Abstract
Both positive and negative interactions among bacteria take place in the environment. We hypothesize that the complexity of the substrate affects the way bacteria interact with greater cooperation in the presence of recalcitrant substrate. We isolated lignocellulolytic bacteria from salt marsh detritus and compared the growth, metabolic activity and enzyme production of pure cultures to those of three-species mixed cultures in lignocellulose and glucose media. Synergistic growth was common in lignocellulose medium containing carboxyl methyl cellulose, xylan and lignin but absent in glucose medium. Bacterial synergism promoted metabolic activity in synergistic mixed cultures but not the maximal growth rate (μ). Bacterial synergism also promoted the production of β-1,4-glucosidase but not the production of cellobiohydrolase or β-1,4-xylosidase. Our results suggest that the chemical complexity of the substrate affects the way bacteria interact. While a complex substrate such as lignocellulose promotes positive interactions and synergistic growth, a labile substrate such as glucose promotes negative interactions and competition. Synergistic interactions among indigenous bacteria are suggested to be important in promoting lignocellulose degradation in the environment.
Keywords: bacterial synergism, lignocellulose degradation, bacterial activity, enzyme production, microbial interaction
Introduction
Bacteria in nature do not live in isolation but interact dynamically with other bacteria in the community. Interactions can be either positive or negative, resulting in different community function outcomes (Newman and Banfield, 2002; Hansen et al., 2007). Factors that mediate the way bacteria interact remain poorly studied.
Competition for resources has been suggested as the prevalent type of interaction among bacteria (Hibbing et al., 2010; Foster and Bell, 2012). As labile nutrients are often limited in the environment, one bacteria might compete with other bacteria by producing antibiotics that inhibit the growth of others (D’Costa et al., 2007; Hibbing et al., 2010). For example, the majority of marine bacterial isolates exhibited antagonistic activity against others from the same environment through production of potential antimicrobial agents (Long and Azam, 2001; Grossart et al., 2004; Rypien et al., 2010).
Although competitive interactions among bacteria are ubiquitous in nature, there are multiple studies showing that using complex microbial communities enhances the rate of bioconversion of lignocellulose to more valuable products (Haruta et al., 2002; Wongwilaiwalin et al., 2010; Jiménez et al., 2014). It is unclear however whether bacterial synergism is directly related to the structural complexity of the substrate and whether bacteria in the consortia switch from synergism to antagonism if labile substrates become available.
Lignocellulose consists of mainly cellulose, hemicellulose and lignin that are strongly cross-linked in plant tissue and thus is recalcitrant to degradation (Lynd et al., 2002). To achieve better degradation, bacteria might form consortia and synergistically degrade lignocellulose (Lynd et al., 2002; Perez et al., 2002). It thus seems likely that the way bacteria interact may depend on substrate complexity with greater cooperation in the presence of recalcitrant substrate.
In this study, we test the hypothesis that the complexity of substrate mediates bacterial interactions. When lignocellulose is the only carbon source, bacteria form consortia and synergistically degrade the complex substrate. However, when labile substrate is available, bacteria compete for the limiting nutrient and antagonistic interaction prevails. To test this hypothesis, we isolated lignocellulolytic bacteria from salt marsh detritus. Bacteria were grown in single and three-species mixed cultures in both lignocellulose medium and glucose medium. We compared the growth, enzyme production and metabolic activity of three-species mixed cultures to those of pure cultures. The specific aims of this study are to determine: 1) the frequency of synergistic degradation among bacteria when grown in lignocellulose medium; 2) whether the occurrence of synergistic growth depends on the complexity of the carbon source; and 3) whether bacterial synergy affects the maximal specific growth rate, enzyme production and metabolic activity of bacteria in mixed cultures.
Materials and Methods
Culture media preparation
Three bacterial isolation media were used. They were composed of Bushnell Haas basal salt medium (Lo et al., 2009) amended with one of the three main components of lignocellulose: carboxymethyl cellulose (CMC) (0.5%), xylan (0.5%) or lignin (0.3%). The basal salt medium was adjusted to pH 7.5 and 1% NaCl, and pre-filtered through G/C filter (1.0 μm) before adding any organic nutrients. Zobell marine medium (HiMedia, Cat# M385) (with 1.5% agar for plates) was used to grow purified bacterial isolates.
Two media, containing either glucose or lignocellulosic compounds, were used to study bacterial interactions. The glucose medium contained basal salt medium with 0.3% glucose and 0.05% yeast extract. The lignocellulose medium, used to simulate recalcitrant carbon substrates found in nature, contained basal salt medium with 0.3% CMC, 0.2% xylan, 0.1% lignin, and 0.05% yeast extract. The relative proportion of CMC, xylan, and lignin, 3:2:1, mimics the typical lignocellulosic composition in grass material (Sun and Cheng, 2002). All components in the lignocellulose medium are water soluble, producing a clear solution that permitted measurement of bacterial growth directly by optical density (OD) in 96-well plates.
