Scientific conferences fulfil many roles, but one of the most important ones is that they help shape the direction in which a scientific discipline grows by promoting person-to-person exchanges of information, ideas and constructive criticisms between scientists from different backgrounds. This interaction also helps to identify areas of controversy and promotes efforts to address and, it is hoped, resolve them. This year is the 30th anniversary of the publication of the first practical description of the polymerase chain reaction [1], arguably one of the simplest and the most widely used molecular technology. It also sees the 7th instalment of the Freising PCR meetings (http://www.qpcr-ngs-2015.net/), which are the longest established, continuous and most influential conferences in this field and have provided a looking glass for conceptual and technical innovation as well as practical applications of PCR-associated methods.
The first of this conference series took place in March 2004, a propitious time for such a conference, eleven years after Russ Higuchi published his paper describing the use of real-time quantitative PCR (qPCR) [2] and nine years after the publication of molecular beacons [3] and a 5′ hydrolysis-based qPCR assay [4]. The meeting was organised, as it still is, by Michael Pfaffl, one of the founding editors of Biomolecular Detection and Quantification (BDQ). Michael had dabbled with competitive reverse transcription (RT)-PCR, preparing synthetic competitive templates and detecting PCR amplification products using HPLC-UV to quantify IGF-1 mRNA levels in bovines [5]. But in 2001 he published his seminal “new mathematical model for relative quantification in real-time RT-PCR” [6], which has been cited over 14,000 times in the peer-reviewed literature. This was followed by another highly cited publication, which introduced the relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in qPCR [7]. Meanwhile another founding editor of BDQ had long since established his reputation as one of the foremost innovators and though leaders in the field. Carl Wittwer described a new type of thermal cycler that was based on heat transfer with air to samples in sealed capillary tubes, similar to a “recirculating hair dryer” [8]. He followed this up with a rapid cycling protocol that introduced the concept of a 15 min PCR [9], something he has recently bested with a 15 s PCR [10]. In fact, by 2004 a considerable amount of Carl's research effort had gone into increasing the efficiency of the PCR and optimising instruments, time and temperature parameters [11], [12], [13], [14], [15] as well as finding suitable applications for all these improvements, for example fusion transcript detection in leukaemia [16], multiplexing with hybridisation probes for genotyping [17] and melt curve analysis for the detection of chromosomal translocations in mantle cell lymphoma [18], to name but a few. Another pioneer of qPCR was Mikael Kubista, who by this time had established his reputation with the invention of light up probes [19], novel fluorescent dyes suitable for qPCR [20] and had also turned his attention to the question of how to deal with PCR assays with different efficiencies in the exponential phase of the reaction [21]. Together with his then student Anders Stahlberg he published two insightful papers on the variability of the reverse transcription step [22], [23], which underlies many molecular assays and was the first empirical demonstration of the enzyme, target and concentration-dependence of cDNA synthesis efficiency. By 2004 Jo Vandesompele had published his ground-breaking GeNorm paper [24], which has been cited over 8000 times and provided an ingenious solution to the problem of how to select appropriate reference genes for the normalisation of RNA data. He had also started to consider approaches to improving and simplifying primer and probe design and announced details of the first public database application for the storage and retrieval of validated qPCR primer and probe sequence records [25].
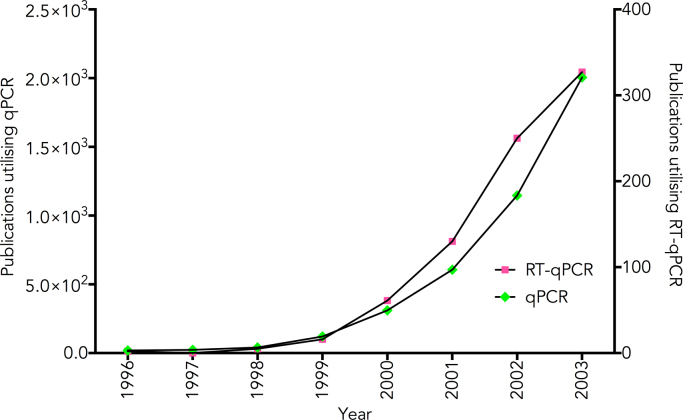
By the time of the first Freising meeting, the advantages of qPCR were beginning to be appreciated, a development reflected in the number of papers published by the end of 2003 that made use of this technology (Fig. 1). The combination of reverse transcription and PCR into RT-qPCR was also utilised from the very beginning and followed the same curve, albeit at a much lower level. However, whilst there were comparatively few technical problems associated with DNA-targeted quantitative PCR, by 2004 it was becoming clear that reliable RT-qPCR assays were a different matter: results depended on RNA quality, consistent RT conditions, appropriate normalisation and the application of suitable statistical methodologies [26], [27]. The uncertainty arising from these four issues with regards to biological or clinical relevance of many RT-qPCR results were discussed at that first meeting and they have continued to play a central role in every one of the subsequent meetings held in September 2005 (2nd), March 2007 (3rd), March 2009 (4th), March/April 2011 (5th), March 2013 (6th) and undoubtedly will do so at the 7th.
