Abstract
Coevolution is relentlessly creating and maintaining biodiversity and therefore has been a central topic in evolutionary biology. Previous theoretical studies have mostly considered coevolution between genetically symmetric traits (i.e. coevolution between two continuous quantitative traits or two discrete Mendelian traits). However, recent empirical evidence indicates that coevolution can occur between genetically asymmetric traits (e.g. between quantitative and Mendelian traits). We examine consequences of antagonistic coevolution mediated by a quantitative predator trait and a Mendelian prey trait, such that predation is more intense with decreased phenotypic distance between their traits (phenotype matching). This antagonistic coevolution produces a complex pattern of bifurcations with bistability (initial state dependence) in a two-dimensional model for trait coevolution. Furthermore, with eco-evolutionary dynamics (so that the trait evolution affects predator–prey population dynamics), we find that coevolution can cause rich dynamics including anti-phase cycles, in-phase cycles, chaotic dynamics and deterministic predator extinction. Predator extinction is more likely to occur when the prey trait exhibits complete dominance rather than semidominance and when the predator trait evolves very rapidly. Our study illustrates how recognizing the genetic architectures of interacting ecological traits can be essential for understanding the population and evolutionary dynamics of coevolving species.
Keywords: coevolution, polygenic continuous trait, major-gene discrete trait, Red Queen dynamics, eco-evolutionary feedbacks, extinction
1. Introduction
Coevolution—reciprocal evolution in interacting species—has been considered a driving force for creating and maintaining biodiversity [1]. Evolutionary ‘arms races’ in predator–prey, parasite–host and exploiter–victim systems [2,3] are thought to create an endless evolutionary chase called Red Queen dynamics [4,5]. In addition to accumulating empirical evidence of coevolution [1,5], recent studies have shown that evolution can be rapid enough to affect contemporary population dynamics [6–8]. Understanding ‘rapid coevolution’ is therefore important for understanding and predicting future ecological dynamics [9–11].
Theoretical studies on coevolution have mostly considered genetically symmetric traits in antagonistic interactions (i.e. coevolution either between continuous quantitative traits or between discrete Mendelian traits) [5]. Continuous quantitative traits are generally modelled using phenotypically based approaches for trait evolutionary dynamics, partly because the genetic basis of ecologically important traits was largely unknown [3]. The approaches include approximate quantitative genetics models for multilocus trait dynamics (e.g. [10–17]), mutation-limited asexual clonal models (Adaptive Dynamics) (e.g. [18–24]) and models for evolutionarily stable strategies (ESS) that cannot be invaded by other strategies (e.g. [25]). These studies have assumed that the coevolving traits are quantitative with a continuum of possible values [26]. Coevolution between Mendelian predator and prey traits has also been studied (e.g. [27–31]). Indeed, there is a large theoretical literature going back to classical population genetics about exploiter–victim coevolution with Mendelian traits (reviewed by Seger [32]).
How do different genetic architectures affect coevolutionary dynamics? One important difference is that Mendelian models have an inherent tendency to cycle, first noted by Haldane [32]. Multiple alleles, multiple loci or multiple species versions of these models readily generate complex cycles or chaotic dynamics [32,33]. A comparison of models with two versus more alleles suggests that cycles would be very likely when prey and predator have continuously varying quantitative traits, but in fact the reverse is true [32]: coevolution of quantitative traits rarely produces cycles. The key difference is that in quantitative trait models, all phenotypes are near the population mean [32]. Cycles in Mendelian models involve switches between extremes: high frequency of one predator type (feeding on its complementary prey genotype) causes increased frequency of the least complementary prey genotype. By contrast, in quantitative trait models, trait distributions change through gradual shifts in the mean of a Gaussian distribution. Coevolution-driven cycles can occur in quantitative trait models [14,19], but these require very strong external stabilizing selection for intermediate trait values [15]. With external stabilizing selection weak or absent, the only possible type of ongoing dynamics is unbounded runaway selection for extreme traits [14,15].
Coevolution models with discrete Mendelian traits have frequently been used for parasite–host coevolution, whereas models for predator–prey coevolution mainly assume continuous quantitative traits [32]. However, empirical studies have found many ecologically important traits controlled by Mendelian inheritance in prey (e.g. [34–36]) or predator (e.g. [37–39]) species [40]. This suggests the potential importance of coevolution between traits with different genetic bases. To the best of our knowledge, no study has examined the outcome of coevolution between quantitative and Mendelian traits, despite empirical evidence for coevolution of this type [36]. In Southeast Asia, some snakes can eat right-handed (dextral) land snails effectively but are not good at eating left-handed (sinistral) snails because of left–right asymmetry in the snakes [36,41]. Within the right-handed snake range, left-handed snail species frequently evolved by speciation from a right-handed ancestor, probably as a counter-adaptation against the biased predation [36,42]. Handedness of snails is determined by two alleles at a single nuclear locus [36,43], whereas the snake traits (teeth number on each side and foraging behaviour) are continuous quantitative traits [41] and therefore they seem to be controlled by many loci with small effects. These observations indicate antagonistic coevolution between quantitative and Mendelian traits. Will the dynamics resemble Mendelian–Mendelian coevolution, quantitative–quantitative coevolution, something in between, or are some new phenomena likely to occur?
In this study, we examine theoretically coevolution between a quantitative predator trait and a Mendelian prey trait. Our model is analogous to matching-allele interaction, in that predators are most able to capture prey whose phenotype matches their own (i.e. a bidirectional axis of vulnerability sensu [3]). We consider both purely coevolutionary dynamics (trait evolution that does not interact with population dynamics) and eco-evolutionary dynamics (traits and population sizes change on the same time scale). Analysis of our models shows that: (i) continuous-discrete coevolution can produce a rich array of dynamics including bistability (initial state dependence), anti-phase cycles, in-phase cycles and chaotic dynamics, (ii) semidominance of the prey trait is more favourable for predator persistence than complete dominance and (iii) increasing heritability of the predator trait can result in deterministic predator extinction. We also present some results for the reverse situation, Mendelian predator and quantitative prey traits.
2. Model
We consider a predator–prey model with phenotype matching (a bidirectional axis) instead of a unidirectional axis of vulnerability (e.g. speed, toxin or armour: [10,12,13,16,17,44]). Phenotype matching means that predators are most successful at capturing prey when their trait value x is close in value to the prey trait value θ, and unsuccessful if |x−θ| is large. Examples of phenotype matching include habitat choice, pattern matching and lock-and-key mechanisms [9,13–15,18–20,45,46]. We assume that per capita attack rate on prey genotype i declines in a Gaussian manner when x differs from θi, 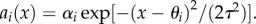 Here αi is the maximum per capita attack rate, and τ determines how steeply the rate decreases as predator–prey mismatch increases [47].
Here αi is the maximum per capita attack rate, and τ determines how steeply the rate decreases as predator–prey mismatch increases [47].
