Abstract
Background
Environmental conditions or chemical agents can interfere with the function of the endoplasmic reticulum, and the resulting endoplasmic reticulum (ER) stress can be toxic to the cell if it is not relieved. The classical compensatory response to ER stress is the unfolded protein response (UPR) that reduces protein load in the ER. However, autophagy may also compensate by removing large insoluble protein aggregates. Agents that stress the ER can have anti-cancer activity, and novel applications of ER stress inducing agents are being investigated. Plant stilbenes are a class of stress responsive molecules that includes resveratrol, which are being investigated as potential therapeutics in humans for conditions such as aging or cancer.
Results
We performed a screen of 1726 small, drug like molecules to identify those that could activate an ER-stress responsive luciferase gene. After secondary screening, we determined that the plant stilbenes pterostilbene and piceatannol were the most potent inducers of ER stress from this group. ER stress can be particularly toxic to cells with high ER load, so we examined their effect on cells expressing the Wnt family of secreted glycoprotein growth factors. Molecular analysis determined that these ER stress-inducing stilbenes could block Wnt processing and also induce autophagy in acute lymphoblastic leukemia cells expressing Wnt16. Combining pterostilbene (to induce ER stress) with chloroquine (to inhibit autophagy) lead to significant cellular toxicity in cells from aggressive acute lymphoblastic leukemia.
Conclusions
Plant stilbenes are potent inducers of ER stress. However, their toxicity is more pronounced in cancer cells expressing Wnt growth factors. The toxicity of stilbenes in these ALL cells can be potentiated by the addition of autophagy inhibitors, suggesting a possible therapeutic application.
Keywords: Stilbenes, Unfolded Protein Response, Wnt growth factors, Stress Responses, high throughput screen
BACKGROUND
The endoplasmic reticulum (ER) is the organelle in the cell responsible for the synthesis of membrane-embedded and secreted proteins. The ER is also responsible for the proper folding and modification of these proteins. If drugs or environmental conditions interfere with the processing of proteins in the ER, an ER stress is elicited and the cell responds with the Unfolded Protein Response (UPR) [1, 2]. The UPR contains three arms: IRE1/XBP1, ATF6, and PERK/ATF4. Together, these three components reduce further protein entry into the ER, assist in folding proteins already in the ER, and remove malfolded proteins to decompress the enlarged ER [3]. If the ER stress persists, or the UPR is insufficient to decompress the ER, prolonged ER stress can result in cell death [4].
We have reported identification of a small molecule that can block the endoribonucease activity of IRE1 and therefore can block activation of the XBP1 arm of the UPR [5]. This molecule was identified through a small molecule screen using an XBP1-responsive luciferase reporter gene. This reporter requires XBP1 splicing to generate a frameshift in the XBP1-luciferase transcript so the luciferase gene is brought into frame and is translated [6]. In this manuscript, we describe the use of this reporter gene to identify novel drug-like agents that induce ER stress, from the library of pharmacologically active compounds (LOPAC) and the NIH clinical collection (NIHCC). We extend these studies to examine the cellular response to the plant stilbenes resveratrol, pterostilbene and piceatannol which all scored as potent ER-stressors in this screen.
Previous work from our group also identified the Wnt family of secreted glycoprotein growth factors as heavily processed in the ER with significant sensitivity to ER-stressing agents like hypoxia or tunicamycin [7]. The processing of Wnt proteins can take up to 18 hours in cells, and serve as a molecular marker of ER function. This extended transit time allows for correct intramolecular bonds from the 23–27 conserved cysteine residues, glycosylation, and lipidation [8]. Interference with Wnt processing can result in proteolysis of the growth factor, and reduction in b-catenin signaling [7].
RESULTS AND DISCUSSION
We decided to screen known drug like molecules for their “off target” ability to induce ER stress. We had constructed a reporter cell in the HT1080 fibrosarcoma background that contained a luciferase gene that was fused to the XBP1 open reading frame. In the event of ER stress, the ER-membrane embedded IRE-1 endoribonuclease performs a precise splicing event in the XBP1 mRNA that removes 26 nucleotides and shifts the open reading frame of the final one third of the protein to produce an active transcription factor [9]. In our XBP1-luciferase reporter gene, the luciferase coding sequence is in frame with the carboxy end of XBP1 open reading frame so that after XBP1 splicing, the luciferase protein is translated and its activity detected [5, 6]. We also used a separate HT1080 clone that contained a constitutively active CMV-driven luciferase as a control for cell death or non-specific inhibition/activation of transcription or translation. These two cell lines were plated in duplicate in 96 well format and individual wells were exposed overnight to 10 µM of the compounds in the library. The next day, luciferase activity was measured and the relative induction of the XBP1 luciferase was calculated for each compound and normalized to any changes in the CMV-luciferase signal. We calculated the fold induction of the XBP1-luciferase activity for each compound and chose those with greater than 1.5 fold induction for secondary analysis. Secondary screen was performed in cells expressing the XBP1-luciferase, the CMV-luciferase, and also in cells expressing an ATF4UTR-luciferase reporter [10]. This reporter is responsive to activation of the PERK kinase and increased translation of the ATF4 gene.