Fig. 1.
Publications utilising qPCR published by the time of the first Freising meeting (1996–12/2003). Results from a PubMed search for the terms (1) “real-time PCR” or “realtime PCR” or “real time PCR” or qPCR and (2) “real-time PCR” or “realtime PCR” or “real time PCR” or qPCR and “reverse transcription” were plotted against the year the publication appeared.
The meeting of minds at the various Freising conferences certainly paved the way for the publication of the minimum information for the publication of quantitative PCR (MIQE) guidelines, which appeared in 2009 [28]. These arose out of the conviction of a worldwide group of PCR practitioners that there were serious issues with the way qPCR experiments in general, but RT-qPCR experiments in particular were being performed. A survey of RT-qPCR practices taken at the 2005 London qPCR meeting had revealed extensive variation in assay design, validation and analysis, with little regard for reporting experimental detail or paying attention to the importance of the four areas mentioned previously [29]. The MIQE guidelines had the aim of establishing best practice guidelines for the design of qPCR experiments and the subsequent transparent reporting of experimental detail. The guidelines have certainly been successful in so far as that there is a universal awareness of their existence, with all major suppliers of qPCR instruments and reagents committed to promoting their use. However, recent surveys of qPCR-based papers continue to find that the vast majority of papers are significantly flawed in the reporting of experimental detail and often use inappropriate methods long since shown to be invalid [30], [31], [32]. The most obvious example is the continued, near-universal use of single reference genes, which for precise applications are not able to generate reliable and biologically meaningful results [24], [33], [34], [35], [36].
Inhibition is one key area that affects significantly the reliability of a (RT)-PCR assay, but is often ignored. Inhibition affects both reverse transcriptases [37] and thermostable DNA polymerases [38] and some polymerases are more susceptible to inhibition than others [39]. This was recognised early on and there have been numerous reports detailing the effects of many different components on the reliability of (RT)-PCR results [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52] and proposing solutions that overcome [21], [49], [53], [54], [55], [56], [57], [58], [59], [60] or at least detect [61] that inhibition. One worrying aspect of inhibition is that it does not affect all PCR reactions to the same extent, i.e. some assays are more susceptible to inhibition than others [62]. This has important implications for any gene expression experiment, as a differential effect of inhibition on target genes of interest as well as the reference genes will result in incorrect results and is also another source of error for PCR-based molecular diagnostic assays. Unfortunately, inhibition testing is not at all common [32] and there is an urgent need to consider inhibition compatibility when conducting PCR analyses [62].
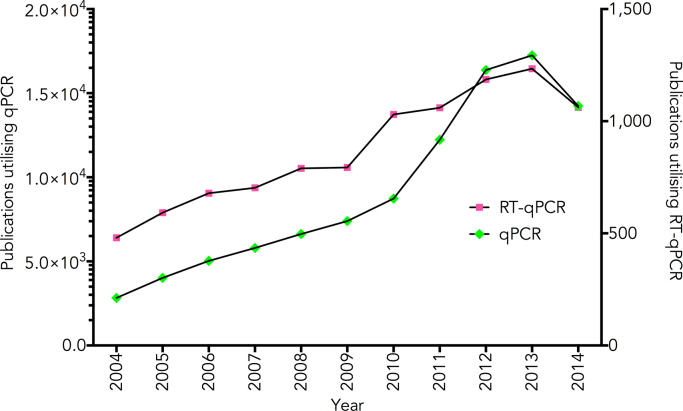
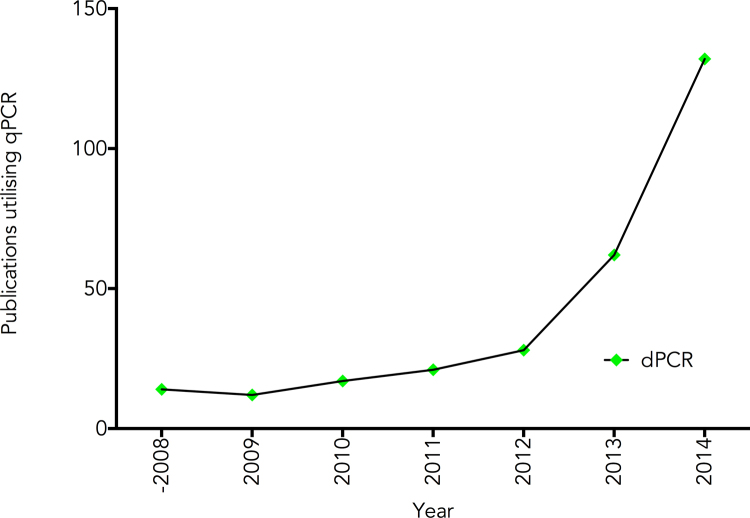
An interesting development is that 2014 is the first year in which there has been a reduction in the number of qPCR as well as RT-qPCR papers published (Fig. 2). It will be interesting to see whether this is a blip or the beginning of an impact exerted by a combination of next generation sequencing and digital PCR. The latter is certainly beginning to take off, but is currently where qPCR was in 2000 (Fig. 3).
