Abstract
Background
G-protein-coupled receptors (GPCRs), in the form of monomers or homodimers that bind heterotrimeric G proteins, are fundamental in the transfer of extracellular stimuli to intracellular signaling pathways. Different GPCRs may also interact to form heteromers that are novel signaling units. Despite the exponential growth in the number of solved GPCR crystal structures, the structural properties of heteromers remain unknown.
Results
We used single-particle tracking experiments in cells expressing functional adenosine A1-A2A receptors fused to fluorescent proteins to show the loss of Brownian movement of the A1 receptor in the presence of the A2A receptor, and a preponderance of cell surface 2:2 receptor heteromers (dimer of dimers). Using computer modeling, aided by bioluminescence resonance energy transfer assays to monitor receptor homomerization and heteromerization and G-protein coupling, we predict the interacting interfaces and propose a quaternary structure of the GPCR tetramer in complex with two G proteins.
Conclusions
The combination of results points to a molecular architecture formed by a rhombus-shaped heterotetramer, which is bound to two different interacting heterotrimeric G proteins (Gi and Gs). These novel results constitute an important advance in understanding the molecular intricacies involved in GPCR function.
Electronic supplementary material
The online version of this article (doi:10.1186/s12915-016-0247-4) contains supplementary material, which is available to authorized users.
Keywords: GPCR, Heterotetramer, Heterotrimeric G protein, Single-particle tracking, BRET, Molecular modeling
Background
G-protein-coupled receptor (GPCR) oligomerization is heavily supported by recent biochemical and structural data [1–6]. Optical-based techniques are instrumental in studying the dynamics and organization of receptor complexes in living cells [7]. For instance, total internal reflection fluorescence microscopy shows that 30 % of muscarinic M1 receptors exist as dimers (with no evidence of higher oligomers) that undergo interconversion with monomers on a timescale of seconds [8]. Similarly, the β1-adrenergic receptors (β1-AR) are expressed as a mixture of monomers and dimers whereas β2-adrenergic receptors (β2-AR) have a tendency to form dimers and higher-order oligomers [9]. Moreover, the monomer-dimer equilibrium of the chemoattractant N-formyl peptide receptor at a physiological level of expression lies within a timescale of milliseconds [10]. Together, these studies in heterologous systems show that a given GPCR is present in a dynamic equilibrium between monomers, dimers, and higher-order oligomers.
Studies in a broad spectrum of GPCRs [11–14] show that these receptors may form heteromers. GPCR heteromers are defined as novel signaling units with functional properties different from homomers and they represent a completely new field of study [15]. Innovative crystallographic techniques have permitted researchers to obtain crystal structures of GPCR families A, B, C, and F, bound to either agonists, antagonists, inverse agonists or allosteric modulators; in the form of monomers or homo-oligomers; and in complex with a G protein or with a ß-arrestin [16]. However, crystal structures of GPCR heteromers have not yet been obtained. Here, we propose a quaternary structure of a heteromer, taking into account the molecular stoichiometry and the interacting G proteins. Adenosine A1-A2A receptor (A1R-A2AR) complexes constitute a paradigm in the GPCR heteromer field because A1R is coupled to Gi and A2AR to Gs; that is, they transduce opposite signals in cyclic adenosine monophosphate (cAMP)-dependent intracellular cascades. First described as a concentration-sensing device in striatal glutamatergic neurons [17], the A1R-A2AR heteromer is thought to function as a Gs/Gi-mediated switching mechanism by which low and high concentrations of adenosine inhibit and stimulate, respectively, glutamate release [17, 18]. The structural basis of this switch is key to understanding heteromer function and the biological advantage behind the GPCR heteromerization phenomenon. Here, we have devised the molecular architecture of the adenosine A1R-A2AR heteromer in complex with G proteins using a combination of microscope-based single-particle tracking, molecular modeling, and energy transfer assays in combination with molecular complementation. The results point to A1 and A2A receptors organizing into a rhombus-shaped heterotetramer that couples to Gi and Gs. The overall structure is very compact and provides interacting interfaces for GPCRs and for G proteins.
Results and discussion
Reciprocal restriction of adenosine receptor motion in the plasma membrane
To examine the dynamics of A1R-A2AR heteromers in the plasma membrane of a living cell, the motion of the receptors tagged with fluorescent proteins (A1R-green fluorescent protein [GFP] or A2AR-mCherry) was measured by real-time single-particle tracking (SPT) (Fig. 1). Examples of fluorescent images and individual particle trajectories are shown in Additional file 1: Figure S1. Analysis of data corresponding to 500 A1R-GFP particles showed a linear relationship between the mean square displacement (MSD) versus time lag in the trajectories of up to 1600 single fluorescent particles (Fig. 1a, c). This is typical for Brownian diffusion, indicating a lack of restrictions in A1R-GFP motion. Co-expression of A2AR-mCherry (Fig. 1b) led to a reduction in the lateral mobility of A1R-GFP, which became confined to plasma membrane regions of 0.461 ± 0.004 μm in diameter. Its diffusion coefficient decreased from 0.381 ± 0.002 μm2/s to 0.291 ± 0.003 μm2/s (p = 0.002, one-tailed t-test). Similarly, A1R-GFP also decreased the A2AR-mCherry diffusion coefficient from 0.317 ± 0.002 μm2/s to 0.143 ± 0.005 μm2/s (p < 0.0001) (Fig. 1d–f). A2AR moved within a confinement zone of 0.941 ± 0.007 μm in diameter that was reduced to 0.360 ± 0.001 μm (p < 0.0001) when both receptors were co-expressed. We conclude from these mobility comparisons that reciprocally restricted motion of the individual receptor particles must be due to A1R-A2AR receptor-receptor interactions.
Fig. 1.
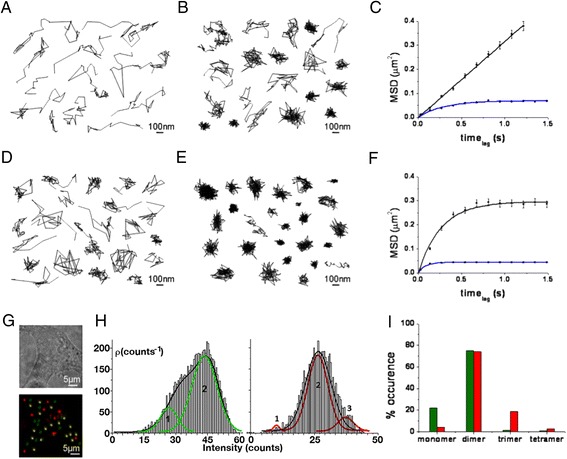
Cell surface mobility of A1R-GFP and A2AR-mCherry. Individual trajectories of particles containing GFP fused to the C-terminus of A1R (A1-GFP) (a and b) or mCherry fused to the C-terminus of A2AR (A2A-mCherry) (d and e) on HEK-293T cells expressing A1-GFP (a), A2A-mCherry (d) or both (b and e). The trajectory and the fluorescence intensity of the individual particles were recorded over time using total internal reflection microscopy (TIRFM) and an electron multiplying charged-coupled device (EMCCD) camera recording. Receptor motion was determined by plotting (versus time lag) the mean square displacement (MSD) of A1-GFP (c) in the absence (black line) or presence of A2A-mCherry (blue line), or A2A-mCherry (f) in the presence (black line) or presence of A1-GFP (blue line). Data sets were fitted to mathematical models of free and confined diffusion for A1R and A2AR respectively. g Co-localization of A1-GFP and A2A-mCherry is observed (yellow dots). Scale bar: 100 nm. h Distribution of the fluorescence signal of A1-GFP (left) and A2A-mCherry (right) within co-localized receptors (yellow dots in g). Curves approximately delineate the number of monomers, dimers, or trimers within the co-localized complex. i Stoichiometry analysis performed for co-localized A1-GFP and A2A-mCherry receptor particles co-expressed in HEK-293T cells (yellow dots in g). Green corresponds to A1-GFP and red to A2A-mCherry
Stoichiometry of A1 and A2A receptor heterocomplexes
The stoichiometry of the fluorescent receptors on the cell surface can be calculated from the brightness distribution of the individual particles [19] (see “Methods”). In cells expressing A1R-GFP, we found the majority of clusters to consist of either two (~47 %) or four (~34 %) receptors, and clusters with one or three receptors were scarce (~10 % and ~9 %, respectively) (Additional file 2: Figure S2A and black bars in Additional file 2: Figure S2C). In the case of A2AR-mCherry, the stoichiometry analysis showed that the clusters mostly expressed trimers (45 %), with dimers (29 %) and tetramers (12 %) the second and third most common populations (Additional file 2: Figure S2D and black bars in Additional file 2: Figure S2F). Remarkably, this stoichiometry for either A1 or A2A receptors was altered when the partner receptor was also expressed. In cells co-expressing A1R-GFP and A2AR-mCherry, the dimer population increased (57 % for A1R-GFP and 49 % for A2AR-mCherry, blue bars in Additional file 2: Figures S2C, F) and became the predominant species (Additional file 2: Figures S2B, C, E, F).
