Abstract
The haemodynamic response function (HRF) is a key component of the blood oxygen level-dependent (BOLD) signal, providing the mapping between neural activity and the signal measured with functional magnetic resonance imaging (fMRI). Most of the time the HRF is associated with task-based fMRI protocols, in which its onset is explicitly included in the design matrix. On the other hand, the HRF also mediates the relationship between spontaneous neural activity and the BOLD signal in resting-state protocols, in which no explicit stimulus is taken into account. It has been shown that resting-state brain dynamics can be characterized by looking at sparse BOLD ‘events’, which can be retrieved by point process analysis. These events can be then used to retrieve the HRF at rest. Crucially, cardiac activity can also induce changes in the BOLD signal, thus affecting both the number of these events and the estimation of the haemodynamic response. In this study, we compare the resting-state haemodynamic response retrieved by means of a point process analysis, taking the cardiac fluctuations into account. We find that the resting-state HRF estimation is significantly modulated in the brainstem and surrounding cortical areas. From the analysis of two high-quality datasets with different temporal and spatial resolution, and through the investigation of intersubject correlation, we suggest that spontaneous point process response durations are associated with the mean interbeat interval and low-frequency power of heart rate variability in the brainstem.
Keywords: resting state, functional magnetic resonance imaging, haemodynamic response, point process, cardiac fluctuations
1. Introduction
Spontaneous fluctuations in the blood oxygen level-dependent (BOLD) signal are correlated with local field potential activity [1]. Analyses of the functional connectivity (FC) between spontaneous low-frequency BOLD signal fluctuations have identified a number of large-scale intrinsic connectivity networks (ICNs). These ICNs are related to sensory, motor, language, social–emotional and cognitive functions, suggesting that spontaneous BOLD fluctuations play a fundamental role in encoding brain function. The above-mentioned patterns are consistent across different levels of consciousness ranging from wakefulness down to sleep and anaesthesia [2], and find correlates with other imaging modalities including electroencephalography (EEG) and magnetoencephalography [3,4].
Recent studies have revealed that these ICNs can be identified by transiently synchronized spontaneous BOLD ‘events’ in the distributed brain regions, which can for example be identified as peaks above the threshold. A growing amount of evidence points to these spontaneous BOLD events governing the brain dynamic at rest [5–7]; these spontaneous BOLD events can be revealed by point process analysis (PPA) [6], the core idea of it being in this context to isolate events in the BOLD time series (for example, peaks in the standardized time series) and to look at their spatial and temporal distribution. Compared with static FC maps constructed from correlations between the whole time series, the FC maps derived by PPA appear to be similar but carry more information on the individual states characterizing brain dynamics [6,8]. Both dynamic FC and PPA are dependent on the variance of BOLD signal, thus they are sensitive to the contribution from non-neuronal fluctuation. A wide range of artefacts may induce changes in BOLD signal, such as thermal noise, hardware limitations [9] and participant's movements inside the scanner [10]. In addition, as the BOLD signal is a measurement of changes in blood flow, oxygenation and volume [11], these changes may be caused by neuronal activity through neurovascular coupling, or alternatively arise from any other physiological processes that affect blood oxygenation or volume [12]. Accordingly, noise and physiological fluctuations may contribute to the rich spatio-temporal information content of dynamic brain activity and connectivity [13]. To reduce confounds deriving from non-neural activity-related processes, plenty of advanced noise cleanup methods have been proposed [14]. Owing to the lack of ground truth in noise removal, most of them focus on improvements in temporal signal-to-noise ratio (tSNR) or reproducibility of FC maps. Yet, no study has explored how and to what extent these confounds affect sparse spontaneous events. As proposed in previous studies, the spontaneous BOLD point process events in resting-state fMRI are assumed to be induced by these spontaneous neural events. Then, it would be possible to retrieve the corresponding haemodynamic response function (HRF) of the spontaneous neural event at rest [15,16]. Apart from the variation of amplitude in BOLD signal, additional temporal characteristics of the haemodynamic response, not available from tSNR and FC maps such as time to peak, could be revealed by statistical analysis of spontaneous point process HRF.
Unlike thermal noise, physiological fluctuations can introduce fluctuations in the fMRI signal that are uncoupled from neural activity, and are among the most important confounds in BOLD signal change [17]. In fact, cardiac mechanisms include changes in cerebral blood flow/volume and arterial pulsatility [18]. Respiration effects include changes in B0 and arterial CO2 partial pressure [19]. Although cardiac and respiratory cycles have relatively high frequencies in contrast to the typical low-frequency (less than 0.1 Hz) BOLD fluctuations, aliasing of physiological components to lower frequency range will inevitably occur owing to lower sampling rate in BOLD fMRI than cardiac and respiratory cycles [20]. Recent studies have shown that these nuisance confounds can significantly alter FC maps of the intrinsic brain networks, such as the default mode network [21–,23]. Nonetheless, ample evidence has been collected to support that resting-state FC does have a neuronal underpinning and cannot purely be the result of physiological noise. To date, it is still not clear to what extent the physiological confounds affect the haemodynamic response retrieved by PPA, and more information on this would be helpful for understanding the physiological foundation of functional coupling among brain regions [24].
A number of methods have been developed to reduce physiological confounds in the BOLD signal [21,22,25–27]. Retrospective image space correction of physiological noise (RETROICOR) is one of the most employed methods to correct the cardiac and respiratory quasi-periodic fluctuations [25]. However, it only filters cyclic effects aliased in the fMRI signal, whereas the physiology-related low-frequency fluctuations remain in the data. The time-shifted respiratory volumes per unit time (RVT) and heart rate (HR) time series were introduced to account for more variance in BOLD signal than that induced by non-periodic fluctuations arising from cardiac and respiratory processes [21,22]. These physiological noise correction models on BOLD have been well validated. In the light of what said so far, we feel it is important to examine their influence on haemodynamic response retrieved from spontaneous point processes.
Unfortunately, the advanced technique to remove the physiological confounds may also remove meaningful variance components reflecting activity in the autonomic nervous system (ANS). ANS activity should be considered as theoretically meaningful information, especially when studying brain areas involved in decision-making, conflict resolution and the experience of emotion [28]. A seed-based static FC analysis of posterior cingulate cortex has shown that general ANS activity is significantly related to spontaneous BOLD activity in default mode network (DMN) and task positive network [29]. The sliding window-based dynamic FC analysis further reveals that heart rate variability (HRV) covaries with temporal changes in dorsal anterior cingulate cortex (dACC) and amygdala connectivity maps [30]. These studies indicate that resting-state BOLD activity also contains both physiology-related spontaneous neuronal activity and non-neural fluctuations [12,21,31,32]. Therefore, it is critical to differentiate BOLD point process from ANS modulation and physiological noise confounding. HRV is a popular non-invasive indicant for assessing the activity in ANS. An analysis reporting how spontaneous point process HRF covaries with HRV could therefore explore the nature of autonomic regulation on brain activity at rest.
