Abstract
Evolutionary biologists since Darwin have hypothesized that closely related species compete more intensely and are therefore less likely to coexist. However, recent theory posits that species diverge in two ways: either through the evolution of ‘stabilizing differences’ that promote coexistence by causing individuals to compete more strongly with conspecifics than individuals of other species, or through the evolution of ‘fitness differences’ that cause species to differ in competitive ability and lead to exclusion of the weaker competitor. We tested macroevolutionary patterns of divergence by competing pairs of annual plant species that differ in their phylogenetic relationships, and in whether they have historically occurred in the same region or different regions (sympatric versus allopatric occurrence). For sympatrically occurring species pairs, stabilizing differences rapidly increased with phylogenetic distance. However, fitness differences also increased with phylogenetic distance, resulting in coexistence outcomes that were unpredictable based on phylogenetic relationships. For allopatric species, stabilizing differences showed no trend with phylogenetic distance, whereas fitness differences increased, causing coexistence to become less likely among distant relatives. Our results illustrate the role of species' historical interactions in shaping how phylogenetic relationships structure competitive dynamics, and offer an explanation for the evolution of invasion potential of non-native species.
Keywords: biogeographic history, coexistence, environmental heterogeneity, phylogenetic relatedness, plant competition, species invasions
1. Introduction
The diversity of ecological interactions on the Earth is the product of approximately 3.5 billion years of evolution, with ongoing extinctions matched by the continual divergence of populations and species. Signatures of this past evolution frequently emerge in the strength of the interactions among current-day species [1] in ways that have potential to further perpetuate divergence and the evolution of interaction strengths [2]. This dynamic feedback between the ecology and evolution of organisms is a central theme in microevolutionary [3,4], macroevolutionary [5] and recent ecological perspectives [6–8], as it promises a more complete picture of the processes that generate and maintain biological diversity.
A long-standing hypothesis in evolutionary biology states that closely related species are more ecologically similar, and that this similarity leads to intense competition and ultimately exclusion [7,9–11]. Despite abundant evidence that closely related species tend to be similar in terms of functional traits and resource requirements [12], the effect of evolutionary relatedness on the outcome of competition tends to be weak or absent in experimental [13] and observational [14,15] tests. This might occur for two reasons: first, contemporary ecological theory [16,17] suggests that species may evolve two types of differences, ‘stabilizing differences’ and ‘fitness differences’, that have opposing effects on competitive outcomes. Stabilizing differences, also known as ‘niche differences’ [17], promote coexistence by causing negative-frequency dependence in interacting species, and act to stabilize diversity by preventing any one species from dominating the community. Fitness differences, by contrast, are inequalities in mean fitness, including intrinsic demographic rates and competitive abilities, which preclude coexistence by favouring a single dominant species [16,17]. Because character displacement may cause species to diverge in both stabilizing differences and fitness differences, the relationship between coexistence and evolutionary time depends on their relative evolutionary trajectories [17–19].
The second reason for inconsistent effects of evolutionary relationships on coexistence and diversity might stem from the high occurrence of non-native species in many contemporary communities. Research on coevolution and adaptive radiations predicts that the divergence of related species depends on a history of competitive interactions, and therefore on whether species have historically occurred in sympatry or in allopatry [20–22]. By occurring in sympatry, we refer to the idea that species have had ample opportunity to interact and potentially influence evolutionary trajectories, rather than a specific mode of speciation. For the vast majority of taxa, evolutionary histories are not sufficiently documented to know when species historically occurred in sympatry or allopatry. However, information on species' native status in a region and current-day distributional overlap can be used to identify species with a history of potential interaction; interactions between pairs of species native to either the same or different regions can therefore be contrasted as a proxy for the influence of coevolutionary history.
We grew 30 Mediterranean annual plant species both alone and in two-species competition to estimate stabilizing and fitness differences [23] in two commonly encountered environments (wet and dry), and used phylogenetic relationships among species pairs to test whether the evolutionary trajectories of stabilizing differences, fitness differences and coexistence depend on coevolutionary history. We predicted that stabilizing differences would increase rapidly and predictably with phylogenetic distance among species pairs that have occurred in sympatry, whereas fitness differences would be constrained (e.g. red queen hypothesis [24] and competitive disarmament [25]). By contrast, we predict a weak or absent relationship between stabilizing differences and phylogenetic distance for allopatric species, due to the lack of coevolutionary history, and no constraints on fitness differences. These predictions would correspond to positive and negative relationships between coexistence and phylogenetic distance, depending on whether species pairs originated from the same or different biogeographic region, respectively.
2. Material and methods
(a). Species selection
Species were selected to meet two criteria: first, species of the same biogeographic origin must have had common affinities for annual grasslands and overlapping geographical distributions across their native ranges, as determined by CalFlora (http://www.calflora.org; California) and Euro + Med (http://www.emplantbase.org; Spain) plant databases, to represent a realistic subset of species that could have potentially interacted over evolutionary time. There were no criteria necessitating the overlap or non-overlap of species of different biogeographic origins in that the species pool was not designed to discriminate species based on invasion history or lack thereof. In total, 30 species were included, 20 of which were native to the Central Valley in California and 10 of which were native to the Mediterranean Basin region of southern Spain (electronic supplementary material, table S1). These regions were selected from among all Mediterranean-climate regions because of their high similarity in climate, especially rainfall [26]. Second, species must have an annual life cycle to estimate lifetime seed production in a single growing season. We constructed a phylogenetic tree using Bayesian methods and the ITS1/5.8S/ITS2 and rbcL sequence regions, as detailed in the electronic supplementary material, Supplementary methods and figure S1. Seeds were acquired from independent donors and commercial suppliers [26], and were tested for per cent germinability under similar soil conditions prior to experimentation. We chose not to cold stratify the seeds because this method is known to induce rather than break dormancy in Mediterranean annuals [27].
