Abstract
Background & objectives:
Amoebiasis is a common parasitic infection caused by Entamoeba histolytica and amoebic liver abscess (ALA) is the most common extraintestinal manifestation of amoebiasis. The aim of this study was to standardise real-time PCR assays (Taqman and SYBR Green) to detect E. histolytica from liver abscess pus and stool samples and compare its results with nested-multiplex PCR.
Methods:
Liver abscess pus specimens were subjected to DNA extraction. The extracted DNA samples were subjected to amplification by nested-multiplex PCR, Taqman (18S rRNA) and SYBR Green real-time PCR (16S-like rRNA assays to detect E. histolytica/E. dispar/E. moshkovskii). The amplification products were further confirmed by DNA sequence analysis. Receiver operator characteristic (ROC) curve analysis was done for nested-multiplex and SYBR Green real-time PCR and the area under the curve was calculated for evaluating the accuracy of the tests to dignose ALA.
Results:
In all, 17, 19 and 25 liver abscess samples were positive for E. histolytica by nested-multiplex PCR, SYBR Green and Taqman real-time PCR assays, respectively. Significant differences in detection of E. histolytica were noted in the real-time PCR assays evaluated (P<0.0001). The nested-multiplex PCR, SYBR Green real-time PCR and Taqman real-time PCR evaluated showed a positivity rate of 34, 38 and 50 per cent, respectively. Based on ROC curve analysis (considering Taqman real-time PCR as the gold standard), it was observed that SYBR Green real-time PCR was better than conventional nested-multiplex PCR for the diagnosis of ALA.
Interpretation & conclusions:
Taqman real-time PCR targeting the 18S rRNA had the highest positivity rate evaluated in this study. Both nested multiplex and SYBR Green real-time PCR assays utilized were evaluated to give accurate results. Real-time PCR assays can be used as the gold standard in rapid and reliable diagnosis, and appropriate management of amoebiasis, replacing the conventional molecular methods.
Keywords: Amoebic liver abscess (ALA), Entamoeba histolytica, nested multiplex PCR, real-time PCR, SYBR Green, Taqman
Amoebiasis, caused by Entamoeba histolytica is the second most common cause of death due to parasitic infections after malaria worldwide and is responsible for approximately 1,00,000 deaths per year occurring mainly in Africa, Indian subcontinent, Central and South America1,2. The clinical features of amoebiasis range from asymptomatic colonization to amoebic dysentery and invasive extraintestinal amoebiasis which is manifested most commonly as amoebic liver abscess (ALA). The extraintestinal manifestations result due to spread of the invasive trophozoites from colon to various organs by haematogenous route. The most commonly affected organs are the liver, pleura, pericardium, brain and genitourinary system3. Although imaging techniques like ultrasonogram (USG)/computerized tomographic (CT) scan/magnetic resonance imaging (MRI) are sensitive for the detection of liver abscess, it is difficult to differentiate ALA from pyogenic liver abscess (PLA)/any other space occupying lesions in liver. Very few patients with ALA have concurrent intestinal disease while most lack bowel symptoms with no findings in stool microscopy4. The most common and serious complication is rupture of the abscess into the pleura or pericardium with pleural rupture being relatively common with good prognosis. With early detection and treatment, mortality of ALA is <1 per cent5.
ALA is usually diagnosed by the demonstration of amoebic antibody-based serological tests combined with absence of bacteria in the abscess pus. The drawback of serological tests is that antibody levels from people residing in endemic areas remain positive for years after infection with E. histolytica, limiting their specificity for the diagnosis of ALA6. Antigen detection based tests performed on serum and liver pus samples, like enzyme-linked immuno sorbent assay (ELISA), counter-current immune-electrophoresis (CIEP) and co-agglutination test usually targeting the Gal/Gal NAc lectin, had been useful in the detection of ALA cases7,8,9.
With advancement in the area of molecular biology based diagnostic tests new techniques with increased sensitivity, specificity and simplicity are available. Specific amplification of E. histolytica DNA has been found to be the most sensitive and specific method of detection of E. histolytica from faeces and liver abscess pus10. The PCR has been widely evaluated in diagnosis of the ALA by demonstration of E. histolytica DNA in the serum as well as in the liver pus, saliva and urine10. But the conventional PCR protocols are time consuming and prone to false positive results due to risk of cross-contamination. Real-time PCR, a closed tube technique that employs fluorescent labels for continuous monitoring of amplicon generation can circumvent the disadvantages of a conventional PCR. The aim of this study was to evaluate real-time PCR in detection of ALA cases by detecting E. histolytica DNA in the liver abscess pus samples and compare its results with the conventional PCR.
Material & Methods
The study was conducted in the department of Microbiology in collaboration with the departments of Medicine and Paediatrics, Jawaharlal Institute of Postgraduate Medical Education and Research (JIPMER), Puducherry from April 2010 to October 2011. The study was approved by the institute's research and human ethics committees.