The predator trait distribution is Gaussian, with mean 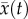 and constant phenotypic variance σ2:
and constant phenotypic variance σ2: 
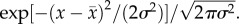 We assume (i) that phenotypic variance has a genetic (heritable) component,
We assume (i) that phenotypic variance has a genetic (heritable) component, 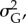 and an environmental component,
and an environmental component, 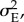 with
with 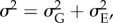 and for simplicity, (ii) that the variance components are constant, which is commonly assumed in dynamic models for quantitative trait evolution [10–12,14,26].
and for simplicity, (ii) that the variance components are constant, which is commonly assumed in dynamic models for quantitative trait evolution [10–12,14,26].
Under our assumptions, the average attack rate by a predator on prey genotype i is
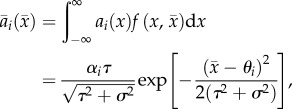 |
2.1 |
where i = 1, 2 and 3 correspond to prey genotypes A1A1, A1A2 and A2A2, respectively. The model represents complete dominance with two discrete prey trait values when θ1 = θ2 ≠ θ3. In semidominance, the heterozygotes have an intermediate trait θ2 = (θ1 + θ3)/2 so there are three distinct prey phenotypes. We assume that θ1 = –θ3 without loss of generality, so that θ2 = 0 under semidominance. The above models represent ends of a spectrum. There is a continuum between these extremes and probably some sort of smooth transition between their dynamics.
(a). Evolutionary dynamics model
Our first model considers purely evolutionary dynamics driven by relative fitness, with total population sizes held constant as in [14]. Let p be the A1 allele frequency. We assume prey genotype fitnesses (in discrete time, with discrete generations) to be 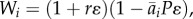 where ɛ is prey generation time, r is the reproduction rate (equal for all genotypes), P is the predator density and
where ɛ is prey generation time, r is the reproduction rate (equal for all genotypes), P is the predator density and 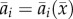 defined by equation (2.1). Thus, the only advantage of one prey phenotype over another is predation risk. We assume that prey start each generation in Hardy–Weinberg equilibrium owing to random mating. To eliminate dynamic behaviours resulting only from the assumption of discrete generations, we pass to a continuous-time approximation in the usual way, by assuming small changes per generation (ɛ ≪ 1) owing to weak selection (e.g. [48], §3.3). In the electronic supplementary material, appendix S1, we show that for small ɛ, the prey evolutionary dynamics are approximated by a continuous-time equation determined by the additive genetic variance and fitness gradient,
defined by equation (2.1). Thus, the only advantage of one prey phenotype over another is predation risk. We assume that prey start each generation in Hardy–Weinberg equilibrium owing to random mating. To eliminate dynamic behaviours resulting only from the assumption of discrete generations, we pass to a continuous-time approximation in the usual way, by assuming small changes per generation (ɛ ≪ 1) owing to weak selection (e.g. [48], §3.3). In the electronic supplementary material, appendix S1, we show that for small ɛ, the prey evolutionary dynamics are approximated by a continuous-time equation determined by the additive genetic variance and fitness gradient,
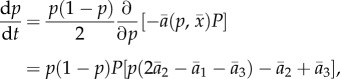 |
2.2 |
where 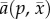 is the per-predator attack rate or average predation rate,
is the per-predator attack rate or average predation rate, 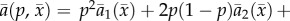

We assume that predator fitness is proportional to per-predator total predation rate, which is determined by the prey abundance N, prey genotype frequencies, and the attack rate on each genotype, 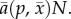 We then adopt a conventional fitness gradient model [26]
We then adopt a conventional fitness gradient model [26]
 |
2.3 |
The speed of adaptive evolution in the predator quantitative trait is determined by the strength of selection and additive genetic variance [49]. The assumption that the trait remains normally distributed was demonstrated to be a good approximation to trait dynamics in various multilocus models numerically [50]. Equations (2.2) and (2.3) are our two-dimensional model for purely evolutionary dynamics. The attack rate is simplified by rescaling predator and prey traits relative to 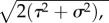 and defining
and defining 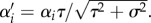 With these, and dropping the prime, the attack rate is
With these, and dropping the prime, the attack rate is 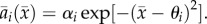 The predator's additive genetic variance on this scale is
The predator's additive genetic variance on this scale is 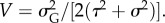
(b). Eco-evolutionary dynamics model
Our second model allows evolutionary and ecological dynamics (changes in predator and prey abundance) to occur on the same time scale. Specifically, we put the predation rates from our evolutionary model into a Rosenzweig–MacArthur-type predator–prey model where the prey has logistic growth, and the predator has a linear (Holling type I) functional response. The dynamics of prey density, N, and predator density, P, are then
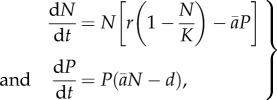 |
2.4 |
where r is the prey intrinsic growth rate, K is the carrying capacity, d is the per capita mortality and 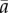 is the average predation rate,
is the average predation rate, 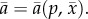 The combination of logistic growth and linear functional response means that with constant parameters, an equilibrium at which predator and prey coexist is always stable [51]. The predator equation often includes conversion efficiency, but we can scale prey abundance so that this equals one.
The combination of logistic growth and linear functional response means that with constant parameters, an equilibrium at which predator and prey coexist is always stable [51]. The predator equation often includes conversion efficiency, but we can scale prey abundance so that this equals one.
The fitnesses implied by equation (2.4) are consistent with our evolutionary model (equations (2.2) and (2.3)). Prey reproduction (r in equation (2.4)) has no effect on equation (2.2) as long as it is the same for all genotypes, so the logistic density-dependence in equation (2.4) does not change the prey trait dynamics. Predator fitness (dP/dt)/P in equation (2.4) equals the per-predator attack rate, 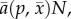 minus the constant d, so the fitness gradient and trait dynamics are still given by equation (2.3).
minus the constant d, so the fitness gradient and trait dynamics are still given by equation (2.3).
Assuming that all prey genotypes are equivalent except for the trait value θi (α = α1 = α2 = α3), we rescale the model as N′ = N/K, P′ = P/K and t′ = rt. With the new parameters 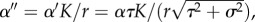 d′ = d/r, and the (double) prime dropped, the full system becomes
d′ = d/r, and the (double) prime dropped, the full system becomes
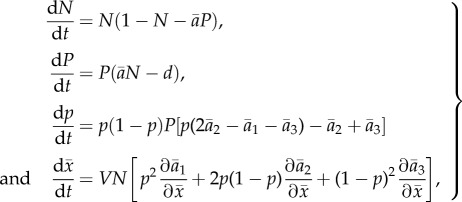 |
2.5 |
with 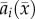 given by
given by  Thus, the model has four parameters (d, α, V, θ1) after rescaling.
Thus, the model has four parameters (d, α, V, θ1) after rescaling.