(b). Greenhouse growing conditions
In January 2012, seeds were sown into 12.7 cm diameter, 23 cm deep treepots filled with a 3 : 2 mixture of sand and screened topsoil, to mimic the sandy loam soils found in annual grasslands. The topsoil was collected locally (Villacci's Garden Depot; Scarborough, Ontario, Canada) to ensure that species were equally naive to the soil microbiome regardless of origin, a requirement for unbiased biogeographic comparisons in a common garden experiment. Each pot was randomly assigned to a position on a bench in the rooftop greenhouse at the University of Toronto (Toronto, Canada). The greenhouse was maintained at day/night temperatures of 14/7°C, which was set to gradually increase to 29/17°C on average by the end of the experiment. High intensity discharge lighting was provided to maintain a 12 h photoperiod. Each pot was watered daily to saturation using a drip irrigation system during a three-week establishment period.
After the establishment period, pots of plants were randomly assigned to either a wet or dry soil moisture regime. Using a drip irrigation system, pots in the wet treatment received 175 ml water twice as often as those in the dry treatment, starting at 1- and 2-day intervals which were extended to 7 and 14 days as the growing season progressed. These two soil moisture regimes were selected to simulate realistic among-year differences in precipitation, or equally, the 30-year average differences between mesic (662 mm) and dry (312 mm) sites across the species ranges (http://www.climate-charts.com). We confirmed that the wet (11.1 ± 0.56%; mean ± s.e. per cent soil moisture content) and dry (5.8 ± 0.42%) treatments were effective using a volumetric water probe (HydroSense™, Campbell Scientific Australia) on empty pots. Prior to flowering (approx. 60 days after planting), each pot was provided with 350 ml of 1500 ppm 20–20–20 NPK fertilizer (Plant Products, Inc., Brampton, Ontario, Canada). Pollination was provided by commercial colonies of the generalist pollinator Bombus impatiens, which were active throughout the flowering period (Biobest Canada, Leamington, Ontario, Canada). The experiment lasted 220 days owing to our inclusion of some summer annual species with relatively long life cycles (Atriplex patula, Chenopodium berlandieri, Crepis capillaris and Madia elegans).
(c). Experimental design
The experiment was designed to parametrize a series of annual plant models [28]. Here we present the model that best fit our data (based on AICc, electronic supplementary material, table S2), the Beverton–Holt model,
| 2.1 |
where Nit and Njt are the numbers of viable seeds of focal species i and j initially planted, λi is the finite rate of increase for species i in the absence of competition, αij and αii are the inter- and intra-specific competition coefficients, respectively, and Nit+1 is the number of viable seeds of focal species i in year t + 1. Because this equation is symmetric, Njt+1 can also be calculated by switching subscripts i and j. To independently estimate λ, we grew each species alone at low densities and enumerated the number of seeds produced per individual; other models (electronic supplementary material, table S2) require a similar parametrization for finite rate of increase in low competition. Specifically, 30 seeds of each species were sown into the pots, and emerged seedlings were thinned to approximately eight maximally spaced individuals that were allowed to mature and produce seed. There were seven replicate pots of this low-density treatment, and these replicates were used to calculate the distribution of log-transformed λ for each combination of species × soil moisture environment (details in the electronic supplementary material) to be used as informative priors in our Bayesian analysis (described below).
To estimate competition coefficients (αii, αjj, αij and αji), we grew species in pairwise competition in pots at an expected density of 70 plants (based on germination rates from pilot experiments). This density is comparable with the seeding density found in annual grasslands (2500 to 5500 plants m−2 [29,30]). Within these pots, we varied the numbers of individuals of the two species (10 : 60, 20 : 50, 30 : 40, 40 : 30, 50 : 20 and 60 : 10 individuals) to create six relative frequency ratios, and had two replicates per frequency ratio. The presence of strong negative-frequency dependence would indicate that stabilizing differences are large between competing species [31].
We used Bayesian modelling (JAGS software v. 3-15, implemented in the ‘rjags' R package) to fit annual plant models to our data, using uninformative priors for all fitted parameter estimates, except log(λ) for which we used the distributions calculated earlier. Similar to previous work [23], all competition coefficients (α parameters in equation (2.1)) were lognormally distributed, and other parameters (such as b in models 4 and 6, electronic supplementary material, table S2) were uniformly distributed but constrained to be positive. We ran four independent Markov chains for 100 000 iterations, with a 10 000 iteration burn-in period based on time to convergence; for each parameter, the mean of the posterior distribution was used as the best estimate. We then compared AICc values for the various models by calculating likelihoods for each model using parameter means. Once we selected the best model (the Beverton–Holt model, as has been commonly fit in other annual plant studies [23,28]), the estimated parameters were used to calculate stabilizing and fitness differences using electronic supplementary material, equations S1 and S2, which are further described below. The entire experiment was replicated in two soil moisture environments (wet and dry, see Greenhouse growing conditions), and included 900 total pots of plants arranged in a completely randomized design.