Fig. 2.
Publications utilising qPCR published from 2004 to 2014. Results from a PubMed search for the terms (1) “real-time PCR” or “realtime PCR” or “real time PCR” or qPCR and (2) “real-time PCR” or “realtime PCR” or “real time PCR” or qPCR and “reverse transcription” were plotted against the year the publication appeared.
Fig. 3.
Publications utilising dPCR. Results from a PubMed search for the term “digital PCR” were plotted against the year the publication appeared.
Ironically, dPCR precedes qPCR [63] but had to await the development of suitable instrumentation to become a serious competitor to qPCR. It allows very precise measurement of DNA molecules by partitioning a limiting dilution of DNA into a succession of individual PCR reactions. DNA templates are randomly distributed into sub-reactions, termed partitions, and as long as there are negative partitions, Poisson statistics can be used to measure the quantities of DNA present for a given proportion of positive partitions (Fig. 4). There is no longer a need for calibration curves [64] and dPCR may even be less susceptible to inhibitors [65]. The publication of the MIQE guidelines for dPCR [66] might just help avoid many of the pitfalls that have opened up for qPCR, especially as it has become clear that many of the issues faced by (RT)-qPCR are also issues for (RT)-dPCR [64], [67], [68], [69].
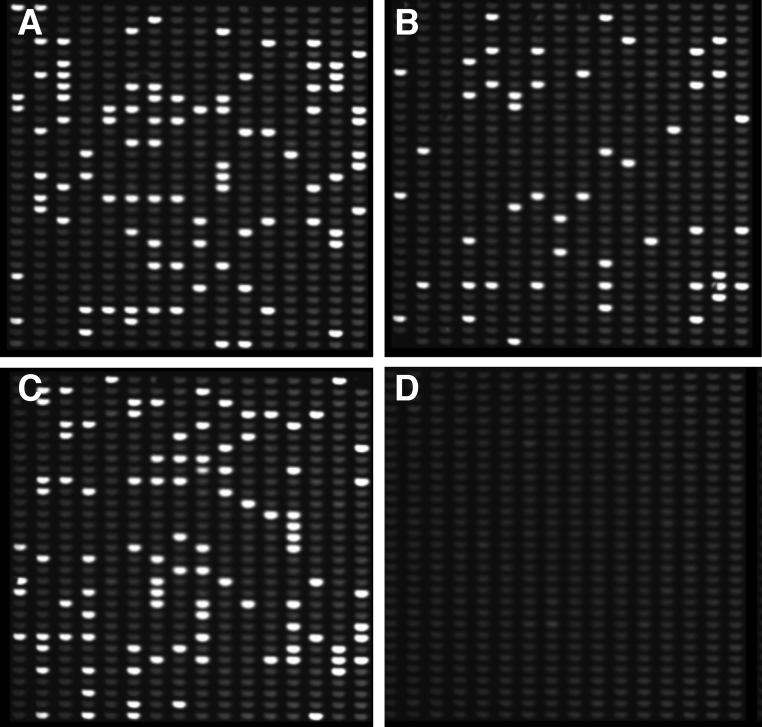
Fig. 4.
dPCR analysis of Aspergillus DNA. DNA concentrations of A. fumigatus, A. terreus and A. flavus preparations were measured on a Nanodrop instrument and samples were diluted to 40 fg/μl, and analysed on a Formulatrix Constellation dPCR instrument following 40 cycles of PCR with Agilent Brilliant III mastermix. Hydrolysis probes and primers targeting the 18S rDNA repeat were used, together with a published protocol [77]. (A) A. fumigatus. (B) A. terreus. (C) A. flavus.
