | – | 5.9 | 1.6 | 2.5 | 1.9 | |||
| Plate 3 | 7.3 | 4.6 | 6.9 | 6.0 | 7.2 | – | 4.3 | 3.4 | 4.0 | |||
| Day 3 | Plate 1 | 2.3 | 0.4 | 1.8 | 0.9 | 2.2 | 5.0 | – | 0.9 | 0.3 | ||
| Plate 2 | 3.2 | 0.5 | 2.7 | 1.8 | 3.1 | 4.1 | 0.9 | – | 0.6 | |||
| Plate 3 | 2.3 | 0.4 | 1.9 | 1.0 | 2.2 | 5.0 | 0.0 | 0.9 | – | |||
| Acceptance criteria | <15% | |||||||||||
Difference in normalized medium signals (% inhibition) between the indicated plates.
4. Discussion
The development of physiologically relevant and statistically robust in vitro assays is a crucial step in the early drug discovery process. Multiple factors contribute to the successful optimization of an assay, including the selection of the adequate assay technology and materials, buffer composition, reaction conditions, enzyme and substrate concentrations, liquid handling equipment, and analytical instrumentation. Screening paradigms with well validated assay systems help in identifying and optimizing clinically relevant leads. The goal of the present study was to provide a complete overview on the critical processes required to successfully develop and validate a cell-free enzymatic assay for small molecule screening and profiling using calf intestine AP as model target.
Initially, the optimal plate type was selected to ensure enzyme stability. Inadequate protein binding to the assay plate may lead to premature loss of enzyme activity during the assay course, shortening of screening window and, most importantly, leading to inaccurate estimations of enzyme activity. In this regard, non-binding plates were shown to sustain AP activity for at least 4 h. Likewise, buffer composition was optimized to mimic physiological conditions. We found that the presence of detergent micelles increased AP activity, presumably by reconstituting its native lipid environment. It is noteworthy that screening the target in non-physiological conditions could bias the hit identification process toward molecules that are inactive in physiological conditions. Beside the optimal plate type and detergents illustrated in this study, some assays may require additional reagents in the reaction buffer, such as carrier proteins, salts, or reducing agents, to ensure sustained target stability.
Subsequent to the optimization of buffer constituents, the kinetic parameters of AP were determined in order to establish the appropriate pH and substrate concentration for screening of inhibitors. For each target class, the most adequate substrate concentration for compound screening and potency assessment depends on the kinetic parameters of the enzyme and the desired modality of inhibition: concentrations below the KM favor the selection of inhibitors that are competitive for the substrate binding site and disfavor the selection of un-competitive inhibitors, and vice versa. If there is no preference for a particular type of inhibitor, conducting the screen or dose–response studies at a substrate concentration around the KM is a good compromise. The kinetic studies using AP demonstrated this enzyme performed well at neutral pH as compared to alkaline pH. More importantly, these studies illustrated the interconnection between pH optima and substrate concentration for AP and the need to optimize these two parameters simultaneously. Kinetic studies also raised the necessity to replace the assay technology by a more sensitive one, which was compatible with the low substrate concentration and neutral pH requirements of the assay.
While optimization of “one-factor-at-a-time” approach may seem a priori a less cost-effective strategy for assay development than optimizing “all-in-one-go” approach, the first one is more practical to optimize independent assay variables whereas the second one is more powerful to co-optimize simultaneously various inter-dependent parameters. For example, the optimization of plate type, pH, and substrate concentration could be a priori established in parallel in an all-in-one experiment by titrating simultaneously substrate and pH in different plate types. While this approach would save time by reducing to one experiment the optimization of three assay components, it would increase reagent consumption and thus assay costs by increasing the number of assay replicates in multiple plate types. In general, as the number of independent parameters to be optimized in a multifactorial assessment increases linearly, the number of conditions to be tested and the consumption of reagents and consumable grow exponentially, making this approach expensive, impractical and error prone, despite the potential time saving benefits. In contrast, the optimization of parameters that are inter-dependent should be conducted solely by a designed all-in-one approach. In this case, a one-factor-at-a-time strategy would identify local instead of global optima, thus leading to select potentially suboptimal assay conditions. For example, the optimization of pH and substrate concentration (two inter-dependent assay parameters) was conducted by simultaneous titration of these two assay variables in a two-dimensional matrix design. This approach allowed to identify not only local pH optima at fixed substrate concentrations or vice versa, but global optima for these two parameters. Therefore, optimizing a combination of one-factor-at-a-time and all-in-one designed experiments is recommended to successfully develop and optimize an in vitro assay for any target of interest.
Upon assay optimization, the suitability of the selected assay conditions for compound screening was verified using reference compounds. Sodium orthovanadate inhibited the AP activity in a dose dependent manner and kinetic studies sodium orthovanadate confirmed the reversibility & mode of action of this inhibitor, providing further support to the appropriateness of the developed assay for mode of action studies. In general, assay conditions that have been optimized adequately should enable to determine the mechanism of action of inhibitors correctly.