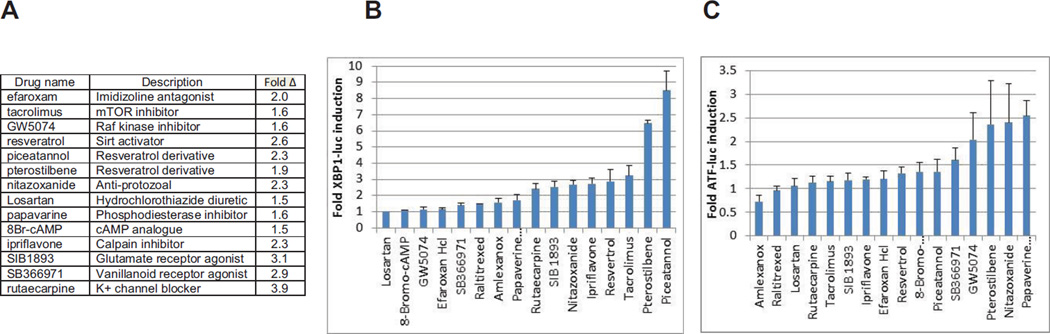
Figure 1A identifies the 13 top inducers, and shows the normalized fold induction of the reporter genes from the primary screen. These compounds resulted in between 1.5 and 3.9 fold induction of the XBP1-luciferse reporter gene. These compounds were then retested manually in a secondary screen with the XBP1-luciferase and the CMV luciferase reporter HT1080 cells and the normalized results show a qualitatively similar, but significantly higher fold induction. Figure 1C shows the effects of the same compounds on the ATF4UTR reporter gene in the second reporter HT1080 cell line. Interestingly, resveratrol and the closely related derivatives piceatannol and pterostilbene were all in the original list of ER-stress inducers, and all were near the top of the most potent inducers in the secondary screen. These results support the idea that there is some structural element(s) in these related compounds that interferes with ER function.
Figure 1.
Identification of small molecules that induce ER stress
Panel A: Results from the primary screen for normalized induction of the XBP1-luciferase reporter gene in cells treated for 24 hours at 10 µM of the indicated drug.
Panel B: Fold induction of the XBP1-luciferase reporter gene after 24 hours of treatment with 20 µM of the indicated compound in manually performed secondary screen of top hits from primary screen.
Panel C: Fold induction of the ATF4-luciferase reporter gene in manually performed secondary screen as described in B.
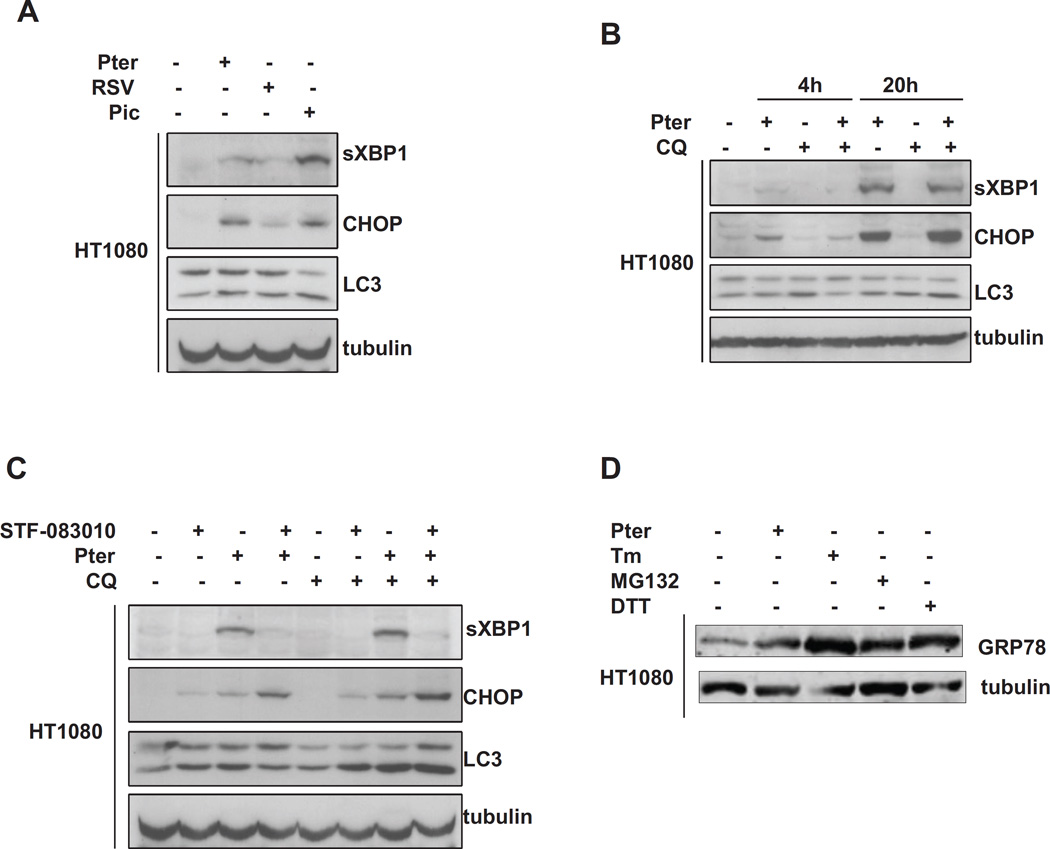
We next decided to molecularly confirm the ability of these compounds to stress the ER using western blot analysis to identify changes in markers of the UPR. Figure 2A shows immunoblot analysis of extracts from parental HT1080 cells treated for 24 hours with 20 µM resveratrol, pterostilbene or piceatannol as indicated. In the cells treated with pterostilbene or piceatannol, we detect both splicing of XBP1 and induction of the ATF4 target gene CHOP. We also find modest increase in LC3 processing, indicating a mild autophagic response. Because pterostilbene appeared to be the most potent of the related molecules in these two responses, we focused on its