Bacterial isolation and classification
Natural detritus was collected from a salt marsh in Ocean Springs, MS, USA (30°23′32″ N 88°47′56″W). Potential lignocellulolytic bacteria were isolated from the detritus using the three isolation media mentioned above. After incubation for 10–14 days at 25°C, representative single colonies were streaked on new agar plates of the same isolation media to obtain well-isolated colonies. Pure single colonies were then transferred and grown on Zobell marine agar. Based on the carbon source used during the initial isolation, the isolates were classified into three groups: cellulose-degrading (C), xylan-degrading (X), or lignin degrading bacteria (L).
To study bacterial interactions, nine bacteria with different colony morphology were randomly selected, three from each group. The nine bacteria were identified by 16S rRNA gene sequencing using universal primers 27F and 1492R (Weisburg et al., 1991; Ciric et al., 2010). The nine sequences have been deposited in GenBank database (Accession Number KJ158195-KJ158203). Putative taxonomic identities of the nine isolates were assigned to genus level using ribosomal database project (RDP) Bayesian classifier (Wang et al., 2007) with minimum bootstrap confidence of 80%. Two of the nine isolates with bootstrap confidence less than 80% at the genus level were assigned to the family level. A phylogenetic analysis of the nine bacteria was conducted by MEGA (Tamura et al., 2011) using neighbor-joining method (Saitou and Nei, 1987).
Culture preparation and growth measurements
The nine isolates were used to determine bacterial interactions in both lignocellulose and glucose media. To prepare the isolates for growth experiments, they were first grown in 3 ml Zobell marine broth in 100 mm × 16 mm polypropylene tubes. After 18 h at 25°C with shaking (250 rpm), 0.2 ml of each culture was used to inoculate 3 ml of either lignocellulose or glucose medium and grown at 25°C with shaking. Those in lignocellulose medium were grown for 30 h and those in glucose medium were grown for 24 h. To conduct growth experiments, seed cultures were prepared by diluting each culture to an optical density (OD595) of 0.01 (ca. 106 CFU/ml) with either lignocellulose or glucose medium.
Growth experiments using nine pure cultures and 27 mixed cultures were conducted to test whether the complexity of carbon source affects bacterial interactions. Each of the 27 mixed cultures were created by combining three pure cultures, one from each of the three groups (C, X, and L) described above. All growth experiments started with cultures at an OD595 of 0.01 in clear flat-bottom 96-well plates. Growth experiments were conducted using 100 μl of glucose medium or lignocellulose medium. For three-species mixed cultures, each species contributed 1/3 of the starting volume. Each culture (pure or mixed) was replicated in four wells in each of three plates thus a total of 12 replicates were used for each pure culture and bacterial combination. Each 96-well plate also included four wells filled with 100 μl sterile lignocellulose or glucose media as blank controls.
Bacterial cultures in growth experiments were grown at 25°C without shaking. Cultures in lignocellulose medium were incubated for 48 h and those in glucose medium for 30 h. Sterile growth media were optically clear at the start of each experiment and growth was determined by measuring the increase in OD595 of each culture (minus the blank controls) using a Synergy 2 microplate reader (BioTek Instruments, Inc., Winooski, VT) over time. To determine maximal growth rates (μ, h−1), optical densities of the cultures during the exponential growth phase were log-transformed and the slope for each culture used.
Definition of synergistic growth
Synergistic growth is defined as having occurred when a mixed culture grew more densely than any of the three corresponding pure cultures. The densest among the three pure cultures is referred to as the reference culture. Thus, a mixed culture is considered to exhibit synergism when it reached significantly higher density (OD595) than its reference culture. No distinction was made whether the higher density resulted from the enhanced growth of all three bacteria or just one or two in the mixed culture. Of interest was the fact that enhanced growth of the mixed cultures indicated more bacterial biomass production and greater degradation of lignocellulose, the ecological process of interest.
Enzyme production assay
The production of lignocellulolytic enzymes was measured using fluorometric assays adopted from widely used methods (Sinsabaugh et al., 1997, 2008; Marx et al., 2001; Saiya-Cork et al., 2002). The assays use enzyme-specific substrates labeled with 4-methylumbelliferone (MUB). The substrates are non-fluorescent, but fluorescence is observed when MUB is released upon substrate hydrolysis by enzymes in the growth medium (Hoppe, 1983; Marx et al., 2001). Enzyme activity is quantified by measuring the amount of fluorescence. 4-MUB-β-D-glucoside, 4-MUB-β-D-cellobioside, and 4-MUB-β-D-xyloside, respectively, were used to test the activities of β-1,4-glucosidase (EC.3.2.1.21), cellobiohydrolase (EC.3.2.1.91), and β-1,4-xylosidase (EC.3.2.1.37). The first two degrade cellulose and the third degrades hemicellulose.