In order to focus the analysis on heteromer complexes, we identified clusters containing both receptors (individual yellow dots in Fig. 1g, displaying both GFP and mCherry fluorescence). In ~1000 analyzed co-localized clusters that consisted of a mixture of A1-GFP and A2A-Cherry (yellow dots in Fig. 1g), we found a similar high amount of dimers of A1R (75 %, left panel in Fig. 1h and green bar in Fig. 1i) and A2AR (74 %, right panel in Fig. 1h and red bar in Fig. 1i). Trimers and tetramers of A1R, and monomers and tetramers of A2AR, were in the minority or negligible (see Fig. 1h, i). In summary, given that the percentage of dimers of either A1R-GFP or A2AR-mCherry in the yellow dots (which show co-localization of the two receptors) was similar and high (~75 %), the heterotetramer containing two A1Rs and two A2ARs must have been the most predominant species. To our knowledge, this is the first stoichiometry data for a GPCR heteromer in living cells.
Arrangement of G proteins interacting with A1 and A2A receptors
Monomeric GPCRs are capable of activating G proteins [20]. However, recent findings suggest that one GPCR homodimer bound to a single G protein may be a common functional unit [21]. Thus, an emerging question is how G proteins couple to GPCR heteromers. Because A1R selectively couples to Gi and A2AR to Gs [22], the working hypothesis was that both Gi and Gs proteins may couple to the A1R-A2AR heterotetramer. To test this hypothesis, we used bioluminescence resonance energy transfer (BRET) assays [23]. In agreement with the SPT experiments (see above), homodimers and heterodimers were detected by BRET assays in cells expressing A1R fused with Renilla luciferase (A1R-Rluc) or yellow fluorescent protein (A1R-YFP) (Fig. 2a), A2AR-Rluc and A2AR-YFP (Fig. 2b), or A1R-Rluc and A2AR-YFP (Fig. 2e). Neither A1R-Rluc nor A2AR-YFP interacted with the ghrelin receptor 1a fused to YFP (GHS1a-YFP), used as a control as a protein unable to directly interact with these adenosine receptors (Fig. 2a, b). In order to test the presence of the two G proteins in the heterotetramer, we transfected cells with minigenes that code for peptides blocking either Gi or Gs binding to GPCRs [24]. In addition, cells were treated with pertussis or cholera toxins that catalyze ADP-ribosylation of Gi or Gs. Clearly, treating cells with pertussis toxin, or expressing the minigene-coded peptide that blocks αi coupling, reduced the value of BRETmax for A1R-A1R homodimers (Fig. 2a) and for A1R-A2AR heterodimers (Fig. 2e) but not for A2AR-A2AR homodimers (Fig. 2b). This indicates that Gi is coupled to A1R in both the homodimer and the heterodimer. Similarly, blocking Gs-receptor interaction using cholera toxin or a minigene-coded peptide that blocks αS coupling reduced BRETmax for A2AR-A2AR homodimers (Fig. 2b) and for A1R-A2AR heterodimers (Fig. 2e) but not for A1R-A1R homodimers (Fig. 2a). Interestingly, BRET curves showed sensitivity to both cholera and pertussis toxins in cells expressing either A1R-Rluc-A1R-YFP and A2AR (Fig. 2c) or A2AR-Rluc-A2AR-YFP and A1R (Fig. 2d). Functionality of constructs and controls in cells expressing minigenes, and in cells expressing the ghrelin GHS1a receptor instead of one of the adenosine receptors, are shown in Additional file 3: Figure S3. To further confirm that Gi binds A2AR in the receptor heteromer, the energy transfer between Rluc fused to the N-terminal domain of the α-subunit of Gi (Gi-Rluc) and A2AR-YFP was analyzed in cells co-expressing or not co-expressing A1R (Fig. 2f). A hyperbolic BRET curve was observed in the presence of A1R, but not in its absence, indicating that Gi and Gs are bound to their respective receptor homodimers within the A1R-A2AR heteromer.
Fig. 2.
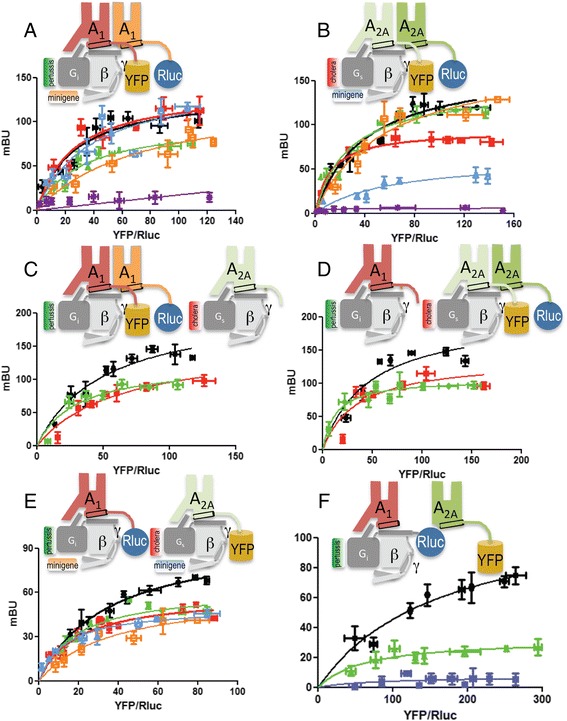
Influence of G proteins on A1R and A2AR homodimerization and heterodimerization. B Bioluminescence resonance energy transfer (BRET) saturation curves were performed in HEK-293T cells 48 h post-transfection with (a, c) 0.3 μg of cDNA corresponding to A1R-Rluc and increasing amounts of A1R-YFP (0.1–1.5 μg cDNA) or GHS1a-YFP (0.25–2 μg cDNA) as negative control (a, purple line), without (a) or with (c) 0.15 μg of cDNA corresponding to A2AR; (b, d) 0.2 μg of cDNA corresponding to A2AR-Rluc and increasing amounts of A2AR-YFP (0.1–1.0 μg cDNA) or GHS1a-YFP (0.25–2 μg cDNA) as negative control (b, purple line), without (b) or with (d) 0.5 μg of cDNA corresponding to A1R; (e) 0.3 μg of cDNA corresponding to A1R-Rluc and increasing amounts of A2AR-YFP (0.1–1.0 μg cDNA); and (f) 0.5 μg of cDNA corresponding to A1R (except control blue curves that were obtained in cells not expressing A1R), 2 μg of cDNA corresponding to Gi-Rluc, and increasing amounts of A2AR-YFP (0.1–0.5 μg cDNA). In panels a, b, and e, cells were also transfected with 0.5 μg of cDNA corresponding to the Gi-related (orange curves) or Gs-related (blue curves) minigenes. Cells were treated for 16 h with medium (black curves), with 10 ng/ml of pertussis toxin (green curves), or with 100 ng/ml of cholera toxin (red curves) prior to BRET determination. To confirm similar donor expressions (approximately 100,000 bioluminescence units) while monitoring the increase in acceptor expression (1000–40,000 fluorescence units), the fluorescence and luminescence of each sample were measured before energy transfer data acquisition. MiliBRET unit (mBU) values are the mean ± standard error of the mean of four to six different experiments grouped as a function of the amount of BRET acceptor. In each panel (top) a cartoon depicts the proteins to which Rluc and YFP were fused and the presence or not of partner receptors and/or Gs or Gi proteins [schemes in c to f are not intended to illustrate on stoichiometry because the predominant form in cells expressing the two receptors was the heterotetramer containing two A1 and two A2A receptors (see “Results”)]
Further, two complementary BRET experiments were performed to determine the orientation of Gi and Gs within the A1R-A2AR heterocomplex. First, Rluc and YFP were respectively fused to the N-terminal domains of the α-subunit of Gi (αi-Rluc) and Gs (αs-YFP) (Fig. 3, bar a); second, they were fused to the N-terminal domain of the γ-subunit (γ-Rluc and γ-YFP) (Fig. 3, bar b). We observed significant energy transfer between γ-Rluc and γ-YFP in cells co-expressing A1R and A2AR (Fig. 3, bar b) but minimal amounts in negative-control cells (Fig. 3, bars c and d). In cells expressing either A1R or A2AR, the energy transfer between γ-Rluc and γ-YFP was also low (Fig. 3, bars e and f), suggesting that dimers but not tetramers were the most prevalent form of surface receptors in single-transfected cells. These results in co-transfected cells corroborate the 2:2 stoichiometry obtained from analysis of the fluorescence in single particles and are consistent with Gi and Gs binding to these A1R-A2AR heterotetramers.