We did not compete all possible pairs of the 30 species included in the experiment. Rather, 10 sympatric (both species native to California) and 10 allopatric (one species native to California, the other to Spain) pairs were selected to represent competitive pairs that were phylogenetically independent (i.e. non-overlapping branch lengths) relative to all other pairs of the same biogeographic history treatment. Additionally, the same 10 Californian focal species were used in both the sympatric and allopatric pairs, competed against 10 other unique species from California or Spain (e.g. Vulpia microstachys versus V. octoflora (sympatric) or V. myuros (allopatric)); this was accounted for in the model fitting, with the parameter estimates of all three species being fit simultaneously (electronic supplementary material, table S3).
(d). Solving for stabilizing differences, fitness differences and coexistence outcomes
Stabilizing (1 − ρ; electronic supplementary material, equation S1) and fitness (κj/κi; electronic supplementary material, equation S2) differences were estimated according to Godoy & Levine [23] by rearranging the parameters from the Beverton–Holt annual plant model described in equation (2.1). When the strength of intra- and inter-specific competition is the same, then 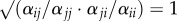 and 1 − ρ = 0, indicating that there are no stabilizing differences between competitors. As the relative strength of intra- to inter-specific competition increases
and 1 − ρ = 0, indicating that there are no stabilizing differences between competitors. As the relative strength of intra- to inter-specific competition increases 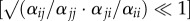 , 1 − ρ approaches 1 indicating that stabilizing differences are large. In five of 40 cases (20 species pairs × two soil moisture environments), 1 − ρ was less than 0, meaning that these species pairs showed evidence of destabilizing effects that are indicative of priority-based competitive outcomes. For simplicity, we followed the convention of setting these values to 1 (complete niche overlap [23]), as this allows comparison of coexistence outcomes. κj/κi is the average fitness difference between species i and j, calculated as the product of the demographic ratio (
, 1 − ρ approaches 1 indicating that stabilizing differences are large. In five of 40 cases (20 species pairs × two soil moisture environments), 1 − ρ was less than 0, meaning that these species pairs showed evidence of destabilizing effects that are indicative of priority-based competitive outcomes. For simplicity, we followed the convention of setting these values to 1 (complete niche overlap [23]), as this allows comparison of coexistence outcomes. κj/κi is the average fitness difference between species i and j, calculated as the product of the demographic ratio (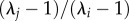 ), and the competitive response ratio (
), and the competitive response ratio (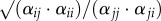 ). Our estimate of fitness differences differs from previous work [23] in that we use K, the larger of κj/κi and κi/κj (electronic supplementary material, equation S3). This was done to simplify the presentation of our results, but does not alter their outcome or interpretation. Mathematical derivations of these equations can be found in the appendices of Godoy & Levine [23].
). Our estimate of fitness differences differs from previous work [23] in that we use K, the larger of κj/κi and κi/κj (electronic supplementary material, equation S3). This was done to simplify the presentation of our results, but does not alter their outcome or interpretation. Mathematical derivations of these equations can be found in the appendices of Godoy & Levine [23].
Species are predicted to coexist locally if both can invade when rare and the other species is at its equilibrium density, a criterion that is met when [16,23]:
| 2.2 |
In the main text, we use the logarithm of the left-hand side of equation (2.2) as our coexistence metric, so that values greater than zero indicate coexistence, whereas those less than zero indicate competitive exclusion. It is important to note that we consider a model of coexistence in which all ungerminated seeds are considered inviable, thereby ignoring the contribution that ungerminated seeds may make to fitness [23] or to inter-annual stabilization through the storage effect [32]. We tested the sensitivity of our results against two alternative models, where the seed bank was set to either realistic species-specific rates (electronic supplementary material, table S4) or 100% viability to determine K, as used previously [23]; our results are qualitatively similar across all three models (electronic supplementary material, figure S2). The species-specific rates for the first alternative model were estimated via a separate germination trial using Petri dishes and gibberellic acid application [26].
(e). Statistical analyses
We analysed stabilizing differences, fitness differences and the coexistence metric using linear mixed effects (LME) models with the R packages ‘lmerTest’ and ‘nlme’. All response variables were tested against a model with phylogenetic distance, biogeographic history, soil moisture treatment and their interactions as fixed effects, and species pair as the random effect since the same pairs of species were grown in each soil moisture treatment. To meet model assumptions (linearity and homoscedasticity of errors), stabilizing differences were logit-transformed and tested with the ‘lmer’ function, whereas fitness differences and the coexistence metric were both log-transformed and coded to include heteroscedastic variance structure (‘weights’ argument in the ‘lme’ function). The results of the LMEs were summarized using function ‘Anova’ (type II analysis of variance) in the ‘car’ package; we report the significant highest-order interactions only in the main text, and the full model outputs in electronic supplementary material, tables S5 and S6. Reported p-values are calculated from χ2-tests of maximum-likelihood ratios.