Next generation high throughput sequencing (NGS) has become an increasingly important element of the Freising meetings, not surprisingly given its companion status with PCR and its impact on biological and clinical applications [70]. There has been rapid progress in sequencing technologies, with less efficient, competitive or more expensive methods dropping out and being replaced with rival faster, cheaper and more accurate technologies. Together with the continuous improvement of bioinformatics tools, this is allowing smaller research groups to consider more routine use of NGS. Indeed, over the course of the next decade this is likely to result in the replacement of rival high-throughput technologies.
As always, of course, there are issues with NGS [71], [72] and, again not surprisingly, these are often associated with whole transcriptome (RNAseq) applications especially when applied to the analysis of low levels of RNA [73], [74].
Unambiguous assembly into a single contig of repetitive elements that are longer than sequencing read-length is another problem with short read NGS technology. Instead, one is left with multiple contigs that leave gaps in whole genome assemblies. Again technology is coming to the rescue and DNA sequencing using nanopore technology shows promise as an alternative method for producing long-read sequence data more cheaply [75]. A recent report uses this technology to solve the structure of a complex antibiotic resistance island in Salmonella typhi [76], achieving median read lengths of 6 kb with an accuracy of 72%. Promisingly, this was achieved without any attempts made to optimise read length, which was determined by the length of the input DNA, not the chemistry. The authors rightly conclude by saying that this type of technology has the potential to create a paradigm shift in genomics, bringing low cost, long-read sequencing to the nonspecialist laboratory.
The increasing focus on accurate as well as precise quantification of nucleic acids, proteins and small molecules requires a forum for the exchange of ideas, concepts, tools and applications that reaches way beyond the scope of a scientific meeting, no matter how well organised, attended and influential. BDQ hopes to provide a platform for precisely this purpose. BDQ is an open access, peer-reviewed international journal dedicated to championing excellence in molecular study design, measurement, data analysis and reporting. Its focus is on the application of qualitative and quantitative molecular methodologies to all areas of clinical and life sciences. The journal has two main aims:
-
•
to provide a forum for discussion and recommendation of guidelines designed to improve the accuracy of molecular measurement, its data analysis and the transparency of its subsequent reporting;
-
•
to publish molecular biology based studies that adhere to best practice guidelines, both current and future.
BDQ was established by a group of scientists based on their experience developing and publishing the MIQE and digital MIQE guidelines. The deliberately broad scope of the journal covers clinical areas such as cancer, epigenetics, metagenomics, and infectious diseases as well non clinical subjects including environmental sciences, microbiology and food science. BDQ revolves around the common theme of promoting excellence in molecular measurement and its data analysis. It will serve as a repository for sharing key findings across what may otherwise be disparate specialties. We look forward to receiving manuscripts from the attendees of this year's conference and hope that BDQ will become the journal of choice for an increasing number of scientific publications dedicated to transparency of reporting, excellence of protocols and significance of results.
Acknowledgements
We thank Drs. Gemma Johnson, Sara Kirvell and Helen Moor for providing the dPCR data shown in Fig. 4.
Contributor Information
Jim F. Huggett, Email: Jim.Huggett@lgcgroup.com.
Justin O’Grady, Email: Justin.OGrady@uea.ac.uk.
Stephen Bustin, Email: Stephen.bustin@anglia.ac.uk.
References