Fig. 3.
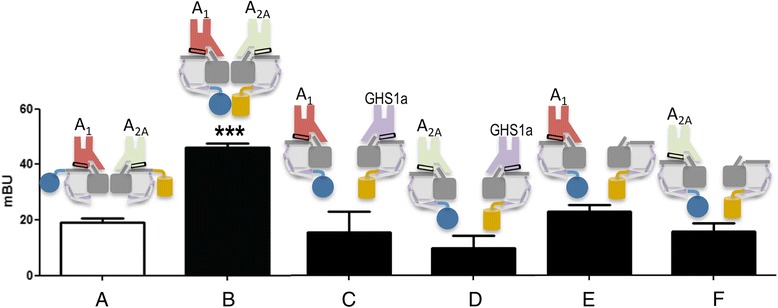
Gs and Gi coupling to adenosine A1R-A2AR heterocomplexes. Bioluminescence resonance energy transfer (BRET) experiments were performed in HEK-293T cells 48 h post-transfection with (a, b) 0.2 μg of cDNA corresponding to A1R and 0.15 μg of cDNA corresponding to A2AR; (c, d) 0.2 μg of cDNA corresponding to A1R or 0.15 μg of cDNA corresponding to A2AR and 0.4 μg of cDNA corresponding to growth hormone secretagogue receptor GHS1a; (e) 0.2 μg of cDNA corresponding to A1R; or (f) 0.15 μg of cDNA corresponding to A2AR. Cells were also transfected with 2 μg of cDNA corresponding to the α-subunit of Gi fused to Rluc and increasing amounts of cDNA corresponding to the α-subunit of Gs fused to YFP (a) or 0.3 μg of cDNA corresponding to the γ-subunit fused to Rluc and increasing amounts of cDNA corresponding to the γ-subunit fused to YFP (b–f). Maximum miliBRET unit (mBU) values are the mean ± standard error of the mean of four different experiments. A scheme showing the protein to which Rluc and YFP were fused is provided (top). ***p < 0.001 by one-way ANOVA with post - hoc Dunnett’s test
Molecular model of Gi and Gs bound to the A1R-A2AR heterotetramer
To identify the orientation of the G protein in the receptor homodimer, we combined energy transfer assays between αs-Rluc (Rluc at the N-terminus of the G protein α-subunit) and A2AR-YFP (Fig. 4a) with information on transmembrane (TM) interfaces based on crystal structures of GPCRs [3, 4], which have been recently summarized [25]. The observed high-energy transfer using αs-Rluc and A2AR-YFP indicated close proximity between the N-tail of the α-subunit of Gs and the C-tail of A2AR. Interestingly, Rluc and YFP in the “monomeric” A2AR-Gs complex (see “Methods”) point toward distant positions in space (Fig. 4b). Therefore, the observed BRET should occur between Rluc in the G protein α-subunit and a second A2AR-YFP protomer. Among all described TM interfaces for receptor homodimerization (see Additional file 4: Figure S4), we propose the TM4/5 interface, which is observed in the oligomeric structure of β1-AR [4] and in structures derived from coarse-grained molecular dynamics (MD) simulations [26]. In fact, this is the only interface that favors BRET between αs-Rluc and a second A2AR-YFP protomer in a homodimer (Fig. 4c). The homologous A1R homodimer was built using the same TM4/5 interface as for A2AR (see Additional file 4: Figure S4 and its legend).
Fig. 4.
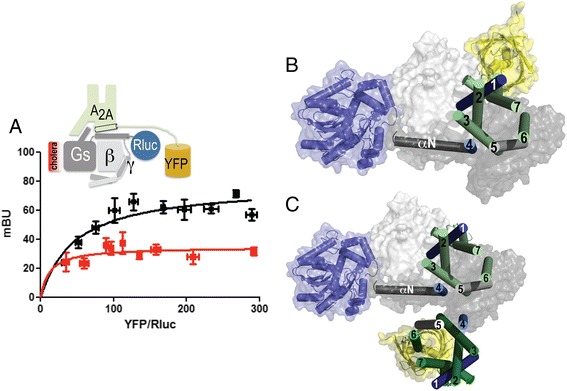
Orientation of a G protein in a receptor homodimer. Bioluminescence resonance energy transfer (BRET) saturation experiments were performed in HEK-293T cells transfected with 2 μg of cDNA corresponding to the α-subunit of Gs fused to Rluc and increasing amounts of A2AR-YFP (0.1–0.5 μg) cDNA. a BRET measurements in cells pretreated for 16 h with medium (black line) or with 100 ng/ml of cholera toxin (red line). Both fluorescence and luminescence of each sample were measured before every experiment to confirm similar donor expressions (approximately 50,000 bioluminescence units) while monitoring the increase in acceptor expression (1000–10,000 fluorescence units). miliBRET unit (mBU) values are the mean ± standard error of the mean of four to five different experiments grouped as a function of the amount of BRET acceptor. A scheme of the placement of donor and acceptor BRET moieties is provided (top). b Molecular model of the A2AR-Gs complex. Rluc (blue) is attached to the N-terminal αN helix of Gs (gray), and YFP (yellow) is attached to the C-terminal domain of A2AR (light green) (see Additional file 9: Figure S9 for details). c Arrangement of A2AR homodimers modeled via the TM4/5 interface as observed in the oligomeric structure of β1-AR [4]. The A2AR protomer bound to αs is shown in light green, whereas the second A2AR-YFP protomer is shown in dark green. The molecular model in panel c (BRET between Rluc in Gs α subunit and YFP in a second A2AR protomer; center-to-center distance between Rluc and YFP of 6.5 nm), in contrast to the model shown in panel B (BRET between Rluc in Gs α subunit and YFP in the G-protein bound A2AR protomer; center-to-center distance between Rluc and YFP of 8.3 nm), would favor the observed high-energy transfer (see panel a) between αs-Rluc and A2AR-YFP
The remaining possible TMs able to form heteromeric interfaces are TM1 and TM5/6 (Fig. 5). Both are possible inter-GPCR interfaces as observed in the structure of the μ-opioid receptor (μ-OR) [3]. To discern between these two possibilities, a bimolecular fluorescence complementation strategy was undertaken. For this purpose, the N-terminal fragment of Rluc8 was fused to A1R (A1R-nRluc8) and its C-terminal domain to A2AR (A2AR-cRluc8), which only upon complementation can act as a BRET donor (Rluc8). The BRET acceptor protein was obtained upon complementation of the N-terminal fragment of YFP Venus protein fused to A1R (A1R-nVenus) and its C-terminal domain fused to A2AR (A2AR-cVenus). When all four receptor constructs were transfected, we obtained a positive and saturable BRET signal (BRETmax of 35 ± 2 mBU and BRET50 of 16 ± 3 mBU) that was not obtained for negative controls (Additional file 5: Figure S5). Figure 5a, b shows that the hemi-donor (A1R-nRluc8 and A2AR-cRluc8) and the hemi-acceptor (A1R-nVenus and A2AR-cVenus) moieties, placed at the C-terminus of the receptors, can only complement if A1R-A2AR heterodimerization occurs via the TM5/6 interface. The TM4/5 interface for homodimerization and the TM5/6 interface for heterodimerization give a rhombus-shaped tetramer organization (Fig. 5a). Remarkably, cell pre-incubation with either pertussis or cholera toxins decreased the BRETmax by 35 % (Fig. 5c), further suggesting that both Gs and Gi proteins bind to the A1R-A2AR heterotetramer.