To perform assays, bacterial cultures growing in lignocellulose medium for 48 h were diluted five-fold with 50 mM MOPS buffer (pH 6.5) and 150 μl of each diluted culture were combined with 50 μl of one of the three 4-MUB-labeled enzyme-specific substrates (200 μM) in black 96-well plates. After incubation at 25°C in the dark for 1 h with shaking at 500 rpm, fluorescence in each well was measured using a Synergy 2 Bio-Tek microplate reader (365ex, 450em). The amount of lignocellulolytic enzyme in each culture was calculated based on the fluorescence of the sample relative to that of a standard. The standard contained 150 μl of dilute bacterial culture and 50 μl of 10 μM MUB. Blank controls contained 150 μl dilute culture and 50 μl buffer. Substrate controls contained 150 μl buffer and 50 μl substrate. The enzyme activity in each sample expressed in nmol/h/ml was calculated using the formula:
where SF is the sample fluorescence, BCF the blank control fluorescence, SCF the substrate control fluorescence, and MF the MUB standard fluorescence. Results were multiplied by five to adjust for the initial five-fold sample dilution. Each sample, standard, blank control and substrate control was replicated four times. The experiment was repeated twice.
Microbial metabolic activity measurement
To test whether bacterial interactions enhance microbial activity, the metabolic activity of bacterial cultures was measured using the 2,3,5-triphenyltetrazolium chloride (TTC) assay (Gabrielson et al., 2002; Burmølle et al., 2006). The colorless TTC is enzymatically reduced by metabolically active bacteria to red 1,3,5-triphenylformazan (TPF) that can be quantified by measuring sample absorbance at 490 nm (Gabrielson et al., 2002). After growth for 30 h in lignocellulose medium, 100 μl of each culture was combined with 25 μl of substrate solution (0.05% TTC and 1.5% glucose) and incubated at 25°C in the dark. Glucose was included to improve sensitivity (da Silva et al., 2008). Sterile culture medium with the same amount of substrate solution served as negative control. Absorbance was measured at 0 and 3 h and the difference in absorbance (ΔA490) was used as a measure of metabolic activity. We also compared the growth-specific metabolic activity (ΔA490 · OD595−1) between mixed and pure cultures. The experiment was repeated twice using eight replicates each time.
Statistical analysis
One-way ANOVA and Tukey HSD test were performed to test whether mixed cultures grew better than their corresponding pure cultures. The effects of synergism on growth, maximal growth rate and metabolic activity were analyzed by one-way ANOVA, followed by Tukey HSD test. Due to the non-normal distribution of enzyme production data, the non-parametric Kruskal-Wallis test was used to test the effect of synergism on enzyme production. All statistical tests were performed in RStudio (www.rstudio.org).
Results
Bacterial isolates classification
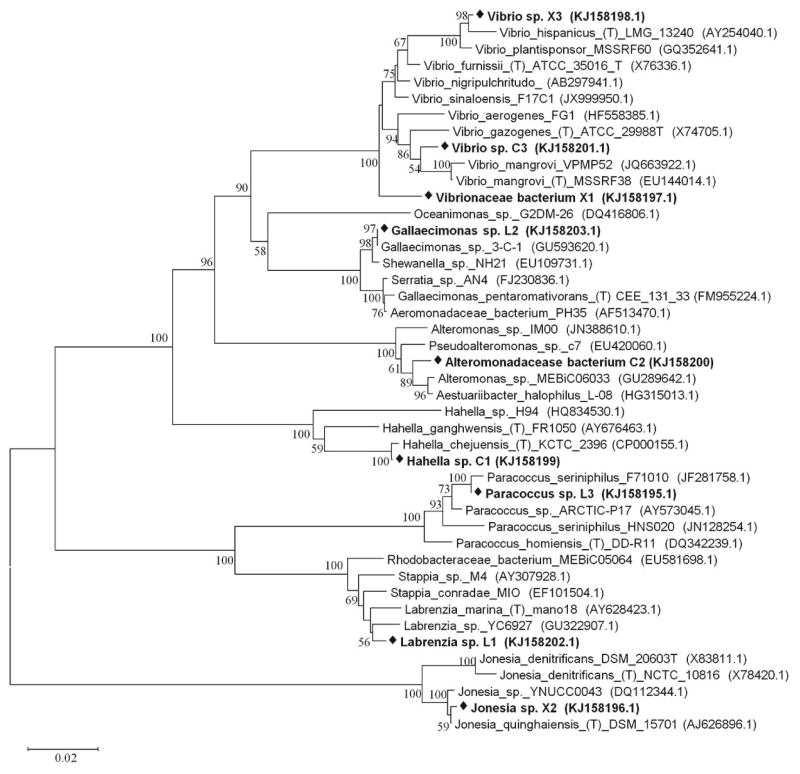
The nine lignocellulolytic bacteria isolated from a natural salt marsh in Ocean Springs, MS, USA were taxonomically diverse (Fig. 1). Two isolates were classified as a-Proteobacteria including Labrenzia sp. (L1) and Paracoccus sp. (L3). Six isolates belonged to -Proteobacteria, including two Vibrio spp. (X3 and C3), one Gallaecimonas sp. (L2), one Hahella sp. (C1) and two presumptively new isolates (X1 and C2) that could not be assigned to the genus level. These two isolates were thus assigned to the family level and classified as a Vibrionaceae bacterium (X1) and a Alteromonadaceae bacterium (C2). The ninth isolate was Jonesia sp. (X2), belonging to Actinobacteria.
