Abstract
Integrity of the mRNA in clinical samples has major impact on the quality of measured expression levels. This is independent of the measurement technique being next generation sequencing (NGS), Quantitative real-time PCR (qPCR) or microarray profiling. If mRNA is highly degraded or damaged, measured data will be very unreliable and the whole study is likely a waste of time and money. It is therefore common strategy to test the quality of RNA in samples before conducting large and costly studies. Most methods today to assess the quality of RNA are ignorant to the nature of the RNA and, therefore, reflect the integrity of ribosomal RNA, which is the dominant species, rather than of mRNAs, microRNAs and long non-coding RNAs, which usually are the species of interest. Here, we present a novel molecular approach to assess the quality of the targeted RNA species by measuring the differential amplification (ΔAmp) of an Endogenous RNase Resistant (ERR) marker relative to a reference gene, optionally combined with the measurement of two amplicons of different lengths. The combination reveals any mRNA degradation caused by ribonucleases as well as physical, chemical or UV damage. ΔAmp has superior sensitivity to common microfluidic electrophoretic methods, senses the integrity of the actual targeted RNA species, and allows for a smoother and more cost efficient workflow.
Keywords: RNA integrity, RNA quality, Endogenous RNase resistant marker, Differential amplicons, ΔAmp, DAmp
1. Introduction
It is today well established that the dominant confounding variation in the analysis of mRNA levels originates from the preanalytical steps comprising sampling, transportation, storage and extraction of the samples rather than from the actual analysis performed with techniques such as reverse transcription quantitative polymerase chain reaction (RT-qPCR), microarray profiling, and next generation sequencing (NGS) [1]. Based on results generated primarily by the European Commission Framework 7 project SPIDIA (www.spidia.eu) [2], the European Standardizing Committee (CEN) initiated in 2013 a project within CEN/TC140 “In vitro diagnostic medical devices” to develop nine new technical specifications for the preanalytical process that are expected to be published autumn 2015, and in 2014 eight projects were approved for ISO (International Organization for Standardization) standards in ISO/TC212 “Clinical laboratory testing and in vitro diagnostic test systems”.
Many factors influence the measurable RNA-levels in biological tissues post sampling. The removal of the tissue/cells from its natural environment can have profound effect on genes’ expressions. Genes may become activated increasing their expression levels manifold or they can be repressed. A system where these problems recently were documented is blood samples collected in EDTA tubes [2]. The presence of EDTA does not kill cells; rather it chelates magnesium resulting in dramatic changes in the ionic composition to which the cells respond. Other complications include RNA degradation by ribonucleases (RNases). RNases are plentiful in biological material and may contaminate water, buffers, vessels, glassware, and plasticware. RNA molecules are damaged by chemicals, such as formalin, which is commonly used to preserve tissue samples for morphological analysis, and by ultraviolet (UV) radiation, which introduces crosslinks. At extreme pH RNA can undergo self-cleavage [3]. All these influences compromise measurements of native gene expression and should be controlled in standard operating procedures (SOPs) for gene expression profiling.
Currently most common test for RNA integrity is to analyze its length distribution using electrophoretic methods on microfluidic platforms such as the Bioanalyzer (Agilent Technologies) or Experion (Bio-Rad), the ScreenTape (Agilent Technologies) or capillary electrophoresis such as the Fragment Analyzer (Advanced Analytical Technologies) and QIAxcel (Qiagen). Bulk RNA is dominated by ribosomal RNAs (rRNA) that make up about 85% of the total amount. Most eukaryotes have two ribosomal subunits composed of four rRNA molecules. In humans these are the 5S (121 bases), 5.8S (156 bases), 18S (1869 bases) and 28S (5070 bases) rRNAs, where S is the Svedberg unit reflecting sedimentation property [4]. The four rRNA molecules are readily separated electrophoretically. The 28S, 18S and 5.8S rRNA molecules are encoded by a single transcriptional unit and therefore produced in equal amounts, resulting in a defined relation between their electropherogram peaks for fully intact material. Because of the much larger sizes of the 28S and 18S rRNAs they dominate the electropherogram and integral RNA is characterized by two distinct peaks. Degradation of RNA is reflected by broadening of the 28S and 18S rRNA peaks toward faster migrating molecules and a general increase of background due to accumulation of degradation fragments of various sizes. Analysis of the electropherogram determines a score that reflects RNA integrity. Different platforms deliver scores calibrated somewhat differently; the most common score is the RNA integrity number, RIN, originally introduced by Schroeder et al. [5]. RIN is a number between 0 and 10, where 10 reflects fully intact RNA and 0 totally degraded RNA. Analoge scores are the RNA Quality Index (RQI) introduced by Bio-Rad, the RNA Quality Number (RQN) introduced by Advanced Analytical, RNA Integrity Number equivalent (RINe) introduced by Agilent, and the RNA Integrity Score (RIS) introduced by Qiagen. Although analyzing length distribution of RNA is today the most common approach to evaluate RNA integrity, its relevance for gene expression analysis has occasionally been called into question [6]. As outlined above the electrophoretic analysis reflects rather the integrity of ribosomal RNAs, which are quite different from the messenger RNAs and microRNAs typically studied. rRNAs have compact structures and are found in tight complexes with proteins, while mRNAs have an 3′ A-tail and 5′-cap that are usually protein bound, which protects from certain nucleolytic degradation. mRNAs may also harbor internal sequences that are cleaved by endonucleases. Indeed, model studies of RNA degradation show rRNAs are degraded at quite different rates from mRNAs [6]. rRNA expression is further regulated differently from mRNA expression and is not expected to be appreciably affected by changes in environmental conditions, which may have profound effect on the expression of mRNAs [7]. The use of RIN to assess samples for miRNA analysis is even more questionable because of the very short length of microRNAs, and empirical data supporting such correlation is limited [8], [9]. The impact of the chemical and structural differences between rRNAs and mRNA/miRNAs may further depend on the actual degradation mechanism. The precision of the electrophoretic assessment of RNA integrity depends on sample quality. While high RIN/RQI values are reproducible and reliable, results obtained on poor quality samples tend to show larger variation due to poor reproducibility of degradation and difficulties in modeling the increased background from the short fragments that form. Our experience is RIN/RQI below 5 are prone to substantial uncertainty and conclusions about quality are ambiguous. This is a major limitation of the electrophoretic approach, since borderline samples often have RIN <5. A more reliable assessment of mRNA integrity in samples of poor quality is therefore desirable to distinguish those were mRNA levels can be measured with reasonable accuracy from those where results would be so uncertain and unreliable that analysis is waste of time and money.