activities. Figure 2B shows that with increasing time up to 20 hours, that there was increasing XBP1 splicing and CHOP induction, indicating continuing ER stress. We also investigated how inhibition of compensatory pathways would affect pterostilbene induced stress. We tested the combination of pterostilbene with either IRE1 inhibitor STF-083010 to block XPB1 splicing [5], or chloroquine to block autophagic turnover. Figure 2C shows that STF-083010 is indeed a good inhibitor of XBP1 splicing, but this leads to an increase in the PERK arm of the UPR and increased CHOP expression. Addition of chloroquine by itself had a modest increase in LC3 processing; however, LC3 processing was dramatically increased by chloroquine with pterostilbene and/or STF-083010. Finally we compared the ER stress induced by pterostilbene with that of classical ER stressing agents. Figure 2D shows the induction of GRP78 in response to 24 hours of either 20 µM pterostilbene, 2.5 µg/ml tunicamycin, 10 nM of the proteasome inhibitor MG-132, or 1.0 mM DTT. This representative western blot shows that pterostilbene is less able to induce ER stress than these classical ER stressing agents. However, ER stress is dose and time dependent, and these doses of classical ER stressors are somewhat more potent than 20 µM pterostilbene.
Figure 2.
Molecular characterizations of the effects of stilbenes on markers of ER stress and autophagy in HT1080 cells.
Panel A: Immunoblot analysis of XBP1 splicing (top), induction of the ATF4 target gene CHOP (middle), and processing of LC3 (bottom) by 20 hours of 20 µM resveratrol, piceatannol and pterostilbene as indicated. Tubulin is used as loading control.
Panel B: Immunoblot analysis of sXBP1, CHOP, and LC3 as performed in A by either 4 or 20 hours of treatment with 20 µM pterostilbene, 20 µM chloroquine, or both.
Panel C: Immunoblot analysis of sXBP1, CHOP and LC3 in cells treated with 20 µM pterostilbene, 10 µM STF-083010, or 20 µM chloroquine alone or in combination.
Panel D: Immunoblot analysis of ER stress comparing 24 hour treatment with either 20 µM pterostilbene, 2.5 µg/ml tunicamycin, 10 nM MG-132, or 1.0 mM DTT. Lysates were probed for GRP78/Bip to indicate the total ER stress of the treatments.