Fig. 1. Phylogenetic relationship of nine bacterial isolates used in study.
Bold text with black diamond indicates the nine bacteria isolates used. The letter and number following the name of the bacterium denote the substrate used in isolating each bacterium, C for cellulose, L for lignin and X for xylan. Bootstrap values are shown as the percentage of 1,000 replicates when greater than 50%. The horizontal bar represents nucleotide substitutions per sequence position. GenBank accession numbers are in parentheses.
Synergistic growth in three-species mixed cultures
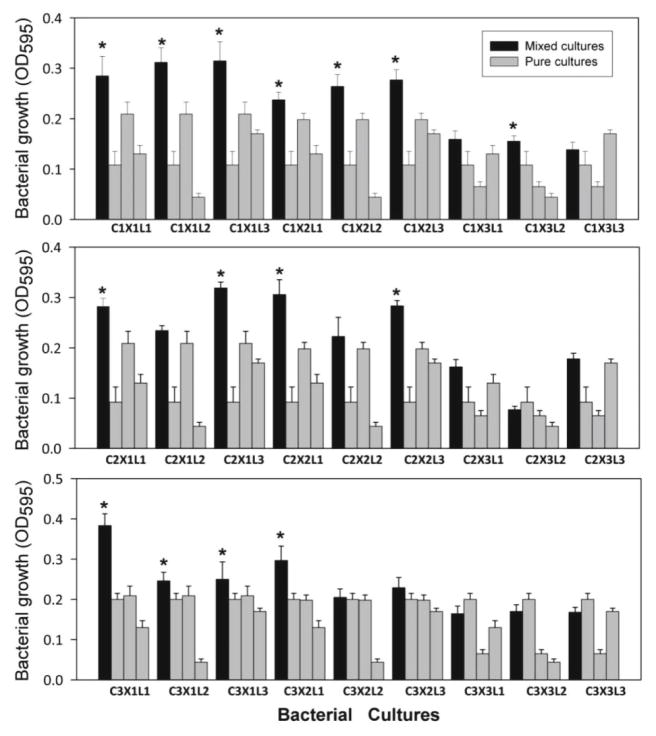
Synergistic growth among three-species mixed cultures was common in lignocellulose medium (Fig. 2). Synergistic growth is defined as having occurred when a mixed culture grew better than any of the three corresponding pure cultures. Fifteen among the 27 possible mixed cultures exhibited significantly greater growth when compared to their corresponding pure cultures (Fig. 2). In fact, all but one of these 15 mixed cultures reached higher densities (OD595 > 0.209) than all the pure cultures. The experiment was repeated twice more with similar results (data not shown). Hereafter, the 15 three-species combinations are designated as synergistic mixed cultures because those mixed cultures showed greater growth than their corresponding cultures that made up each of them. The remaining 12 mixed cultures are designated as non-synergistic mixed cultures.
Fig. 2. Growth of mixed cultures compared to pure cultures in lignocellulose medium.
Growth of 27 mixed cultures (black bars) in relationship to the growth of their three corresponding pure cultures (gray bars). N = 12. Error bar is one standard deviation. Asterisks indicate significantly greater growth compared to pure cultures. One-way ANOVA (F35, 396 = 151.77, P < 0.001) followed by Tukey HSD test (P < 0.05).
Carbon source dependent synergistic interaction
To determine whether the synergistic growth was substrate dependent, we repeated the mixed-culture experiment using glucose as the sole carbon source. For comparing bacterial growth in glucose medium and lignocellulose medium, we plotted the growth of each mixed culture against the growth of its reference culture (the pure culture with the greatest growth among the three that made up the mixed culture) (Fig. 3). Although cultures grown in glucose medium reached higher densities than the same cultures in lignocellulose medium, none of the 27 mixed cultures in glucose medium reached higher density than their reference cultures. All had optical densities below the isometric line that indicates equal growth between mixed and reference cultures (Fig. 3). This result suggests the dominance of negative interaction or competition among three species in mixed cultures when grown in glucose medium. In contrast, most of the mixed cultures (21 in 27) reached densities above the isometric line when grown in lignocellulose medium (Fig. 3). Our results show that bacterial synergism occurred frequently in lignocellulose medium but never in glucose medium, suggesting that bacterial synergistic growth was dependent on the structural complexity of the carbon source.
Fig. 3. Comparison of mixed culture growth in lignocellulose and glucose medium.