As alternative to the electrophoretic approach molecular methods have been proposed to assess RNA integrity. One is the 3′/5′ assay, which exploits the proximity of the targeted sequence to the A-tail at the mRNA 3′-end [10]. Reverse transcribing the mRNA using oligo(T) primer most of the cDNA should contain the complement to the mRNA 3′-end, while only full length cDNAs reverse transcribed from intact mRNAs should harbor the complement to the mRNA 5′-end. Hence, by quantifying the presence of the 5′-end and 3′-end sequences of an mRNA using oligo(T) primed RT-qPCR, the ratio should indicate the fraction of full lengths mRNA and, hence, reflect mRNA integrity. Although the 3′/5′ assay has been reported to work well in some model systems [10], [11] it has not been particularly useful for biological samples. One complication is that degraded samples contain DNA and RNA fragments that may prime the reverse transcription and the required specific oligo(T) priming is lost [12].
A more recent molecular method employs amplicon length to assess mRNA integrity. It is based on using paired qPCR assays that produce amplicons of different length from the same target [13], [14]. For intact mRNA, using probe based assays, the Cqs of the short and long amplicons should be virtually the same (for SYBR based assays Cqs depend on amplicon lengths, which has to be considered), while for fragmented RNA the Cq of the longer amplicon is higher than that of the shorter due to a lower yield [15].
In this work we evaluate the approach of measuring differential amplicons, ΔAmp = CqAmplicon1–CqAmplicon2, and the amplicon ratio amplicon1/amplicon2 = 2ΔAmp (assuming 100% PCR efficiency and same fluorescence intensity per amplicon) as indicator of RNA integrity. We also present a new RNA marker that is virtually resistant to RNases and found in most eukaryotic cells [16]. This new Endogenous RNase Resistant (ERR) marker can be combined with an mRNA marker, such as a reference gene, that has normal sensitivity to RNases, to assess physical and chemical degradation of the mRNA in addition to damage caused by post sampling enzymatic degradation. We validate the ΔAmp approach by measuring mRNA degradation of unprotected biological material exposed to air at ambient temperature, under conditions were RNases are active, upon exposure to heat, UV light, and formalin.
2. Material and methods
2.1. Modelling of enzymatic RNA degradation
2.1.1. RNA degradation in lysed HeLa cells by added RNase I
A pellet of frozen HeLa cells (8 × 105) was re-suspended in 250 μl of RNase free H2O (Life Technologies, Inc.) to fully lyse the cells. The re-suspended cells where divided into six aliquots (6 × 40 μl) and mixed with 40 μl of tris-HCl buffer (10 mM) containing different amounts of RNase I (final concentration: 0.005–0.125 U/μl, Cat# EN0601, Thermo Scientific Inc.). The aliquots were incubated at room temperature for 30 s followed by the addition of 320 μl β-mercaptoethanol and immediately extracted using QIAshredder columns and RNeasy® Mini kit (Qiagen GmbH, Germany) according to the manufacturer’s instructions. RNA concentration and integrity were assessed spectroscopically (NanoDrop 1000, Thermo Scientific, Inc.) and using automated gel electrophoresis (Experion system, Bio-Rad, Inc.).
Extracted RNA was diluted to 50 ng/μl. 500 ng of RNA from each aliquot was reversed transcribed into cDNA in 20 μl reactions using iScript™ cDNA Synthesis Kit (Bio-Rad, Inc.) on a Mx3005P qPCR system (Agilent Technologies, Inc.) according to the manufacturer’s instructions. Negative (RNase free H2O) and no-RT (equal mixture of all six samples) controls were included to test for gDNA background and contamination. After reverse transcription the cDNA was diluted 10X and stored at −20 °C until further analysis.
All cDNA was analyzed using validated qPCR assays targeting human ERR marker [16] (Supplementary material 1) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Supplementary material 1). qPCR was performed in 20 μl reactions with 10 μl PerfeCTa® SYBR® Green FastMix® (2X, Quanta Bioscience, Inc.), 0.3 μl(ERR marker)/0.6 μl(GAPDH) primer mix (forward and reverse, 10 μM), 7.7 μl(ERR marker)/7.4 μl(GAPDH) RNase free H2O and 2 μl of cDNA template (10X diluted) on a MX 3005 P qPCR system (Agilent Technologies, Inc.) according to the manufacturer’s instructions. Cycling conditions; activation: 3 min 95 °C, cycling: 20 s 95 °C, 20 s 58 °C + fluorescence acquisition, 20 s 72 °C for 45 cycles, melt curve: 20 s 95 °C, 20 s 58 °C and continuous fluorescence acquisition between 58 and 95 °C.
2.2. Degradation by ribonucleases
To study enzymatic degradation in tissue two frozen mouse livers were used. One liver was sectioned into six pieces of similar sizes in the presence of dry ice to prevent thawing and put at −80 °C overnight. The other liver was crushed into fine powder in the presence of dry ice to prevent thawing using a frozen mortar and pestle. The tissue powder was divided into eight aliquots and stored at −80 °C overnight. The frozen liver pieces and the powder were exposed to room temperature (pieces: 0, 10, 20, 40, 60, 120 min; powder: 0, 1, 5, 10, 20, 40, 60, 90, 120 min) and then homogenized (Tissuelyzer, Qiagen GmbH, Germany) in ice cold QiaZol (Qiagen GmbH, Germany) and extracted for RNA with a miRNeasy® Mini Kit (Qiagen GmbH, Germany) according to the manufacturer’s instructions. RNA concentration and integrity were estimated spectroscopically (NanoDrop 1000, Thermo Scientific, Inc.) and using automated gel electrophoresis (Experion system, Bio-Rad, Inc.). Samples were normalized to 50 ng/μl of RNA and 500 ng from each was reversed transcribed into cDNA in a 20 μl reaction using SuperScript® III First-Strand Synthesis SuperMix (Life Technologies, Inc.) with a mix of random hexamer and OligodT priming according to the manufacturer’s instructions on a CFX96 qPCR instrument (Bio-Rad, Inc.). Negative (RNase free H2O) and no-RT (equal mixture of all samples for tissue pieces or powder) controls were also included to test for gDNA and contamination. The cDNA was stored in aliquots at −20 °C until further analysis.