In this study, we investigate to what extent the estimation of the spontaneous point process haemodynamic response is affected by changes in physiological noise, rather than solely by central processes such as neural or astrocytic control. The combination of RETROICOR, RVT and HR is employed to deconvolve the physiological fluctuation influence. As HRV is estimated from cardiac activity, we explore only physiological noise correction effect of the quasi-periodic and non-periodic cardiac fluctuations. Then, spontaneous point process HRF maps are retrieved from the residual BOLD signal. Quantitative analysis on HRF map affected by cardiac fluctuation is performed. Finally, correlation analysis between HRV and spontaneous point process HRF is further explored.
2. Material and methods
Two different resting-state (rs) fMRI datasets are included in this study. The first dataset is the enhanced Nathan Kline Institute-Rockland Sample (NKI-RS), acquired from 3 T Siemens scanners [33]. Here we focus on two different TRs (TR=0.645 s, TE=30 ms, FA=60°, 3 mm isotropic voxels, 900 volumes; and TR=2.5 s, TE=30 ms, FA=80°, 3 mm isotropic voxels, 120 volumes) sequentially collected by multiband (acceleration factor=4) and conventional echo-planar imaging (EPI) sequence. Anatomical images were obtained using an MPRAGE sequence with a resolution of 1 mm3 isotropic. Right-handed subjects in release 4 with complete demographic information were employed in our analysis (n=67, 17 females, age: 12–85 with mean 50.6 and standard deviation (s.d.) 20.8 years). They were instructed to keep their eyes open and fixate a crosshair.
The 7 T rs fMRI test–retest dataset used in this study has been publicly released by the Consortium for Reliability and Reproducibility (CoRR) project [34]. Twenty-two participants (10 females) were scanned during two sessions spaced one week apart. The subjects were instructed to stay awake, keep eyes open and focus on a cross. Their age ranged from 21 to 30 years with mean 25.1 and s.d. 2.2, one left-handed subject was excluded, resulting in all right-handed subjects. Each session included two 1.5 mm isotropic whole-brain resting-state scans (TR=3.0 s, TE=17 ms, FA=70°, 1.5 mm isotropic voxels, 300 volumes, GRAPPA acceleration with iPAT factor of 3) and gradient echo field map. Structural images were acquired by three-dimensional MP2RAGE sequence with a resolution of 0.7 mm isotropic.
(a). Physiological noise models and heart rate variability
Physiological data (respiratory and cardiac traces) were simultaneously recoded for each rs-fMRI scan. The original data in 7 T dataset (5000 Hz) were down-sampled to 100 Hz. The data in NKI-RS dataset are recorded at a sample rate of 62.5 Hz. Two cardiac fluctuation correction models were constructed to account for components related to (i) cardiac phases (CP) and (ii) heart rate (HR). The respiration fluctuations are also included to account for the physiological noise influences: (i) respiratory phases (RP) and the interaction effects between CP and RP (InterCRP) and (ii) respiratory volume per unit time (RVT). Models for cardiac and respiratory phases and their interaction effects were based on RETROICOR [25] and its extension [35]. Cardiac and respiratory response functions were employed to model HR and RVT onto physiological process of the fMRI time series [21,22,26,27]. For each subject, a set of 20 physiological regressors (i.e. fourth-order Fourier expansion for RP, third-order Fourier expansion for CP, second-order Fourier expansion for InterCRP, RVT and HR) was created using the Matlab PhysIO toolbox (http://www.translationalneuromodeling.org/tnu-checkphysretroicor-toolbox/) for each slice in each fMRI run. Cardiac fluctuation correction based on different combinations of these regressors was studied to investigate the effect of cardiac pulse, performing by a generalized linear model (GLM). The combinations are
(1) RP & RVT (RPV-model),
(2) RP & RVT & CP & InterCRP (RPVC-model),
(3) RP & RVT & HR (RPVH-model), and
(4) RP & RVT & CP & InterCRP & HR, i.e. all models (RPVCH-model).
HRV analysis was performed on the interbeat interval (IBI) time series in each resting-state session scan, using the HRV analysis software (HRVAS, https://github.com/jramshur/HRVAS). The IBI time series were calculated as the peak-to-peak interval of photoplethysmography signal. IBI outliers in each session were removed. The outliers were defined as intervals deviating 20% from the previous interval. To alleviate any non-stationarities within IBI time series, wavelet packet detrending was used before HRV analysis. Finally, time domain and frequency-domain measures were derived from IBI series, including: mean IBI; the standard deviation of the normal-to-normal (NN) interval series, SDNN; the root mean square of successive differences of the IBI series, RMSSD; spectral power of low-frequency (LF: 0.04–0.15 Hz) and high-frequency (HF: 0.15–0.4 Hz) band power and LF/HF ratio, which represents a measure of sympathovagal balance.
(b). Magnetic resonance imaging data processing
All structural images in both datasets were manually reoriented to the anterior commissure and segmented into grey matter, white matter and cerebrospinal fluid (CSF), using the standard segmentation option in SPM 12 [36]. Resting-state fMRI data pre-processing was subsequently carried out using both AFNI and SPM12 package with default parameters [36,37], including slice timing correction (T), registration (R), physiological noise model correction (C), despiking (D) and normalization (N). To examine the pre-processing procedure effect on point process acquisition, three commonly used orders of pre-processing steps were applied to the dataset: (i) DCTRN, (ii) DRCTN, and (iii) DTRCN. The raw volumes were despiked using AFNI's 3dDespike algorithm to mitigate the impact of outliers. In slice timing step, the EPI volumes of each run were corrected for the temporal difference in acquisition among different slices to match the middle time slice or half TR (for TR=0.645 s); in the registration step, the images were realigned to the first volume of the first run, the gradient echo field map was processed to create a voxel displacement map and used to correct the realigned images for geometrical distortion (only for 7 T dataset), then the generated mean image across all realigned volumes was coregistered with the structural image, and the resulting warps applied to all the realigned volumes; in physiological noise correction step, physiological noise models were regressed as the covariates, the physiological model regressors from middle time slice were used for DTRCN, whereas each slice data were regressed by physiological model regressors constructed from different time acquisition for DCTRN and DRCTN. Finally, all the processed BOLD images were spatially normalized into MNI space. A conjunction mask was then created to sufficiently cover for all participants in each dataset.
Six head motion parameters obtained in the realigning step, Legendre polynomials up to second order were included in a linear regression to remove possible spurious variances from the data. Then, the residual time series were temporally band-pass filtered (0.008–0.1 Hz) and submitted for further HRF retrieval and statistical analysis, including the s.d. and coefficient of variation (CV, i.e. s.d./mean).