A total of 50 liver abscess pus specimens collected during the study period with written informed consent were included in the study. The inclusion criteria for case selection (confirmed cases) included liver abscess pus specimens from nested-multiplex PCR confirmed cases of amoebiasis (n=17), suspected cases of ALA (solitary hypoechoic lesion detected by ultrasonogram of liver with thick brownish aspirate) negative by nested - multiplex PCR (n=22) and pyogenic liver abscess (PLA) pus specimens (control group) (n=11). Liver abscess pus samples were collected in sterile capped containers and immediately transported to the microbiology laboratory. The samples were stored at -20°C until further use.
DNA extraction: The DNA from the samples were extracted by modified cetyl trimethyl ammonium bromide (CTAB) method10. All the extracted samples were tested in parallel for Entamoeba DNA by nested-multiplex PCR, SYBR Green and Taqman real-time PCR methods.
Standard strains used: E. histolytica HM1:IMSS, E. dispar SAW 760 and E. moshkovskii Laredo were the standard strains used as positive control in the present study.
Nested-multiplex PCR: The nested-multiplex PCR protocol was adapted from earlier studies from our laboratory10,11,12. The primer sequences targeted the 16S-like rRNA gene.
Genus - specific primers. First round PCR).
Entamoeba genus: E-1 5’ TAAGATGCACGAGAGCGAAA 3’(forward primer).
E-2 5’GTACAAAGGGCAGGGACGTA3’(reverse primer).
Species - specific primers (Second round nested- multiplex PCR).
E. histolytica: Eh - 1 5’AAGCATTGTTTCTAGATCTGAG 3’(forward primer).
Eh - 2 5’AAGAGGTCTAACCGAAATTAG 3’ (reverse primer).
E. dispar: Ed - 1 5’TCTAATTTCGATTAGAACTCT 3’(forward primer).
Ed - 2 5’TCCCTACCTATTAGACATAGC 3’(reverse primer).
E. moshkovskii: Em- 1 5’GAAACCAAGAGTTTCACAAC 3’(forward primer).
Em- 2 5’CAATATAAGGCTTGGATGAT 3’(reverse primer).
PCR conditions: A 25 μl amplification mixture containing 10 μl of master mix (Bangalore Genei, Bangalore), 0.3 μM of forward and reverse primers. First round PCR: Genus-specific primers used, Second Nested Multiplex PCR: Species - specific primers used) with 2.5 μl of the extracted template DNA and the remaining of the 25 μl volume Milli-Q water was used. The reaction was carried out in an Eppendorf Master cycler (Eppendorf, Germany). For genus - specific PCR, the PCR mix was subjected to an initial denaturation at 96°C for 2 min, followed by 30 cycles of amplification - each at 92°C for 60 sec (denaturation), 56°C for 60 sec (annealing), and 72°C for 90 sec (extension). Finally, one cycle of extension at 72°C for seven minutes was performed. In the species - specific nested multiplex PCR (which had multiple primer sets in the same tube), only the annealing temperature was changed to 48°C, leaving the other parameters of the amplification cycles unchanged.
The amplification products (3 μl) were separated by electrophoresis through 1.8 per cent agarose gel (Agarose Low EEO, Bangalore Genei products, Bengaluru, India) in 0.5 x Tris-borate-EDTA at 120V for 45 min and were visualized by ethidium bromide staining under UV light for bands of DNA of appropriate sizes (Eh-439 bp, Ed-174 bp and Em-553 bp), ascertained in comparison to a standard 100 bp DNA ladder used as a molecular weight marker. Positive (E. histolytica HM1:IMSS strain DNA) and negative control reactions (Milli-Q water added instead of DNA) were included with each batch of samples analysed by nested-multiplex PCR (Fig. 1).
Fig. 1.
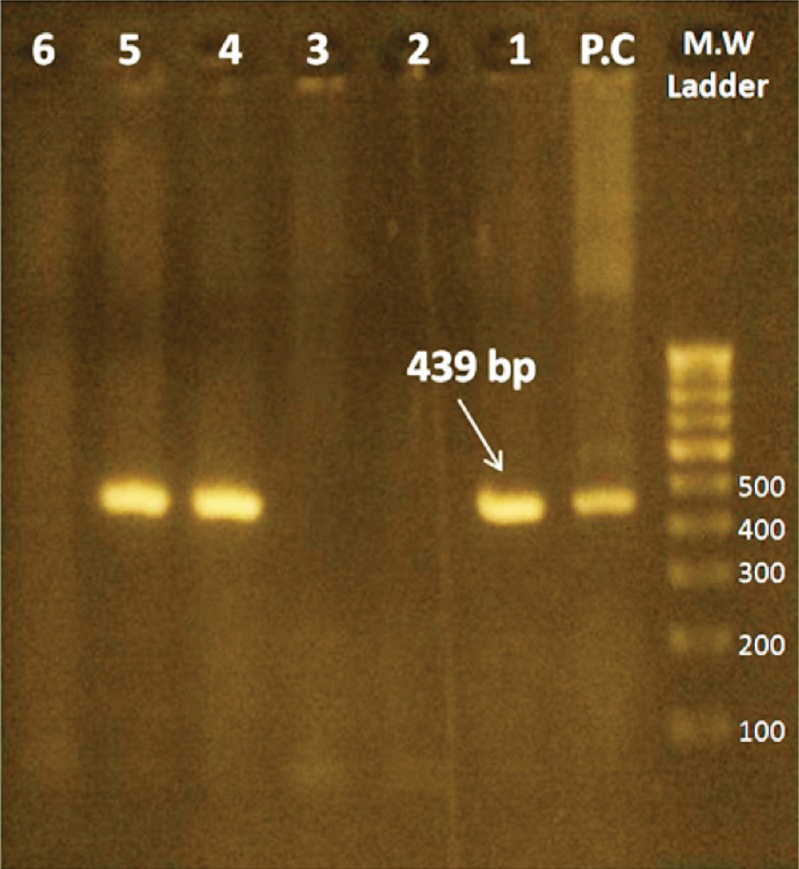
Agarose gel electrophoresis of amplicons from nested-multiplex PCR, showing M.W. marker-DNA molecular weight marker [represented in base pairs (bp)], P.C - Positive control (Eh - HM1:IMSS strain), Lanes 1, 4 and 5-Positive test samples, Lanes 2, 3 and 6 - Negative test samples.