3. Results
(a). Purely evolutionary dynamics
With predator and prey abundance constant, we can eliminate more parameters by rescaling 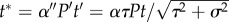 in the last two lines of equation (2.5), giving (with the asterisk dropped)
in the last two lines of equation (2.5), giving (with the asterisk dropped)
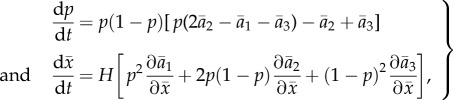 |
3.1 |
where 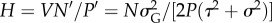 and
and 
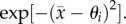 This leaves only two parameters, the scaled predator genetic variation (H) and prey trait difference (θ1).
This leaves only two parameters, the scaled predator genetic variation (H) and prey trait difference (θ1).
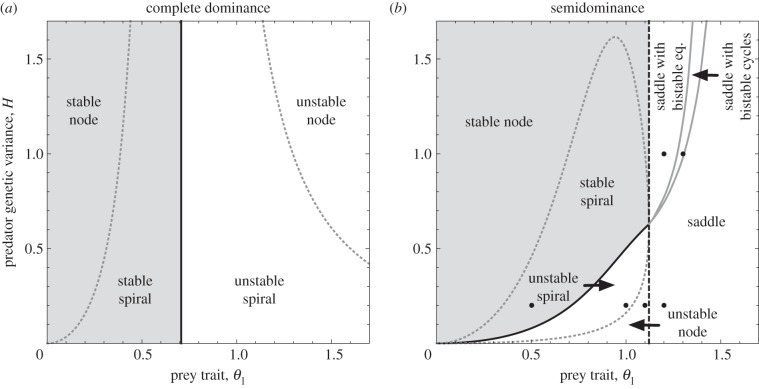
We found that the complete dominance model is stable when the prey trait θ1 is smaller than the threshold (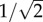 ), and is unstable otherwise, by local stability analyses (figure 1a; the mathematical analysis is in the electronic supplementary material, appendix S2). The speed of predator evolution, H, does not affect local stability and the prey trait distance is the sole determinant of stability. At the stable equilibrium, the predator trait is between the prey homozygotes (
), and is unstable otherwise, by local stability analyses (figure 1a; the mathematical analysis is in the electronic supplementary material, appendix S2). The speed of predator evolution, H, does not affect local stability and the prey trait distance is the sole determinant of stability. At the stable equilibrium, the predator trait is between the prey homozygotes (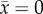 ), because the prey phenotypes are similar enough for predators to exploit both of them simultaneously.
), because the prey phenotypes are similar enough for predators to exploit both of them simultaneously.
Figure 1.
Phase diagrams for the internal equilibrium of the two-dimensional evolutionary system. Horizontal axis is the prey trait (θ1) and vertical axis is the predator genetic variance (H). (a) Complete dominance (θ1 = θ2 =–θ3) and (b) semidominance (θ1 =–θ3 and θ2 = 0). Light grey regions: the stable internal equilibrium. White regions: the unstable internal equilibrium. Solid black line/curve: Hopf bifurcations. Dashed black line: a saddle-node bifurcation. Solid grey curves: Hopf bifurcations of the two additional interior equilibria and the condition for two bistable cycles to merge obtained by numerical simulations. Dashed grey curves: boundaries between nodes and spirals (real and complex eigenvalues, respectively). Black points in figure 1b indicate parameter values for figures 2 and 3.
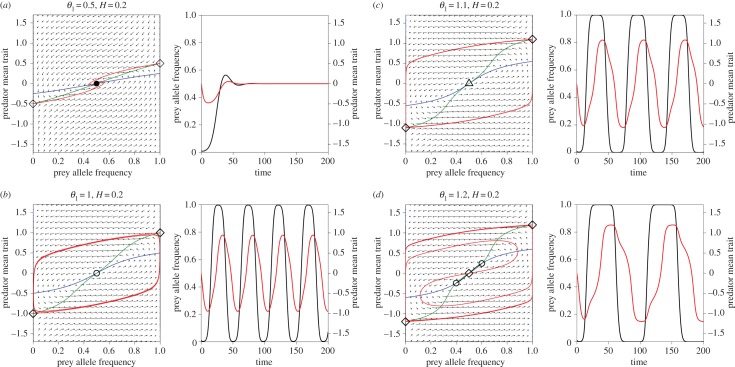
By contrast, the parameter region in which the semidominance model is stable decreases gradually as θ1 increases (figure 1b). Thus, stability results from simultaneously increasing predator evolution speed and decreasing the prey trait difference (figure 1b). When the prey trait difference is smaller than about 1.12, slowing predator evolution can create limit cycles through a Hopf bifurcation (a black curve in figure 1b). When the predator evolution speed is smaller than about 0.628, increasing the prey trait difference also results in a Hopf bifurcation. This is illustrated in figure 2, using parameter values shown in figure 1b (black points with H = 0.2). We used an R [52] implementation of the pplane program (math.rice.edu/∼dfield/dfpp.html) to compute and plot fixed points, nullclines, phase arrows, trajectories and local stable and unstable manifolds for saddle points (see the electronic supplementary material for our pplane R scripts). Increasing the prey trait distance first changes the equilibrium from a stable node to a stable focus (figure 2a). Then, a Hopf bifurcation occurs and the system shows a limit cycle (figure 2b). The cycle amplitude at first increases with the prey trait distance, but as prey allele frequency is bounded (0 ≤ p ≤ 1), trajectories spend longer periods of time near p = 0 and 1 (figure 2c,d). The dynamics in figure 2c,d look like heteroclinic (saddle-to-saddle) trajectories, but in fact they are more extreme versions of figure 2b: the unstable manifolds of the saddles on the boundary (red curves) quickly converge onto a limit cycle that passes very close to the saddles, approaching one saddle almost along its stable manifolds (the boundaries p = 0 and 1), then leaving almost along the unstable manifold and approaching the other saddle. This process repeats ad infinitum, so that the prey allele frequency has recurrent jumps between p = 0 and 1 (figure 2c,d).
Figure 2.
Bifurcation patterns with increasing value of the prey phenotypic difference (θ1) in the two-dimensional evolutionary model with semidominance when H = 0.2. Phase planes and trait dynamics trajectories are shown. (a) θ1 = 0.5, (b) θ1 = 1, (c) θ1 = 1.1 and (d) θ1 = 1.2. In the phase plane diagrams, horizontal axis is the prey allele frequency (p) and vertical axis is the predator mean trait (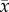 ). Black circle, stable spiral; white circle, unstable spiral; white triangle, unstable node; diamond, saddle. Blue curves, p nullclines; green curves,
). Black circle, stable spiral; white circle, unstable spiral; white triangle, unstable node; diamond, saddle. Blue curves, p nullclines; green curves, 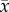 nullclines. Black lines/curves (sometimes overlapping with p = 0 and 1 lines): local stable manifolds for saddle points; red curves: local unstable manifolds for saddle points. In the trait dynamics plots, black curves are the prey allele frequency (p) and red curves are the predator mean trait (
nullclines. Black lines/curves (sometimes overlapping with p = 0 and 1 lines): local stable manifolds for saddle points; red curves: local unstable manifolds for saddle points. In the trait dynamics plots, black curves are the prey allele frequency (p) and red curves are the predator mean trait (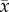 ).