3. Results and discussion
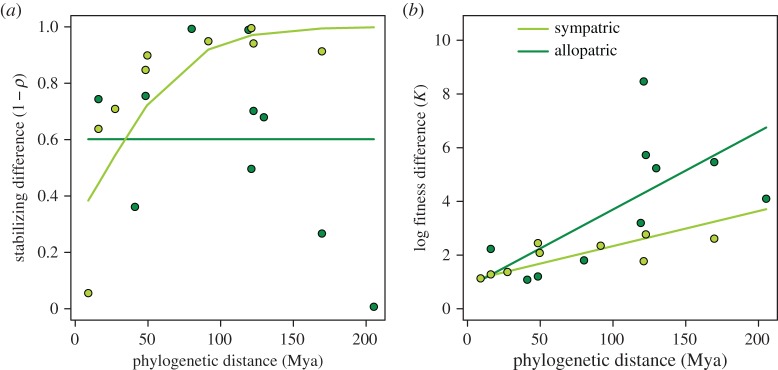
Consistent with evolutionary theory, our results support the hypothesis that the macroevolutionary trajectories of stabilizing differences are mediated by biogeographic history (phylogenetic distance by biogeographic history interaction 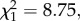 p = 0.003). Species that have had potential to evolve in sympatry rapidly accumulated stabilizing differences with phylogenetic distance, resulting in nearly complete ecological separation by 90 Myr (figure 1a, light shade). This was not observed in species pairs that have been evolving in allopatry (i.e. in California versus Spain); stabilizing differences showed no relationship with phylogenetic distance (figure 1a, dark shade). These lines of phylogenetic evidence are consistent with theory that competition imposes selective pressure for divergence in resource use among sympatrically evolving species [22]. A relationship would not be expected between allopatric species pairs because they are evolutionarily naive to each other; species native to separate regions have experienced distinct coevolutionary trajectories, such that stabilizing differences in the native range would not be predictive of those in the introduced range.
p = 0.003). Species that have had potential to evolve in sympatry rapidly accumulated stabilizing differences with phylogenetic distance, resulting in nearly complete ecological separation by 90 Myr (figure 1a, light shade). This was not observed in species pairs that have been evolving in allopatry (i.e. in California versus Spain); stabilizing differences showed no relationship with phylogenetic distance (figure 1a, dark shade). These lines of phylogenetic evidence are consistent with theory that competition imposes selective pressure for divergence in resource use among sympatrically evolving species [22]. A relationship would not be expected between allopatric species pairs because they are evolutionarily naive to each other; species native to separate regions have experienced distinct coevolutionary trajectories, such that stabilizing differences in the native range would not be predictive of those in the introduced range.
Figure 1.
Biogeographic history alters the evolutionary trajectory of stabilizing and fitness differences. (a) Stabilizing differences rapidly increase among sympatric species pairs (light shade), whereas allopatric species pairs (dark shade) show no relationship. (b) Fitness differences, by contrast, increase over evolutionary time in both sympatric and allopatric pairs, but are larger on average among allopatric pairs. Stabilizing differences have a maximum of one (electronic supplementary material, equation S1; lines are fitted from the logit-transformed data), whereas fitness differences have no upper limit (electronic supplementary material, equation S2). Because soil moisture had no effect on stabilizing or fitness differences, each point is a fitted average across soil moisture environments for each species pair. (Online version in colour.)
Our results are striking and appear to contradict the few existing experimental studies that test whether stabilizing differences are explained by phylogenetic relatedness, and find no relationship [18,19]. This apparent contradiction might be explained by two methodological differences between our study and those that precede it. First, our study is the first to our knowledge to incorporate information on historical species distributions. In rerunning our analysis without accounting for differences in biogeographic history, we find that phylogenetic distance fails to explain variation in stabilizing differences (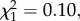 p = 0.756), a result more in line with previous studies. Second, our competitive pairs were selected to be phylogenetically independent (see Material and methods). This approach is necessary because any time there is overlap in evolutionary history among contrasted pairs, the number of comparisons made will be greater than the number of independent observations from a phylogenetic perspective. This limits the inferential power of many observational and experimental tests [33].
p = 0.756), a result more in line with previous studies. Second, our competitive pairs were selected to be phylogenetically independent (see Material and methods). This approach is necessary because any time there is overlap in evolutionary history among contrasted pairs, the number of comparisons made will be greater than the number of independent observations from a phylogenetic perspective. This limits the inferential power of many observational and experimental tests [33].
Similar to stabilizing differences, we found that the pattern of past evolution of fitness differences depended on biogeographic history. Specifically, fitness differences increased as an accelerating function of phylogenetic distance (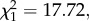 p < 0.001) and were greater overall among allopatric species pairs (
p < 0.001) and were greater overall among allopatric species pairs (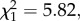 p = 0.016; figure 1b). In other words, just as coevolutionary dynamics lead to a greater probability of stabilizing coexistence, they appear to constrain the degree of fitness differences that lead to competitive exclusion across the entire phylogeny.
p = 0.016; figure 1b). In other words, just as coevolutionary dynamics lead to a greater probability of stabilizing coexistence, they appear to constrain the degree of fitness differences that lead to competitive exclusion across the entire phylogeny.
The effect of biogeographic history on fitness differences could arise from factors other than coevolutionary dynamics if, for example, the growing conditions in our experiment were more similar to the ambient environment in either California or Spain. If this were the case, we would expect absolute fitness differences (κj/κi) to be biased towards species from a particular region. Further investigation suggests that this is not the case; a post hoc test showed no consistent fitness advantage for a particular region (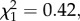 p = 0.515). It appears that the effect of biogeographic history we observe reflects differences in historical interactions, rather than experimental conditions favouring species from one region.
p = 0.515). It appears that the effect of biogeographic history we observe reflects differences in historical interactions, rather than experimental conditions favouring species from one region.