Real-time PCR
(i) Taqman-based chemistry - The Taqman-based real-time PCR protocol was adapted from earlier studies13,14,15. The primer sequences utilized targeted the 18S rRNA (amplicon size -231 bp) and were as follows: Ehd- F- 5’ ATTGTCGTGGCATCCTAACTCA 3’; Ehd- R- 5’ GCGGACGGCTCATTATAACA 3’.
The probe sequences utilized for detection of E. histolytica and E. dispar were as follows: histolytica-96T VIC -UCAUUGAAUGAAUUGGC CAUUU-NFQ; dispar-96T FAM-UUACUUACAUAAAUUGGCCACUUUG-NFQ.
PCR conditions - A 25 μl reaction volume constituting 12.5 μl of 2X Taqman gene expression master mix, 1.25 μl of 20X primer-probe mix (0.5 μM of each primer/0.1 μM of each probe), 2.5 μl of 10X extracted sample DNA and 8.75 μl of milli-Q water was used. The concentrations used were according to the recommendations in Step-one software (Applied Biosystems). The reaction was carried out in an Applied Biosystems RT-PCR thermal cycler. The cycling conditions were as follows, an initial holding stage of 95°C for three minutes, followed by 40 cycles of 95°C for 15 sec, 60°C for 30 sec and 72°C for 30 sec (cycling stage). The fluorescence was detected at the end of each extension plateau. Positive (E. histolytica HM1: IMSS strain DNA) and negative control (Milli-Q water added instead of DNA) reactions were included with each batch of samples analysed (Fig. 2).
Fig. 2.
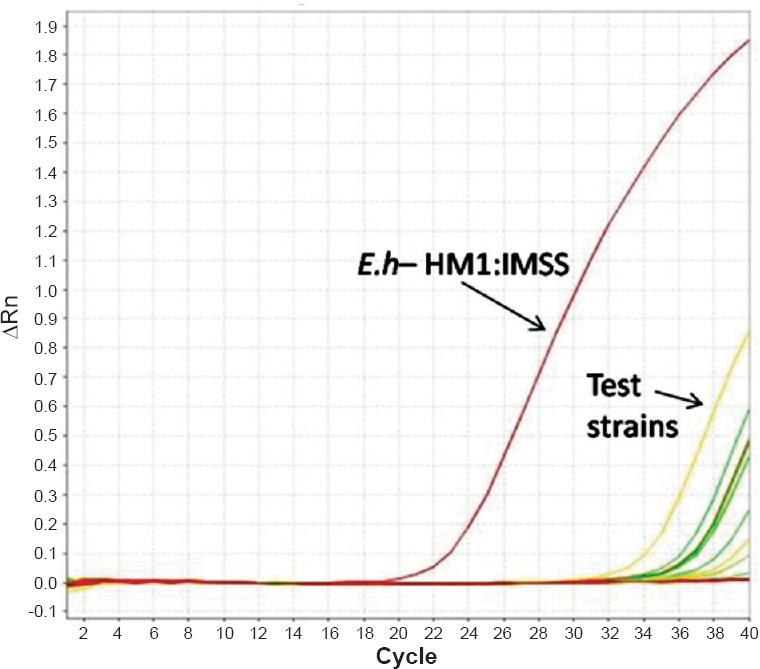
Amplification curve obtained in Taqman real-time PCR assay. Eh-HM1:IMSS:- Reference strain used as positive control. A test sample positive for E. histolytica (fluorescence signal) is indicated with an arrow.
(ii) SYBR Green based chemistry - The conventional nested-multiplex PCR was adapted for SYBR Green real-time PCR assay.The primers utilized were the same as nested - multiplex PCR (16S-like rRNA gene target).