(c). Spontaneous point process event and haemodynamic response function retrieval
We employed a blind deconvolution technique to retrieve spontaneous point process HRF from resting-state BOLD fMRI signal [15]. A linear time-invariant model for the observed resting-state BOLD response is assumed. We hypothesize that a common HRF is shared across the various spontaneous point process events at a given voxel, resulting in a more robust estimation. After cardiac fluctuation correction, the BOLD signal y(t) at a particular voxel is given by
| 2.1 |
where x(t) is a sum of time-shifted delta functions centred at the onset of each spontaneous point process event and h(t) is the haemodynamic response to these events, c is a constant term indicating the baseline magnitude of the BOLD response, ε(t) represents additive noise and ⊗ denotes convolution. The noise errors are not independent in time owing to aliased biorhythms and unmodelled neural activity, and are accounted for using an AR(p) model during the parameter estimation (we set p=1 in this study). Although no explicit external inputs exist in rs fMRI acquisitions, we still could retrieve the timing of these spontaneous events by means of the blind deconvolution technique [15]. The lag between the peak of neural activation and the peak of BOLD response is presumed to be k×(TR/N) seconds (where 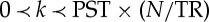 , N=3, PST is the peristimulus time, in the resting-state sense, where the ‘stimulus’ is the neural event resulting in a BOLD signature). The timing set S of these resting-state BOLD transients is defined as the time points exceeding a given threshold around a local peak, is built in the following way: s{i}=ti, y(ti)≥θ and y(ti)≻y(ti−τ) and y(ti)≻y(ti+τ), where we set τ=1,2 and θ=σ (i.e. the s.d.) in this study. The exact time lag can be obtained by minimizing the mean-squared error of equation (2.1), i.e. solving the optimization problem
, N=3, PST is the peristimulus time, in the resting-state sense, where the ‘stimulus’ is the neural event resulting in a BOLD signature). The timing set S of these resting-state BOLD transients is defined as the time points exceeding a given threshold around a local peak, is built in the following way: s{i}=ti, y(ti)≥θ and y(ti)≻y(ti−τ) and y(ti)≻y(ti+τ), where we set τ=1,2 and θ=σ (i.e. the s.d.) in this study. The exact time lag can be obtained by minimizing the mean-squared error of equation (2.1), i.e. solving the optimization problem
| 2.2 |
In order to avoid pseudo-point process events induced by motion artefacts, a temporal mask with framewise displacement (FD)<0.3 was added to exclude these bad pseudo-event onsets from timing set S by means of data scrubbing [38]. A smoothed finite impulse response (sFIR) model is employed to retrieve the spontaneous point process HRF shape [39].
To characterize the shape of the haemodynamic response, three parameters, namely response height (including non-normalized and normalized by baseline magnitude c, i.e. percent signal change, hereafter referred to as response height, and response height-PSC), time to peak and full width at half maximum (FWHM), were estimated, which could be interpretable in terms of potential measures for response magnitude, latency and duration of neuronal activity [40].
After we retrieved the resting-state HRF for each cardiac fluctuation correction model, response height (-PSC) outlier was rejected by the Grubbs test, then the corresponding HRF parameters for each subject were spatially smoothed (8 mm FWHM), finally individually entered into a random-effects analysis (one-way ANOVA within subjects, with three covariates (age, gender and mean FD) to identify regions which showed significant haemodynamic differences after cardiac fluctuation correction; subjects with mean FD>0.3 were excluded in the statistical analysis). Correlation between each HRV indicator and HRF parameter was analysed by multiple regressions with three covariates (age, gender and mean FD). Type I error owing to multiple comparisons across voxels was controlled by familywise error rate (FWE, voxel-wise correction, p<0.05, cluster size 20).
3. Results
(a). Variance explained by cardiac fluctuations
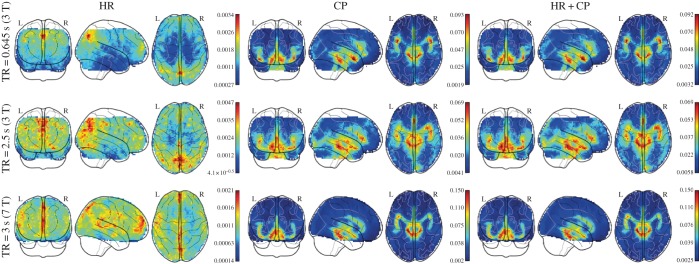
Four regressors of physiological noise correction models are entered into mass univariate GLM analysis, and the adjusted R-square of cardiac fluctuations are estimated by nested model. To include head motion parameters (obtained in the realigning step), here we report only the results after DTRCN pre-processing. Figure 1 shows the averaged fraction of variance explained by quasi-periodic and non-periodic cardiac fluctuation regressors at voxel level over subjects. Most higher adjusted R-square values for quasi-periodic cardiac fluctuation are distributed on the brainstem (the anatomical locations were obtained using the maximum probability tissue atlas from the OASIS project (http://www.oasis-brains.org/) included in SPM12 and provided by Neuromorphometrics, Inc., under academic subscription (http://neuromorphometrics.com/)). For HR, the adjusted R-square value distribution is much lower and more homogeneous, and higher explained variance can also be found in cortical networks, such as DMN.
Figure 1.
Spatial distribution of voxelwise adjusted R-squared values for different cardiac fluctuations in different TRs. First column: 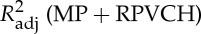 –
–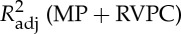 . Second column:
. Second column: 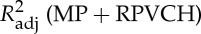 –
–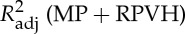 . Third column:
. Third column: 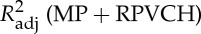 –
–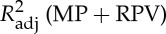 . MP, motion parameter. (Online version in colour.)
. MP, motion parameter. (Online version in colour.)
(b). Spatial distributions of resting-state blood oxygen level-dependent statistical map and haemodynamic response function
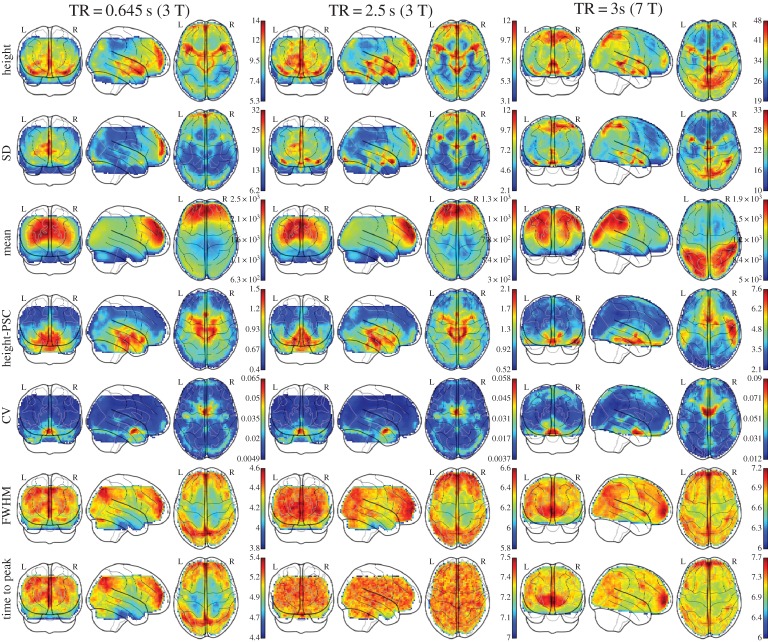
HRF parameters of each voxel are estimated and mapped on a brain template (figure 2). The median maps of each HRF parameter exhibit spatial heterogeneity across different physiological noise correction models (figure 3). They present similar spatial distributions: higher response height/FWHM/time to peak is present in the occipital/frontal lobe and precuneus, higher response height-PSC is distributed in the brainstem and surrounding areas. The baseline amplitudes in different MRI scanners exhibit different spatial distributions. The interested reader can find spatial maps of these parameters in other datasets [16].