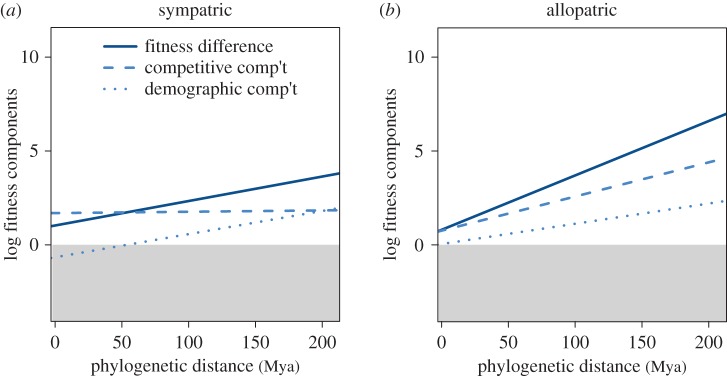
Fitness differences arise from the combined effects of differences in demographic rates and differences in competitive abilities, both of which have been hypothesized to lead to competitive exclusion by invasive species [34]. Despite reports that many invasive species differ in demographic characteristics, such as seed production [35], we found that allopatric species pairs significantly diverged in competitive ability but the trend in demographic rates was not significant (figure 2b, dashed versus dotted line; electronic supplementary material, table S6). By contrast, sympatric species pairs significantly diverged in demographic rates but not competitive ability (figure 2a, dotted versus dashed line; electronic supplementary material, table S6), with divergence in overall fitness differences matching divergence in demographic rates. In other words, species that have evolved in sympatry or allopatry both diverge in fitness differences over evolutionary time, but the fitness component responsible for this divergence is distinct.
Figure 2.
Fitness differences arise through alternate mechanisms in sympatric and allopatric species pairs. In log-space, fitness differences (solid line) are the sum of competitive (dashed line) and demographic (dotted line) components (see Material and methods), shown here as fitted relationships. (a) In sympatric species pairs, fitness differences are minimal relative to allopatric species pairs, and appear to accumulate over macroevolutionary time primarily through differences in demographic rates. (b) In allopatric species pairs, the more rapid increase in fitness differences over evolutionary time is driven solely by divergence in competitive ability, as the apparent divergence in demographic rates is non-significant. Values in the shaded area indicate that the species with the highest fitness had the lowest fitness component. (Online version in colour.)
The relationship between species coexistence and phylogenetic distance ultimately depends on biogeographic history (phylogenetic distance and biogeographic history interaction 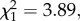 p = 0.049), due to differing evolutionary trajectories of stabilizing and fitness differences. For species pairs that occur in allopatry, coexistence is most likely to occur between close relatives, and becomes increasingly unlikely over macroevolutionary time (figure 3, dark shade). In sympatric species pairs, by contrast, the past evolution of stabilizing and fitness differences has counteractive effects on coexistence that are nearly perfectly matched, resulting in local coexistence outcomes that are random with respect to phylogeny (figure 3, light shade; slope = −0.02 ± 0.01 s.e.). Interestingly, our coexistence metric was less variable among sympatric than among allopatric species pairs, and generally straddled the threshold between coexistence and exclusion (figure 3, light shade); in three cases, the soil moisture treatment alone was enough to cause sympatric species pairs to cross this threshold so that species were predicted to coexist in at least one environment. Our results highlight the importance of environmental variation in maintaining species diversity among native species, but suggest that this variation may be less successful in maintaining diversity among species from different regions.
p = 0.049), due to differing evolutionary trajectories of stabilizing and fitness differences. For species pairs that occur in allopatry, coexistence is most likely to occur between close relatives, and becomes increasingly unlikely over macroevolutionary time (figure 3, dark shade). In sympatric species pairs, by contrast, the past evolution of stabilizing and fitness differences has counteractive effects on coexistence that are nearly perfectly matched, resulting in local coexistence outcomes that are random with respect to phylogeny (figure 3, light shade; slope = −0.02 ± 0.01 s.e.). Interestingly, our coexistence metric was less variable among sympatric than among allopatric species pairs, and generally straddled the threshold between coexistence and exclusion (figure 3, light shade); in three cases, the soil moisture treatment alone was enough to cause sympatric species pairs to cross this threshold so that species were predicted to coexist in at least one environment. Our results highlight the importance of environmental variation in maintaining species diversity among native species, but suggest that this variation may be less successful in maintaining diversity among species from different regions.
Figure 3.

The effect of evolutionary history on coexistence outcomes depends on biogeographic history. Species coexistence is not influenced by the phylogenetic distances of species pairs that occur in sympatry (light shade, slope not significantly different from zero), but the probability of coexistence decreases with the phylogenetic distance of allopatric pairs (dark shade). Species pairs were grown in wet (squares) and dry (triangles) environments, but soil moisture does not alter the effect of evolutionary history on coexistence (fitted lines are averaged across environments). The dashed line indicates the threshold between coexistence (positive values) and competitive exclusion (negative values); the coexistence metric is given by equation (2.2) in Material and methods. Points connected by a solid line represent cases in which a species pair was predicted to coexist in one but not both environments (3 of 10 sympatric pairs, 0 of 10 allopatric pairs). (Online version in colour.)
The use of two sets of environmental conditions in deconstructing the components of species' competitive dynamics is a strength of our experiment, and is the first to do so to our knowledge; the sensitivity of these components to the underlying environment is heretofore unknown [36]. We found that the soil moisture environment did not affect stabilizing differences, fitness differences or coexistence (all p > 0.15), and did not influence how these variables responded to biogeographic history or phylogenetic distance. The presence of an effect would have indicated that certain environments cause species to overlap more or less in resource use (affecting stabilizing differences) or to have stronger or weaker competitive asymmetries (affecting fitness differences [37]). Instead, the varied responses observed across species pairs likely reflect species-specific differences in responses to soil moisture limitation [26]; in a dry environment, for example, some species pairs might experience more overlap in resource use, whereas others might experience less overlap. Although we do not have the data to identify the exact mechanism of species-specific differences, it likely has to do with whether or not traits relevant to competition converge or diverge between species in different environments. Whether the same results would be obtained in response to other environmental conditions that, unlike soil moisture, are not also an essential resource, warrants further investigation.