PCR conditions - A 25 μl reaction volume constituting 12.5 μl of 2X Power SYBR Green PCR master mix, 0.25 μl of forward and reverse primers (0.1 μM concentration each), 2.5 μl of 10X extracted sample DNA and 11 μl of mill-Q water was used. Primers targeting different species were run in separate tubes. The concentrations used were according to the recommendations in Step-one software (Applied Biosystems). The reaction was carried out in an Applied Biosystems real-time-PCR thermal cycler. The cycling conditions were as follows, an initial holding stage of 95°C for 15 min followed by 50 cycles of 95°C for 15 sec, 60°C for one minute, 72°C for 1.5 min and 80°C for 30 sec (cycling stage). The cycling stage was followed by melting curve analysis to determine the specific melting curve temperature. This stage constituted rising of temperature from 40 to 95°C at 1°C rise per minute. The fluorescence was detected at the end of the 80°C incubation plateau and during the melting curve analysis (melting temperature, Tm- 78.96°C) (Fig. 3). The amplified product were confirmed by running agarose gel electrophoresis (1.8%) without ethidium bromide staining under UV light for bands of DNA of appropriate sizes (Eh – 439 bp, Ed – 174 bp and Em – 553 bp), ascertained in comparison to a standard 100 bp DNA ladder used as a molecular weight marker. SYBR Green was added to the DNA molecular weight marker as a DNA intercalating dye (Fig. 4). Positive and negative control reactions were included with each batch of samples analysed.
Fig. 3.
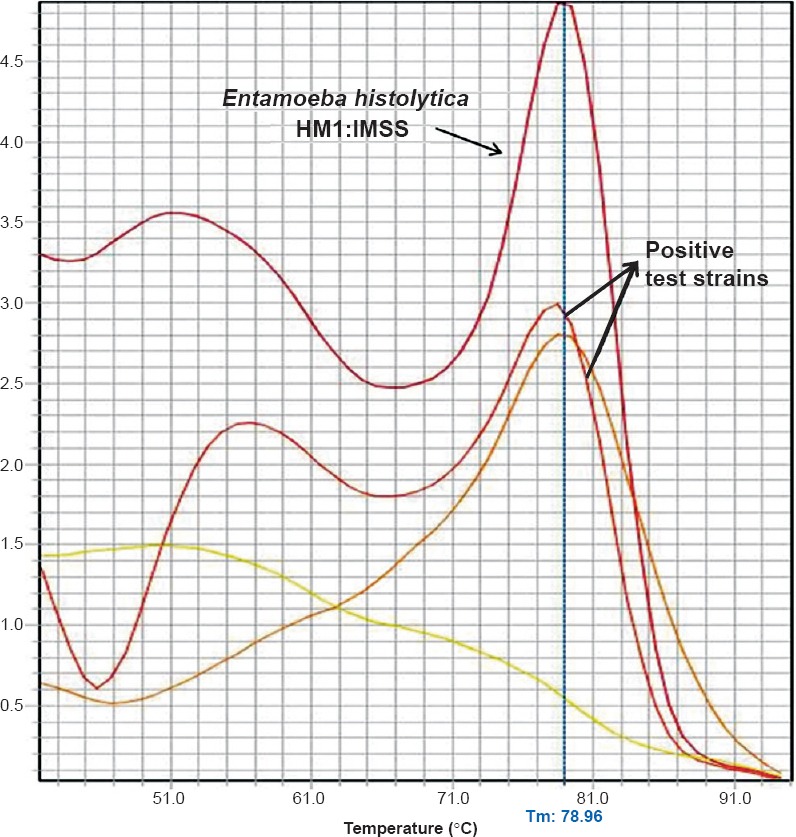
Melting curve analysis of the amplified products obtained in the SYBR Green real-time PCR assay. Entamoeba histolytica HM1:IMSS:- Reference strain used as positive control (Tm-78.96°C); Tm-Melting temperature of the amplicon. Melting temperature curves of the amplicons from positive test samples are indicated with an arrow.
Fig. 4.
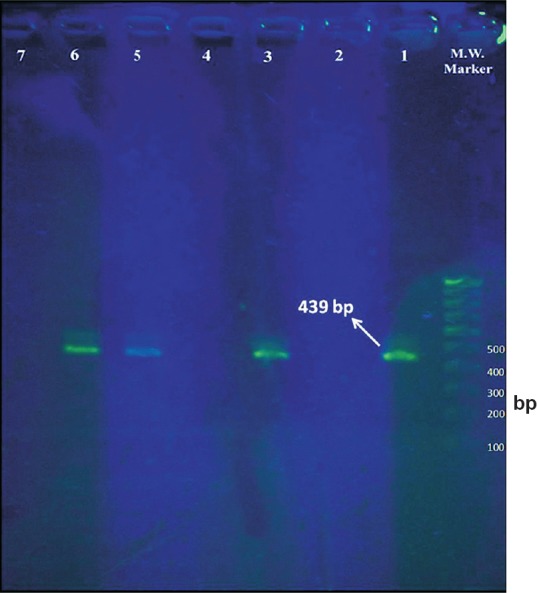
Agarose gel electrophoresis of amplified products from SYBR Green real-time PCR assay (without ethidium bromide staining). M.W. marker - 100 bp DNA molecular weight marker, Lane 1 - Positive control (amplicon size - 439 bp), Lane 2 - Negative control, Lanes 3, 5 and 6 - Positive test samples (439 bp - E. histolytica), Lanes 4 and 7 - Negative test samples.
Sequence analysis: Amplified products from the positive control strain, E. histolytica HM1: IMSS and five positive samples were sequenced by Peak Trace Basecaller Software (Sanger's DNA sequencing) using ABI 3730xls sequencer (Macrogen Inc., South Korea) and were found to have maximum identity with E. histolytica small subunit RNA sequence with accession number- AB608092.1.