Fig. 5.
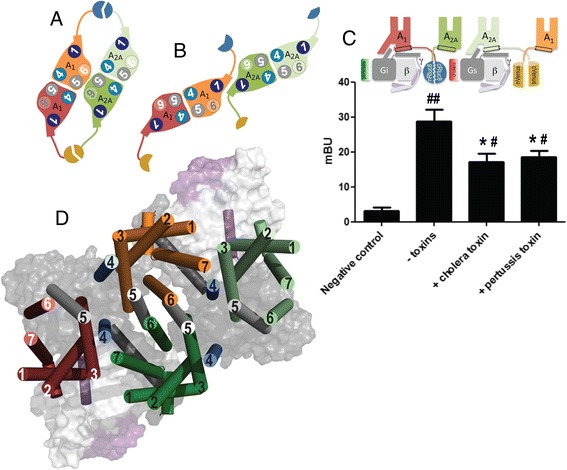
Bioluminescence resonance energy transfer (BRET)-aided construction of a model consisting of Gi and Gs bound to an A1R-A2AR heterotetramer. a, b A1R-A2AR tetramer built using TM5/6 (a) or TM1 (b) inter-receptor interfaces modeled as in the structure of the μ opioid receptor [3]. TM helices 1, 4, and 5, involved in receptor dimerization, are highlighted in dark blue, light blue, and gray, respectively. nRluc8 and cRluc8 are shown in blue and nVenus and cVenus in dark yellow. c BRET and bimolecular fluorescence complementation experiments were performed in HEK-293T cells transfected with 1.5 μg of cDNA corresponding to A1R-cRluc8 and A2AR-nRluc8, and 1.5 μg of cDNA corresponding to A1R-nVenus and A2AR-cVenus. As the negative control, cells were transfected with 1 μg of cDNA corresponding to nRluc8 and 1.5 μg of cDNA corresponding to A2AR-nRluc8, A1R-nVenus, and A2AR-cVenus. Cells were treated for 16 h with medium (– toxins), 10 ng/ml of pertussis toxin (+ pertussis), or 100 ng/ml of cholera toxin (+ cholera) prior to BRET determination. The relative amount of BRET is given as in Fig. 4 and values are the mean ± standard error of the mean of three different experiments. Student’s t-test showed statistically significant differences with respect to the control (# p < 0.05, ## p < 0.01) and a significant effect in the presence of either toxin over BRET in the absence of toxins (*p < 0.05). A schematic representation at the top shows the protein to which the hemi luminescent or fluorescent proteins were fused. d Molecular model of the A1R-A2AR tetramer in complex with Gi and Gs. A1R bound to Gi is shown in red, Gi-unbound A1R is shown in orange, A2AR bound to Gs is shown in dark green, Gs-unbound A2AR is shown in light green, and the α, β-, and γ-subunits of Gi and Gs are shown in dark gray, light gray, and purple, respectively. Transmembrane helices 4 and 5 are highlighted in light blue and gray, respectively
We next evaluated, using computational tools, whether the proposed A1R-A2AR heterotetramer could couple to both Gi and Gs proteins. Clearly, the external protomers of the proposed A1R-A2AR heterotetramer can bind to Gi and Gs proteins (Fig. 5d). This model positions the α-subunits of Gi and Gs in close contact, facing the interior of the tetrameric complex, while the N-terminal α-helices of αi and αs point outside the complex. The N-terminal α-helices of the γ-subunits are in close proximity, facing the inside (Additional file 6: Figure S6), which explains the significant energy transfer observed between γ-Rluc and γ-YFP (Fig. 3, bar b). The model provides experimental insights into the structural arrangement of heteromers consisting of two GPCRs and coupled to two G proteins, the possibility of which has recently been discussed [25]. We used MD simulations to study the stability of this complex. Additional file 7: Figure S7 shows root-mean-square deviations (rmsd) on protein α-carbons throughout the MD simulation, as well as key intermolecular distances among protomers and G proteins. Clearly, both the A1R protomer bound to Gi and the A1R protomer that does not interact with it maintained a close structural similarity (rmsd ≈ 0.3 nm) relative to the initial structures. Similar results were obtained for the A2AR protomers (bound and unbound to Gs) (Additional file 7: Figure S7A). The fact that rmsd values of the whole system, formed by the A1R-A2AR heterotetramer bound to Gi and Gs, are of the order of 0.6 nm indicates that the initial structural model is maintained during the MD simulation (Additional file 7: Figure S7A). As a consequence, selected intermolecular distances among protomers and G proteins remain constant during the MD simulation (Additional file 7: Figure S7B). A key aspect in the assembly of the heterotetramer is the TM interfaces for homodimerization (TM4/5) and heterodimerization (TM5/6). Additional file 8: Figure S8B shows rmsd values of the four-helix bundle forming the TM4/5 and TM5/6 interfaces, the initial and final snapshots of these bundles, and the evolution of the A1R-A2AR heterotetramer during the MD simulation. Clearly, the rather small structural variations of these four-helix bundles, also reflected by rmsd <0.3 nm, suggest a stable complex. Notably, the TM5/6 four-helix bundle seems more stable than the TM4/5 bundles, as shown by its lower rmsd value. Additional file 8: Figure S8B, C depicts contact maps of the TM4/5 and TM5/6 interfaces, as well as the evolution of the network of hydrophobic interactions within these interfaces during the MD simulation.
Conclusions
For more than a decade, experimental evidence has supported the occurrence of homo-oligomers and hetero-oligomers of GPCRs [21]. However, our basic understanding of what makes heteromers different from homomers remains unknown. Our results, studying adenosine receptors as a model heteromer, point to three important new findings. First, the predominant stoichiometry in cells expressing A1R-A2AR heteromers is 2:2; that is, a dimer of dimers (tetramer). Second, two different heterotrimeric G proteins can couple to heteromers, the overall complex constituting a functional unit. Third, the molecular orientation within the heteromer complex affords various qualitatively different interfaces; the two more relevant are the inter-protomer heteromeric interface and the inter-G-protein interface. Presumably, the two interfaces provide the key characteristic of heteromers: the ability of one protomer/G-protein complex to influence the signaling of the other. Surely, allosteric effects occurring between heteroreceptors and between Gs and Gi proteins are due to conformational changes transmitted along the intimately interacting molecules in the complex. In our controlled cell transfection system, which expressed a low density of receptors, minor species formed by monomers and trimers were found in addition to a predominance of tetramers in the plasma membrane, strongly supporting the occurrence of an in vivo dynamic distribution of receptors.
Adenosine was, from an evolutionary point of view, one of the first extracellular regulators given that it is involved in energy and nucleic acid metabolisms. Adenosine A1 and A2A receptors are expressed in almost every mammalian organ and tissue. In the heart, where adenosine plays a key role in both inotropic and chronotropic regulation, A1R-mediated cardioprotection did not occur in A2AR knockout mice, suggesting an interaction between A1 and A2A receptors. In neurons, A1 and A2A receptors show co-localization, leading to inter-receptor interactions unveiled by pharmacological treatments. For instance, Okada et al. [27] showed that cAMP-dependent protein kinase A plays a role in the regulation of hippocampal serotonin release mediated by both A1 and A2A receptors. Similarly, the control of γ-amino butyric acid transport in astrocytes was attributed to the expression of A1R-A2AR heteromers and to a specific mechanism by which the heteromer signals via Gi or Gs depending on the concentration of adenosine [28]. The structural basis of the differential signaling by the heteromer/G-protein macromolecular complex likely implies communication at the receptor-receptor level but also between Gs and Gi. Because the binding of two G proteins to a heterodimer is not feasible due to steric clashes [25], our finding that the A1R-A2AR heterotetramer may bind to both Gs and Gi provides a structural framework to interpret experimental data.