).
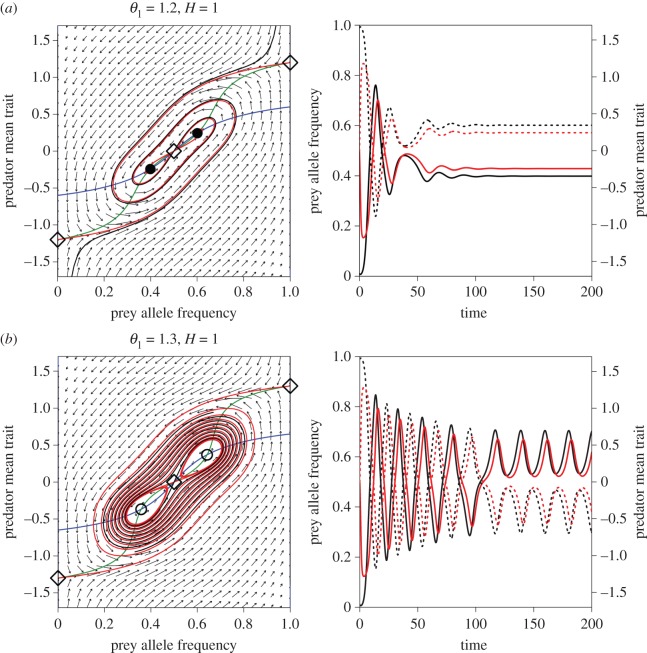
As the prey trait distance increases further, the unstable focus changes to an unstable node (figure 2c), and then (when an eigenvalue passes through zero) the node becomes a saddle and at the same time splits into three equilibria (figure 2d; electronic supplementary material, appendix S2). The two new equilibria (figure 2d) are initially unstable when H is small, but they are locally stable when H is large and predators evolve rapidly. In that case, increasing the prey trait difference results in a bifurcation to bistable equilibria (figure 3a) and then to bistable limit cycles (figure 3b; electronic supplementary material, figures S1 and S2: a dashed black line and grey curve in figure 1b). Further increase in the prey trait difference causes the two bistable cycles to grow and then merge.
Figure 3.
Bistability in the two-dimensional evolutionary model with semidominance when H = 1. (a) θ1 = 1.2, (b) θ1 = 1.3. Figure legends are the same as figure 2.
Bistability means that the outcome of coevolution depends on the initial condition (solid and dotted lines in figure 3). In the semidominance model (figure 1b), it is possible for rapid predator evolution to constrain predator trait dynamics between the heterozygote (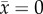 ) and either one of homozygotes (
) and either one of homozygotes (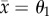 or –θ1) depending on the initial state, when predator evolution is rapid (H is large) and the prey trait difference is intermediate (θ1 is around 1.2), as follows. If the predator mean trait is near one of the homozygote phenotypes, heterozygotes increase in frequency in response to predation. Once heterozygotes are common enough, the predator trait evolves rapidly (owing to large H) towards the heterozygote phenotype (
or –θ1) depending on the initial state, when predator evolution is rapid (H is large) and the prey trait difference is intermediate (θ1 is around 1.2), as follows. If the predator mean trait is near one of the homozygote phenotypes, heterozygotes increase in frequency in response to predation. Once heterozygotes are common enough, the predator trait evolves rapidly (owing to large H) towards the heterozygote phenotype ( ); as a result, the predator phenotype is closer to zero than the prey mean phenotype, and this reverses the direction of prey evolution back towards the original homozygote (figure 3). When the prey trait difference is small, predators can exploit the three prey phenotypes simultaneously and the system is stabilized. When the prey trait difference is large, the homozygotes are so different from the heterozygote that it becomes difficult for the predator trait to get ahead of the prey phenotype. Thus, large H and intermediate θ1 are necessary for bistability to occur.
); as a result, the predator phenotype is closer to zero than the prey mean phenotype, and this reverses the direction of prey evolution back towards the original homozygote (figure 3). When the prey trait difference is small, predators can exploit the three prey phenotypes simultaneously and the system is stabilized. When the prey trait difference is large, the homozygotes are so different from the heterozygote that it becomes difficult for the predator trait to get ahead of the prey phenotype. Thus, large H and intermediate θ1 are necessary for bistability to occur.
(b). Eco-evolutionary dynamics
By local stability analyses (electronic supplementary material, appendix S3) and numerical simulations, we found that coevolutionary dynamics in the four-dimensional eco-evolutionary system shows stable equilibria, limit cycles, bistable equilibria, bistable cycles or deterministic predator extinction depending on the prey trait (θ1) and predator mortality (d) (figure 4). Limit cycles are a consequence of coevolution, because the ecological subsystem is always stable with prey logistic growth and predator linear functional response (electronic supplementary material, appendix S3). The bifurcations of the complete dominance model are not affected by the speed of predator evolution, V, but increasing V stabilizes the semidominance model (electronic supplementary material, figure S3).
Figure 4.
Phase diagrams of the four-dimensional eco-evolutionary system. Horizontal axis is the prey trait (θ1) and vertical axis is the predator mortality (d). (a) Complete dominance and (b) semidominance. Figure legends are the same as figure 1 except for dark grey regions: deterministic predator extinction. Other parameter values are V = 0.01 and α = 10. Black points in figure 4a,b indicate parameter values for figures 5 and 6, respectively.
Deterministic predator extinction can occur when the predator equilibrium density becomes negative at the internal equilibrium (‘extinction’ regions in figure 4). Here the prey phenotypes are so different that predators cannot exploit them to invade with the intermediate trait ( ) even when the prey density is at its equilibrium. In addition, coevolution can drive deterministic extinction with distinct prey phenotypes, as gradual predator evolution cannot keep up with jumping prey evolution (figure 5b: ‘extinction by coevolution’ regions in figure 4). This occurs because of the genetic asymmetry between prey and predator: while predator evolution is gradual, the prey trait distribution can shift from one extreme phenotype to the other by changing allele frequencies. Therefore, the complete dominance model (figure 4a) causes predator extinction more easily than the semidominance model (figure 4b) because the heterozygous phenotype works as a bridge between the two extreme phenotypes of homozygotes in the semidominance model.
) even when the prey density is at its equilibrium. In addition, coevolution can drive deterministic extinction with distinct prey phenotypes, as gradual predator evolution cannot keep up with jumping prey evolution (figure 5b: ‘extinction by coevolution’ regions in figure 4). This occurs because of the genetic asymmetry between prey and predator: while predator evolution is gradual, the prey trait distribution can shift from one extreme phenotype to the other by changing allele frequencies. Therefore, the complete dominance model (figure 4a) causes predator extinction more easily than the semidominance model (figure 4b) because the heterozygous phenotype works as a bridge between the two extreme phenotypes of homozygotes in the semidominance model.