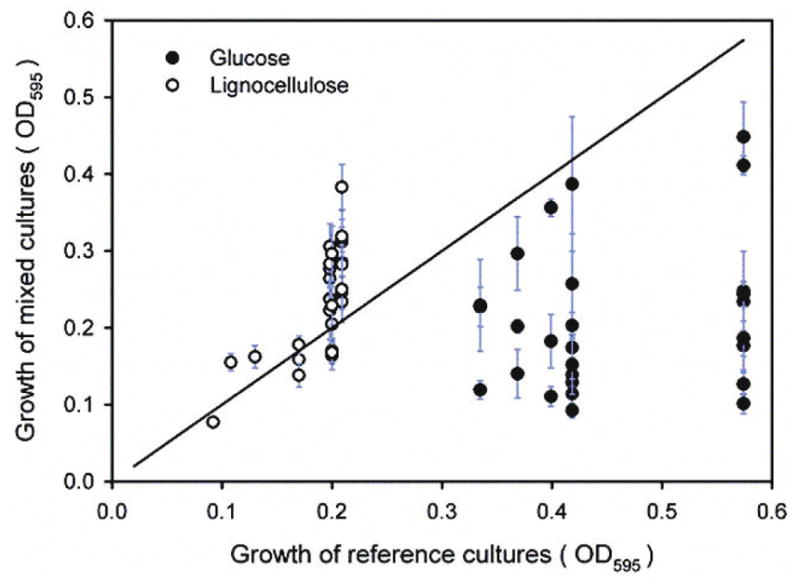
Each data point is the OD595 of one mixed culture (mean ± SD, n =12) plotted against the OD595 of its reference culture (the pure culture with the greatest growth among the three that made up the mixed culture). The isometric line represents equal growth between mixed cultures and their reference cultures.
Bacterial growth and activity during lignocellulose degradation
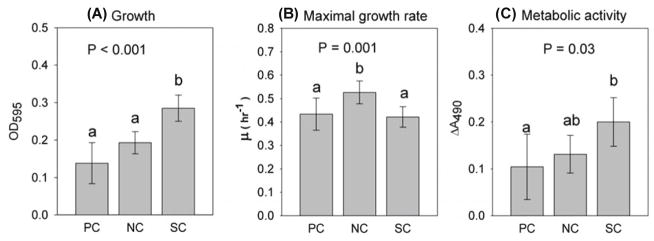
To explore the potential mechanism for bacterial synergism in lignocellulose medium, we compared the maximal growth rate and metabolic activity among the three culture groups: synergistic mixed cultures (the 15 mixed cultures that exhibited synergistic growth), non-synergistic mixed cultures and pure cultures. Synergistic mixed cultures reached the highest cell density among the three groups (F2, 33 = 26.78, P < 0.001) (Fig. 4A) but did not grow faster during the exponential phase (Fig. 4B). The mean growth density of synergistic mixed cultures (OD595 = 0.280) was more than twice that of pure cultures (OD595 = 0.135) and 1.6 times that of non-synergistic mixed cultures (OD595 = 0.176) (Fig. 4A). In terms of maximal growth rate, there was, however, no significant difference between synergistic mixed cultures (mean = 0.429 h−1) and pure cultures (mean = 0.430 h−1) (Fig. 4B). Interestingly, although they grew less than synergistic mixed cultures (Fig. 4A), non-synergistic mixed cultures had higher maximal growth rate (mean = 0.542 h−1) than synergistic mixed cultures and pure cultures (F2, 33 = 8.29, P = 0.001) (Fig. 4B).
Fig. 4. Effect of synergy on bacterial growth, specific growth rate and metabolic activity.
Gray bar is the mean value of each group. Error bars represent 95% confidence interval. Different letters indicate significant difference (P < 0.05). NC = non-synergistic mixed cultures, n = 12; PC = pure cultures, n = 9; SC = synergistic mixed cultures, n = 15.
Bacteria in synergistic mixed cultures had higher metabolic activity (mean ΔA490 = 0.186) than those in pure cultures (mean ΔA490 = 0.096) (F2, 33 = 3.79, P = 0.033) but not significantly higher than non-synergistic mixed cultures (mean ΔA490 = 0.132) (Fig. 4C). Although synergistic mixed cultures had higher metabolic activity than pure cultures, there was no significant difference in the growth-specific metabolic activity (ΔA490 OD595−1) among the three groups after normalizing for cell density (data not shown).
Bacterial production of lignocellulolytic enzymes
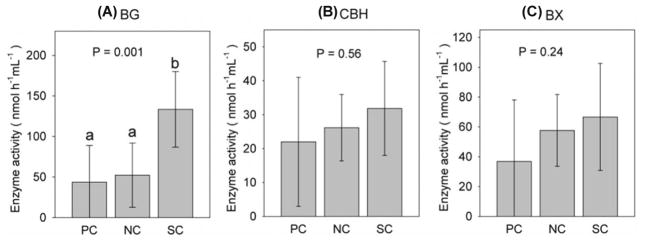
Bacterial synergism promoted the production of β-1,4-glucosidase (BG) but not the production of cellobiohydrolase (CBH) or β-1,4-xylosidase (BX) (Fig. 5). The mean BG activity among synergistic mixed cultures (133.5 nmol/h/ml) was over three times that of pure cultures (43.7 nmol/h/ml), and 2.6 times that of non-synergistic mixed cultures (52.3 nmol/h/ml) ( 2 =13.87, df = 2, P = 0.001) (Fig. 6A). Activities of CBH and BX for synergistic group were slightly higher than two other groups but not significantly different from them (Fig. 5B and C). When adjusted for cell density, there was no significant difference in production of the three enzymes tested (nmol/h/ml/OD595) among synergistic mixed cultures, non-synergistic growth and pure culture groups (data not shown).