Figure 2.
Median maps of HRF parameters (first, fourth, sixth, seventh rows) and BOLD s.d./mean/CV (second, third, fifth rows) across subjects (pre-processed by DRCTN). (Online version in colour.)
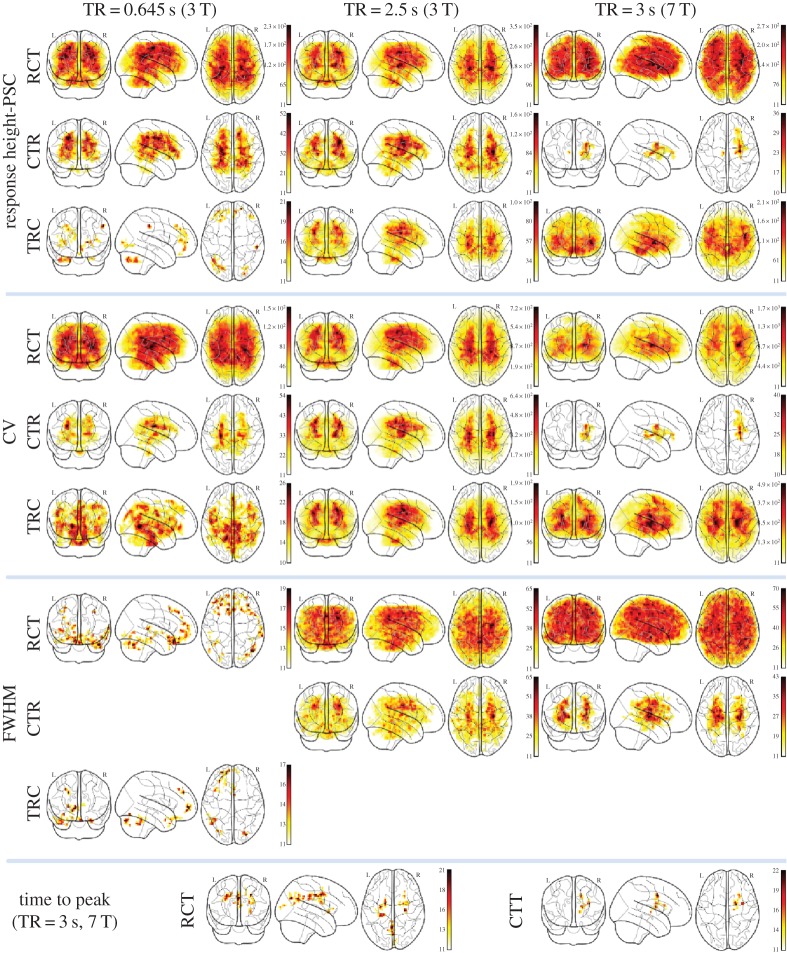
Figure 3.
Main effect on HRF parameters of four cardiac fluctuation correction models with different pre-processing procedures (repeated-measures ANOVA F-value, p<0.05, FWE correction). CTR, DCTRN; RCT, DRCTN; TRC, DTRCN. (Online version in colour.)
(c). Group difference among the cardiac fluctuation correction models
Repeated-measures ANOVA reveals that HRF response height is significantly different across models. The main effect of the cardiac fluctuation correction model on response height is mainly located in the brainstem and the surrounding pulsatile CSF regions and cortex (figure 3; p<0.05, FWE-corrected). The results are strongly affected by different pre-processing procedures and magnetic field strengths. The post hoc analyses suggest that HR gives a limited contribution to the variance of the point process response height (and PSC), whereas the significant magnitude increase is caused by cardiac cycle. The main effect maps of response height show highly similar spatial distribution with CV, s.d. and the response height-PSC (s.d. and response height are not shown in figure 3). The other HRF parameters are less sensitive to cardiac fluctuation correction. The cardiac fluctuation correction on FWHM exhibits high sensitivity to different pre-processing procedures. While the post hoc analyses further indicate that cardiac cycle extends the response duration. The only significant differences found in time to peak are in the 7 T dataset (TR=3 s); the post hoc tests show quasi-periodic cardiac fluctuation extends the time latency in precuneus.
(d). Correlation between haemodynamic response function and heart rate variability
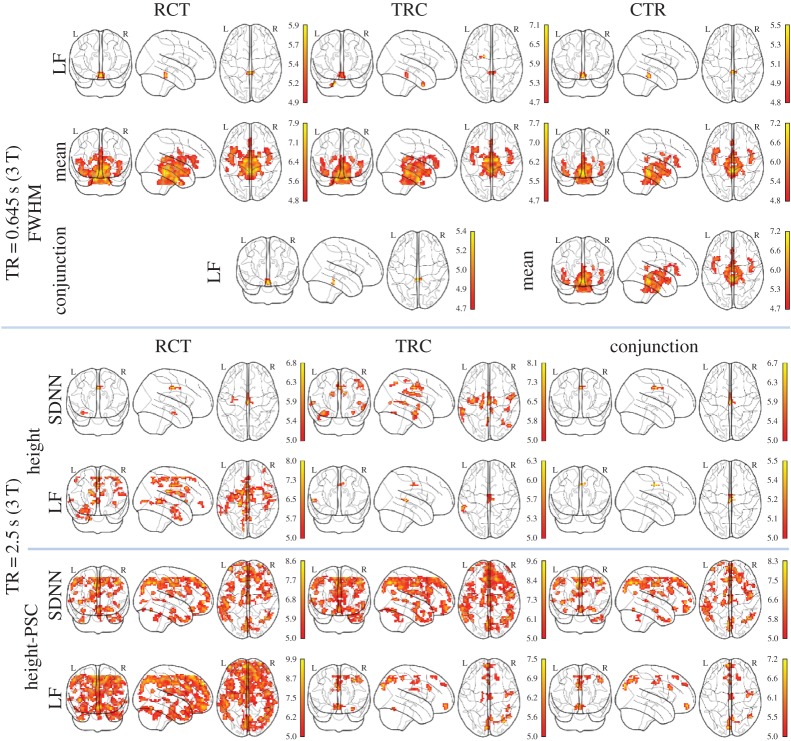
Only the correlation maps that passed the conjunction analysis obtained from two or three pre-processing procedures are reported. The correlation map reveals that the results appear to be dependent on the TR (figure 4; p<0.05, FWE-corrected). For TR=0.645 s, FWHM appears to be the only HRF parameter significantly linearly correlated with two HRV indicators. One is the mean IBI in midbrain, pons and surrounding areas: culmen, parahippocampal gyrus, thalamus, insula, superior temporal gyrus and dorsal anterior cingulate; the other is LF power in midbrain and cerebellum anterior lobe (figure 4, top). These positive correlations are also significant without cardiac fluctuation correction in all pre-processing procedures. For TR=2.5 s, only the response height and PSC are significantly correlated with some HRV indicators: LF power and SDNN (figure 4, middle and bottom). The positive linear relationship with LF power/SDNN in response magnitude map is mainly distributed in midcingulate cortex (MCC). More regions are found to be also correlated between LF power and response magnitude-PSC, namely cuneus, precuneus, inferior parietal lobule, angular, precentral gyrus, anterior cingulate cortex (ACC), medial/superior frontal gyrus and superior parietal lobule. The positive correlation between response magnitudes PSC and SDNN shows a spatial pattern similar to the one of LF power, apart from above-reported regions, but extends to include hippocampus, parahippocampal gyrus, caudate, middle/inferior/superior temporal gyrus, supramarginal gyrus, postcentral gyrus and inferior/middle frontal.