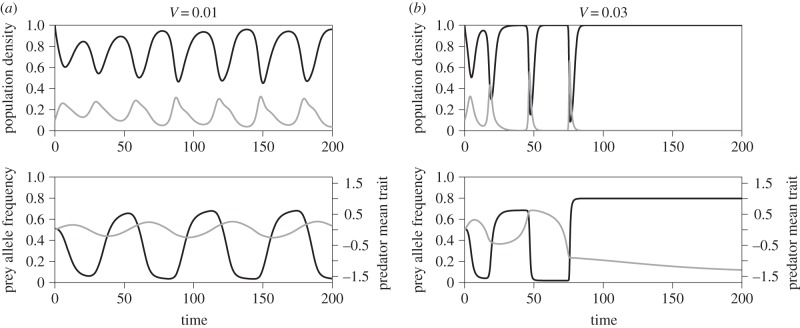
Figure 5.
Rapid coevolution can cause extinction with complete dominance. (a) V = 0.01, (b) V = 0.03. Black curves are prey and grey curves are predator. Other parameter values are θ1 = 1.4, d = 1 and α = 10.
Predator extinction depends on initial conditions as well as the predator trait heritability, 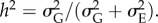 Increasing the heritability (and the speed of predator adaptation, H and V) tends to stabilize purely coevolutionary cycles (figure 1; electronic supplementary material, figure S3), but it can also result in deterministic predator extinction (figure 5). When the speed of evolution is slow, the predator stays in between the prey homozygotes and behaves as a generalist that exploits both homozygotes simultaneously (figure 5a). However, fast evolution can cause overreaction of the predator trait to specialize on one of the homozygotes. After the prey approaches one extreme (homozygote) phenotype, the predator phenotype follows and gets close enough to the homozygote, and then the other prey allele starts to become common. The predator continues to evolve in the direction of the mean prey phenotype until the mean prey phenotype (evolving now in the opposite direction) crosses the predator phenotype. As the prey phenotype escapes, predator density drops until the predator no longer affects prey evolution and prey allele frequency stops changing. At this point, the homozygote that was previously common is rare, but it still exists. The predator is then trapped in a local basin around the now-rare homozygote (an evolutionary trap, sensu [53]) and cannot return to the intermediate trait value (figure 5b). Because predator abundance is a continuous variable it never becomes exactly zero in our model, so the predator trait continues to evolve even when predator density is extremely low (figure 5b). Therefore, increasing the speed of predator evolution expands the ‘extinction by coevolution’ regions in the phase diagrams (figure 4; electronic supplementary material, figure S3).
Increasing the heritability (and the speed of predator adaptation, H and V) tends to stabilize purely coevolutionary cycles (figure 1; electronic supplementary material, figure S3), but it can also result in deterministic predator extinction (figure 5). When the speed of evolution is slow, the predator stays in between the prey homozygotes and behaves as a generalist that exploits both homozygotes simultaneously (figure 5a). However, fast evolution can cause overreaction of the predator trait to specialize on one of the homozygotes. After the prey approaches one extreme (homozygote) phenotype, the predator phenotype follows and gets close enough to the homozygote, and then the other prey allele starts to become common. The predator continues to evolve in the direction of the mean prey phenotype until the mean prey phenotype (evolving now in the opposite direction) crosses the predator phenotype. As the prey phenotype escapes, predator density drops until the predator no longer affects prey evolution and prey allele frequency stops changing. At this point, the homozygote that was previously common is rare, but it still exists. The predator is then trapped in a local basin around the now-rare homozygote (an evolutionary trap, sensu [53]) and cannot return to the intermediate trait value (figure 5b). Because predator abundance is a continuous variable it never becomes exactly zero in our model, so the predator trait continues to evolve even when predator density is extremely low (figure 5b). Therefore, increasing the speed of predator evolution expands the ‘extinction by coevolution’ regions in the phase diagrams (figure 4; electronic supplementary material, figure S3).
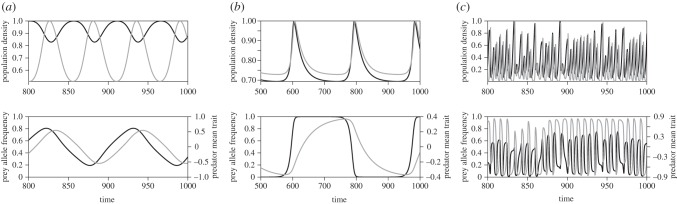
Adding population densities to the purely evolutionary model allows a greater range of dynamics to occur including anti-phase cycles (figure 6a), in-phase cycles (figure 6b) and chaotic dynamics (figure 6c). Anti-phase cycles are characterized by out-of-phase oscillations in predator and prey densities (i.e. predator maxima coinciding with prey minima and vice versa), and arise when the predator mortality and prey trait difference are large. In-phase cycles have a longer cycle period and occur when predator mortality and prey trait difference are both small (black points in figure 4b). Chaotic dynamics arises when the attack rate and heritability are large enough (electronic supplementary material, figure S4).
Figure 6.
Examples of anti-phase cycles (a) and in-phase cycles (b) with semidominance and chaotic cycles with complete dominance (c). Population dynamics are scaled so that the maximum value is one. Black curves are prey and grey curves are predator. (a) θ1 = 1 and d = 5.5, (b) θ1 = 0.4 and d = 1, and other parameter values are the same as figure 4 (V = 0.01 and α = 10). (c) θ1 = 0.9, d = 2, V = 0.3 and α = 20.
4. Discussion
(a). Comparing discrete–continuous with continuous–continuous coevolution
Our results show that genetic asymmetry is potentially important for understanding and predicting ecological and evolutionary dynamics. We explored the consequences of antagonistic coevolution between quantitative predator and Mendelian prey traits. As we did not include stabilizing selection towards an intermediate optima [14,45] or the Holling type II functional response, a continuous–continuous analogue of our model will result in either stable equilibrium or runaway escalation towards infinite trait values [15]. By contrast, we found a wide variety of possible dynamics in discrete–continuous coevolution, including coevolutionary cycles (figure 2), bistable equilibria and bistable cycles (figure 3) in the purely evolutionary system (figure 1). When we couple population and evolutionary dynamics, deterministic predator extinction (figure 5), anti-phase cycles, in-phase cycles and chaotic dynamics (figure 6) are also possible (figure 4). Therefore, continuous–discrete coevolution is more similar to discrete–discrete coevolution in terms of stability, as it can produce coevolutionary cycles without stabilizing selection. Furthermore, the rich dynamics from simple models demonstrate that traits' genetic architectures can determine the dynamics of coevolution [15,45,54].