cDNA was analyzed with qPCR assays targeting mouse ERR marker, cyclophilin A (PPIA), Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Tyrosine 3-/Tryptophan 5-monooxygenase activation protein (YWHAZ) (Supplementary material 1). qPCR was performed in 10 μl with 5 μl iQ™ SYBR® Green Supermix (2X, BioRad, Inc.), 0.4 μl primer mix (forward and reverse, 10 μM), 2.6 μl RNase free H2O and 2 μl of cDNA template (5X diluted) on a LightCycler 480 qPCR instrument (Roche, Inc.) according to the manufacturer’s instructions. Cycling conditions; activation: 3 min 95 °C, cycling: 20 s 95 °C, 20 s 58 °C + fluorescence acquisition, 20 s 72 °C for 45 cycles, melt curve: 15 s 95 °C, 15 s 58 °C and continuous fluorescence acquisition from 58 to 95 °C.
2.3. Thermal and radiation damage
2.3.1. Thermal and radiation induced degradation of purified human RNA
Two 100 μl aliquots of purified human RNA (aliquot 1 = 90 ng/μl RQN = 9, aliquot 2 = 70 ng/μl RQN = 8.1) were exposed to ultraviolet (UV) light (aliquot 1) or heat (95 °C, aliquot 2) for 60 min. 10 μl fractions were removed from each aliquot at 0 (before exposure), 1, 10, 20, 40 and 60 min. All fractions were immediately stored at −20 °C after collection. RNA integrity was assessed with automated gel electrophoresis (Experion system, Bio-Rad, Inc.).
Five microliter of each fraction (UV or heat treated) was reverse transcribed in 20 μl reactions using SuperScript® III First-Strand Synthesis SuperMix (Life Technologies, Inc.) and random hexamer priming according to manufacturer’s instructions on a CFX96 qPCR instrument (Bio-Rad, Inc.). Negative (RNase free H2O) and no-RT (equal mixture of all six samples) controls were included. After reverse transcription cDNA was stored at −20 °C.
cDNA from all the samples and controls were analyzed using validated qPCR assays targeting human ERR marker, human beta-2-microglobulin (B2 M) and human 18 s rRNA (18S) producing multiple amplicon lengths for each target (Supplementary Material 1). qPCR was performed in 10 μl with 5 μl PerfeCTa® SYBR® Green FastMix® (2X, Quanta Bioscience, Inc.), 0.2 μl primer mix (forward and reverse, 10 μM), 2.8 μl RNase free H2O and 2 μl of cDNA template (5X diluted) on a LightCycler 480 qPCR instrument (Roche, Inc.) according to manufacturer’s instructions. Cycling conditions: activation: 3 min 95 °C, cycling: 10 s 95 °C, 10 s 58 °C + fluorescence acquisition, 35 s 72 °C for 45 cycles, melt curve: 15 s 95 °C, 15 s 58 °C and continuous fluorescence acquisition from 58 to 95 °C.
2.4. Chemical degradation
Total RNA from rat liver (1 μg/sample) was degraded in technical replicates (n = 2) in 17 μl of formalin solution (10%), buffered to neutral pH (Sigma–Aldrich) at room temperature for 0, 1, 10, 20, 40, 60, 120 and 240 min. The RNA was then purified to remove formalin using a miRNeasy FFPE kit (Qiagen) according to the manufacturer’s instructions starting from step 15 in the protocol. The RNA was eluted with 20 μl RNase free water and reverse transcribed using GrandScript Reverse Transcription kit (TATAA Biocenter AB) in a ViiA™ 7 instrument (Life Technologies, Inc.) following manufacturer’s instructions using 10 μl volume with 100 ng total RNA per reaction. The cDNA was diluted 10× in DNase/RNase free H2O and stored at −20 °C.
cDNA from all the samples and controls was analyzed using validated qPCR assays targeting rat beta-2-microglobulin (B2 M), rat 18 s rRNA (18S), and the Endogenous RNase Resistant (ERR) [16] marker [Supplementary Material 1] producing multiple amplicon lengths for each target. qPCR was performed in 10 μl with 5 μl 2X TATAA SYBR® GrandMaster® Mix (TATAA Biocenter AB), 400 nM primer and DNase/RNase free H2O (Life Technologies, Inc.) and 4 μl of template on a ViiA™ 7 instrument (Life Technologies, Inc.) according to the manufacturer’s instructions. For NTCs 4 μl of LPA-H2O (50 ng/μl) was added instead of template. Cycling conditions; activation: 1 min 95 °C, Cycling: 3 s 95 °C, 20 s 60 °C + fluorescence acquisition, 10 s 72 °C for 50 cycles, melt curve: 15 s 95 °C, 15 s 60 °C and continuous fluorescence acquisition from 60 to 95 °C.