- 1.Saiki R.K., Scharf S., Faloona F., Mullis K.B., Horn G.T., Erlich H.A. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985;230:1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- 2.Higuchi R., Fockler C., Dollinger G., Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (NY) 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 3.Tyagi S., Kramer F.R. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 4.Heid C.A., Stevens J., Livak K.J., Williams P.M. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- 5.Pfaffl M., Meyer H.H., Sauerwein H. Quantification of insulin-like growth factor-1 (IGF-1) mRNA: development and validation of an internally standardised competitive reverse transcription-polymerase chain reaction. Exp Clin Endocrinol Diabetes. 1998;106:506–513. doi: 10.1055/s-0029-1212025. [DOI] [PubMed] [Google Scholar]
- 6.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:E45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pfaffl M.W., Horgan G.W., Dempfle L. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002;30:e36. doi: 10.1093/nar/30.9.e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wittwer C.T., Fillmore G.C., Hillyard D.R. Automated polymerase chain reaction in capillary tubes with hot air. Nucleic Acids Res. 1989;17:4353–4357. doi: 10.1093/nar/17.11.4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wittwer C.T., Fillmore G.C., Garling D.J. Minimizing the time required for DNA amplification by efficient heat transfer to small samples. Anal Biochem. 1990;186:328–331. doi: 10.1016/0003-2697(90)90090-v. [DOI] [PubMed] [Google Scholar]
- 10.Farrar J.S., Wittwer C.T. Extreme PCR: efficient and specific DNA amplification in 15–60 seconds. Clin Chem. 2015;61:145–153. doi: 10.1373/clinchem.2014.228304. [DOI] [PubMed] [Google Scholar]
- 11.Wittwer C.T., Garling D.J. Rapid cycle DNA amplification: time and temperature optimization. Biotechniques. 1991;10:76–83. [PubMed] [Google Scholar]
- 12.Wittwer C.T., Marshall B.C., Reed G.H., Cherry J.L. Rapid cycle allele-specific amplification: studies with the cystic fibrosis delta F508 locus. Clin Chem. 1993;39:804–809. [PubMed] [Google Scholar]
- 13.Swerdlow H., Jones B.J., Wittwer C.T. Fully automated DNA reaction and analysis in a fluidic capillary instrument. Anal Chem. 1997;69:848–855. doi: 10.1021/ac961104o. [DOI] [PubMed] [Google Scholar]
- 14.Wittwer C.T., Ririe K.M., Andrew R.V., David D.A., Gundry R.A., Balis U.J. The LightCycler: a microvolume multisample fluorimeter with rapid temperature control. Biotechniques. 1997;22:176–181. doi: 10.2144/97221pf02. [DOI] [PubMed] [Google Scholar]
- 15.Bernard P.S., Lay M.J., Wittwer C.T. Integrated amplification and detection of the C677T point mutation in the methylenetetrahydrofolate reductase gene by fluorescence resonance energy transfer and probe melting curves. Anal Biochem. 1998;255:101–107. doi: 10.1006/abio.1997.2427. [DOI] [PubMed] [Google Scholar]
- 16.Hussey C.E., Lyon E., Millson A., Lay M.J., Wittwer C.T., Segal G.H. A rapid practical RT-PCR-based approach for the detection of the PML/RAR alpha fusion transcript in acute promyelocytic leukemia. Am J Clin Pathol. 1999;112:256–262. doi: 10.1093/ajcp/112.2.256. [DOI] [PubMed] [Google Scholar]
- 17.Bernard P.S., Pritham G.H., Wittwer C.T. Color multiplexing hybridization probes using the apolipoprotein E locus as a model system for genotyping. Anal Biochem. 1999;273:221–228. doi: 10.1006/abio.1999.4217. [DOI] [PubMed] [Google Scholar]
- 18.Bohling S.D., Wittwer C.T., King T.C., Elenitoba-Johnson K.S. Fluorescence melting curve analysis for the detection of the bcl-1/JH translocation in mantle cell lymphoma. Lab Invest. 1999;79:337–345. [PubMed] [Google Scholar]
- 19.Svanvik N., Stahlberg A., Sehlstedt U., Sjoback R., Kubista M. Detection of PCR products in real time using light-up probes. Anal Biochem. 2000;287:179–182. doi: 10.1006/abio.2000.4824. [DOI] [PubMed] [Google Scholar]
- 20.Bengtsson M., Karlsson H.J., Westman G., Kubista M. A new minor groove binding asymmetric cyanine reporter dye for real-time PCR. Nucleic Acids Res. 2003;31:e45. doi: 10.1093/nar/gng045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bar T., Stahlberg A., Muszta A., Kubista M. Kinetic Outlier Detection (KOD) in real-time PCR. Nucleic Acids Res. 2003;31:e105. doi: 10.1093/nar/gng106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stahlberg A., Kubista M., Pfaffl M. Comparison of reverse transcriptases in gene expression analysis. Clin Chem. 2004;50:1678–1680. doi: 10.1373/clinchem.2004.035469. [DOI] [PubMed] [Google Scholar]
- 23.Stahlberg A., Hakansson J., Xian X., Semb H., Kubista M. Properties of the reverse transcription reaction in mRNA quantification. Clin Chem. 2004;50:509–515. doi: 10.1373/clinchem.2003.026161. [DOI] [PubMed] [Google Scholar]