Figure 4.
Correlation maps between HRV and HRF parameters (p<0.05, FWE correction). Top: TR=0.645 s. Middle and bottom: TR=2.5 s (SDNN, LF). The warm (cool) colour denotes positive (negative) T-value. (Online version in colour.)
4. Discussion
We investigated how cardiac fluctuations affect the resting-state point process haemodynamic response estimation. These quasi-periodic fluctuations appear to influence the point process HRF magnitude and duration mainly in the brainstem and surrounding cortical. In addition, our results suggest that HRF parameters are sensitive to pre-processing procedures. However, we found a robust correlation between spontaneous point process response duration and mean interbeat interval (higher IBI correspond to slower heart rate)/low-frequency power in brainstem at short TR dataset (TR=0.645). Such positive correlations are persistent and not affected by cardiac fluctuation correction.
Previous neuroimaging studies aiming to elucidate brain–heart interactions have looked at the regional cerebral blood flow, derived from positron emission tomography (PET) or arterial spin labelling (ASL) [31,41], at brain activity in PET/task fMRI or brain connectivity in rs fMRI [12,22,31]. Our results show for the first time, to the best of our knowledge, that the reconstruction of haemodynamic response at rest is affected by cardiac activity confounds, especially affecting the temporal information on the duration and latency of the BOLD signature of cortical events. Our findings are consistent with the PET and above-mentioned fMRI studies, involving the brainstem and surrounding areas, insula and dorsal anterior cingulate. The brainstem, including the medulla oblongata, pons and midbrain, is the most important integrative control centre for ANS function and plays an important role in the regulation of cardiac and respiratory function [42,43]. There are more potential sources of signal variance in the brainstem than in any other part of the brain, owing to its anatomical structure: it is highly vascularized with arteries and veins in midbrain, is surrounded by the pulsatile flow of the CSF and it is more connected to the lungs. These factors have been reported to induce stronger changes in the magnetic field B0 [44,45]. The well-established physiological noise correction methods sharply regress out a very large proportion of spurious variation in the brainstem signal. Nonetheless, the linear correlation analysis across subjects still shows that brainstem activity is associated with HRV. The simple mean value of heart rate reveals such relationship with spontaneous point process response durations. In addition, LF power, which is generally thought to be modulated by both sympathetic and parasympathetic activity, is robustly correlated with the response duration in the midbrain. These phenomena are not affected by different processing pipelines, but cannot be evidenced with longer TRs. This may be explained by the fact that a more precise estimation of haemodynamic response duration requires a higher sample rate. To further confirm the effect from different magnitude field strength, a short TR acquisition with a 7 T MRI scanner would be a great resource.
With longer TRs, the associations between HRF parameters and HRV are more sensitive to the processing steps. No significant correlation could be found after performing the physiological noise correction first. Moreover, the spontaneous point process response magnitude and its normalization (PSC) are the only indexes that are correlated with HRV parameters. Apart from LF power, SDNN is also significantly correlated with brain areas involved in autonomic activity. SDNN also reflects both sympathetic and parasympathetic activity, providing an index of total HRV [46,47]. Our results reveal that LF and SDNN share regions in MCC that are correlated with response magnitude and its normalization. They are canonical brain areas associated with sympathetic regulation [43]. Other regions showing significant associations in the current analysis have been reported to be related to autonomic activity in previous studies [43]. In fact, the insular cortex is posited to act as an integrator on the brain–heart axis [48]: it has a prominent role in limbic–autonomic integration and is involved in the perception of emotional significance [49]; it also participates in visceral motor regulation, including blood pressure control, in cooperation with subcortical autonomic centres [50–52]. The dACC is also involved in autonomic control [30,53]; the network consisting of insula, dACC and amygdala has been described as crucial in the regulation of central ANS [54]. A human neuroimaging meta-analysis on electrodermal activity and high-frequency HRV revealed that midbrain, insula and supramarginal gyrus are associated with sympathetic and parasympathetic regulation; ACC, thalamus and primary somatosensory cortex are associated with sympathetic regulation, whereas precuneus, superior temporal gyri and angular are associated with parasympathetic regulations [43]. The precuneus and angular gyrus are also among the key nodes of DMN. Several studies have shown that FC maps of the DMN are modulated by heart rate and RVT [12,22]. Intriguingly, the spatial extent of the correlation map with normalized response magnitude is larger than when raw response magnitude is considered. It is worth mentioning that there is no significant correlation between HRF estimation and HRV after conjunction analysis in the 7 T dataset. Apart from stringent thresholds for significance, magnetic field strength and age distributions are different in the two datasets: a study has shown that SDNN index exhibits a linearly correlated pattern of decline with ageing for both genders [47].
A recent study reported a significantly decreased test–retest reliability in FC by physiological noise correction techniques [23]. These results were explained by assuming that these physiological fluctuations are similar and reproducible within a subject across sessions, but to a lesser extent than between subjects. Another explanation given in the same study posited that physiological noise correction could also remove the signal of interest.
Physiological fluctuations have been shown to be proportional to magnetic field strength [55]: the physiological processes may therefore contribute much more to variance in BOLD signal when data are acquired with a strong field. Apart from cardiac fluctuations, respiration is another physiological fluctuation that has also been found to strongly modulate the rs fMRI BOLD signal [26,27]. Respiration fluctuations will induce variations in arterial level of CO2, then cause either validations or vasoconstriction, resulting in blood flow and oxygenation changes [56]. HRF magnitude variation is intrinsically related to the CO2 centration owing to vascular reactivity [19,57,58]. In this study, a vascular modulator such as a breath-holding task was not present in all datasets; RVT is used as a surrogate for arterial CO2 concentration to capture breathing rate and depth from respiratory belt measurements suggested by [26]. In addition, as respiration and cardiac pulsations are tightly correlated [59,60], we first partial out respiratory fluctuations before investigating the impact of cardiac fluctuations on the estimation of resting-state point process HRF. The spontaneous events retrieved by point process may include actual neural events, autonomic activities and their interactions. However, in the estimation of the HRF, we hypothesize that they result in a common shape. This may intrinsically limit the disambiguation of the two.