Methods
Total internal reflection microscopy and single-particle data analysis
Single-particle imaging and tracking were performed on a Nikon Total Internal Reflection Fluorescence (TIRF) system, as detailed in Additional file 11: Supplementary Methods. Typically, 500 readouts of a 512 × 512-pixel region, the full array of the CCD chip, were acquired. For single-particle data analysis, parameters were calculated by applying the equations described in Additional file 11: Supplementary Methods.
Cell culture and transient transfection
HEK-293T cells were grow at 37 °C in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Thermo Fischer Scientific, Madrid, Spain) supplemented with 2 mM L-glutamine, 100 U/ml penicillin/streptomycin, and 5 % (v/v) heat-inactivated fetal bovine serum (FBS) (all supplements were from Invitrogen, Paisley, UK). Cells were transiently transfected with cDNA corresponding to receptors, fusion proteins, A2AR mutants, or G-protein minigene vectors obtained as detailed in an expanded view by the polyethylenimine (PEI; SigmaAldrich, Cerdanyola del Vallès, Spain) method. Sample protein concentration was determined using a Bradford assay kit (Bio-Rad, Munich, Germany) using bovine serum albumin dilutions as standards. For single-particle imaging, cells were seeded into six-well plates containing glass coverslips (No. 1, round, 24 mm; Assistent, Sondheim, Germany) or into the Lab-Tek Chambered #1.0 Borosilicate Coverglass System (Nunc, Thermo Fisher Scientific, Schwerte, Germany). Cell transient transfections were performed with Lipofectamine™ 2000 (Invitrogen, Life Technologies, Darmstadt, Germany) or FuGENE 6 (Roche Applied Science, Indianapolis, IN, USA) and the application of 0.1–0.2 μg plasmid DNA per well. Before each experiment, cells were washed three times with 200 μL phenol red-free DMEM.
Plasmids
DNA sequences encoding amino acid residues 1–155 and 155–238 of YFP Venus protein, and amino acids residues 1–229 and 230–311 of RLuc8 protein were subcloned in the pcDNA3.1 vector to obtain the YFP Venus and RLuc8 hemi-truncated proteins. The human cDNAs for adenosine receptors, A2AR and A1R, cloned into pcDNA3.1, were amplified without their stop codons using sense and antisense primers harboring unique EcoRI and BamHI sites to clone receptors into the pcDNA3.1RLuc vector (pRLuc-N1; PerkinElmer, Wellesley, MA, USA), and EcoRI and KpnI to clone A2AR, A1R, or GHS1a into the pEYFP-N1 vector (enhanced yellow variant of GFP; Clontech, Heidelberg, Germany). Gαs cloned into the SFV1 vector, Gαi cloned into the pcDNA3.1 vector, or Gγ cloned into the pEYFP-C1 vector were amplified without their stop codons using sense and antisense primers harboring unique HindIII and BamHI sites to clone them into the pcDNA3.1-Rluc vector, or EcoRI and KpnI to clone Gαs into the pEYFP-N1 vector. The amplified fragments were subcloned to be in-frame with restriction sites of the pcDNA3.1RLuc or pEYFP-N1 vectors to give plasmids that expressed proteins fused to RLuc or YFP on the N-terminal end (Gαs-RLuc, Gαi-RLuc, Gγ-RLuc, Gαs-YFP, and Gγ-YFP) or the C-terminal end (A1R-RLuc, A2AR-RLuc, A1R-YFP, A2AR-YFP, and GHS1a-YFP). The human cDNAs for A1R or GHS1a were subcloned into pcDNA3.1-nRLuc8 or pcDNA3.1-nVenus to give plasmids that expressed A1R or GHS1a fused to either nRLuc8 or nYFP Venus on the C-terminal end of the receptor (A1R-nRLuc8 and A1R-nVenus or GHS1a-nRLuc8 and GHS1a-nVenus). The cDNAs for human A2A or GHS1a receptors were subcloned into pcDNA3.1-cRLuc8 or pcDNA3.1-cVenus to give plasmids that expressed receptors fused to either cRLuc8 or cYFP Venus on the C-terminal end of the receptor (A2AR-cRLuc8 and A2AR-cVenus or GHS1a-cRLuc8 and GHS1a-cVenus). Expression of constructs was tested by confocal microscopy and the receptor-fusion protein functionality by measuring ERK1/2 phosphorylation and cAMP production, as described previously [13, 14, 17, 29].
“Minigene” plasmid vectors are constructs designed to express relatively short polypeptide sequences following their transfection into mammalian cells. Here, we used minigene constructs encoding the carboxyl-terminal 11-amino acid residues from Gα subunits of Gi1/2 (Gi minigene) or Gs (Gs minigene) G proteins; the resulting peptides inhibit G-protein coupling to the receptor and consequently inhibit the receptor-mediated cellular responses as previously described [24]. The cDNA encoding the last 11 amino acids of human Gα subunit corresponding to Gi1/2 (I K N N L K D C G L F) or Gs (Q R M H L R Q Y E L L), inserted in a pcDNA3.1 plasmid vector, were generously provided by Dr Heidi Hamm.
Energy transfer assays
For BRET and complementation BRET assays, HEK-293T cells were transiently cotransfected with a constant amount of cDNA encoding for proteins fused to RLuc, nRLuc8, or cRLuc8, and with increasing amounts of the cDNA corresponding to proteins fused to YFP, nYFP Venus, or cYFP Venus (see figure legends). To quantify protein-YFP expression or protein-reconstituted YFP Venus expression, cells (20 μg protein) were distributed in 96-well microplates (black plates with a transparent bottom) and fluorescence was read in a FLUOstar OPTIMA Fluorimeter (BMG Labtechnologies, Offenburg, Germany) equipped with a high-energy xenon flash lamp, using a 10 nm bandwidth excitation filter at 400 nm reading. Protein fluorescence expression was determined as the fluorescence of the sample minus the fluorescence of cells expressing the BRET donor alone. For BRET measurements, the equivalent of 20 μg of cell suspension were distributed in 96-well microplates (Corning 3600, white plates; Sigma) and 5 μM coelenterazine h (Molecular Probes, Eugene, OR, USA) was added. After 1 min for BRET or after 5 min for BRET with bimolecular fluorescence complementation, the readings were collected using a Mithras LB 940 that allows the integration of the signals detected in the short-wavelength filter at 485 nm (440–500 nm) and the long-wavelength filter at 530 nm (510–590 nm). To quantify protein-RLuc or protein-reconstituted RLuc8 expression, luminescence readings were also performed 10 min after adding 5 μM coelenterazine h. The net BRET was defined as [(long-wavelength emission)/(short-wavelength emission)] – Cf, where Cf corresponds to [(long-wavelength emission)/(short-wavelength emission)] for the donor construct expressed alone in the same experiment. BRET is expressed as miliBRET units (mBU; net BRET × 1000).