A caveat to our interpretation of our findings is that we can only infer past histories of interaction from present-day distributional data [38], given that macroevolutionary change takes place on timescales that are not directly observable. For this reason, it is important that we weigh our results against explanations other than competition for the patterns we observed, such as neutral evolution or specialization on different environment types. The predictions expected given these alternative mechanisms differ from those under competition in two key ways. First, if environmental specialization alone was responsible for divergence, then species would show clear differences in habitat association, or in their abilities to persist in a common environment in the absence of competition; neither was true for the species in our experiment. Second, under either neutral evolution or environmental specialization, stabilizing differences would not be expected to evolve differently between sympatric and allopatric species pairs, nor would they be constrained to the narrow range of high stabilization that we observed among sympatric pairs. For these reasons, we argue that although we cannot definitively rule out alternative mechanisms, historical interactions likely constrain the evolution of competitive similarities and dissimilarities among species.
Regardless of the mechanism of divergence, the simultaneous evolution of differences that promote and prevent coexistence, as inferred through phylogenetic relationships, provides new insight into the diverse patterns of evolutionary relatedness found in natural communities. Specifically, we find no evidence that closely related species are less likely to coexist; for species that occur in sympatry, we show that the effect of evolutionary relatedness on coexistence is unpredictable even though the effects of evolution on the underlying determinants of coexistence are well understood. This result contradicts common interpretations of over a decade of observational work in ecology, where patterns of phylogenetic dissimilarity (i.e. ‘overdispersion’) in communities are typically considered evidence of competitive filtering [7]. Although this interpretation has been called into question repeatedly in recent years [17,39], our evidence is consistent with other recent work [18,19] that competition does not likely result in phylogenetic overdispersion, at least in our annual plant system. Instead, competition is most likely to generate patterns of phylogenetic similarity (i.e. ‘underdispersion’) in communities containing mixed-provenance species, and indeed a survey of the literature [15] finds evidence of phylogenetic underdispersion in approximately 60% of published studies.
Our experiment has implications for understanding whether interactions among species from different regions are fundamentally different from interactions among species from a common region. Observational studies of plant invasions have produced seemingly contradictory results; distantly related plants are less likely to establish upon introduction [40] but become noxious invaders more frequently if they do establish [41], compared with close relatives which tend to naturalize [40,41]. In our study, distant relatives from Spain had much lower or much higher mean fitness than competitors from California, a result that reconciles previous work [40,41]. Specifically, our results suggest that at the earliest stage of invasion, divergence in fitness may generally predict why some species fail to establish (distant relatives of lower fitness [40]) while others have spectacularly negative impacts on native diversity (distant relatives of higher fitness [41]). Although this result does not establish plant characteristics that make some species noxious invaders [42], it supports the general finding that species have a greater potential to become noxious invaders when they are naive to a region [43].
Our patterns of rapid predictable divergence in fitness differences but not stabilizing differences among species from different floras has been hypothesized [44], but has remained untested until now. Future work is needed to identify the traits that underlie stabilizing and fitness differences among non-native competitors. Although specific traits have been implicated as contributors to species' competitive differences [45] and invasion success [35], the potential for traits to differentially contribute to stabilizing and fitness differences depending on biogeographic history is heretofore unexplored.
The intricate interplay between the ecology and evolution of organisms remains an important area of research for understanding diversity and its response to global changes, such as species invasions. In this study, we have tested one of the most long-standing hypotheses about the relationships between evolutionary relatedness, competition and coexistence. Our work highlights the role that historical interactions play in determining the stability of current-day interactions and the impacts of non-native species.
Supplementary Material
Acknowledgements
We thank M. Cadotte and members of the Gilbert laboratory for their feedback, as well as two anonymous reviewers for their comments improving the manuscript. Greenhouse assistance was provided by B. Hall, A. Petri and many undergraduate assistants, most notably C. Blackford. Bees were provided by J. Thomson.
Data accessibility
The raw data used to fit our annual plant models and our nexus tree file have been made publically available on Dryad: http://dx.doi.org/10.5061/dryad.v2211; the fitted parameter estimates, seed viability data and GenBank sequence accessions are in the electronic supplementary material.
Authors' contributions
R.M.G., B.G. and J.W. designed the experiment, R.M.G. collected the data, R.M.G. and B.G. analysed the data, R.M.G. and J.T.W. constructed the phylogeny, R.M.G. and B.G. wrote the manuscript.
Competing interests
We have no competing interests.
Funding
Funding was provided by NSERC Discovery (research; B.G. [402355-2011] and J.T.W. [402013-2011]) and CGSD (personal; R.M.G.).