Fig. 5. Effect of synergy on bacterial production of lignocellulolytic enzymes.
BG, β-1,4-glucosidase; CBH, cellobiohydrolase; BX, β-1,4-xylosidase. Gray bar is the mean enzyme activity of each group. Error bars represent 95% confidence interval. Different letters indicate significant difference (P < 0.05). NC = non-synergistic mixed cultures, n = 12; PC = pure cultures, n = 9; SC = synergistic mixed cultures, n = 15.
Discussion
We found that three-species mixed cultures composed of taxonomically diverse bacteria frequently exhibited greater growth during lignocellulose degradation. The results suggest that bacterial synergism may be important to detritus degradation in the salt marsh ecosystem where those bacteria were isolated. Whether bacterial synergism takes place appeared to depend on the chemical complexity of the carbon source. In lignocellulose medium most of the mixed cultures exhibited synergistic growth but none exhibited synergistic growth in glucose medium. Our results support the hypothesis that the complexity of carbon source plays a role in determining bacterial interactions in the environment.
Several mechanisms that promote synergistic growth have been proposed. For example, synergistic growth can result when multiple species produce complementary enzymes and take part in metabolite cross feeding (Wintermute and Silver, 2010; Kostylev and Wilson, 2012). Due to the complexity and recalcitrance of lignocellulosic substrate, the complete degradation of lignocellulose requires multiple enzymes (Kostylev and Wilson, 2012; Van Dyk and Pletschke, 2012). Furthermore, enzyme cocktails containing cellulases, xylanases and lignin peroxidases that are produced by multiple species can significantly enhance lignocellulose degradation rate (Lynd et al., 2002; Guevara and Zambrano, 2006; Kostylev and Wilson, 2012; Van Dyk and Pletschke, 2012). In addition to making enzyme cocktails where enzymes act synergistically, mixed cultures can also promote production of enzymes that produce simple sugars promoting bacterial growth. In our study, β-1,4-glucosidase (BG) activity was more than three folds higher in synergistic mixed cultures than in pure cultures (Fig. 5A). The higher β-1,4-glucosidase activity likely produced more glucose that contributed to the enhanced growth of mixed cultures. Thus, it appears that mixed cultures in lignocellulose medium not only produced enzymes that were complementary but also produced more active enzymes so that refractory substrates can be degraded more effectively compared to pure cultures.
Another mechanism that promotes bacterial synergism is metabolite cross feeding that allows bacteria to utilize complex substrate in a cooperative manner (Flint et al., 2007; Wintermute and Silver, 2010). During late growth, some species in a mixed culture may produce metabolites that are toxic to themselves but are used by others. In this case, mixed cultures can alleviate problems of feedback regulation and metabolite repression present in pure cultures (Zuroff and Curtis, 2012). In the present study, synergistic mixed cultures reached higher densities than pure cultures but their maximal growth rate during exponential growth was not higher (Fig. 4A and B). One possible explanation is that after exponential growth when the primary nutrients are depleted, bacteria in mixed cultures can continue to grow by utilizing metabolites produced by others. As a result, mixed cultures reached higher growth density than pure cultures that had no partners to exchange metabolites with. This was supported by observations of higher metabolic activity in the synergistic mixed cultures after the exponential growth (Fig. 4C).
Although most mixed cultures (15 of 27) in lignocellulose medium exhibited synergistic growth and degraded more lignocellulose than pure cultures, some (12 of 27) did not under the same culture condition. This suggests that the specific combination of bacteria may be important. The physiological or metabolic response of each bacteria may depend on the presence of others in the mixed culture. Some combinations achieved enhanced growth while others exhibited competition perhaps due to resource overlap or competing metabolic capabilities of neighboring bacteria (Hibbing et al., 2010; Freilich et al., 2011; Elias and Banin, 2012).
It was striking that none of the 27 mixed cultures exhibited synergistic growth in glucose medium. The likely explanation is that cooperation serves no purpose in the presence of a labile carbon source such as glucose and thus competition is prevalent. The concept that substrate complexity regulates the type of bacterial interaction is supported by published studies. For example, Long and Azam (2001) and Grossart et al. (2004) reported that antagonism was common among bacteria found in marine ecosystems. A possible explanation lies in the marine (ZoBell) agar used in the laboratory studies. The limited amounts of peptone and yeast extract in the medium may have spurred competition among bacteria to result in the common antagonism observed. Similarly, both Nielsen et al. (2000) and Breugelmans et al. (2008) showed that when citrate was used as the carbon source, bacteria competed for the labile nutrient and usually formed separated biofilm colonies. In contrast, when complex benzyl compounds were the only carbon source in the medium, bacteria degraded the refractory substrate more efficiently than pure cultures and usually formed mixed-species biofilm. This suggests the possibility that bacteria grown together in glucose-limiting medium in the present study might compete for the labile substrate by producing antimicrobial chemicals to inhibit competitors, resulting in the predominance of antagonistic interactions.