Statistical analysis: Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were calculated using Graph Pad Instat3 Software (University of California, San Diego, USA). Receiver operator characteristic (ROC) curve analysis was done using SPSS 16.0 for Windows (SPSS Inc., Chicago, IL, USA) for nested-multiplex and SYBR Green real-time PCR and the area under the curve was calculated for evaluating the accuracy of the tests to dignose ALA.
Results
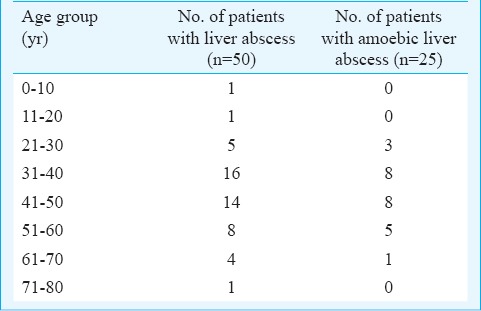
Among the 50 patients with liver abscess (study population), 48 were adults and two were in paediatric age group. The median age group of the study population was 43.5 years. Twenty one of 25 ALA cases diagnosed were in the age range of 30-60 yr. The two paediatric cases included in the study were negative for amoebic aetiology. The age distribution of the study population and the ALA cases is shown in Table I. Males and females were affected in the ratio of 11.5 : 1. Among the PLA cases, the predominant bacterial isolate was Escherichia coli, seen in six (55%) of 11 patients. In one case of PLA, ß-haemolytic streptococci was isolated along with E. coli. The other isolates obtained were Enterobacter spp.(2), Klebsiella pneumoniae (2) and Chromobacterium violaceum (1). Bacterial co-infection of confirmed ALA cases was seen in two (8%) of 25 patients in this study. In one case, E. coli and in another Enterobacter spp. was isolated.
Table I.
Age distribution of the study population (liver abscess cases) and those with amoebic liver abscess
Nested-multiplex PCR: Seventeen of the 50 samples were positive for E. histolytica and were included in the confirmed case group. The remaining 22 liver abscess samples were in the suspected ALA case group. Two samples which were positive for E. histolytica by nested - multiplex PCR had co-existent bacterial infection. The ROC curve of nested-multiplex PCR and the area corresponding area under the curve is shown in Fig. 5.
Fig. 5.
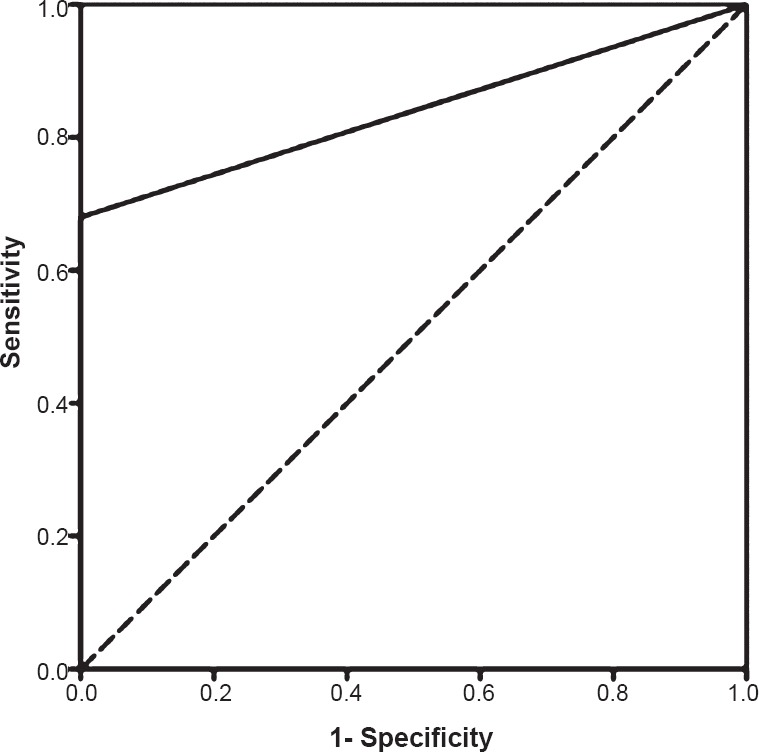
Receiver operator characteristic (ROC) curve of nested-multiplex PCR. Area under the curve: 0.84.
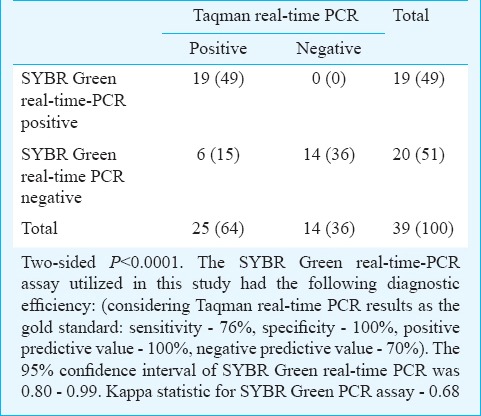
SYBR Green real-time PCR: Real-time PCR assay using the SYBR Green methodology, targeting the 16S-like rRNA of E. histolytica was performed for the 50 liver abscess pus samples. Nineteen samples were found to be positive for E. histolytica. Concordant positive results (both nested-multiplex PCR and SYBR Green RT-PCR positive) were seen in 15 liver abscess pus samples. Two samples positive for E. histolytica by nested-multiplex PCR were not detected by the SYBR Green real-time PCR assay, and four samples detected positive by SYBR Green assay were negative for E. histolytica by nested-multiplex PCR. The ROC curve of SYBR Green real-time PCR and the corresponding area under the curve is shown in Fig. 6.