However, caution must be taken while using the screening campaign assay conditions to establish the inhibition modality and binding affinity of the diverse chemical series identified in the screening campaign. In general, the first step in the characterization of the hits could be the reversibility assessment described in Section 3.2.1 and depicted in Fig. 4B. The methodology described in Section 3.2.2 can in-general be followed to determine compound potency and mechanism-of-action, and guide SAR optimization of reversible inhibitors. However, for time-dependent (slow binding, slow dissociation, and tight binding) and irreversible (covalent binding) inhibitors, modifications to the assay methodology and analysis are required in order to correctly establish compound potency and prioritize candidates. Although tight binding and irreversible inhibition may be a priori undesirable mechanisms for target attenuation due to long-term or permanent inactivation of the target enzyme, in some instances molecules with these characteristics may possess clinical advantages by providing high selectivity and sustained pharmacological responses [11].
The AP assay protocol was further miniaturized and automated to reduce costs, increase throughput, and ensure day-to-day reproducibility. The suitability of the assay for small molecule screening and profiling was validated by quantitative assessment of the signal variability within plates, between plates, and between days, and comparison of performance indicators with recommended cut-off values for assay acceptance.
The overall scheme of assay development, validation, quantitative assessment and screening paradigm described herein could be used as a general guide of assay development for any mammalian or microbial enzymatic target class. Besides the primary screening described herein, it is desirable to counter-screen the hits against a panel of structurally and functionally related enzyme classes. In the case of mammalian targets, these secondary screenings serve the purpose of determining compound selectivity and predict potential off-target effects. In the case of microbial targets (bacterial, protozoan, or fungal proteins), these assessments explore the potential spectrum of the identified antimicrobial candidates. In both cases, assay conditions for each secondary target in the selectivity panel must individually be optimized following similar process described for the primary target.
In conclusion, our study illustrates the comprehensive process of developing and validating an enzymatic assay for discovery and profiling of small molecule inhibitors. The most critical parameters for assay development and validation, quantitative assessments, bottlenecks, and the corrective measures to overcome them were discussed in detail using AP as a model target.
Acknowledgements
We thank Dr. Srividya Swaminathan, Dr. Chitkala Satyanarayana, Dr. Jonathan Wierschke, and Dr. Andrew Mhyre for useful discussions and suggestions.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bdq.2015.03.001.
Contributor Information
Vicente Sancenon, Email: VicenteEnrique.Sancenon-Galarza@amriglobal.com.
Saravanakumar Dhakshinamoorthy, Email: Saravanakumar.Dhakshinamoorthy@amriglobal.com.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Williams K.P., Scott J.E. Enzyme assay design for high-throughput screening. Methods Mol Biol. 2009;565:107–126. doi: 10.1007/978-1-60327-258-2_5. [DOI] [PubMed] [Google Scholar]
- 2.Klink T.A., Staeben M., Twesten K., Kopp A.L., Kumar M., Dunn R.S. Development and validation of a generic fluorescent methyltransferase activity assay based on the transcreener AMP/GMP assay. J Biomol Screen. 2012;17:59–70. doi: 10.1177/1087057111421624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sachsenmeier K.F., Hay C., Brand E., Clarke L., Rosenthal K., Guillard S. Development of a novel ectonucleotidase assay suitable for high-throughput screening. J Biomol Screen. 2012;17:993–998. doi: 10.1177/1087057112443987. [DOI] [PubMed] [Google Scholar]
- 4.Fosset M., Chappelet-Tordo D., Lazdunski M. Intestinal alkaline phosphatase. Physical properties and quaternary structure. Biochemistry. 1974;13:1783–1788. doi: 10.1021/bi00706a001. [DOI] [PubMed] [Google Scholar]
- 5.Chappelet-Tordo D., Fosset M., Iwatsubo M., Gache C., Lazdunski M. Intestinal alkaline phosphatase. Catalytic properties and half of the sites reactivity. Biochemistry. 1974;13:1788–1795. doi: 10.1021/bi00706a002. [DOI] [PubMed] [Google Scholar]
- 6.Millan J.L. Alkaline phosphatases. Structure, substrate specificity and functional relatedness to other members of a large superfamily of enzymes. Purinergic Signal. 2006;2:335–341. doi: 10.1007/s11302-005-5435-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akcakaya H., Aroymak A., Gokce S. A quantitative colorimetric method of measuring alkaline phosphatase activity in eukaryotic cell membranes. Cell Biol Int. 2007;31:186–190. doi: 10.1016/j.cellbi.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 8.Evans D.F., Pye G., Bramley R., Clark A.G., Dyson T.J., Hardcastle J.D. Measurement of gastrointestinal pH profiles in normal ambulant human subjects. Gut. 1988;29:1035–1041. doi: 10.1136/gut.29.8.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross M.H., Ely J.O., Archer J.G. Alkaline phosphatase activity and pH optima. J Biol Chem. 1951;192:561–568. [PubMed] [Google Scholar]
- 10.Gee K.R., Sun W.C., Bhalgat M.K., Upson R.H., Klaubert D.H., Latham K.A. Fluorogenic substrates based on fluorinated umbelliferones for continuous assays of phosphatases and beta-galactosidases. Anal Biochem. 1999;273:41–88. doi: 10.1006/abio.1999.4202. [DOI] [PubMed] [Google Scholar]
- 11.Copeland R.A. Evaluation of enzyme inhibitors in drug discovery: a guide for medicinal chemists and pharmacologists. John Wiley & Sons Inc.; Hoboken, New Jersey: 2008. Lead optimization and structure-activity relationships for reversible inhibitors; pp. 111–140. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.