- 24.Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:0034. doi: 10.1186/gb-2002-3-7-research0034. 0034.1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pattyn F., Speleman F., De Paepe A., Vandesompele J. RTPrimerDB: the real-time PCR primer and probe database. Nucleic Acids Res. 2003;31:122–123. doi: 10.1093/nar/gkg011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bustin S.A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–193. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 27.Bustin S.A. Quantification of mRNA using real-time reverse transcription PCR (RT-PCR): trends and problems. J Mol Endocrinol. 2002;29:23–39. doi: 10.1677/jme.0.0290023. [DOI] [PubMed] [Google Scholar]
- 28.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 29.Bustin S.A. Real-time, fluorescence-based quantitative PCR: a snapshot of current procedures and preferences. Expert Rev Mol Diagn. 2005;5:493–498. doi: 10.1586/14737159.5.4.493. [DOI] [PubMed] [Google Scholar]
- 30.Abdel Nour A.M., Azhar E., Damanhouri G., Bustin S.A. Five years MIQE guidelines: the case of the Arabian countries. PLOS ONE. 2014;9:e88266. doi: 10.1371/journal.pone.0088266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dijkstra J.R., van Kempen L.C., Nagtegaal I.D., Bustin S.A. Critical appraisal of quantitative PCR results in colorectal cancer research: can we rely on published qPCR results? Mol Oncol. 2014;8:813–888. doi: 10.1016/j.molonc.2013.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bustin S.A., Benes V., Garson J., Hellemans J., Huggett J., Kubista M. The need for transparency and good practices in the qPCR literature. Nat Methods. 2013;10:1063–1067. doi: 10.1038/nmeth.2697. [DOI] [PubMed] [Google Scholar]
- 33.Tricarico C., Pinzani P., Bianchi S., Paglierani M., Distante V., Pazzagli M. Quantitative real-time reverse transcription polymerase chain reaction: normalization to rRNA or single housekeeping genes is inappropriate for human tissue biopsies. Anal Biochem. 2002;309:293–300. doi: 10.1016/s0003-2697(02)00311-1. [DOI] [PubMed] [Google Scholar]
- 34.Dheda K., Huggett J.F., Chang J.S., Kim L.U., Bustin S.A., Johnson M.A. The implications of using an inappropriate reference gene for real-time reverse transcription PCR data normalization. Anal Biochem. 2005;344:141–143. doi: 10.1016/j.ab.2005.05.022. [DOI] [PubMed] [Google Scholar]
- 35.Huggett J., Dheda K., Bustin S., Zumla A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 2005;6:279–284. doi: 10.1038/sj.gene.6364190. [DOI] [PubMed] [Google Scholar]
- 36.Dheda K., Huggett J.F., Bustin S.A., Johnson M.A., Rook G., Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. 2004;37:112–119. doi: 10.2144/04371RR03. [DOI] [PubMed] [Google Scholar]
- 37.Imai H., Yamada O., Morita S., Suehiro S., Kurimura T. Detection of HIV-1 RNA in heparinized plasma of HIV-1 seropositive individuals. J Virol Methods. 1992;36:181–184. doi: 10.1016/0166-0934(92)90149-8. [DOI] [PubMed] [Google Scholar]
- 38.Cone R.W., Hobson A.C., Huang M.L. Coamplified positive control detects inhibition of polymerase chain reactions. J Clin Microbiol. 1992;30:3185–3189. doi: 10.1128/jcm.30.12.3185-3189.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Abu Al-Soud W., Radstrom P. Capacity of nine thermostable DNA polymerases to mediate DNA amplification in the presence of PCR-inhibiting samples. Appl Environ Microbiol. 1998;64:3748–3753. doi: 10.1128/aem.64.10.3748-3753.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tichopad A., Didier A., Pfaffl M.W. Inhibition of real-time RT-PCR quantification due to tissue-specific contaminants. Mol Cell Probes. 2004;18:45–50. doi: 10.1016/j.mcp.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 41.Thornton C.G., Passen S. Inhibition of PCR amplification by phytic acid, and treatment of bovine fecal specimens with phytase to reduce inhibition. J Microbiol Methods. 2004;59:43–52. doi: 10.1016/j.mimet.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 42.Al-Soud W.A., Radstrom P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39:485–493. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Morre S.A., van Dijk R., Meijer C.J., van den Brule A.J., Kjaer S.K., Munk C. Pooling cervical swabs for detection of Chlamydia trachomatis by PCR: sensitivity, dilution, inhibition, and cost-saving aspects. J Clin Microbiol. 