To reduce the computational cost and the bias in the linear estimation framework, we employ canonical functions for HR and RVT haemodynamic response [22,27]. Moreover, the lagged RVT and HR regressors are not included in our analysis. These may reduce the contribution of HR in the regression model. However, increasing the number of regressors may induce more bias in GLM especially for short time series (120 volumes in TR=2.5). The more flexible sFIR model could then minimize the risk of assumptions about the spontaneous point process HRF shape [39]. In addition, the sFIR model may also include the components related to cardiac fluctuation in the haemodynamic response, when the latter is not eliminated from the BOLD signal. It has been shown that different processing steps could dramatically change the tSNR in fMRI BOLD signal. In particular, volume registration before physiological noise correction and not performing slice timing correction before physiological noise correction will result in the greatest reduction of temporal noise [61]. Such effect on estimation of spontaneous point process HRF has not been investigated. Resting-state point process is dependent on the variance of BOLD signal; repeated-measures ANOVA results show that HRF magnitudes are similar to BOLD CV and SD in most cases. Our results confirm that different processing steps affect the HRF estimation, not only of its magnitude but also of its temporal parameters (latency and duration). In addition, we find that the effects of HR on HRF estimation are more evident when the DTRCN processing procedure is used together with 3 T dataset, and when DRCTN is used with short TR (0.645 s). Apart from the differences owing to processing, the sFIR model is essentially more sensitive to temporal noise [40].
This study has some limitations that should be noted. First, the proposed method to retrieve the HRF at rest only uses BOLD data. In [16], we have started to explore some validation strategies involving models, ASL, PET and simultaneous EEG-fMRI data. The most convincing validation would nonetheless involve data where the neural activity and the BOLD signal are both extracted, for instance an experiment with simultaneous BOLD signal and intracortical recordings of neural signals [62]. It is worth noting that the processing order ‘DTRCN’ may not capture the aliased physiological perturbation owing to the placement of RETROICOR after slice-timing correction, especially for long TR datasets. Performing despiking before physiological noise correction could improve the regression model fitting by removing large spikes. The despiking procedure was skipped in [38,61]; nonetheless, it appears to improve the results of volume registration over time as illustrated in [63]. However, each procedure has potential disadvantages: temporal correction (slice-timing correction and despking) before realignment may interpolate signals from different brain regions if there is significant head movement; temporal correction after realignment, on the other hand, may shift voxels to adjacent slices and hence disturb temporal order: this problem is especially relevant for interleaved and multiband acquisitions such as those used in this study. To cope with the latter problem, motion-modified RETROICOR has been proposed to include slice contribution to every voxel [61]. This procedure, however, might induce higher computational costs and a bias in the regression model. Therefore, the influences of different pre-processing procedures on HRF estimation should be further explored. In addition, we did not find any significant association between amygdala with HRV parameters. Moreover, no significant correlation with HF power, RMSSD or LF/HF was found after conjunction analysis.
It is well known that head motion is an unavoidable source of noise in the BOLD signal [10]. To avoid motion-related artefact contribution to point process detection, in addition to adding motion parameters as a nuisance regressor in the GLM, data scrubbing was performed [38], and mean FD of each subject was included as a covariate for further statistical analysis [64]. This procedure ensures that our findings are unlikely to be affected by motion artefact.
This study has demonstrated the impact of physiological noise correction on resting-state HRF estimation, validated at different TRs and magnetic field strength. Several processing pipelines are employed to explore the sensitivity in estimation of resting-state HRF. Intersubject correlation analyses between HRF and HRV parameters suggest that ANS fluctuations modulate the estimation of spontaneous point process response in brainstem.
Data accessibility
NKI Rockland data are available and described at http://dx.doi.org/10.3389/fnins.2012.00152. CoRR 7 T TRT data are available and described at http://dx.doi.org/10.1038/sdata.2014.54.
Author' contributions
G.R.W. and D.M. designed the research; G.R.W. analysed the data; G.R.W. and D.M. wrote the paper.
Competing interests
We declare we have no competing interests.
Funding
G.R.W. was supported by the Natural Science Foundation of China (grant no. 61403312), and the Fundamental Research Funds for the Central Universities (grant no. 2362014xk04).
References
- 1.Raichle ME. 2010. Two views of brain function. Trends Cogn. Sci. 14, 180–190. ( 10.1016/j.tics.2010.01.008) [DOI] [PubMed] [Google Scholar]
- 2.Barkhof F, Haller S, Rombouts SA. 2014. Resting-state functional MR imaging: a new window to the brain. Radiology 272, 29–49. ( 10.1148/radiol.14132388) [DOI] [PubMed] [Google Scholar]
- 3.Mantini D, Perrucci MG, Del Gratta C, Romani GL, Corbetta M. 2007. Electrophysiological signatures of resting state networks in the human brain. Proc. Natl Acad. Sci. USA 104, 13 170–13 175. ( 10.1073/pnas.0700668104) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Pasquale F. et al. 2010. Temporal dynamics of spontaneous MEG activity in brain networks. Proc. Natl Acad. Sci. USA 107, 6040–6045. ( 10.1073/pnas.0913863107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deco G, Jirsa VK. 2012. Ongoing cortical activity at rest: criticality, multistability, and ghost attractors. J. Neurosci. 32, 3366–3375. ( 10.1523/JNEUROSCI.2523-11.2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tagliazucchi E, Balenzuela P, Fraiman D, Chialvo DR. 2012. Criticality in large-scale brain FMRI dynamics unveiled by a novel point process analysis. Front. Physiol. 