3. Results
3.1. Degradation by RNases
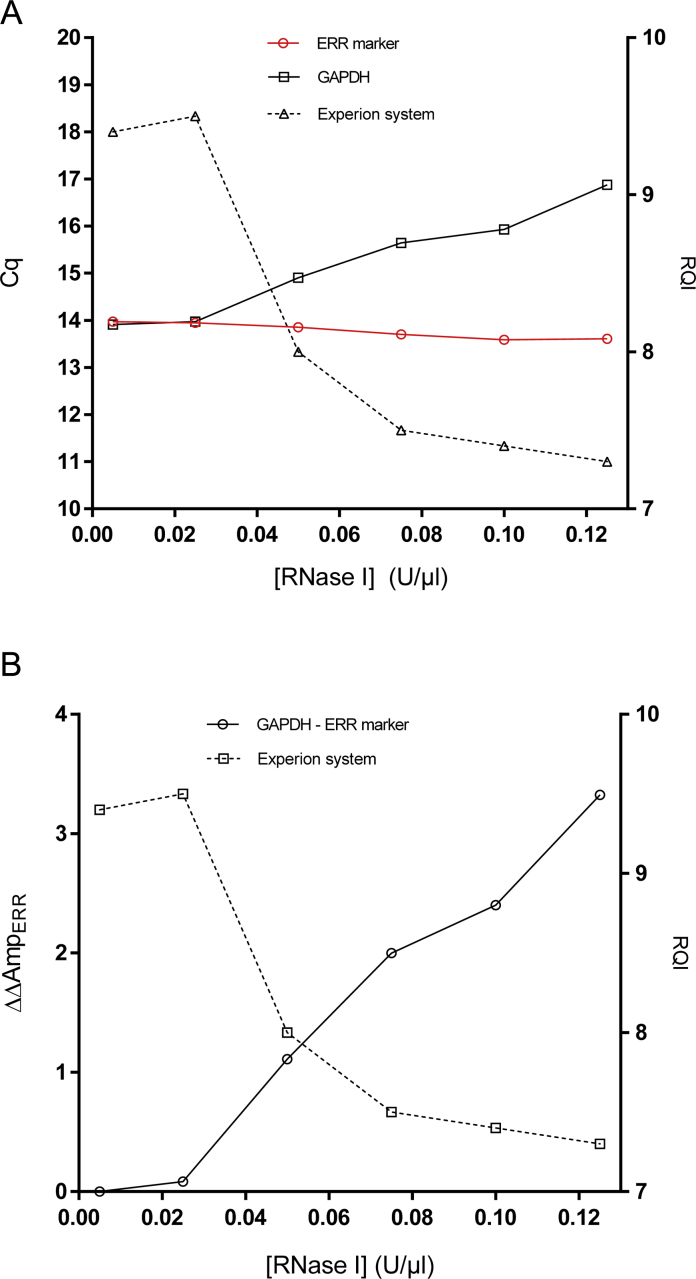
Integrity of RNA from lysed HeLa cells incubated in the presence of RNase I was monitored electrophoretically measuring RQI and with RT-qPCR measuring one normal transcript, which is the common reference gene (RG) glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and the stability ERR marker (Fig. 1a). RNase I concentrations below 0.025 U/μl have no discernible effect on the RNA integrity as reflected by the electrophoresis (RQI = 9.4–9.5) and the RT-qPCR (both Cqs are around 14). With RNase I concentrations above 0.025 units the Cq of the GAPDH transcript as well as the RQI decrease steadily, while the Cq of the ERR marker remains unchanged. At the controlled conditions used in this experiment the input amount of RNA for cDNA synthesis was the same for all samples and the measured Cqs are comparable over time. Hence, the increase in Cq of GAPDH with time reflects degradation of mRNA. However, for field samples of unknown composition the Cq of a reference genes alone could not be interpreted in terms of RNA integrity, since the total amount of native transcript as well as post sampling degradation may vary across samples. However, the Cq of the ERR marker is not affected by the incubation with RNase I and reflects its native level. The effect of RNase I degradation is therefore evident from the comparison of the measured level of the reference gene transcript (here GAPDH) with that of the ERR marker as ΔAmpERR:ΔAmpERR = CqRG − CqERR, which here can be further normalized to the initial amount of intact mRNA as: ΔΔAmpERR (RNase) = ΔAmpERR([RNase]) − ΔAmpERR ([RNase] = 0) (Fig. 1b). Comparing with the effect on RQI shown in the same graph, we see that while RQI decreases from about nine to six, ΔAmpERR increases with over three cycles. ΔAmpERR of three cycles reflects degradation of almost 90% of the GAPDH transcripts to below 151 bases, which is the length of the GAPDH amplicon. This difference is readily measured and reflects the nucleolytic degradation of mRNA, in difference to the change in RQI, which reflects degradation of rRNA.
Fig. 1.
RNase I treated HeLa cells. Cq and RQI plotted against [RNase I] (A). ΔΔAmpERR(RNase) = ΔAmpERR ([RNase]) − ΔERR ([RNase] = 0) and RQI plotted against [RNase I] (B).
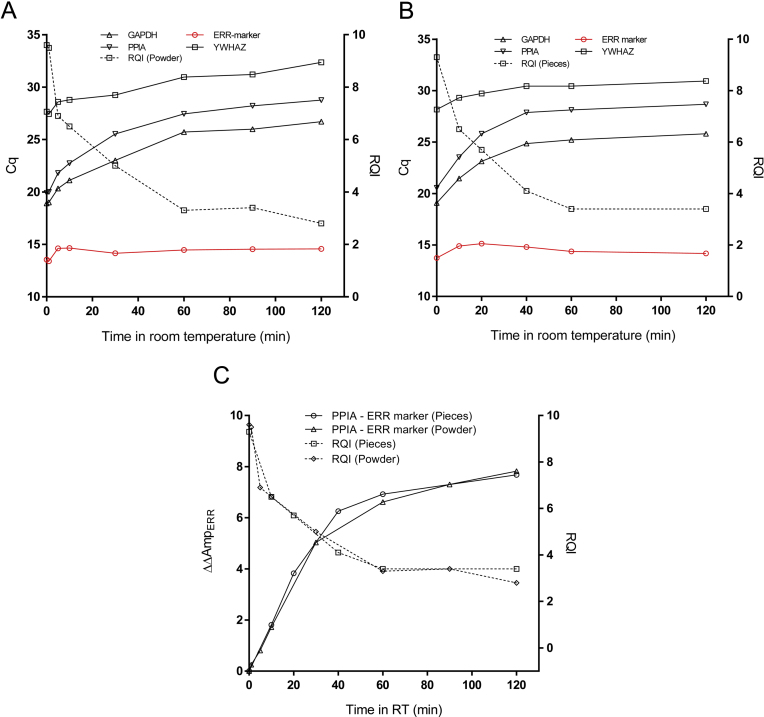
Fig. 2 shows that RNA in small pieces and in powder of liver tissue at room temperature degrades at similar rates as reflected by RQI measured over time. The RQI decreases initially steeply, then slower and after 60 min it levels off at a level of about 3–4. This might suggest to the investigator that most degradation occurs initially and after 60 min either all RNA is degraded or degradation has ceased. However, measuring the presence of transcripts of three commonly used reference genes by RT-qPCR we find degradation continuous for at least the 120 min we studied the material. The Cqs of the three reference genes are different, because the transcripts are present at different levels, but the changes of their Cqs over time are very similar indicating similar degradation rates. The disagreement between the RQI time course and the changes in Cqs of the common reference genes could be due to different mechanisms of degradation of rRNA, which is reflected by the RQI, and by the Cqs, which reflects degradation of the three targeted mRNAs. Indeed, differences in the degradation of rRNA and mRNA by RNases was recently reported [6]. Another reason could be that the electrophoretic approach becomes insensitive to further degradation at a certain level and RQI values of 3–4 are too low to reliably reflect the integrity of the RNA present, if any. In fact, the RQI, like the other indicators based on electrophoretic traces, do not have absolute defined scales, since they were calibrated using artificially degraded RNA samples, for which there were no independent means to assess the integrity of the RNA.