References
- 1.Peay KG, Belisle M, Fukami T. 2012. Phylogenetic relatedness predicts priority effects in nectar yeast communities. Proc. R. Soc. B 279, 749–758. ( 10.1098/rspb.2011.1230) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer JR, Kassen R. 2007. The effects of competition and predation on diversification in a model adaptive radiation. Nature 446, 432–435. ( 10.1038/nature05599) [DOI] [PubMed] [Google Scholar]
- 3.Gravel D, Bell T, Barbera C, Bouvier T, Pommier T, Venail P, Mouquet N. 2011. Experimental niche evolution alters the strength of the diversity–productivity relationship. Nature 469, 89–92. ( 10.1038/nature09592) [DOI] [PubMed] [Google Scholar]
- 4.Zuppinger-Dingley D, Schmid B, Petermann JS, Yadav V, De Deyn GB, Flynn DFB. 2014. Selection for niche differentiation in plant communities increases biodiversity effects. Nature 515, 108–111. ( 10.1038/nature13869) [DOI] [PubMed] [Google Scholar]
- 5.Mahler DL, Ingram T, Revell LJ, Losos JB. 2013. Exceptional convergence on the macroevolutionary landscape in island lizard radiations. Science 341, 292–295. ( 10.1126/science.1232392) [DOI] [PubMed] [Google Scholar]
- 6.Johnson MT, Stinchcombe JR. 2007. An emerging synthesis between community ecology and evolutionary biology. Trends Ecol. Evol. 22, 250–257. ( 10.1016/j.tree.2007.01.014) [DOI] [PubMed] [Google Scholar]
- 7.Webb CO, Ackerly DD, McPeek MA, Donoghue MJ. 2002. Phylogenies and community ecology. Annu. Rev. Ecol. Syst. 33, 475–505. ( 10.1146/annurev.ecolsys.33.010802.150448) [DOI] [Google Scholar]
- 8.Schoener TW. 2011. The newest synthesis: understanding the interplay of evolutionary and ecological dynamics. Science 331, 426–429. ( 10.1126/science.1193954) [DOI] [PubMed] [Google Scholar]
- 9.Darwin C. 1859. On the origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. London, UK: John Murray. [PMC free article] [PubMed] [Google Scholar]
- 10.Elton C. 1946. Competition and the structure of ecological communities. J. Anim. Ecol. 15, 54–68. ( 10.2307/1625) [DOI] [Google Scholar]
- 11.Harper JL, Clatworthy JN, McNaughton IH, Sagar GR. 1961. The evolution and ecology of closely related species living in the same area. Evolution 15, 209–227. ( 10.2307/2406081) [DOI] [Google Scholar]
- 12.Violle C, Nemergut DR, Pu Z, Jiang L. 2011. Phylogenetic limiting similarity and competitive exclusion. Ecol. Lett. 14, 782–787. ( 10.1111/j.1461-0248.2011.01644.x) [DOI] [PubMed] [Google Scholar]
- 13.Cahill JF, Kembel SW, Lamb EG, Keddy PA. 2008. Does phylogenetic relatedness influence the strength of competition among vascular plants? Perspect. Plant Ecol. Evol. Syst. 10, 41–50. ( 10.1016/j.ppees.2007.10.001) [DOI] [Google Scholar]
- 14.Anderson TM, Shaw J, Olff H. 2011. Ecology's cruel dilemma, phylogenetic trait evolution and the assembly of Serengeti plant communities. J. Ecol. 99, 797–806. ( 10.1111/j.1365-2745.2011.01795.x) [DOI] [Google Scholar]
- 15.Vamosi SM, Heard SB, Vamosi JC, Webb CO. 2009. Emerging patterns in the comparative analysis of phylogenetic community structure. Mol. Ecol. 18, 572–592. ( 10.1111/j.1365-294X.2008.04001.x) [DOI] [PubMed] [Google Scholar]
- 16.Chesson P. 2000. Mechanisms of maintenance of species diversity. Annu. Rev. Ecol. Syst. 31, 343–366. ( 10.1146/annurev.ecolsys.31.1.343) [DOI] [Google Scholar]
- 17.Mayfield MM, Levine JM. 2010. Opposing effects of competitive exclusion on the phylogenetic structure of communities. Ecol. Lett. 13, 1085–1093. ( 10.1111/j.1461-0248.2010.01509.x) [DOI] [PubMed] [Google Scholar]
- 18.Narwani A, Alexandrou MA, Oakley TH, Carroll IT, Cardinale BJ. 2013. Experimental evidence that evolutionary relatedness does not affect the ecological mechanisms of coexistence in freshwater green algae. Ecol. Lett. 16, 1373–1381. ( 10.1111/ele.12182) [DOI] [PubMed] [Google Scholar]
- 19.Godoy O, Kraft NJB, Levine JM. 2014. Phylogenetic relatedness and the determinants of competitive outcomes. Ecol. Lett. 17, 836–844. ( 10.1111/ele.12289) [DOI] [PubMed] [Google Scholar]
- 20.Abrams PA. 1986. Character displacement and niche shift analyzed using consumer–resource models of competition. Theor. Popul. Biol. 29, 107–160. ( 10.1016/0040-5809(86)90007-9) [DOI] [PubMed] [Google Scholar]
- 21.Thompson JN. 1999. The evolution of species interactions. Science 284, 2116–2118. ( 10.1126/science.284.5423.2116) [DOI] [PubMed] [Google Scholar]
- 22.Dayan T, Simberloff D. 2005. Ecological and community-wide character displacement: the next generation. Ecol. Lett. 8, 875–894. ( 10.1111/j.1461-0248.2005.00791.x) [DOI] [Google Scholar]
- 23.Godoy O, Levine JM. 2014. Phenology effects on invasion success: insights from coupling field experiments to coexistence theory. Ecology 95, 726–736. ( 10.1890/13-1157.1) [DOI] [PubMed] [Google Scholar]