Fig. 6.
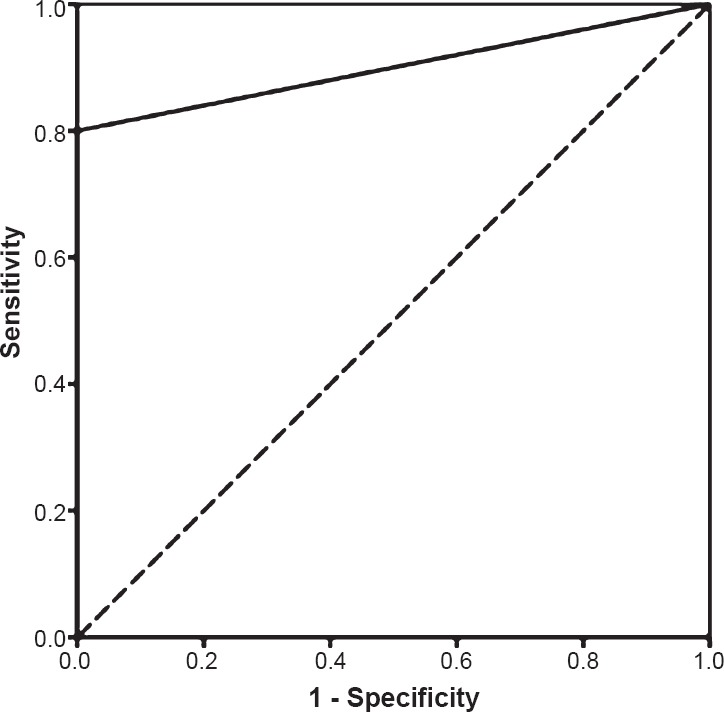
Receiver operator characteristic (ROC) curve of SYBR Green real-time PCR. Area under the curve: 0.9.
Taqman-based real-time PCR: Real-time PCR using Taqman assay targeting the 18S rRNA of E. histolytica was performed for the 50 liver abscess pus samples. Twenty five samples were positive for E. histolytica, which included all the positive cases detected by nested-multiplex PCR and SYBR Green real-time PCR assays. The positivity rate of Taqman real-time PCR assay was 50 per cent (25 positive of 50 liver abscess pus samples). Comparison of nested-multiplex PCR and SYBR Green real-time PCR assay results was done with Taqman real-time PCR assay (Tables II, III).
Table II.
Comparison of results of nested multiplex - PCR and Taqman real-time PCR assays
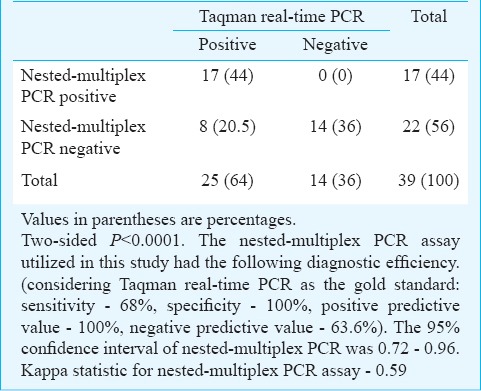
Table III.
Comparison of results of SYBR Green and Taqman real-time PCR assays
Discussion
Real-time PCR methods combine PCR chemistry with amplicon detection by fluorescence. There are various methodologies available, of which the Taqman assay (5’nuclease assay), molecular beacons, FRET (flourescence resonance energy transfer) probes and SYBR Green chemistries are well studied. Real-time PCR has been developed for the diagnosis of various parasitic diseases like malaria, amoebiasis, toxoplasmosis, leishmaniasis, trypanosomiasis, babesiosis, cryptosporidiosis, giardiasis, etc16. In this study we utilized Taqman and SYBR Green based real-time PCR for the diagnosis of amoebiasis.