Fig. 2.
Room temperature incubated frozen mouse liver tissue. Cq and RQI plotted against time in room temperature for tissue powder (A) and pieces (B). ΔΔAmpERR(t) = ΔAmpERR(t)- ΔAmpERR (t = 0) and RQI plotted against time in room temperature for tissue powder and pieces (C).
In contrast to the reference genes the ERR marker is virtually resistant to the degradation by RNase I. Fig. 2c compares ΔΔAmpERR (t) = ΔAmpERR (t = 0) − ΔAmpERR (t) to the RQI. While ΔΔAmpERR increases throughout the entire time course of the experiment, RQI reflects the degradation only during the first 60 min. Throughout the experiment ΔΔAmpERR increases by 8 cycles, which corresponds to 256-fold (=28) difference. Hence, the sensitivity of ΔΔAmpERR is sufficient to detect a loss of 99.7% (=1 − 1/256) of the amount of the initial transcripts post sampling. The initial drop in RQI from close to 10 to about 7 corresponds to an increase of 1 cycle in ΔΔAmpERR, suggesting that RQI is very sensitive to the initial degradation, when 50% of the transcripts are lost. RQI then changes from 7 down to about 3.5, while ΔΔAmpERR increases from 1 to 6 cycles, and then there are no more changes in the RQI while ΔΔAmpERR continues to increase to at least 8 cycles. Clearly, ΔΔAmpERR reflects degradation over a wider range than RQI and it is also more sensitive when degradation is extensive.
To test if the ERR resistance is limited to the particular sequence amplified, which might be due to an unusual secondary or tertiary structure or tight binding to proteins, we repeated the experiment amplifying other regions of the ERR and also using amplicons of different lengths (Supplementary material 2). The formation of all those amplicons was virtually ignorant to the RNase treatment suggesting that the entire ERR RNA is resistant to RNases.
3.2. Degradation by heat
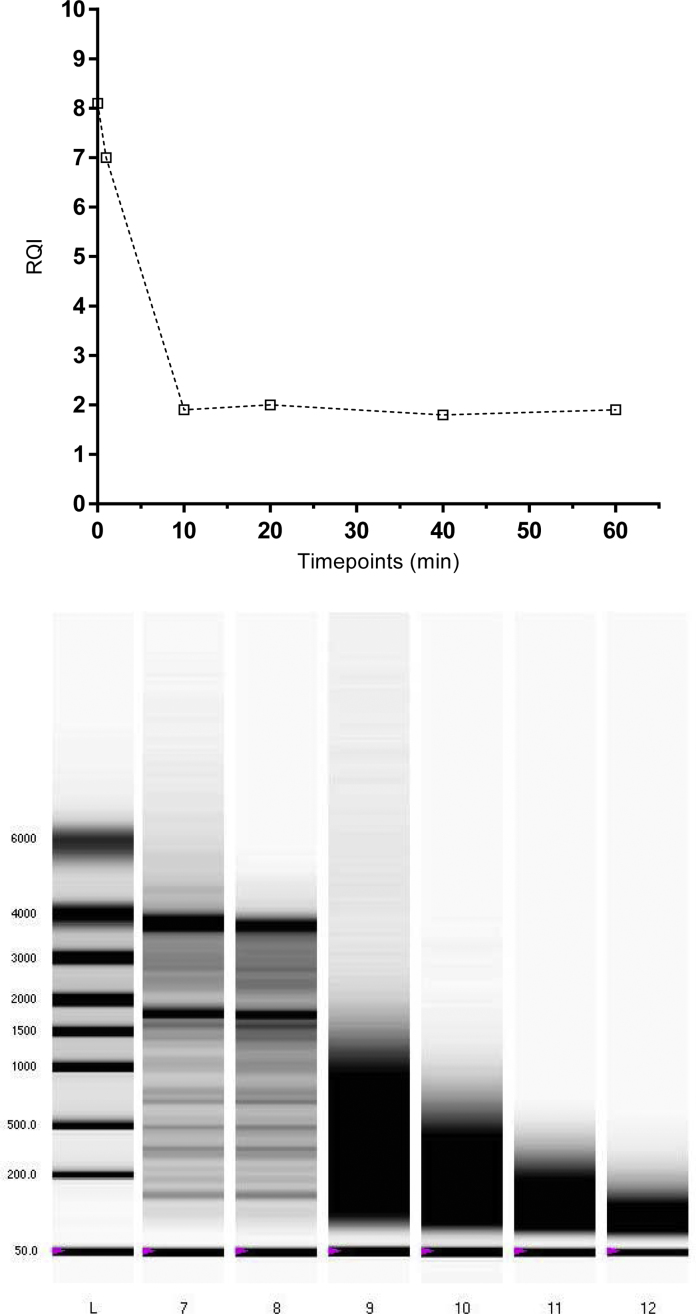
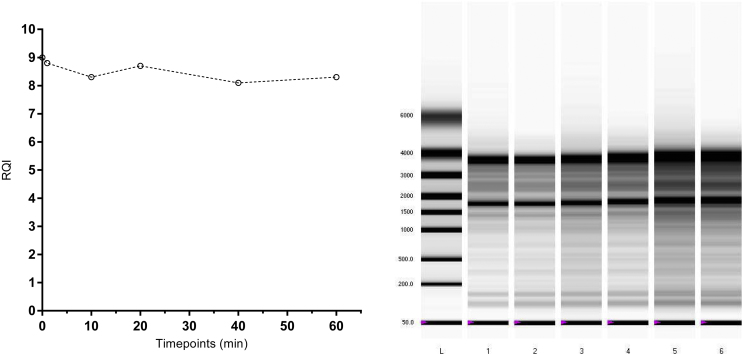
Exposure of RNA to 95 °C leads to degradation that is reflected by a smear in the electrophoretic traces and a drop of RQI from over 8 to below 2 in 10 min, where after no further changes are observed (Fig. 3). The degradation was also monitored by qPCR assessing the cDNA produced by reverse transcription with assays producing short (S ∼100 bp), medium length (M, 100–200 bp), and long (L, >300 bp) amplicons comparing the amounts produced pair-wise.