2001;39(6):2375–2376. doi: 10.1128/jcm.39.6.2375-2376.2001. [Letter] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mahony J., Chong S., Jang D., Luinstra K., Faught M., Dalby D. Urine specimens from pregnant and nonpregnant women inhibitory to amplification of Chlamydia trachomatis nucleic acid by PCR, ligase chain reaction, and transcription-mediated amplification: identification of urinary substances associated with inhibition and removal of inhibitory activity. J Clin Microbiol. 1998;36:3122–3126. doi: 10.1128/jcm.36.11.3122-3126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belec L., Authier J., Eliezer-Vanerot M.C., Piedouillet C., Mohamed A.S., Gherardi R.K. Myoglobin as a polymerase chain reaction (PCR) inhibitor: a limitation for PCR from skeletal muscle tissue avoided by the use of Thermus thermophilus polymerase. Muscle Nerve. 1998;21:1064–1067. doi: 10.1002/(sici)1097-4598(199808)21:8<1064::aid-mus11>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 46.Toye B., Woods W., Bobrowska M., Ramotar K. Inhibition of PCR in genital and urine specimens submitted for Chlamydia trachomatis testing. J Clin Microbiol. 1998;36:2356–2358. doi: 10.1128/jcm.36.8.2356-2358.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gibb A.P., Wong S. Inhibition of PCR by agar from bacteriological transport media. J Clin Microbiol. 1998;36:275–276. doi: 10.1128/jcm.36.1.275-276.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schwartz I., Varde S., Nadelman R.B., Wormser G.P., Fish D. Inhibition of efficient polymerase chain reaction amplification of Borrelia burgdorferi DNA in blood-fed ticks. Am J Trop Med Hyg. 1997;56:339–342. doi: 10.4269/ajtmh.1997.56.339. [DOI] [PubMed] [Google Scholar]
- 49.Chen J.T., Lane M.A., Clark D.P. Inhibitors of the polymerase chain reaction in Papanicolaou stain. Removal with a simple destaining procedure. Acta Cytol. 1996;40:873–877. doi: 10.1159/000333994. [DOI] [PubMed] [Google Scholar]
- 50.Wiedbrauk D.L., Werner J.C., Drevon A.M. Inhibition of PCR by aqueous and vitreous fluids. J Clin Microbiol. 1995;33:2643–2646. doi: 10.1128/jcm.33.10.2643-2646.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akane A., Matsubara K., Nakamura H., Takahashi S., Kimura K. Identification of the heme compound copurified with deoxyribonucleic acid (DNA) from bloodstains, a major inhibitor of polymerase chain reaction (PCR) amplification. J Forensic Sci. 1994;39:362–372. [PubMed] [Google Scholar]
- 52.Rossen L., Norskov P., Holmstrom K., Rasmussen O.F. Inhibition of PCR by components of food samples, microbial diagnostic assays and DNA-extraction solutions. Int J Food Microbiol. 1992;17:37–45. doi: 10.1016/0168-1605(92)90017-w. [DOI] [PubMed] [Google Scholar]
- 53.Burkhart C., Norris M., Haber M. A simple method for the isolation of genomic DNA from mouse tail free of real-time PCR inhibitors. J Biochem Biophys Methods. 2002;52:145–149. doi: 10.1016/s0165-022x(02)00052-0. [DOI] [PubMed] [Google Scholar]
- 54.Ogunjimi A.A., Choudary P.V. Adsorption of endogenous polyphenols relieves the inhibition by fruit juices and fresh produce of immuno-PCR detection of Escherichia coli O157:H7. FEMS Immunol Med Microbiol. 1999;23:213–220. doi: 10.1111/j.1574-695X.1999.tb01241.x. [DOI] [PubMed] [Google Scholar]
- 55.Jung R., Lubcke C., Wagener C., Neumaier M. Reversal of RT-PCR inhibition observed in heparinized clinical specimens. Biotechniques. 1997;23(24):26. doi: 10.2144/97231bm03. 28. [DOI] [PubMed] [Google Scholar]
- 56.Kamatchiammal S., Saravanakumar D., Kumarasamy N., Solomon S., Sritharan M., Sritharan V. A simple method for inhibition free PCR amplification of target DNA directly from clinical specimens. Indian J Clin Biochem. 1997;12:78–80. doi: 10.1007/BF02867961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kreader C.A. Relief of amplification inhibition in PCR with bovine serum albumin or T4 gene 32 protein. Appl Environ Microbiol. 1996;62:1102–1106. doi: 10.1128/aem.62.3.1102-1106.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gilgen M., Hofelein C., Luthy J., Hubner P. Hydroxyquinoline overcomes PCR inhibition by UV-damaged mineral oil. Nucleic Acids Res. 1995;23:4001–4002. doi: 10.1093/nar/23.19.4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.De Boer S.H., Ward L.J., Li X., Chittaranjan S. Attenuation of PCR inhibition in the presence of plant compounds by addition of BLOTTO. Nucleic Acids Res. 1995;23:2567–2568. doi: 10.1093/nar/23.13.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Widjojoatmodjo M.N., Fluit A.C., Torensma R., Verdonk G.P., Verhoef J. The magnetic immuno polymerase chain reaction assay for direct detection of salmonellae in fecal samples. J Clin Microbiol. 1992;30:3195–3199. doi: 10.1128/jcm.30.12.3195-3199.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nolan T., Hands R.E., Ogunkolade B.W., Bustin S.A. SPUD: a qPCR assay for the detection of inhibitors in nucleic acid preparations. Anal Biochem. 2006;351:308–310. doi: 10.1016/j.ab.2006.01.051. [DOI] [PubMed] [Google Scholar]