In conclusion, our results show that certain combinations of indigenous bacteria from a salt marsh exhibit growth synergism that is dependent on the complexity of the substrate. When lignocellulose is the only carbon source, bacteria tend to interact synergistically to degrade the complex substrate. When glucose is the only carbon source and in limited amounts, bacteria compete for the labile substrate resulting in the predominance of antagonistic interactions. To better understand the mechanism, additional studies on the relationship between the chemical complexity of the substrate and bacterial compositions in the consortia are needed. Currently, it is unclear how different species contribute in the consortia. Some may produce enzymes that degrade a particular component of lignocellulose more effectively while others may produce stimulatory exudates or quorum sensing molecules that coordinate interactions among bacteria.
Acknowledgments
We gratefully acknowledge the financial support received from the Atlantic Oceanographic and Meteorological Laboratory, NOAA (Award # NA11OAR4320199) via the Northern Gulf Institute (Award # 191001-363405-01/TO 009).
References
- Breugelmans P, Barken KB, Tolker-Nielsen T, Hofkens J, Dejonghe W, Springael D. Architecture and spatial organization in a triple-species bacterial biofilm synergistically degrading the phenylurea herbicide linuron. FEMS Microbiol Ecol. 2008;64:271–282. doi: 10.1111/j.1574-6941.2008.00470.x. [DOI] [PubMed] [Google Scholar]
- Burmølle M, Webb JS, Rao D, Hansen LH, Sørensen SJ, Kjelleberg S. Enhanced biofilm formation and increased resistance to antimicrobial agents and bacterial invasion are caused by synergistic interactions in multispecies biofilms. Appl Environ Microbiol. 2006;72:3916–3923. doi: 10.1128/AEM.03022-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciric L, Philp JC, Whiteley AS. Hydrocarbon utilization within a diesel-degrading bacterial consortium. FEMS Microbiol Lett. 2010;303:116–122. doi: 10.1111/j.1574-6968.2009.01871.x. [DOI] [PubMed] [Google Scholar]
- D’Costa VM, Griffiths E, Wright GD. Expanding the soil antibiotic resistome: exploring environmental diversity. Curr Opin Microbiol. 2007;10:481–489. doi: 10.1016/j.mib.2007.08.009. [DOI] [PubMed] [Google Scholar]
- Da Silva WJ, Seneviratne J, Parahitiyawa N, Rosa EA, Samaranayake LP, Del Bel Cury AA. Improvement of XTT assay performance for studies involving Candida albicans biofilms. Braz Dent J. 2008;19:364–369. doi: 10.1590/s0103-64402008000400014. [DOI] [PubMed] [Google Scholar]
- Elias S, Banin E. Multi-species biofilms: living with friendly neighbors. FEMS Microbiol Rev. 2012;36:990–1004. doi: 10.1111/j.1574-6976.2012.00325.x. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Duncan SH, Scott KP, Louis P. Interactions and competition within the microbial community of the human colon: links between diet and health. Environ Microbiol. 2007;9:1101–1111. doi: 10.1111/j.1462-2920.2007.01281.x. [DOI] [PubMed] [Google Scholar]
- Foster KR, Bell T. Competition, not cooperation, dominates interactions among culturable microbial species. Curr Biol. 2012;22:1845–1850. doi: 10.1016/j.cub.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Freilich S, Zarecki R, Eilam O, Segal ES, Henry CS, Kupiec M, Gophna U, Sharan R, Ruppin E. Competitive and cooperative metabolic interactions in bacterial communities. Nat Commun. 2011;2:589. doi: 10.1038/ncomms1597. [DOI] [PubMed] [Google Scholar]
- Gabrielson J, Hart M, Jarelov A, Kuhn I, McKenzie D, Mollby R. Evaluation of redox indicators and the use of digital scanners and spectrophotometer for quantification of microbial growth in microplates. J Microbiol Methods. 2002;50:63–73. doi: 10.1016/s0167-7012(02)00011-8. [DOI] [PubMed] [Google Scholar]
- Grossart HP, Schlingloff A, Bernhard M, Simon M, Brinkhoff T. Antagonistic activity of bacteria isolated from organic aggregates of the German Wadden Sea. FEMS Microbiol Ecol. 2004;47:387–396. doi: 10.1016/S0168-6496(03)00305-2. [DOI] [PubMed] [Google Scholar]
- Guevara C, Zambrano MM. Sugarcane cellulose utilization by a defined microbial consortium. FEMS Microbiol Lett. 2006;255:52–58. doi: 10.1111/j.1574-6968.2005.00050.x. [DOI] [PubMed] [Google Scholar]
- Hansen SK, Haagensen JAJ, Gjermansen M, Jørgensen TM, Tolker-Nielsen T, Molin S. Characterization of a Pseudomonas putida rough variant evolved in a mixed-species biofilm with Acinetobacter sp strain C6. J Bacteriol. 2007;189:4932–4943. doi: 10.1128/JB.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta S, Cui Z, Huang Z, Li M, Ishii M, Igarashi Y. Construction of a stable microbial community with high cellulose-degradation ability. Appl Microbiol Biotechnol. 2002;59:529–534. doi: 10.1007/s00253-002-1026-4. [DOI] [PubMed] [Google Scholar]