Fig. 3.
Purified human RNA incubated at 95 °C. RQI plotted against incubation time and gel picture of all time points.
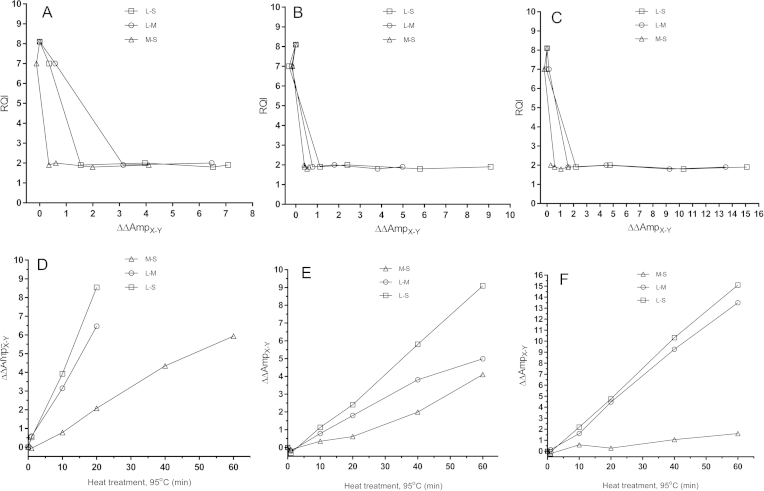
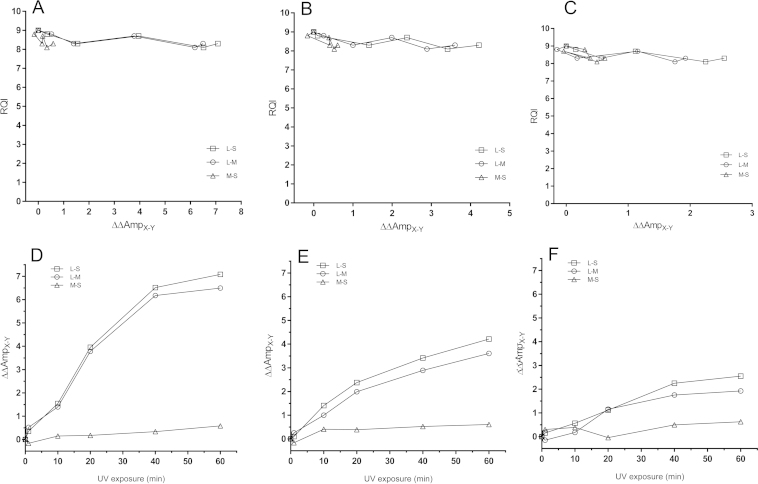
For systems with different amplicon lengths we define ΔAmp as ΔAmpX–Y = CqX − CqY (where X, is either L = long or M = medium amplicon, and Y is either M = medium or S = short amplicon), and ΔΔAmpX/Y = ΔAmpX–Y (t = 0) − ΔAmpX–Y(t) (Fig. 4d–f) to relate to the amount of fully intact mRNA present before heat treatment. We measured ΔΔAmpX–Y for all three combinations of amplicon lengths (L–S, L–M, M–S) for three different targets: a typical reference gene (B2 M), 18S rRNA and the ERR marker. ΔΔAmpX–Y depends on the target and also on the selected amplicons’ lengths. Generally, ΔΔAmpX–Y is largest for the L–S combination (i.e., ΔΔAmpL–S), where the amplicons’ length differ the most, and it reaches values well over 8 cycles (Fig. 4a–c). For the 18S rRNA ΔΔAmpL–S reaches even 14 cycles reflecting exceeding sensitivity to heat degradation. Notably, the ERR marker, which was resistant to degradation by RNases, is sensitive to heat, and all three markers tested: 18S rRNA, the reference gene B2 M, and the ERR marker show similar responses. This is expected, as heat treatment should be ignorant to the chemical nature of the RNA. Reaching the extreme ΔΔAmpL–S levels, however, is only possibly for the 18S rRNA, because of its very high abundance. However, for all three markers, and all three lengths combinations, ΔΔAmpX–Y shows superior sensitivity and dynamic range to the RQI (Fig. 4d–f) when RNA is degraded by heat.
Fig. 4.
Purified human RNA incubated at 95 °C. RQI plotted against ΔΔAmpERR for B2 M (A), ERR marker (B) and 18S (C). Incubation time plotted against ΔΔAmpERR for B2 M (D), ERR marker (E) and 18S (F).
3.3. Damage by UV light
We see no effect on RQI or electrophoretic traces of purified human RNA exposed to UV light for up to 60 min (Fig. 5), suggesting it is intact. Applying ΔAmp qPCR, however, we find pronounced aberrations induced by the UV for all three targets: the reference gene B2 M, 18S rRNA, and the ERR marker (Fig. 6). Although the ΔΔAmpX–Y values vary, which they should because of the different amplicon lengths, we observe changes of several cycles; the highest being 7.1. Clearly, the RNA is extensively damaged by the UV irradiation and amplification is severely compromised. This damage, however, is not sensed by the electrophoretic analysis and it is not reflected by the RQI. To validate this is not an effect of the particular Experion (Bio-Rad) electrophoretic system, the experiment was repeated using the Bioanalyzer (Agilent Technologies), and also extending the UV exposure to 120 min (Supplementary material 3). Even after 120 min of UV exposure, degradation is not obvious from the electrophoretic traces, while ΔAmp qPCR analysis reflects severe damage. By increasing the UV intensity we eventually could measure degradation by electrophoresis (Supplementary material 3) [9], but at this stage the damage to the RNA was so severe we could no longer amplify the material after the initial time points (Supplementary Material 3). Clearly, UV light induces damage to RNA that severely interferes with its quantification by RT-qPCR, but is not always reflected by electrophoretic analysis and RIN/RQI numbers.