Computational model of the A1R-A2AR tetramer in complex with Gi and Gs
The crystal structure of inactive A2AR [PDB:4EIY] [30] was used for the construction of human A2AR [UniProt:P29274] and A1R [UniProt:P30542] homology models using Modeller 9.12 [31]. These receptors share 51 % of sequence identity and 62 % of sequence similarity, excluding the C-terminal after helix 8. Intracellular loop 3 (ICL3) of A2AR (Lys209–Gly218) and A1R (Asn212–Ser219) were modeled using Modeller 9.12 [31] using ICL3 of squid rhodopsin [PDB:2Z73] as a template. The C-terminus tails of A1R, containing 16 amino acids (Pro311–Asp326), and of A2AR, containing 102 amino acids (Gln311–Ser412), were modeled as suggested for the oxoeicosanoid receptor (OXER) [32] (see Additional file 9: Figure S9 for details). The N-terminus of A1R and A2AR were not included in the model. The “active” conformations of A1R bound to Gi and A2AR bound to Gs were modeled using the crystal structure of β2-AR in complex with Gs [PDB:3SN6] [33]. The globular α-helical domain of the α-subunit was modeled in the “closed” conformation [34], using the crystal structure of [AlF4−]-activated Gi [PDB:1AGR]. The location of YFP [PDB:2RH7] attached to the C-tail of A2AR was determined as suggested for the OXER [32] (see Additional file 9: Figure S9 for details). Rluc [PDB:2PSD] and YFP were fused to the to the N-terminus of the α-subunits and γ-subunits of Gi and Gs by a covalent bond. The structures of adenosine receptor oligomers were modeled via the TM4/5 interface for homodimerization, using the oligomeric structure of the β1-AR [PDB:4GPO] [4], or via the TM5/6 interface for heterodimerization, using the structure of the μ-OR [PDB:4DKL] [3]. The Gi-bound A1R and Gs-bound A2AR protomers were rotated 10° to avoid the steric clash of the N-terminal helix of Gi and Gs with the C-terminal helix (Hx8) of Gs-unbound A2AR and Gi-unbound A1R, respectively. This computational model, without Rluc and YFP, was placed in a rectangular box containing a lipid bilayer (814 molecules of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine - POPC -) with explicit solvent (102,973 water molecules) and a 0.15 M concentration of Na+ and Cl− (1762 ions). This initial complex was energy-minimized and subsequently subjected to a 10 ns MD equilibration, with positional restraints on protein coordinates. These restraints were released and 500 ns of MD trajectory were produced at constant pressure and temperature (see Additional file 10: Movie M1). Computer simulations were performed with the GROMACS 4.6.3 simulation package [35], using the AMBER99SB force field as implemented in GROMACS and Berger parameters for POPC lipids. This procedure has been previously validated [36].
Availability of data and materials
The crystal structures 4EIY, 2Z73, 3SN6, 1AGR, 2RH7, 2PSD, 4GPO, and 4DKL are available from PDB (http://www.rcsb.org). All other relevant data are within the paper and its Additional files.
Acknowledgments
We acknowledge the technical help provided by Jasmina Jiménez (CIBERNED, University of Barcelona). This study was supported by grants from the Spanish Ministerio de Ciencia y Tecnología (SAF2009-07276, SAF2010-18472, SAF2011-23813, SAF2013-48271-C2-2-R; those grants may include FEDER funds), the Max Planck Society, the German Cancer Research Center, and the German Ministry for Education and Research (BMBF). PJM and LP participate in the European COST Action CM1207 (GLISTEN). Authors gratefully acknowledge the computer resources provided by the Barcelona Supercomputing Center - Centro Nacional de Supercomputación.
Additional files
Examples of receptor trajectories in HEK-293T cells. Images of cells expressing A1R-GFP (A) and of particular trajectories of A1R-GFP-containing (B) or A2AR-mCherry-containing (C) particles. (TIF 1164 kb)
Graphical description of the stoichiometry of A1R-GFP, A2AR-mCherry or both A1-GFP and A2A-mCherry. The fluorescence intensity signal distribution (gray area) detected for more than 7000 independent observations is given for HEK-293T cells expressing A1-GFP (A), A2A-mCherry (D), or both A1-GFP and A2A-mCherry (B, E). The stoichiometry analysis was performed for A1-GFP (A, B) and A2A-mCherry (D, E). Curves approximately delineating the amount of monomers, dimers, trimers, and tetramers are displayed in green for A1-GFP (A, B) and in red for A2A-mCherry (D-E). The occurrence on the cell surface of monomers, dimers, trimers, and tetramers for A1-GFP (C) expressed alone (black bars) or in the presence of A2A-mCherry (blue bars) and for A2A-mCherry (F) expressed alone (black bars) or in the presence of A1-GFP (blue bars) was calculated by stoichiometry analysis from results shown in A, B, D, and E. (TIF 455 kb)
Controls of cAMP production and BRET assays in cells expressing minigenes and in cells expressing the ghrelin GHS1a receptor instead of one of the adenosine receptors. (A,B) cAMP determination in HEK-293T cells transfected with (A) 0.3 μg of cDNA corresponding to A1R or (B) with 0.2 μg of cDNA corresponding to A2AR with (control) or without 0.5 μg of cDNA corresponding to minigenes coding for peptides blocking either Gi or Gs binding. Cells were stimulated with the A1R agonist N6-Cyclopentyladenosine (CPA) (10 nM, red bars) in the presence of 0.5 μM forskolin (Fk) or with the A2AR agonist 4-[2-[[6-Amino-9-(N-ethyl-β-D-ribofuranuronamidosyl)-9H-purin-2-yl]amino]ethyl]benzenepropanoic acid hydrochloride (CGS-21680) (200 nM, blue bars). Values expressed as % of the forskolin-treated cells (CPA reduces forskolin-induced cAMP levels, red bars) or of the basal (CGS 21680 per se enhances cAMP levels, blue bars) are given as mean ± SD (n = 4–8). One-way ANOVA followed by a Bonferroni post - hoc test showed a significant effect of CPA when compared with that of forskolin (red bars, ***p < 0.001) or of CGS 21680 when compared to basal cAMP levels (blue bars, ## p < 0.01, ### p < 0.001). (C, D) BRET saturation curves were performed in HEK-293T cells transfected with (C) 0.3 μg cDNA coding for A1R-Rluc, increasing amounts of cDNA coding for A1R-YFP (0.1–1.5 μg cDNA), and 0.4 μg cDNA coding for GHS1a, or (D) with 0.2 μg of cDNA coding for A2AR-Rluc, increasing amounts of cDNA coding for A2AR-YFP (0.1–1.0 μg cDNA), and 0.5 μg cDNA coding for to GHS1a. Prior to BRET determination, cells were treated for 16 h with medium (black curves), with 10 ng/ml of pertussis toxin (green curves), or with 100 ng/ml of cholera toxin (red curves). mili BRET units (mBU) are given as the mean ± SD (n = 4–6 different experiments grouped as a function of the amount of BRET acceptor). (TIF 1418 kb)