3, 15 ( 10.3389/fphys.2012.00015) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Petridou N, Gaudes CC, Dryden IL, Francis ST, Gowland PA. 2013. Periods of rest in fMRI contain individual spontaneous events which are related to slowly fluctuating spontaneous activity. Hum. Brain Mapp. 34, 1319–1329. ( 10.1002/hbm.21513) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu X, Duyn JH. 2013. Time-varying functional network information extracted from brief instances of spontaneous brain activity. Proc. Natl Acad. Sci. USA 110, 4392–4397. ( 10.1073/pnas.1216856110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Renvall V, Hari R. 2009. Transients may occur in functional magnetic resonance imaging without physiological basis. Proc. Natl Acad. Sci. USA 106, 20 510–20 514. ( 10.1073/pnas.0911265106) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Power JD, Mitra A, Laumann TO, Snyder AZ, Schlaggar BL, Petersen SE. 2014. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage 84, 320–341. ( 10.1016/j.neuroimage.2013.08.048) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ogawa S, Lee TM, Kay AR, Tank DW. 1990. Brain magnetic resonance imaging with contrast dependent on blood oxygenation. Proc. Natl Acad. Sci. USA 87, 9868–9872. ( 10.1073/pnas.87.24.9868) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Birn RM. 2012. The role of physiological noise in resting-state functional connectivity. Neuroimage 62, 864–870. ( 10.1016/j.neuroimage.2012.01.016) [DOI] [PubMed] [Google Scholar]
- 13.Hutchison RM. et al. 2013. Dynamic functional connectivity: promise, issues, and inter- pretations. Neuroimage 80, 360–378. ( 10.1016/j.neuroimage.2013.05.079) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murphy K, Birn RM, Bandettini PA. 2013. Resting-state fMRI confounds and cleanup. Neuroimage 80, 349–359. ( 10.1016/j.neuroimage.2013.04.001) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu GR, Liao W, Stramaglia S, Ding JR, Chen H, Marinazzo D. 2013. A blind deconvolution approach to recover effective connectivity brain networks from resting state fMRI data. Med. Image Anal. 17, 365–374. ( 10.1016/j.media.2013.01.003) [DOI] [PubMed] [Google Scholar]
- 16.Wu G-R, Marinazzo D. 2015. Retrieving the Hemodynamic Response Function in resting state fMRI: methodology and applications. PeerJ PrePrints 3, e1621 ( 10.7287/peerj.preprints.1317v1) [DOI] [PubMed] [Google Scholar]
- 17.Logothetis NK. 2008. What we can do and what we cannot do with fMRI. Nature 453, 869–878. ( 10.1038/nature06976) [DOI] [PubMed] [Google Scholar]
- 18.Greitz D, Franck A, Nordell B. 1993. On the pulsatile nature of intracranial and spinal CSF-circulation demonstrated by MR imaging. Acta Radiol. 34, 321–328. ( 10.1177/028418519303400403) [DOI] [PubMed] [Google Scholar]
- 19.Murphy K, Harris AD, Wise RG. 2011. Robustly measuring vascular reactivity differences with breath-hold: normalising stimulus-evoked and resting state BOLD fMRI data. Neuroimage 54, 369–379. ( 10.1016/j.neuroimage.2010.07.059) [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharyya PK, Lowe MJ. 2004. Cardiac-induced physiologic noise in tissue is a direct observation of cardiac-induced fluctuations. Magn. Reson. Imaging 22, 9–13. ( 10.1016/j.mri.2003.08.003) [DOI] [PubMed] [Google Scholar]
- 21.Shmueli K, van Gelderen P, de Zwart JA, Horovitz SG, Fukunaga M, Jansma JM, Duyn JH. 2007. Low-frequency fluctuations in the cardiac rate as a source of variance in the resting-state fMRI BOLD signal. Neuroimage 38, 306–320. ( 10.1016/j.neuroimage.2007.07.037) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chang C, Cunningham JP, Glover GH. 2009. Influence of heart rate on the BOLD signal: the cardiac response function. Neuroimage 44, 857–869. ( 10.1016/j.neuroimage.2008.09.029) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Birn RM. et al. 2014. The influence of physiological noise correction on test-retest reliability of resting-state functional connectivity. Brain Connect. 4, 511–522. ( 10.1089/brain.2014.0284) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Valdes-Sosa PA, Roebroeck A, Daunizeau J, Friston K. 2011. Effective connectivity: influence, causality and biophysical modeling. Neuroimage 58, 339–361. ( 10.1016/j.neuroimage.2011.03.058) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glover GH, Li TQ, Ress D. 2000. Image-based method for retrospective correction of physiological motion effects in fMRI: RETROICOR. Magn. Reson. Med. 44, 162–167. () [DOI] [PubMed] [Google Scholar]
- 26.Birn RM, Diamond JB, Smith MA, Bandettini PA. 2006. Separating respiratory-variation-related fluctuations from neuronal-activity-related fluctuations in fMRI. Neuroimage 31, 1536–1548. ( 10.1016/j.neuroimage.2006.02.048) [DOI] [PubMed] [Google Scholar]
- 27.Birn RM, Smith MA, Jones TB, Bandettini PA. 2008. The respiration response function: the temporal dynamics of fMRI signal fluctuations related to changes in respiration. Neuroimage 40, 644–654. ( 10.1016/j.neuroimage.2007.11.059) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Iacovella V, Hasson U. 2011. The relationship between BOLD signal and autonomic nervous system functions: implications for processing of ‘physiological noise’. Magn. Reson. Imaging 29, 1338–1345. ( 10.1016/j.mri.2011.03.006) [DOI] [PubMed] [Google Scholar]
- 29.Fan J, Xu P, Van Dam NT, Eilam-Stock T, Gu X, Luo YJ, Hof PR. 2012. Spontaneous brain activity relates to autonomic arousal. J. Neurosci. 32, 11 176–11 186. ( 10.1523/JNEUROSCI.1172-12.2012) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chang C, Metzger CD, Glover GH, Duyn JH, Heinze HJ, Walter M. 2013. Association between heart rate variability and fluctuations in resting-state functional connectivity. Neuroimage 68, 93–104. ( 10.1016/j.neuroimage.2012.11.038) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thayer JF, Ahs F, Fredrikson M, Sollers JJ 3rd, Wager TD. 2012. A meta-analysis of heart rate variability and neuroimaging studies: implications for heart rate variability as a marker of stress and health. Neurosci. Biobehav. Rev. 36, 747–756. ( 10.1016/j.neubiorev.2011.11.009) [DOI] [PubMed] [Google Scholar]
- 32.Garfinkel SN, Minati L, Gray MA, Seth AK, Dolan RJ, Critchley HD. 2014. Fear from the heart: sensitivity to fear stimuli depends on individual heartbeats. J. Neurosci. 34, 6573–6582. ( 10.1523/JNEUROSCI.3507-13.2014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nooner KB. et al. 2012. The NKI-Rockland sample: a model for accelerating the pace of discovery science in psychiatry. Front. Neurosci. 6, 152 ( 10.3389/fnins.2012.00152) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gorgolewski KJ. et al. 2015. A high resolution 7-Tesla resting-state fMRI test-retest dataset with cognitive and physiological measures. Sci. Data 2, 140054 ( 10.1038/sdata.2014.54) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harvey AK, Pattinson KT, Brooks JC, Mayhew SD, Jenkinson M, Wise RG. 2008. Brainstem functional magnetic resonance imaging: disentangling signal from physiological noise. J. Magn. Reson. Imaging 28, 1337–1344. ( 10.1002/jmri.21623) [DOI] [PubMed] [Google Scholar]