The nested-multiplex PCR utilized in this study detected 17 cases of ALA amongst the 39 suspected cases of extraintestinal amoebiasis. The specificity of the nested-multiplex PCR was found to be 100 per cent as none of the samples in the control group (11 liver abscess pus samples from pyogenic liver abscess) yielded any positive results. The positivity rate of nested-multiplex PCR in this study was 34 per cent (17 of 50 liver abscess pus samples, tested positive for amoebic aetiology). There are very few studies in literature comparing the results of conventional PCR with real-time PCR. Roy et al17 compared conventional PCR targeting small subunit rRNA with molecular-beacon based real-time PCR assay in detection of E. histolytica in ALA patients; the sensitivity and specificity for the conventional PCR were 72 and 99 per cent. Our conventional nested-multiplex PCR, targeting the 16S-like rRNA had a sensitivity and specificity of 68 and 100 per cent, respectively. The SYBR Green real-time PCR optimized in this study could detect 19 cases of ALA of the 39 suspected cases of amoebiasis. Fifteen liver abscess pus samples were positive for amoebic aetiology by both nested-multiplex PCR and SYBR Green assay. It was observed that two liver abscess pus samples which were positive for E. histolytica by nested-multiplex PCR and Taqman real-time PCR assay were negative by SYBR Green real-time PCR assay, and also the SYBR Green assay could detect four cases of ALA which were negative by nested-multiplex PCR. The specificity of SYBR Green real-time PCR assay was 100 per cent since none of the control group samples yielded any positive results. The positivity rate of SYBR Green assay optimized in this study was 38 per cent (19 of 50 liver abscess samples were positive for amoebiasis). Qvanstrom et al13 reported the use of SYBR Green real-time PCR (targeting 18S rRNA) in detection of E. histolytica from liver abscess samples. The assay detected five of nine confirmed amoebic liver abscess pus samples. Rahman et al18 used SYBR Green assay targeting the SREHP gene in genotyping of E. histolytica. The Taqman-based real-time PCR assay optimized in this study detected 25 of the 39 suspected cases of amoebiasis. This assay could detect all the positive cases detected by nested-multiplex PCR and SYBR Green real-time PCR assay. This assay also had 100 per cent specificity since none of the control group samples (pyogenic liver abscess) yielded positive results. Othman et al19 using real-time PCR detected E. histolytica DNA in 23 of 30 liver abscess pus samples. The positivity rate of the optimized Taqman real-time PCR assay in this study was 50 per cent (25 of 50 liver abscess samples were found to be positive for amoebic aetiology).
Among the 39 liver abscess pus samples from the suspected cases of amoebiasis, 25 were positive for E. histolytica. In the remaining 14 cases, no aetiology was found. The possible explanation in these cases could be anaerobic organisms, tubercular liver abscess, fungal abscess, antibiotic use prior to admission and lower levels of genome copies which could not be detected by the real-time PCR assays utilized.
The positive cases of ALA detected by SYBR Green and Taqman real-time PCR assay were confirmed to be E. histolytica by DNA sequencing analysis. Considering the optimized Taqman real-time PCR assay as the gold standard, the diagnostic efficiency of nested-multiplex PCR and SYBR Green assay were calculated. The nested-multiplex PCR had a sensitivity and specificity of 68 and 100 per cent, respectively. In this study, five liver abscess samples were initially falsely identified to be positive for E. histolytica by nested-multiplex PCR. This was detected when these five samples tested negative in Taqman real-time PCR assay and were confirmed by repeat DNA extraction and nested-multiplex PCR testing which gave negative results for E. histolytica. This may be attributed to cross-contamination of DNA, which is a disadvantage of conventional nested-multiplex PCR technique since the extracted DNA is handled several times (during the two cycles of amplification and while performing agarose gel electrophoresis). Such a disadvantage is absent in real-time PCR assay since there is no post-PCR handling of DNA. The nested-multiplex PCR and SYBR Green real-time PCR assays utilized in this study had an average post-extraction processing time of seven and six hours, respectively, whereas the post-extraction processing time for the optimized Taqman real-time PCR assay was one and a half hours. Qvanstrom et al13 reported an estimated time from reception of specimen to final result as 7 h for conventional nested-multiplex PCR and SYBR Green real-time PCR assays. The Taqman assays utilized in their study had an estimated time of four hours which comparatively was a shorter duration than our study (six hours) since they utilized commercial DNA extraction kit (QIAGEN Inc, California, USA).
In addition to the above mentioned advantages of real-time PCR over conventional PCR, quantitative analysis can also be performed with real-time PCR assays. In amoebiasis, the parasite content in the sample varies tremendously between or even within specimens from the same patient. Hence quantitative analysis does not play much role in the estimation of parasite burden in amoebiasis patients. But this analysis would be useful in food, water and environmental samples in assessing the parasite burden. Although real-time PCR has numerous advantages compared to the conventional PCR, the limitation of high cost equipment and laboratory set-up exists.
The limitation of our study was that the performance characteristics of the real-time PCR assays like the limit of detection (cysts or trophozoites/ml of sample) and the amplification efficiency (slope) were not evaluated. Hence comparison of amplification efficacy with other published real-time PCR assays could not be performed. The difference in the positivity rate between the two real-time PCR assays optimized (Taqman and SYBR Green assays) in this study might be due to (i) difference in the target amplified in the two assays (18S rRNA in Taqman assay and 16S-like rRNA in the SYBR Green assay; (ii) suboptimal amplification efficiency in Taqman and SYBR Green real-time PCR. Since the amplicon size recommended for optimal amplification efficiency is 50-150 bp, and the amplicon sizes of Taqman and SYBR Green assays optimised in this study were larger, which would have led to less efficient amplification during the PCR cycle; and (iii) utilization of primers with low melting temperature (51°C) in SYBR Green real-time PCR assay which ideally should be 58-60°C for optimal amplification.