Possible interfaces in A2AR homodimers in complex with Gs. In A–E, the A2AR homodimer was modeled through TM4 using the H1-receptor structure as template (A), through TM5 using the structure of squid rhodopsin (B), through TM4/5 using the β1-receptor structure (C), and via TM5/6 (D) and TM1 (E) using the μ-OR structure. TM helices 1, 4, and 5 involved in receptor dimerization are highlighted in dark blue, light blue, and gray, respectively. A2AR protomers bound to Gs (in gray) are shown in light green, whereas Gs-unbound A2AR protomers are shown in dark green. Rluc (blue) is attached to the N-terminal αN helix of Gs, and YFP (yellow) is attached to the C-terminal domain of the Gs-unbound A2AR protomer (light green). It is important to note that the position of YFP is highly dependent on the orientation of the long and highly flexible C-tail of A2AR (102 amino acids, Gln311–Ser412), which was modeled as described for the OXER [32] (see Additional file 9: Figure S9 for details). Despite these limitations, we can crudely estimate the approximate distances between the center of mass of Rluc and YFP as 4.6, 10.1, 6.5, 11.6, and 8.3 nm for panels A–E, respectively. Thus, among all these possible dimeric interfaces, only the molecular models depicted in panels A (TM4 interface) and C (TM4/5 interface) would favor the observed high-energy transfer between Gs-Rluc and A2AR-YFP (Fig. 4a in main paper). However, there is a steric clash between the N-terminal helix of Gs and the dark-green protomer in the TM4 interface. Accordingly, we have modeled A2AR homodimerization via the TM4/5 interface. Unfortunately, similar experiments with cells transfected with Gi-Rluc and A1R-YFP could not be accomplished because of a lack of receptor expression (not shown); it is likely that the shorter C-tail of A1R (16 amino acids, Pro311–Asp326) could not accommodate YFP in the presence of Gi in the right three-dimensional structure. The A1R homodimer was built using the same TM4/5 interface as for A2AR. (TIF 3135 kb)
BRET assays in cells expressing fusion proteins containing hemi-Rluc8 and hemi-Venus moieties fused to adenosine receptors or containing the ghrelin GHS1a receptor instead of one of the adenosine receptors. (A) Saturation BRET curve in HEK-293T co-transfected with 1.5 μg of the two cDNAs corresponding to A1R-cRLuc8 and A2AR-nRLuc8 and with increasing amounts of cDNAs corresponding to A1R-nVenus and A2AR-cVenus (equal amounts of the two cDNAs). BRETmax was 35 ± 2 mBU and BRET50 was 16 ± 3 mBU. BRET in cells expressing cRluc8 instead of A1R-cRluc8 gave a linear, non-saturable signal. (B) Comparison of BRET responses using complementary and non-complementary pairs, or replacing one adenosine receptor with the ghrelin GHS1a (gn) receptor. Data are mean ± SD of three different experiments grouped as a function of the amount of BRET acceptor. ***p < 0.001 with respect to BRET in cells expressing adenosine receptors and hemi-Rluc8 and hemi-Venus proteins. (TIF 398 kb)
Details of the relative position of Rluc and YFP in a receptor heterotetramer interacting with two G proteins. Computational-based model of Gs and Gi bound to the adenosine A1R-A2AR heterotetramer. Rluc and YFP fused to the N-terminal domain of the Gα-subunits point toward different positions in space (A), whereas Rluc and YFP fused to Gγ-subunits are close (B). The color code of the proteins is depicted in the adjacent schematic representations (TM4 and TM5 of GPCR protomers are in light blue and gray, respectively). (TIF 6445 kb)
Molecular dynamics (MD) simulation of the adenosine A1R-A2AR heterotetramer in complex with Gi and Gs. (A) Root-mean-square deviations (rmsd) on protein α-carbons of the whole system (black solid line), of the two A1Rs (orange and red solid lines), of the two A2ARs (light and dark green solid lines), of Gi (gray solid line), and of Gs (gray dotted line) throughout the MD simulation. This color scheme matches with the color of the different proteins depicted in the two adjacent schematic representations. (B) Intermolecular distances between the N-terminal helices of the γ-subunit of Gi and Gs (magenta line), the N-terminal helices of the α-subunit of Gi and Gs (gray line), the N-terminal helix of the α-subunit of Gi and the C-terminal helix (Hx8) of inactive A1R (orange line), the N-terminal helix of the α-subunit of Gs and the C-terminal Hx8 of inactive A2AR (green line), the C-terminal Hx8 of A1R and A2AR (blue lines). These computed intermolecular distances are depicted as double arrows in the two adjacent schematic representations. (TIF 6973 kb)
Evolution of TM4/5 and TM5/6 interfaces as devised from MD simulations of the adenosine A1R-A2AR heterotetramer in complex with Gi and Gs. (A) Representative snapshots (20 structures collected every 25 ns) of the TM domains of A1R bound to Gi (red), Gi-unbound A1R (orange), A2AR bound to Gs (dark green), and Gs-unbound A2AR (light green). TM helices 4 and 5 are highlighted in light blue and gray, respectively. Initial (at 0 ns, transparent cylinders) and final (at 500 ns, solid cylinders) snapshots of TM interfaces are shown for homodimerization (TM4/5, within rectangles) and heterodimerization (TM5/6, within a circle) bundles. TM helices 4 (light blue), 5 (gray), and 6 (orange and green) are highlighted. (B) Root-mean-square deviations (rmsd) on protein α-carbons of the four-helix bundles forming the TM5/6 interface (orange solid line), TM4/5 interface of A1R (blue dotted line), and TM4/5 interface of A2AR (blue solid line) throughout the MD simulation. (C) Contact maps of the TM4/5 interface (rectangles in panel A) in the A1R or A2AR homodimer (left and right panels) and of the TM5/6 interface (circle in panel A) in the A1R-A2AR heterodimer (middle panel). Darker dots show more frequent contacts. (D) Detailed view of the extensive network of hydrophobic interactions (mainly of aromatic side chains) within the TM4/5 (left and right panels) and TM5/6 (middle panel) interfaces. The amino acids are numbered following the generalized numbering scheme of Ballesteros and Weinstein [37, 38]. This allows easy comparison among residues in the 7TM segments of different receptors. (TIF 4004 kb)
Positioning YFP in the C-tail of A2AR. The complex between the A2AR protomer (in light green) and Gs (α-subunit in dark grey and yellow, β-subunit in light gray, and γ-subunit in purple) was constructed from the crystal structure of β2 in complex with Gs [33]. Although the exact conformation of the A2AR C-tail (102 amino acids, Gln311–Ser412) cannot unambiguously be determined, its orientation was modeled as in the C-tail of squid rhodopsin [39], which contains the conserved amphipathic helix 8 that runs parallel to the membrane and an additional cytoplasmic helix 9. Thus, the C-tail of A2AR expands (see solid light green line) and points intracellularly toward the N-termini of the γ-subunit as suggested for OXER [32]. The laboratory of Kostenis has shown that the C-terminal of OXER, labeled with Rluc (OXER-Rluc), gets close to the N-terminal of the γ-subunit, labeled with GFP (γ-GFP) [32]. Analogously, we propose that YFP attached to the C-tail of A2AR is positioned near the N-termini of the γ-subunit (in purple). (TIF 2395 kb)
Additional file 10: Movie M1.
Assembly of adenosine A1 and A2A receptors in complex with two G proteins and MD simulation of the system. The assembly of Gs and Gi bound to the adenosine A1R-A2AR heterotetramer was subjected to 500 ns of MD simulation in a rectangular box containing the system, the lipid bilayer, explicit solvent, and ions. A1R protomers are in orange and red, A2AR protomers in light and dark green, Gα in white, Gβ in gray, and Gγ in purple. For easier visualization of protomer-protomer interfaces, TMs 4 and 5 are highlighted in blue and white, respectively. (MPEG 87870 kb)
Supplementary methods. (DOCX 72 kb)
Footnotes
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GN performed the molecular biology. GN, MB, EM, and DA perfomed BRET experiments. MZ-F performed single-particle tracking experiments. AC and LP-B performed molecular modeling studies. AC, VC, JM, and EIC analyzed the data. CL, LP, AJG-S, PJM, and RF designed the experiments, supervised the work in the respective laboratories and wrote the manuscript.. All authors read and approved the final manuscript.
Contributor Information
Peter J. McCormick, Email: p.mccormick@uea.ac.uk
Rafael Franco, Email: rfranco123@gmail.com.