(b). Semidominance versus complete dominance
We found that semidominance can promote trait cycling when the prey trait difference is small, stabilize dynamics when the prey trait difference is large (figure 1), and prevent predator extinction (figure 4) compared to the complete dominance model, as heterozygotes work as a bridge between the two extreme homozygotes. Interestingly, predators mostly exploit heterozygotes in stable coexistence, and heterozygotes are maintained by subsidy from homozygotes via sexual reproduction (phenotypic subsidy) [55]. As stable coexistence of three asexual genotypes sharing the same predator and resource is impossible [56], sexual reproduction is an important factor for coevolution with semidominance.
(c). Effects of increasing heritability
Increasing the predator trait heritability (h2) intuitively appears beneficial for the predator because it accelerates adaptive responses to prey phenotypic changes. Indeed, increasing heritability promotes population persistence under directional environmental change [8]. However, larger heritability can cause the predator trait to overreact to prey frequency change, and eventually results in predator extinction (figure 5). This phenomenon was previously discussed in the context of periodic abiotic environmental change [57]; evolution of prey traits has a similar effect on the predator in discrete–continuous coevolution.
Using a different parameter rescaling (electronic supplementary material, appendix S4), we can examine the effects of heritability and trait variance separately. In the complete dominance model, the system is always stable when the prey trait difference is small (electronic supplementary material, figure S5a), but large trait variance stabilizes the system otherwise (electronic supplementary material, figure S5b). In the semidominance model, on the other hand, both heritability and variance are necessary for the system to be stable (electronic supplementary material, figures S5c,d). This underlines that the components of trait variation matter, not just the total amount of trait variation [47,55].
(d). Complicated cycles
Anti-phase cycles were observed in predator–prey microcosm experiments with rapid prey evolution [7,58]. Our findings suggest that in-phase and chaotic cycles might also occur as a result of eco-evolutionary feedbacks in experimental studies. The possibility of in-phase cycles [59] and chaotic cycles [24,45] of (co)evolving predator–prey systems has been theoretically suggested, but our study illustrates another possible mechanism.
Our model's in-phase cycles (figure 6b) are similar to the canard cycles in models for prey evolution with multiple predators [60]. A canard is a trajectory that spends a long time near an unstable object. In the in-phase cycles, prey allele frequency spends a long time near each extreme (p = 0 or 1) after the direction of selection has shifted to favour the opposite extreme, because of low trait variance, and then suddenly moves to the other extreme. As a result, the lag between traits is larger than that in anti-phase cycles (figure 6).
(e). Comparison of empirical systems: snake–snail and cichlid systems
Our model is a simple representation of coevolution between quantitative and Mendelian traits, not a specific model for snake–snail coevolution mediated by handedness [36]. For example, snail coiling direction is determined by their maternal genotypes [42], and this delayed inheritance can stabilize coevolutionary cycles (electronic supplementary material, appendix S5, and figure S6). In addition, the reproductive incompatibility between snails with opposite coils [43] will affect coevolutionary dynamics, because rare genotypes have low fitness [42]; even with complete dominance, reproductive incompatibility causes bistability [42]. Therefore, the combination of delayed inheritance and reproductive incompatibility in land snails makes cyclic dynamics unlikely compared to our model.
Scale-eating cichlid fish and their prey are a classical example of negative frequency-dependent selection [37]. There are right- and left-handed scale eaters, with ‘handedness’ determined by one locus with two alleles [38]. Because prey fish behaviourally adapt to more frequent phenotypes [37] and learning can be described by a quantitative trait model [61], this may be regarded as coevolution between discrete predator and continuous prey traits. This reversed system also results in coevolutionary bistability with the reversed effects of parameters: smaller predator trait difference and larger prey evolution speed destabilize dynamics (electronic supplementary material, appendix S6 and figure S7). As behavioural dynamics are probably fast relative to evolution, the cichlid system may become unstable (electronic supplementary material, figure S7).
5. Conclusion
Quantitative genetics models for trait change along a fitness gradient are commonly used to model evolution, phenotypic plasticity and even learning by changing the time scale of adaptation [61]. On the other hand, recent studies showed that different mechanisms of rapid adaptation can produce distinct ecological dynamics. For example, phenotypic plasticity, rapid evolution and evolution of plasticity can cause distinct eco-evolutionary dynamics [62]. The number of loci that control ecologically important traits also affects ecological speciation [42], evolutionary rescue [63] and coevolution [14,28,45]. Our study suggests that different genetic architectures can result in distinct eco-evolutionary dynamics. Functional genomic approaches (e.g. [34,35]) and further studies on coevolution with different forms of genetic asymmetry such as ploidy [31], epigenetics [64] and recombination [65] may produce insights on contemporary eco-evolutionary dynamics with diverse genetic architectures.
Supplementary Material
Acknowledgements
We thank A. Sasaki, N. G. Hairston Jr and M. Hoso for discussion and valuable comments on the earlier manuscript. We also thank two anonymous reviewers and members of the Ellner lab for their helpful comments.
Data accessibility
Supplementary R scripts: Dryad http://dx.doi.org/10.5061/dryad.7jq44.
Authors' contributions
M.Y. conceived the study. M.Y. and S.P.E. constructed and analysed the models. M.Y. wrote the initial manuscript and S.P.E. substantially contributed to manuscript revisions.
Competing interests
We declare we have no competing interests.
Funding
M.Y. was supported by the Japan Society for the Promotion of Science (JSPS) Postdoctoral Fellowship for Research Abroad (24-869) and is supported by Hakubi Center for Advanced Research and John Mung Program of Kyoto University. S.P.E. was supported by National Science Foundation (NSF) DEB-1256719 and DEB-1353039.