- 36.Ashburner J. 2012. SPM: a history. Neuroimage 62, 791–800. ( 10.1016/j.neuroimage.2011.10.025) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox RW. 1996. AFNI: software for analysis and visualization of functional magnetic resonance neuroimages. Comput. Biomed. Res. 29, 162–173. ( 10.1006/cbmr.1996.0014) [DOI] [PubMed] [Google Scholar]
- 38.Power JD, Barnes KA, Snyder AZ, Schlaggar BL, Petersen SE. 2012. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage 59, 2142–2154. ( 10.1016/j.neuroimage.2011.10.018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Goutte C, Nielsen FA, Hansen LK. 2000. Modeling the haemodynamic response in fMRI using smooth FIR filters. IEEE Trans. Med. Imag. 19, 1188–1201. ( 10.1109/42.897811) [DOI] [PubMed] [Google Scholar]
- 40.Lindquist MA, Wager TD. 2007. Validity and power in hemodynamic response modeling: a comparison study and a new approach. Hum. Brain Mapp. 28, 764–784. ( 10.1002/hbm.20310) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Restom K, Behzadi Y, Liu TT. 2006. Physiological noise reduction for arterial spin labeling functional MRI. Neuroimage 31, 1104–1115. ( 10.1016/j.neuroimage.2006.01.026) [DOI] [PubMed] [Google Scholar]
- 42.Mendelowitz D. 1999. Advances in parasympathetic control of heart rate and cardiac function. News Physiol. Sci. 14, 155–161. [DOI] [PubMed] [Google Scholar]
- 43.Beissner F, Meissner K, Bar KJ, Napadow V. 2013. The autonomic brain: an activation likelihood estimation meta-analysis for central processing of autonomic function. J. Neurosci. 33, 10 503–10 511. ( 10.1523/JNEUROSCI.1103-13.2013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brooks JC, Faull OK, Pattinson KT, Jenkinson M. 2013. Physiological noise in brainstem FMRI. Front. Hum. Neurosci. 7, 623 ( 10.3389/fnhum.2013.00623) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barry RL, Coaster M, Rogers BP, Newton AT, Moore J, Anderson AW, Zald DH, Gore JC. 2013. On the origins of signal variance in FMRI of the human midbrain at high field. PLoS ONE 8, e62708 ( 10.1371/journal.pone.0062708) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Task Force of the European Society of Cardiology and North American Society of Pacing and Electrophysiology. 1996. Heart rate variability: standards of measurement, physiological interpretation and clinical use. Circulation 93, 1043–1065. ( 10.1161/01.CIR.93.5.1043) [DOI] [PubMed] [Google Scholar]
- 47.Umetani K, Singer DH, McCraty R, Atkinson M. 1998. Twenty-four hour time domain heart rate variability and heart rate: relations to age and gender over nine decades. J. Am. College Cardiol. 31, 593–601. ( 10.1016/S0735-1097(97)00554-8) [DOI] [PubMed] [Google Scholar]
- 48.Nagai M, Hoshide S, Kario K. 2010. The insular cortex and cardiovascular system: a new insight into the brain–heart axis. J. Am. Soc. Hypertens. 4, 174–182. ( 10.1016/j.jash.2010.05.001) [DOI] [PubMed] [Google Scholar]
- 49.Augustine JR. 1996. Circuitry and functional aspects of the insular lobe in primates including humans. Brain Res. Rev. 22, 229–244. ( 10.1016/S0165-0173(96)00011-2) [DOI] [PubMed] [Google Scholar]
- 50.Lane RD, McRae K, Reiman EM, Chen K, Ahern GL, Thayer JF. 2009. Neural correlates of heart rate variability during emotion. Neuroimage 44, 213–222. ( 10.1016/j.neuroimage.2008.07.056) [DOI] [PubMed] [Google Scholar]
- 51.Napadow V, Dhond R, Conti G, Makris N, Brown EN, Barbieri R. 2008. Brain correlates of autonomic modulation: combining heart rate variability with fMRI. Neuroimage 42, 169–177. ( 10.1016/j.neuroimage.2008.04.238) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gianaros PJ, Jennings JR, Sheu LK, Derbyshire SW, Matthews KA. 2007. Heightened functional neural activation to psychological stress covaries with exaggerated blood pressure reactivity. Hypertension 49, 134–140. ( 10.1161/01.HYP.0000250984.14992.64) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Critchley HD. et al. 2003. Human cingulate cortex and autonomic control: converging neuroimaging and clinical evidence. Brain 126, 2139–2152. ( 10.1093/brain/awg216) [DOI] [PubMed] [Google Scholar]
- 54.Critchley HD. 2005. Neural mechanisms of autonomic, affective, and cognitive integration. J. Comp. Neurol. 493, 154–166. ( 10.1002/cne.20749) [DOI] [PubMed] [Google Scholar]
- 55.Kruger G, Glover GH. 2001. Physiological noise in oxygenation-sensitive magnetic resonance imaging. Magn. Reson. Med. 46, 631–637. ( 10.1002/mrm.1240) [DOI] [PubMed] [Google Scholar]
- 56.Van den Aardweg JG, Karemaker JM. 2002. Influence of chemoreflexes on respiratory variability in healthy subjects. Am. J. Respir. Crit. Care Med. 165, 1041–1047. ( 10.1164/ajrccm.165.8.2104100) [DOI] [PubMed] [Google Scholar]
- 57.Kannurpatti SS, Biswal BB. 2008. Detection and scaling of task-induced fMRI-BOLD response using resting state fluctuations. Neuroimage 40, 1567–1574. ( 10.1016/j.neuroimage.2007.09.040) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Handwerker DA, Gazzaley A, Inglis BA, D'Esposito M. 2007. Reducing vascular variability of fMRI data across aging populations using a breathholding task. Hum. Brain Mapp. 28, 846–859. ( 10.1002/hbm.20307) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pitzalis MV, Mastropasqua F, Massari F, Passantino A, Forleo C, Luzzi G, Totaro P, De Nicolò M, Rizzon P. 1997. Dependency of premature ventricular contractions on heart rate. Am. Heart J. 133, 153–161. ( 10.1016/S0002-8703(97)70203-5) [DOI] [PubMed] [Google Scholar]
- 60.Princi T, Accardo A, Peterec D. 2006. Linear and non-linear assessment of heart rate variability in congenital central hypoventilation syndrome. Biomed. Sci. Instrum. 42, 434–439. [PubMed] [Google Scholar]
- 61.Jones TB, Bandettini PA, Birn RM. 2008. Integration of motion correction and physiological noise regression in fMRI. Neuroimage 42, 582–590. ( 10.1016/j.neuroimage.2008.05.019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Logothetis NK. 2003. The underpinnings of the BOLD functional magnetic resonance imaging signal. J. Neurosci. 23, 3963–3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jo HJ, Gotts SJ, Reynolds RC, Bandettini PA, Martin A, Cox RW, Saad ZS. 2013. Effective preprocessing procedures virtually eliminate distance-dependent motion artifacts in resting state FMRI. J. Appl. Math 2013, 935154 ( 10.1155/2013/935154) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Van Dijk KR, Sabuncu MR, Buckner RL. 2012. The influence of head motion on intrinsic functional connectivity MRI. Neuroimage 59, 431–438. ( 10.1016/j.neuroimage.2011.07.044) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
NKI Rockland data are available and described at http://dx.doi.org/10.3389/fnins.2012.00152. CoRR 7 T TRT data are available and described at http://dx.doi.org/10.1038/sdata.2014.54.