References
- 1.Fung JJ, Deupi X, Pardo L, Yao XJ, Velez-Ruiz GA, Devree BT, et al. Ligand-regulated oligomerization of beta(2)-adrenoceptors in a model lipid bilayer. EMBO J. 2009;28:2384–2392. doi: 10.1038/emboj.2009.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albizu L, Cottet M, Kralikova M, Stoev S, Seyer R, Brabet I, et al. Time-resolved FRET between GPCR ligands reveals oligomers in native tissues. Nat Chem Biol. 2010;6:587–594. doi: 10.1038/nchembio.396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, et al. Crystal structure of the micro-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang J, Chen S, Zhang JJ, Huang XY. Crystal structure of oligomeric β1-adrenergic G protein-coupled receptors in ligand-free basal state. Nat Struct Mol Biol. 2013;20:419–425. doi: 10.1038/nsmb.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lane JR, Donthamsetti P, Shonberg J, Draper-Joyce CJ, Dentry S, Michino M, et al. A new mechanism of allostery in a G protein-coupled receptor dimer. Nat Chem Biol. 2014;10:745–752. doi: 10.1038/nchembio.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vinals X, Moreno E, Lanfumey L, Cordomi A, Pastor A, de La Torre R, et al. Cognitive impairment induced by delta9-tetrahydrocannabinol occurs through heteromers between cannabinoid cb1 and serotonin 5-ht2a receptors. PLoS Biol. 2015;13:e1002194. doi: 10.1371/journal.pbio.1002194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasai RS, Kusumi A. Single-molecule imaging revealed dynamic GPCR dimerization. Curr Opin Cell Biol. 2014;27:78–86. doi: 10.1016/j.ceb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 8.Hern JA, Baig AH, Mashanov GI, Birdsall B, Corrie JE, Lazareno S, et al. Formation and dissociation of M1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc Natl Acad Sci U S A. 2010;107:2693–2698. doi: 10.1073/pnas.0907915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calebiro D, Rieken F, Wagner J, Sungkaworn T, Zabel U, Borzi A, et al. Single-molecule analysis of fluorescently labeled G-protein-coupled receptors reveals complexes with distinct dynamics and organization. Proc Natl Acad Sci U S A. 2013;110:743–748. doi: 10.1073/pnas.1205798110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kasai RS, Suzuki KG, Prossnitz ER, Koyama-Honda I, Nakada C, Fujiwara TK, et al. Full characterization of GPCR monomer-dimer dynamic equilibrium by single molecule imaging. J Cell Biol. 2011;192:463–480. doi: 10.1083/jcb.201009128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vilardaga JP, Nikolaev VO, Lorenz K, Ferrandon S, Zhuang Z, Lohse MJ. Conformational cross-talk between alpha2A-adrenergic and mu-opioid receptors controls cell signaling. Nat Chem Biol. 2008;4:126–131. doi: 10.1038/nchembio.64. [DOI] [PubMed] [Google Scholar]
- 12.Fribourg M, Moreno JL, Holloway T, Provasi D, Baki L, Mahajan R, et al. Decoding the signaling of a GPCR heteromeric complex reveals a unifying mechanism of action of antipsychotic drugs. Cell. 2011;147:1011–1023. doi: 10.1016/j.cell.2011.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzalez S, Moreno-Delgado D, Moreno E, Perez-Capote K, Franco R, Mallol J, et al. Circadian-related heteromerization of adrenergic and dopamine D(4) receptors modulates melatonin synthesis and release in the pineal gland. PLoS Biol. 2012;10:e1001347. doi: 10.1371/journal.pbio.1001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navarro G, Ferre S, Cordomi A, Moreno E, Mallol J, Casado V, et al. Interactions between intracellular domains as key determinants of the quaternary structure and function of receptor heteromers. J Biol Chem. 2010;285:27346–27359. doi: 10.1074/jbc.M110.115634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferre S, Baler R, Bouvier M, Caron MG, Devi LA, Durroux T, et al. Building a new conceptual framework for receptor heteromers. Nat Chem Biol. 2009;5:131–134. doi: 10.1038/nchembio0309-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–194. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- 17.Ciruela F, Casado V, Rodrigues RJ, Lujan R, Burgueno J, Canals M, et al. Presynaptic control of striatal glutamatergic neurotransmission by adenosine A1-A2A receptor heteromers. J Neurosci. 2006;26:2080–2087. doi: 10.1523/JNEUROSCI.3574-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orru M, Bakesova J, Brugarolas M, Quiroz C, Beaumont V, Goldberg SR, et al. Striatal pre- and postsynaptic profile of adenosine A(2A) receptor antagonists. PLoS One. 2011;6:e16088. doi: 10.1371/journal.pone.0016088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harms GS, Cognet L, Lommerse PH, Blab GA, Kahr H, Gamsjager R, et al. Single-molecule imaging of l-type Ca(2+) channels in live cells. Biophys J. 2001;81:2639–2646. doi: 10.1016/S0006-3495(01)75907-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Whorton MR, Bokoch MP, Rasmussen SG, Huang B, Zare RN, Kobilka B, et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci U S A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferre S, Casado V, Devi LA, Filizola M, Jockers R, Lohse MJ, et al. G protein-coupled receptor oligomerization revisited: functional and pharmacological perspectives. Pharmacol Rev. 2014;66:413–434. doi: 10.1124/pr.113.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fredholm BB, IJzerman AP, Jacobson KA, Linden J, Muller CE. International Union of Basic and Clinical Pharmacology. LXXXI. Nomenclature and classification of adenosine receptors--an update. Pharmacol Rev. 2011;63:1–34. doi: 10.1124/pr.110.003285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carriba P, Navarro G, Ciruela F, Ferre S, Casado V, Agnati L, et al. Detection of heteromerization of more than two proteins by sequential BRET-FRET. Nat Methods. 2008;5:727–733. doi: 10.1038/nmeth.1229. [DOI] [PubMed] [Google Scholar]
- 24.Gilchrist A, Li A, Hamm HE. G alpha COOH-terminal minigene vectors dissect heterotrimeric G protein signaling. Sci STKE. 2002;2002:pl1. doi: 10.1126/stke.2002.118.pl1. [DOI] [PubMed] [Google Scholar]
- 25.Cordomi A, Navarro G, Aymerich MS, Franco R. Structures for G-protein-coupled receptor tetramers in complex with G proteins. Trends Biochem Sci. 2015;40:548–551. doi: 10.1016/j.tibs.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Mondal S, Johnston JM, Wang H, Khelashvili G, Filizola M, Weinstein H. Membrane driven spatial organization of GPCRs. Sci Rep. 2013;3:2909. doi: 10.1038/srep02909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okada M, Nutt DJ, Murakami T, Zhu G, Kamata A, Kawata Y, et al. Adenosine receptor subtypes modulate two major functional pathways for hippocampal serotonin release. J Neurosci. 2001;21:628–640. doi: 10.1523/JNEUROSCI.21-02-00628.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cristovao-Ferreira S, Navarro G, Brugarolas M, Perez-Capote K, Vaz SH, Fattorini G, et al. A1R-A2AR heteromers coupled to Gs and G i/0 proteins modulate GABA transport into astrocytes. Purinergic Signal. 2013;9:433–449. doi: 10.1007/s11302-013-9364-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Canals M, Marcellino D, Fanelli F, Ciruela F, de Benedetti P, Goldberg SR, et al. Adenosine A2A-dopamine D2 receptor-receptor heteromerization: qualitative and quantitative assessment by fluorescence and bioluminescence energy transfer. J Biol Chem. 2003;278:46741–46749. doi: 10.1074/jbc.M306451200. [DOI] [PubMed] [Google Scholar]
- 30.Liu W, Chun E, Thompson AA, Chubukov P, Xu F, Katritch V, et al. Structural basis for allosteric regulation of GPCRs by sodium ions. Science. 2012;337:232–236. doi: 10.1126/science.1219218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marti-Renom MA, Stuart AC, Fiser A, Sanchez R, Melo F, Sali A. Comparative protein structure modeling of genes and genomes. Annu Rev Biophys Biomol Struct. 2000;29:291–325. doi: 10.1146/annurev.biophys.29.1.291. [DOI] [PubMed] [Google Scholar]
- 32.Blattermann S, Peters L, Ottersbach PA, Bock A, Konya V, Weaver CD, et al. A biased ligand for OXE-R uncouples Gα and Gβγ signaling within a heterotrimer. Nat Chem Biol. 2012;8:631–8. doi: 10.1038/nchembio.962. Epub 2012 May 27. [DOI] [PubMed]
- 33.Rasmussen SG, DeVree BT, Zou Y, Kruse AC, Chung KY, Kobilka TS, et al. Crystal structure of the β2 adrenergic receptor-Gs protein complex. Nature. 2011;477:549–555. doi: 10.1038/nature10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chung KY, Rasmussen SG, Liu T, Li S, DeVree BT, Chae PS, et al. Conformational changes in the G protein Gs induced by the β2 adrenergic receptor. Nature. 2011;477:611–615. doi: 10.1038/nature10488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pronk S, Pall S, Schulz R, Larsson P, Bjelkmar P, Apostolov R, et al. GROMACS 4.5: a high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics. 2013;29:845–854. doi: 10.1093/bioinformatics/btt055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cordomi A, Caltabiano G, Pardo L. Membrane protein simulations using AMBER force field and Berger lipid parameters. J Chem Theory Comput. 2012;8:948–958. doi: 10.1021/ct200491c. [DOI] [PubMed] [Google Scholar]
- 37.Ballesteros JA, Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure-function relations in G-protein coupled receptors. Methods Neurosci. 1995;25:366–428. doi: 10.1016/S1043-9471(05)80049-7. [DOI] [Google Scholar]
- 38.Isberg V, de Graaf C, Bortolato A, Cherezov V, Katritch V, Marshall FH, et al. Generic GPCR residue numbers - aligning topology maps while minding the gaps. Trends Pharmacol Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murakami M, Kouyama T. Crystal structure of squid rhodopsin. Nature. 2008;453:363–367. doi: 10.1038/nature06925. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The crystal structures 4EIY, 2Z73, 3SN6, 1AGR, 2RH7, 2PSD, 4GPO, and 4DKL are available from PDB (http://www.rcsb.org). All other relevant data are within the paper and its Additional files.