References
- 1.Thompson JN. 2013. Relentless evolution. Chicago, IL: The University of Chicago Press. [Google Scholar]
- 2.Dawkins R, Krebs JR. 1979. Arms races between and within species. Proc. R. Soc. Lond. B 205, 489–511. ( 10.1098/rspb.1979.0081) [DOI] [PubMed] [Google Scholar]
- 3.Abrams PA. 2000. The evolution of predator–prey interactions: theory and evidence. Annu. Rev. Ecol. Syst. 31, 79–105. ( 10.1146/annurev.ecolsys.31.1.79) [DOI] [Google Scholar]
- 4.Van Valen L. 1973. A new evolutionary law. Evol. Theory 1, 1–30. [Google Scholar]
- 5.Brockhurst MA, Chapman T, King KC, Mank JE, Paterson S, Hurst GDD. 2014. Running with the Red Queen: the role of biotic conflicts in evolution. Proc. R. Soc. B 281, 20141382 ( 10.1098/rspb.2014.1382) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schoener TW. 2011. The newest synthesis: understanding the interplay of evolutionary and ecological dynamics. Science 331, 426–429. ( 10.1126/science.1193954) [DOI] [PubMed] [Google Scholar]
- 7.Ellner SP. 2013. Rapid evolution: from genes to communities, and back again? Funct. Ecol. 27, 1087–1099. ( 10.1111/1365-2435.12174) [DOI] [Google Scholar]
- 8.Gonzalez A, Ronce O, Ferriere R, Hochberg ME. 2013. Evolutionary rescue: an emerging focus at the intersection between ecology and evolution. Phil. Trans. R. Soc. B 368, 20120404 ( 10.1098/rstb.2012.0404) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calcagno V, Dubosclard M, de Mazancourt C. 2010. Rapid exploiter–victim coevolution: the race is not always to the swift. Am. Nat. 176, 198–211. ( 10.1086/653665) [DOI] [PubMed] [Google Scholar]
- 10.Mougi A, Iwasa Y. 2010. Evolution towards oscillation or stability in a predator–prey system. Proc. R. Soc. B 277, 3163–3171. ( 10.1098/rspb.2010.0691) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Northfield TD, Ives AR. 2013. Coevolution and the effects of climate change on interacting species. PLoS Biol. 11, e1001685 ( 10.1371/journal.pbio.1001685) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saloniemi I. 1993. A coevolutionary predator–prey model with quantitative characters. Am. Nat. 141, 880–896. ( 10.1086/285514) [DOI] [PubMed] [Google Scholar]
- 13.Abrams PA, Matsuda H. 1997. Fitness minimization and dynamic instability as a consequence of predator–prey coevolution. Evol. Ecol. 11, 1–20. ( 10.1023/a:1018445517101) [DOI] [Google Scholar]
- 14.Gavrilets S. 1997. Coevolutionary chase in exploiter–victim systems with polygenic characters. J. Theor. Biol. 186, 527–534. ( 10.1006/jtbi.1997.0426) [DOI] [PubMed] [Google Scholar]
- 15.Nuismer SL, Doebeli M, Browning D. 2005. The coevolutionary dynamics of antagonistic interactions mediated by quantitative traits with evolving variances. Evolution 59, 2073–2082. ( 10.1111/j.0014-3820.2005.tb00918.x) [DOI] [PubMed] [Google Scholar]
- 16.Tien RJ, Ellner SP. 2012. Variable cost of prey defense and coevolution in predator–prey systems. Ecol. Monogr. 82, 491–504. ( 10.1890/11-2168.1) [DOI] [Google Scholar]
- 17.Cortez MH, Weitz JS. 2014. Coevolution can reverse predator–prey cycles. Proc. Natl Acad. Sci. USA 111, 7486–7491. ( 10.1073/pnas.1317693111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marrow P, Law R, Cannings C. 1992. The coevolution of predator–prey interactions: ESSs and Red Queen dynamics. Proc. R. Soc. B 250, 133–141. ( 10.1098/rspb.1992.0141) [DOI] [Google Scholar]
- 19.Dieckmann U, Marrow P, Law R. 1995. Evolutionary cycling in predator–prey interactions: population dynamics and the Red Queen. J. Theor. Biol. 176, 91–102. ( 10.1006/jtbi.1995.0179) [DOI] [PubMed] [Google Scholar]
- 20.Van der Laan JD, Hogeweg P. 1995. Predator–prey coevolution: interactions across different timescales. Proc. R. Soc. B 259, 35–42. ( 10.1098/rspb.1995.0006) [DOI] [Google Scholar]
- 21.Weitz JS, Hartman H, Levin SA. 2005. Coevolutionary arms races between bacteria and bacteriophage. Proc. Natl Acad. Sci. USA 102, 9535–9540. ( 10.1073/pnas.0504062102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Best A, White A, Boots M. 2009. The implications of coevolutionary dynamics to host–parasite interactions. Am. Nat. 173, 779–791. ( 10.1086/598494) [DOI] [PubMed] [Google Scholar]
- 23.Carval D, Ferriere R. 2010. A unified model for the coevolution of resistance, tolerance, and virulence. Evolution 64, 2988–3009. ( 10.1111/j.1558-5646.2010.01035.x) [DOI] [PubMed] [Google Scholar]
- 24.Dercole F, Ferriere R, Rinaldi S. 2010. Chaotic Red Queen coevolution in three-species food chains. Proc. R. Soc. B 277, 2321–2330. ( 10.1098/rspb.2010.0209) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown JS, Vincent TL. 1992. Organization of predator–prey communities as an evolutionary game. Evolution 46, 1269–1283. ( 10.2307/2409936) [DOI] [PubMed] [Google Scholar]
- 26.Abrams PA. 2001. Modelling the adaptive dynamics of traits involved in inter- and intraspecific interactions: an assessment of three methods. Ecol. Lett. 4, 166–175. ( 10.1046/j.1461-0248.2001.00199.x) [DOI] [Google Scholar]
- 27.Levin SA, Udovic JD. 1977. A mathematical model of coevolving populations. Am. Nat. 111, 657–675. ( 10.1086/283198) [DOI] [Google Scholar]
- 28.Gavrilets S, Hastings A. 1998. Coevolutionary chase in two-species systems with applications to mimicry. J. Theor. Biol. 191, 415–427. ( 10.1006/jtbi.1997.0615) [DOI] [PubMed] [Google Scholar]
- 29.Waltman P, Braselton J, Braselton L. 2002. A mathematical model of a biological arms race with a dangerous prey. J. Theor. Biol. 218, 55–70. ( 10.1006/jtbi.2002.3057) [DOI] [PubMed] [Google Scholar]
- 30.Mostowy R, Engelstädter J. 2011. The impact of environmental change on host–parasite coevolutionary dynamics. Proc. R. Soc. B 278, 2283–2292. ( 10.1098/rspb.2010.2359) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Switkes JM, Moody ME. 2001. Coevolutionary interactions between a haploid species and a diploid species. J. Math. Biol. 42, 175–194. ( 10.1007/s002850100071) [DOI] [PubMed] [Google Scholar]
- 32.Seger J. 1992. Evolution of exploiter–victim relationships. In Natural enemies: the population biology of predators, parasites and diseases (ed. Crawley MJ.), pp. 3–25. Oxford, UK: Blackwell. [Google Scholar]
- 33.Seger J. 1988. Dynamics of some simple host–parasite models with more than two genotypes in each species. Phil. Trans. R. Soc. Lond. B 319, 541–555. ( 10.1098/rstb.1988.0064) [DOI] [PubMed] [Google Scholar]