Fig. 5.
Purified human RNA exposed to UV radiation. RQI plotted against incubation time and gel picture of all time points.
Fig. 6.
Purified human RNA exposed to UV radiation. RQI plotted against ΔΔAmpERR for B2 M (A), ERR marker (B) and 18S (C). Exposure time plotted against ΔΔAmpERR for B2 M (D), ERR marker (E) and 18S (F).
3.4. Degradation by fixatives
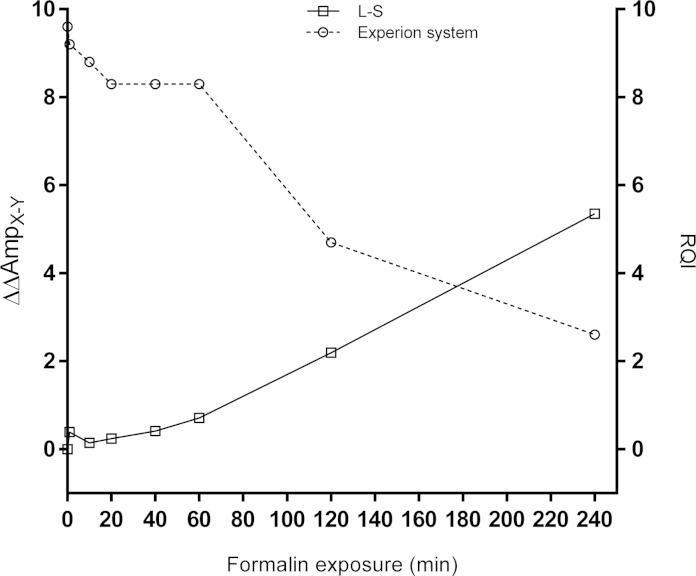
To test the effect of formalin fixation extracted RNA was treated with formalin at room temperature for various length of times and analyzed with ΔAmp qPCR using Differential Amplicons and by automated electrophoresis determining RQI (Fig. 7). The ΔΔAmpX–Y and RQI show similar (though inverse) time profiles. This is expected if the formalin treatment induces breaks in the RNA, in addition to various chemical modifications that do not cause strand breaks, as rRNA dominates the electropherogram. Other RNAs tested show similar degradation profiles, suggesting that all RNA species are damaged by formalin to similar extent (Supplementary material 2).
Fig. 7.
RQI and ΔΔAmpX–Y for 18S rRNA as function of the time exposed to formalin.
4. Discussion
RNases in live cells are contained and degradation of mRNAs is strictly controlled as part of the cell’s RNA metabolism. However, post sampling, during the preanalytical process, cells die, become permeabilized and/or lysed, which brings endogenous RNases into contact with RNA and exogenous RNases may enter leading to mRNA degradation. This can severely compromise quantitative gene expression analysis. In established workflows in pathology tissues are treated with formalin and embedded in paraffin (FFPE), which preserves samples’ morphology but is deleterious to the RNA. These factors may profoundly influence measured expression levels leading to unreliable and in worst cases fatally incorrect results [17]. mRNA degradation is a particular severe problem when analysing tissues rich in RNases, such as pancreas and spleen. It may also be a problem when tissue or cell samples are frozen and then thawed, since freezing damages the cells, releasing RNases giving them access to the RNA. Equipment and reagents may also be contaminated with RNases leading to serious degradation. It is therefore critical to test the integrity of the RNA before performing serious analyses [18]. Traditionally this is done using electrophoresis, which works very well under many conditions, but has limitations.
Here we have developed a new molecular approach based on measuring differential amplicons (ΔAmp) to assess specifically the integrity of mRNA for expression profiling studies and we have evaluated it on test systems, where RNA was degraded or damaged by exposure to ribonuclease, heat, ultraviolet radiation, and formalin. The approach is based on an Endogenous RNase Resistant (ERR) marker that is virtually resistant to RNases, which is assayed by RT-qPCR producing two amplicons of different length and also compared to a representative reference gene. ΔAmp is compared to the current method based on assessing the integrity of RNA by means of electrophoresis using the Experion (Bio-Rad) or Bioanalyzer (Agilent Technologies).
The degree of damage caused by RNases is reflected by the differential expression of a reference gene(s) and the ERR marker. The reference genes are assumed to have been selected carefully and validated, such that their expression is stable across samples and invariant to the studied conditions. The differential expression ΔAmpERR = CqRG − CqERRthen reflects the integrity of the mRNA in the sample. The sensitivity of ΔAmpERR to the damage caused by RNases is much greater than that of the RQI and related indexes obtained from analysis of electropherograms. Furthermore, the ΔAmpERR reflects the integrity of mRNA rather than of rRNA that dominate the electropherogram and has quite different resistance to RNases. We do not know the mechanism behind the stability of the ERR marker, but it is independent of the targeted sequence within the ERR, and the marker is stable in human (Fig. 1), mouse (Fig. 2) and rat (not shown).
Degradation by heat is expected to affect all RNA molecules similarly and is reflected by measuring ΔAmp, expressed as ΔAmpX–Y = CqX − CqY. Essentially any RNA that is reasonably abundant can be targeted. The lengths of the amplicons can be chosen to reflect the degree of degradation that is most relevant for each experiment; the shorter the amplicons the higher the degree of degradation can be assessed. Biological samples are usually not exposed to heat and heat degradation is rarely a problem. One exception is during the paraffination and de-paraffination of FFPE samples. The non-specific degradation by heat is also expected to cause similar damage as the non-specific degradation that occurs during chemical fixation of tissues. Electrophoretic analysis measuring indicators such as RQI are relevant to assess non-specific damage, which affect mRNAs and rRNAs the same way. ΔAmpX–Y is, however, a much more sensitive quality indicator that is also more relevant for gene expression measurements.