Footnotes
Conflicts of Interest: None.
References
- 1.Stanley SL. Amoebiasis. Lancet. 2003;361:1025–34. doi: 10.1016/S0140-6736(03)12830-9. [DOI] [PubMed] [Google Scholar]
- 2.Haque R, Huston CD, Hughes M, Houpt E, Petri WA., Jr Amebiasis. N Engl J Med. 2003;348:1565–73. doi: 10.1056/NEJMra022710. [DOI] [PubMed] [Google Scholar]
- 3.Garcia LS. Diagnostic medical parasitology. 5th ed. Washington DC: ASM Press; 2006. [Google Scholar]
- 4.Thompson JE, Jr, Forlenza S, Verma R. Amebic liver abscess: a therapeutic approach. Rev Infect Dis. 1985;7:171–9. doi: 10.1093/clinids/7.2.171. [DOI] [PubMed] [Google Scholar]
- 5.Anesi JA, Gluckman S. Amebic liver abscess: Amebic liver abscess Anesi and Gluckman. Clin Liver Dis. 2015;6:41–3. doi: 10.1002/cld.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haque R, Kabir M, Noor Z, Rahman SM, Mondal D, Alam F, et al. Diagnosis of amebic liver abscess and amebic colitis by detection of Entamoeba histolytica DNA in blood, urine, and saliva by a real-time PCR assay. J Clin Microbiol. 2010;48:2798–801. doi: 10.1128/JCM.00152-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karki BM, Parija SC. Co-agglutination test for the detection of circulating antigen in amebic liver abscess. Am J Trop Med Hyg. 1999;60:498–501. doi: 10.4269/ajtmh.1999.60.498. [DOI] [PubMed] [Google Scholar]
- 8.Parija SC, Karki BM. Detection of circulating antigen in amoebic liver abscess by counter-current immunoelectrophoresis. J Med Microbiol. 1999;48:99–101. doi: 10.1099/00222615-48-1-99. [DOI] [PubMed] [Google Scholar]
- 9.Haque R, Neville LM, Hahn P, Petri WA., Jr Rapid diagnosis of Entamoeba infection by using Entamoeba and Entamoeba histolytica stool antigen detection kits. J Clin Microbiol. 1995;33:2558–61. doi: 10.1128/jcm.33.10.2558-2561.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khairnar K, Parija SC. A novel nested multiplex polymerase chain reaction (PCR) assay for differential detection of Entamoeba histolytica, E. moshkovskii and E. dispar DNA in stool samples. BMC Microbiol. 2007;7:47. doi: 10.1186/1471-2180-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Parija SC, Khairnar K. Detection of excretory Entamoeba histolytica DNA in the urine, and detection of E. histolytica DNA and lectin antigen in the liver abscess pus for the diagnosis of amoebic liver abscess. BMC Microbiol. 2007;7:41. doi: 10.1186/1471-2180-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Khairnar K, Parija SC. Detection of Entamoeba histolytica DNA in the saliva of amoebic liver abscess patients who received prior treatment with metronidazole. J Health Popul Nutr. 2008;26:418–25. doi: 10.3329/jhpn.v26i4.1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qvanstrom Y, James C, Xayavong M, Holloway BP, Visvesvara GS, Sriram R, et al. Comparison of real-time PCR protocols for differential laboratory diagnosis of amebiasis. J Clin Microbiol. 2005;43:5491–7. doi: 10.1128/JCM.43.11.5491-5497.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verweij JJ, Blangé RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, et al. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol. 2004;42:1220–3. doi: 10.1128/JCM.42.3.1220-1223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kebede A, Verweij JJ, Endeshaw T, Messele T, Tasew G, Petros B, et al. The use of real-time PCR to identify Entamoeba histolytica and E. dispar infections in prisoners and primary-school children in Ethiopia. Ann Trop Med Parasitol. 2004;98:43–8. doi: 10.1179/000349804225003082. [DOI] [PubMed] [Google Scholar]
- 16.Espy MJ, Uhl JR, Sloan LM, Buckwalter SP, Jones MF, Vetter EA, et al. Real-time PCR in clinical microbiology: applications for routine laboratory testing. Clin Microbiol Rev. 2006;19:165–256. doi: 10.1128/CMR.19.1.165-256.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Roy S, Kabir M, Mondal D, Ali IK, Petri WA, Jr, Haque R. Real-time-PCR assay for diagnosis of Entamoeba histolytica infection. J Clin Microbiol. 2005;43:2168–72. doi: 10.1128/JCM.43.5.2168-2172.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rahman S, Haque R, Roy S, Mondal M. Genotyping of Entamoeba histolytica by real-time polymerase chain reaction with Sybr Green I and melting curve analysis. Bangladesh Vet Med. 2006;4:53–60. [Google Scholar]
- 19.Othman N, Mohamed Z, Verweij JJ, Huat LB, Olivos-García A, Yeng C, et al. Application of real-time polymerase chain reaction in detection of Entamoeba histolytica in pus aspirates of liver abscess patients. Foodborne Pathog Dis. 2010;7:637–41. doi: 10.1089/fpd.2009.0427. [DOI] [PubMed] [Google Scholar]