- 34.Colosimo PF, et al. 2005. Widespread parallel evolution in sticklebacks by repeated fixation of Ectodysplasin alleles. Science 307, 1928–1933. ( 10.1126/science.1107239) [DOI] [PubMed] [Google Scholar]
- 35.Rosenblum EB, Römpler H, Schöneberg T, Hoekstra HE. 2010. Molecular and functional basis of phenotypic convergence in white lizards at White Sands. Proc. Natl Acad. Sci. USA 107, 2113–2117. ( 10.1073/pnas.0911042107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hoso M, Kameda Y, Wu SP, Asami T, Kato M, Hori M. 2010. A speciation gene for left–right reversal in snails results in anti-predator adaptation. Nat. Commun. 1, 133 ( 10.1038/ncomms1133) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hori M. 1993. Frequency-dependent natural selection in the handedness of scale-eating cichlid fish. Science 260, 216–219. ( 10.1126/science.260.5105.216) [DOI] [PubMed] [Google Scholar]
- 38.Hori M, Ochi H, Kohda M. 2007. Inheritance pattern of lateral dimorphism in two cichlids (a scale eater, Perissodus microlepis, and an herbivore, Neolamprologus moorii) in Lake Tanganyika. Zool. Sci. 24, 486–492. ( 10.2108/zsj.24.486) [DOI] [PubMed] [Google Scholar]
- 39.Hedrick PW, Ritland K. 2012. Population genetics of the white-phased ‘Spirit’ black bear of British Columbia. Evolution 66, 305–313. ( 10.1111/j.1558-5646.2011.01463.x) [DOI] [PubMed] [Google Scholar]
- 40.Bell G. 2010. Fluctuating selection: the perpetual renewal of adaptation in variable environments. Phil. Trans. R. Soc. B 365, 87–97. ( 10.1098/rstb.2009.0150) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoso M, Asami T, Hori M. 2007. Right-handed snakes: convergent evolution of asymmetry for functional specialization. Biol. Lett. 3, 169–172. ( 10.1098/rsbl.2006.0600) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamichi M, Sasaki A. 2013. Single-gene speciation with pleiotropy: effects of allele dominance, population size, and delayed inheritance. Evolution 67, 2011–2023. ( 10.1111/evo.12068) [DOI] [PubMed] [Google Scholar]
- 43.Ueshima R, Asami T. 2003. Single-gene speciation by left-right reversal: a land-snail species of polyphyletic origin results from chirality constraints on mating. Nature 425, 679 ( 10.1038/425679a) [DOI] [PubMed] [Google Scholar]
- 44.Sasaki A, Godfray HCJ. 1999. A model for the coevolution of resistance and virulence in coupled host–parasitoid interactions. Proc. R. Soc. Lond. B 266, 455–463. ( 10.1098/rspb.1999.0659) [DOI] [Google Scholar]
- 45.Kopp M, Gavrilets S. 2006. Multilocus genetics and the coevolution of quantitative traits. Evolution 60, 1321–1336. ( 10.1111/j.0014-3820.2006.tb01212.x) [DOI] [PubMed] [Google Scholar]
- 46.Khibnik AI, Kondrashov AS. 1997. Three mechanisms of Red Queen dynamics. Proc. R. Soc. Lond. B 264, 1049–1056. ( 10.1098/rspb.1997.0145) [DOI] [Google Scholar]
- 47.Schreiber SJ, Bürger R, Bolnick DI. 2011. The community effects of phenotypic and genetic variation within a predator population. Ecology 92, 1582–1593. ( 10.1890/10-2071.1) [DOI] [PubMed] [Google Scholar]
- 48.Otto SP, Day T. 2007. A biologist's guide to mathematical modeling in ecology and evolution. Princeton, NJ: Princeton University Press. [Google Scholar]
- 49.Lande R. 1976. Natural selection and random genetic drift in phenotypic evolution. Evolution 30, 314–334. ( 10.2307/2407703) [DOI] [PubMed] [Google Scholar]
- 50.Turelli M, Barton NH. 1994. Genetic and statistical analyses of strong selection on polygenic traits: what, me normal? Genetics 138, 913–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murdoch WW, Briggs CJ, Nisbet RM. 2003. Consumer–resource dynamics. Princeton, NJ: Princeton University Press. [Google Scholar]
- 52.R Development Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 53.Ferrière R, Dieckmann U, Couvet D. 2004. Evolutionary conservation biology. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 54.Doebeli M. 1996. Quantitative genetics and population dynamics. Evolution 50, 532–546. ( 10.2307/2410829) [DOI] [PubMed] [Google Scholar]
- 55.Bolnick, et al. 2011. Why intraspecific trait variation matters in community ecology. Trends Ecol. Evol. 26, 183–192. ( 10.1016/j.tree.2011.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jones LE, Becks L, Ellner SP, Hairston Jr NG, Yoshida T, Fussmann GF. 2009. Rapid contemporary evolution and clonal food web dynamics. Phil. Trans. R. Soc. B 364, 1579–1591. ( 10.1098/rstb.2009.0004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bürger R, Krall C. 2004. Quantitative-genetic models and changing environments. In Evolutionary conservation biology (eds Ferrière R, Dieckmann U, Couvet D), pp. 171–187. Cambridge, UK: Cambridge University Press. [Google Scholar]
- 58.Yoshida T, Jones LE, Ellner SP, Fussmann GF, Hairston NG Jr. 2003. Rapid evolution drives ecological dynamics in a predator–prey system. Nature 424, 303–306. ( 10.1038/nature01767) [DOI] [PubMed] [Google Scholar]
- 59.Cortez MH, Ellner SP. 2010. Understanding rapid evolution in predator–prey interactions using the theory of fast–slow dynamical systems. Am. Nat. 176, E109–E127. ( 10.1086/656485) [DOI] [PubMed] [Google Scholar]
- 60.Hiltunen T, Ellner SP, Hooker G, Jones LE, Hairston Jr NG. 2014. Eco-evolutionary dynamics in a three-species food web with intraguild predation: intriguingly complex. Adv. Ecol. Res. 50, 41–73. ( 10.1016/B978-0-12-801374-8.00002-5) [DOI] [Google Scholar]
- 61.Abrams PA, Matsuda H, Harada Y. 1993. Evolutionarily unstable fitness maxima and stable fitness minima of continuous traits. Evol. Ecol. 7, 465–487. ( 10.1007/BF01237642) [DOI] [Google Scholar]
- 62.Yamamichi M, Yoshida T, Sasaki A. 2011. Comparing the effects of rapid evolution and phenotypic plasticity on predator–prey dynamics. Am. Nat. 178, 287–304. ( 10.1086/661241) [DOI] [PubMed] [Google Scholar]
- 63.Orr HA, Unckless RL. 2008. Population extinction and the genetics of adaptation. Am. Nat. 172, 160–169. ( 10.1086/589460) [DOI] [PubMed] [Google Scholar]
- 64.Mostowy R, Engelstädter J, Salathé M. 2012. Non-genetic inheritance and the patterns of antagonistic coevolution. BMC Evol. Biol. 12, 93 ( 10.1186/1471-2148-12-93) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bell G, Maynard Smith J. 1987. Short-term selection for recombination among mutually antagonistic species. Nature 328, 66–68. ( 10.1038/328066a0) [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Supplementary R scripts: Dryad http://dx.doi.org/10.5061/dryad.7jq44.