- 24.Stenseth NC. 1979. Where have all the species gone? On the nature of extinction and the Red Queen Hypothesis. Oikos 33, 196–227. ( 10.2307/3543998) [DOI] [Google Scholar]
- 25.Kisdi E, Geritz SAH. 2001. Evolutionary disarmament in interspecific competition. Proc. R. Soc. Lond. B. 268, 2589–2594. ( 10.1098/rspb.2001.1842) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Germain RM, Gilbert B. 2014. Hidden responses to environmental variation: maternal effects reveal species niche dimensions. Ecol. Lett. 17, 662–669. ( 10.1111/ele.12267) [DOI] [PubMed] [Google Scholar]
- 27.Baskin CC, Baskin JM. 2001. Seeds: ecology, biogeography, and evolution of dormancy and germination. San Diego, CA: Academic Press. [Google Scholar]
- 28.Levine JM, HilleRisLambers J. 2009. The importance of niches for the maintenance of species diversity. Nature 461, 254–257. ( 10.1038/nature08251) [DOI] [PubMed] [Google Scholar]
- 29.Harrison S. 1999. Local and regional diversity in a patchy landscape: native, alien, and endemic herbs on serpentine. Ecology 80, 70–80. ( 10.2307/176980) [DOI] [Google Scholar]
- 30.Bartolome JW. 1979. Germination and seedling establishment in California annual grassland. J. Ecol. 67, 273–281. ( 10.2307/2259350) [DOI] [Google Scholar]
- 31.Adler PB, HilleRisLambers J, Levine JM. 2007. A niche for neutrality. Ecol. Lett. 10, 95–104. ( 10.1111/j.1461-0248.2006.00996.x) [DOI] [PubMed] [Google Scholar]
- 32.Chesson P, Huntly N. 1989. Short-term instabilities and long-term community dynamics. Trends Ecol. Evol. 4, 293–298. ( 10.1016/0169-5347(89)90024-4) [DOI] [PubMed] [Google Scholar]
- 33.Harmon LJ, Glor RE. 2010. Poor statistical performance of the Mantel test in phylogenetic comparative analyses. Evolution 64, 2173–2178. ( 10.1111/j.1558-5646.2010.00973.x) [DOI] [PubMed] [Google Scholar]
- 34.Daehler CC. 2003. Performance comparisons of co-occurring native and alien invasive plants: implications for conservation and restoration. Annu. Rev. Ecol. Evol. Syst. 34, 183–211. ( 10.1146/annurev.ecolsys.34.011802.132403) [DOI] [Google Scholar]
- 35.van Kleunen M, Weber E, Fischer M. 2010. A meta-analysis of trait differences between invasive and non-invasive plant species. Ecol. Lett. 13, 235–245. ( 10.1111/j.1461-0248.2009.01418.x) [DOI] [PubMed] [Google Scholar]
- 36.Kraft NJB, Adler PB, Godoy O, James EC, Fuller S, Levine JM. 2014. Community assembly, coexistence and the environmental filtering metaphor. Funct. Ecol. 29, 592–599. ( 10.1111/1365-2435.12345) [DOI] [Google Scholar]
- 37.HilleRisLambers J, Adler PB, Harpole WS, Levine JM, Mayfield MM. 2012. Rethinking community assembly through the lens of coexistence theory. Annu. Rev. Ecol. Evol. Syst. 43, 227–248. ( 10.1146/annurev-ecolsys-110411-160411) [DOI] [Google Scholar]
- 38.Connell JH. 1980. Diversity and the coevolution of competitors, or the ghost of competition past. Oikos 35, 131–138. ( 10.2307/3544421) [DOI] [Google Scholar]
- 39.Kraft NJB, Cornwell WK, Webb CO, Ackerly DD. 2007. Trait evolution, community assembly, and the phylogenetic structure of ecological communities. Am. Nat. 170, 271–283. ( 10.1086/519400) [DOI] [PubMed] [Google Scholar]
- 40.Diez JM, Sullivan JJ, Hulme PE, Edwards G, Duncan RP. 2008. Darwin's naturalization conundrum: dissecting taxonomic patterns of species invasions. Ecol. Lett. 11, 674–681. ( 10.1111/j.1461-0248.2008.01178.x) [DOI] [PubMed] [Google Scholar]
- 41.Strauss SY, Webb CO, Salamin N. 2006. Exotic taxa less related to native species are more invasive. Proc. Natl Acad. Sci. USA 103, 5841–5845. ( 10.1073/pnas.0508073103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Kleunen M, Dawson W, Schlaepfer D, Jeschke JM, Fischer M. 2010. Are invaders different? A conceptual framework of comparative approaches for assessing determinants of invasiveness. Ecol. Lett. 13, 947–958. ( 10.1111/j.1461-0248.2010.01503.x) [DOI] [PubMed] [Google Scholar]
- 43.Simberloff D, Souza L, Nunez MA, Barrios-Garcia MN, Bunn W. 2012. The natives are restless, but not often and mostly when disturbed. Ecology 93, 598–607. ( 10.1890/11-1232.1) [DOI] [PubMed] [Google Scholar]
- 44.MacDougall AS, Gilbert B, Levine JM. 2009. Plant invasions and the niche. J. Ecol. 97, 609–615. ( 10.1111/j.1365-2745.2009.01514.x) [DOI] [Google Scholar]
- 45.Kraft NJB, Godoy O, Levine JM. 2015. Plant functional traits and the multidimensional nature of species coexistence. Proc. Natl Acad. Sci. USA 112, 797–802. ( 10.1073/pnas.1413650112) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data used to fit our annual plant models and our nexus tree file have been made publically available on Dryad: http://dx.doi.org/10.5061/dryad.v2211; the fitted parameter estimates, seed viability data and GenBank sequence accessions are in the electronic supplementary material.