- 62.Huggett J.F., Novak T., Garson J.A., Green C., Morris-Jones S.D., Miller R.F. Differential susceptibility of PCR reactions to inhibitors: an important and unrecognised phenomenon. BMC Res Notes. 2008;1:70. doi: 10.1186/1756-0500-1-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sykes P.J., Neoh S.H., Brisco M.J., Hughes E., Condon J., Morley A.A. Quantitation of targets for PCR by use of limiting dilution. Biotechniques. 1992;13:444–449. [PubMed] [Google Scholar]
- 64.Huggett J.F., Whale A. Digital PCR as a novel technology and its potential implications for molecular diagnostics. Clin Chem. 2013;59:1691–1693. doi: 10.1373/clinchem.2013.214742. [DOI] [PubMed] [Google Scholar]
- 65.Hoshino T., Inagaki F. Molecular quantification of environmental DNA using microfluidics and digital PCR. Syst Appl Microbiol. 2012;35:390–395. doi: 10.1016/j.syapm.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 66.Huggett J.F., Foy C.A., Benes V., Emslie K., Garson J.A., Haynes R. The digital MIQE guidelines: minimum information for publication of quantitative digital PCR experiments. Clin Chem. 2013;59:892–902. doi: 10.1373/clinchem.2013.206375. [DOI] [PubMed] [Google Scholar]
- 67.Sanders R., Mason D.J., Foy C.A., Huggett J.F. Considerations for accurate gene expression measurement by reverse transcription quantitative PCR when analysing clinical samples. Anal Bioanal Chem. 2014 doi: 10.1007/s00216-014-7857-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nixon G., Garson J.A., Grant P., Nastouli E., Foy C.A., Huggett J.F. Comparative study of sensitivity, linearity, and resistance to inhibition of digital and nondigital polymerase chain reaction and loop mediated isothermal amplification assays for quantification of human cytomegalovirus. Anal Chem. 2014;86:4387–4394. doi: 10.1021/ac500208w. [DOI] [PubMed] [Google Scholar]
- 69.Sanders R., Mason D.J., Foy C.A., Huggett J.F. Evaluation of digital PCR for absolute RNA quantification. PLOS ONE. 2013;8:e75296. doi: 10.1371/journal.pone.0075296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Del Chierico F., Ancora M., Marcacci M., Camma C., Putignani L., Conti S. Choice of next-generation sequencing pipelines. Methods Mol Biol. 2015;1231:31–47. doi: 10.1007/978-1-4939-1720-4_3. [DOI] [PubMed] [Google Scholar]
- 71.DeWoody J.A., Abts K.C., Fahey A.L., Ji Y., Kimble S.J., Marra N.J. Of contigs and quagmires: next-generation sequencing pitfalls associated with transcriptomic studies. Mol Ecol Resour. 2013;13:551–558. doi: 10.1111/1755-0998.12107. [DOI] [PubMed] [Google Scholar]
- 72.Xuan J., Yu Y., Qing T., Guo L., Shi L. Next-generation sequencing in the clinic: promises and challenges. Cancer Lett. 2013;340:284–295. doi: 10.1016/j.canlet.2012.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.McIntyre L.M., Lopiano K.K., Morse A.M., Amin V., Oberg A.L., Young L.J. RNA-seq: technical variability and sampling. BMC Genomics. 2011;12:293. doi: 10.1186/1471-2164-12-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bhargava V., Head S.R., Ordoukhanian P., Mercola M., Subramaniam S. Technical variations in low-input RNA-seq methodologies. Sci Rep. 2014;4:3678. doi: 10.1038/srep03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Maitra R.D., Kim J., Dunbar W.B. Recent advances in nanopore sequencing. Electrophoresis. 2012;33:3418–3428. doi: 10.1002/elps.201200272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ashton P.M., Nair S., Dallman T., Rubino S., Rabsch W., Mwaigwisya S. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nat Biotechnol. 2014 doi: 10.1038/nbt.3103. [DOI] [PubMed] [Google Scholar]
- 77.Johnson G.L., Bibby D.F., Wong S., Agrawal S.G., Bustin S.A. A MIQE-compliant real-time PCR assay for Aspergillus detection. PLOS ONE. 2012;7:e40022. doi: 10.1371/journal.pone.0040022. [DOI] [PMC free article] [PubMed] [Google Scholar]