- Hibbing ME, Fuqua C, Parsek MR, Peterson SB. Bacterial competition: surviving and thriving in the microbial jungle. Nat Rev Microbiol. 2010;8:15–25. doi: 10.1038/nrmicro2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe H. Significance of exoenzymatic activities in the ecology of brackish water: measurements by means of methylum-belliferyl-substrates. Mar Ecol Prog Ser. 1983;11:299–308. [Google Scholar]
- Jiménez D, Korenblum E, van Elsas J. Novel multi-species microbial consortia involved in lignocellulose and 5-hydroxymethylfurfural bioconversion. Appl Microbiol Biotechnol. 2014;98:2789–2803. doi: 10.1007/s00253-013-5253-7. [DOI] [PubMed] [Google Scholar]
- Kostylev M, Wilson D. Synergistic interactions in cellulose hydrolysis. Biofuels. 2012;3:61–70. [Google Scholar]
- Lo YC, Saratale GD, Chen WM, Bai MD, Chang JS. Isolation of cellulose-hydrolytic bacteria and applications of the cellulolytic enzymes for cellulosic biohydrogen production. Enzym Microb Technol. 2009;44:417–425. [Google Scholar]
- Long RA, Azam F. Antagonistic Interactions among Marine Pelagic Bacteria. Appl Environ Microbiol. 2001;67:4975–4983. doi: 10.1128/AEM.67.11.4975-4983.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynd LR, Weimer PJ, van Zyl WH, Pretorius IS. Microbial cellulose utilization: fundamentals and biotechnology. Microbiol Mol Biol Rev. 2002;66:506–577. doi: 10.1128/MMBR.66.3.506-577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marx MC, Wood M, Jarvis SC. A microplate fluorimetric assay for the study of enzyme diversity in soils. Soil Biol Biochem. 2001;33:1633–1640. [Google Scholar]
- Newman DK, Banfield JF. Geomicrobiology: how molecular-scale interactions underpin biogeochemical systems. Science. 2002;296:1071–1077. doi: 10.1126/science.1010716. [DOI] [PubMed] [Google Scholar]
- Nielsen AT, Tolker-Nielsen T, Barken KB, Molin S. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environ Microbiol. 2000;2:59–68. doi: 10.1046/j.1462-2920.2000.00084.x. [DOI] [PubMed] [Google Scholar]
- Perez J, Munoz-Dorado J, de la Rubia T, Martinez J. Biodegradation and biological treatments of cellulose, hemicellulose and lignin: an overview. Int Microbiol. 2002;5:53–63. doi: 10.1007/s10123-002-0062-3. [DOI] [PubMed] [Google Scholar]
- Rypien KL, Ward JR, Azam F. Antagonistic interactions among coral-associated bacteria. Environ Microbiol. 2010;12:28–39. doi: 10.1111/j.1462-2920.2009.02027.x. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Saiya-Cork KR, Sinsabaugh RL, Zak DR. The effects of long term nitrogen deposition on extracellular enzyme activity in an Acer saccharum forest soil. Soil Biol Biochem. 2002;34:1309–1315. [Google Scholar]
- Sinsabaugh RL, Findlay S, Franchini P, Fischer D. Enzymatic analysis of riverine bacterioplankton production. Limnol Oceanogr. 1997;42:29–38. [Google Scholar]
- Sinsabaugh RL, Lauber CL, Weintraub MN, Ahmed B, Allison SD, Crenshaw C, Contosta AR, Cusack D, Frey S, Gallo ME, et al. Stoichiometry of soil enzyme activity at global scale. Ecol Lett. 2008;11:1252–1264. doi: 10.1111/j.1461-0248.2008.01245.x. [DOI] [PubMed] [Google Scholar]
- Sun Y, Cheng J. Hydrolysis of lignocellulosic materials for ethanol production: a review. Bioresour Technol. 2002;83:1–11. doi: 10.1016/s0960-8524(01)00212-7. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Van Dyk JS, Pletschke BI. A review of lignocellulose bioconversion using enzymatic hydrolysis and synergistic cooperation between enzymes--Factors affecting enzymes, conversion and synergy. Biotechnol Adv. 2012;30:1458–1480. doi: 10.1016/j.biotechadv.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintermute EH, Silver PA. Emergent cooperation in microbial metabolism. Mol Syst Biol. 2010;6:407. doi: 10.1038/msb.2010.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wongwilaiwalin S, Rattanachomsri U, Laothanachareon T, Eurwilaichitr L, Igarashi Y, Champreda V. Analysis of a thermophilic lignocellulose degrading microbial consortium and multi-species lignocellulolytic enzyme system. Enzyme Microb Technol. 2010;47:283–290. [Google Scholar]
- Zuroff TR, Curtis WR. Developing symbiotic consortia for lignocellulosic biofuel production. Appl Microbiol Biotechnol. 2012;93:1423–1435. doi: 10.1007/s00253-011-3762-9. [DOI] [PubMed] [Google Scholar]