RNA damage by UV radiation is found to have small impact on the RQI/RIN. Considering that UV radiation primarily induces intramolecular crosslinking of thymines and rarely induces strand breaks, the insensivity of the electrophoretic approach to this kind of damage is reasonable. The UV induced damage is, however, readily, sensed by ΔAmp and reflected by the ΔAmpX–Y.
The new approach presented here has important advantages to the current strategy based on automated electrophoretic systems determining quality indicators such as RIN and RQI values. The electrophoretic approaches are not particularly sensitive to more seriously degraded RNA; they reflect quality of rRNA rather than mRNA, and to UV damage. In contrast ΔAmp assesses directly the quality of mRNA and can be designed with essentially any sensitivity and dynamic range by carefully optimizing the amplicons’ lengths and choosing appropriate targets. Here we used amplicons that were about 75 bp, 150 bp and 300 bp and a typical reference gene, 18S rRNA and an ERR marker, which allowed us to detect over 99% degradation by several different mechanisms. Using three amplicon lengths and three targets is unnecessary for routine applications. For regular studies two amplicon lengths of e.g., the ERR marker combined with one or several reference genes is sufficient.
Acknowledgement
This work was supported by EU grants FP7-PEOPLE-ITN-2008-237956: EduGlia (Innovative Techniques and Models to Study Glia-Neuron interactions) and FP7-HEALTH-2007-B-222916: SPIDIA (Standardisation and improvement of generic pre-analytical tools and procedures for in vitro diagnostics), research projects AV0Z50520701, Grant Agency of the Czech Republic [P303/13/02154S]; BIOCEV CZ.1.05/1.1.00/02.0109 and the program for perspective researchers of the Academy of the Czech Republic.
Footnotes
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bdq.2015.09.002.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Tichopad A., Kitchen R., Riedmaier I., Becker C., Stahlberg A., Kubista M. Design and optimization of reverse-transcription quantitative pcr experiments. Clin. Chem. 2009;55:101816–101823. doi: 10.1373/clinchem.2009.126201. [DOI] [PubMed] [Google Scholar]
- 2.Pazzagli M., Malentacchi F., Simi L., Orlando C., Wyrich R., Günther K. SPIDIA-RNA: first external quality assessment for the pre-analytical phase of blood samples used for RNA based analyses. Methods. 2013;59:20–31. doi: 10.1016/j.ymeth.2012.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Li Y., Breaker R.R. Kinetics of RNA degradation by specific base catalysis of transesterification involving the 2′-hydroxyl group. J. Am. Chem. Soc. 1999;121(23):5364–5372. [Google Scholar]
- 4.Larson D.E., Zahradka P., Sells B.H. Control points in eukaryotic ribosome biogenesis. Biochem. Cell Biol. 1991;69:5–22. doi: 10.1139/o91-002. [DOI] [PubMed] [Google Scholar]
- 5.Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sidova M., Tomankova S., Abaffy P., Kubista M., Sindelka R. Effects of post-mortem and physical degradation on RNA integrity and quality. Biomol. Detect. Quantif. 2015 doi: 10.1016/j.bdq.2015.08.002. http://dx.doi.org/10.1016/j.bdq.2015.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rainen L., Oelmueller U., Jurgensen S., Wyrich R., Ballas C., Schram J. Stabilization of mRNA expression in whole blood samples. Clin. Chem. 2002;48(11):1883–1890. [PubMed] [Google Scholar]
- 8.Ibberson D., Benes V., Muckenthaler M., Castoldi M. RNA degradation compromises the reliability of microRNA expression profiling. BMC Biotechnol. 2009;9:102. doi: 10.1186/1472-6750-9-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becker C., Hammerle-Fickinger C., Riedmaier I., Pfaffl M.W. mRNA and microRNA quality control for RT-qPCR analysis. Methods. 2010;50(4):237–243. doi: 10.1016/j.ymeth.2010.01.010. [DOI] [PubMed] [Google Scholar]
- 10.Nolan T., Hands R.E., Bustin S.A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 2006;1(3):1559–1582. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 11.Die J.V., Roman B. RNA quality assessment: a view from plant qPCR studies. J. Exp. Bot. 2012;63:6069–6077. doi: 10.1093/jxb/ers276. [DOI] [PubMed] [Google Scholar]
- 12.Ståhlberg A., Håkansson J., Xian X., Semb H., Kubista M. Properties of the reverse transcription reaction in mRNA quantification. Clin. Chem. 2004;50(3):509–515. doi: 10.1373/clinchem.2003.026161. [DOI] [PubMed] [Google Scholar]
- 13.Viertler C., Groelz D., Gündisch S., Kashofer K., Reischauer B., Riegman P. A new technology for stabilization of biomolecules in tissues for combined histological and molecular analyses. J. Mol. Diagn. 2012;14(5):458–466. doi: 10.1016/j.jmoldx.2012.05.002. [DOI] [PubMed] [Google Scholar]
- 14.Kubista M., Björkman J., Svec D., Sjöback S. RNA quality matters. Eur. Pharmaceut. Rev. 2012;17(6):63–67. [Google Scholar]
- 15.Kashofer K., Viertler C., Pichler M., Zatloukal K. Quality control of RNA preservation and extraction from paraffin-embedded tissue: implications for RT-PCR and microarray analysis. PLoS One. 2013;8(7):e70714. doi: 10.1371/journal.pone.0070714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.J. Björkman, M. Kubista. Methods for assessing rna quality. Patent No. 13710858. 5-1404, 2015.
- 17.Kap M., Sieuwerts A.M., Kubista M., Oomen M., Arshad S., Riegman P. The influence of tissue procurement procedures on RNA integrity, gene expression, and morphology in porcine and human liver tissue. Biopreserv. Biobank. 2015;13(3):200–206. doi: 10.1089/bio.2014.0076. [DOI] [PubMed] [Google Scholar]
- 18.CEN/TC 140 “In vitro diagnostic medical devices”.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.