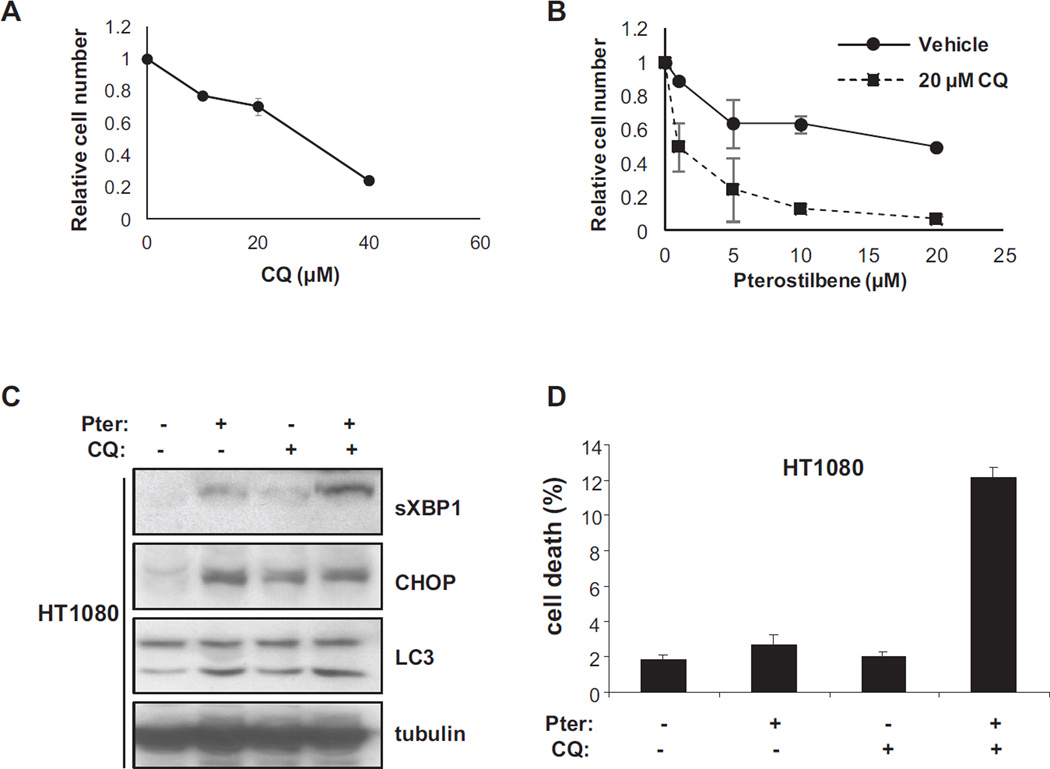
Pterostilbene has been reported to be toxic to cancer cells [11, 12], so we measured its antiproliferative effect alone and in combination with chloroquine. We first measured the growth inhibition by chloroquine alone to establish a relatively non-toxic dose. Figure 3A shows that 20 µM chloroquine has only a 30% reduction in viable cells at 72 hours. We then tested pterostilbene alone and in combination with 20 µM chloroquine for growth inhibition at 72 hours. Figure 3B shows that the concentration to achieve 50% inhibition is 19.7 µM for pterostilbene alone, but only 0.98 µM for pterostilbene with 20 µM chloroquine. In combination, these curves indicate a more than additive effect of the two agents on the inhibition of growth of the HT1080 cells.
Figure 3.
Treatment with pterostilbene is toxic in the presence of autophagy inhibitors.
Panel A: Growth inhibition of chloroquine alone on HT1080 cells as measured by the MTT assay after 72 hours of treatment. Data reported is average of two experiments performed in triplicate.
Panel B: Growth inhibition of pterostilbene alone and in combination with 20 µM chloroquine on HT1080 cells measured by MTT assay after 72 hours. Data reported is average of two experiments performed in triplicate.
Panel C: Western blot analysis of molecular changes in sXBP1, CHOP and LC3 in HT1080 cells treated for 72 hours with 20uM pterostilbene, chloroquine or both.
Panel D: Toxicity of the treatments described in panel C. Percent dead cells reported as measured by trypan blue exclusion assay.
The MTT assay is sensitive to reduced proliferation as well as cell death so we therefore measured cell death directly. We first treated HT1080 cells with 20 µM pterostilbene, 20 µM chloroquine and the combination for 72 hours and examined cell lysates for markers of stress and quantitated toxicity by trypan blue exclusion. Figure 3C shows that the markers of ER stress and autophagy are similar to what was determined in figure 2 at earlier time points. We also measured the toxicity of the drugs using trypan blue exclusion and this showed an even more dramatic interaction of the two drugs. In figure 3D we find that pterostilbene alone was only very mildly toxic after 72 hours and chloroquine treatment showed essentially no toxicity. However, the combination of drugs resulted in very statistically significant amount of cell death (P<0.01).
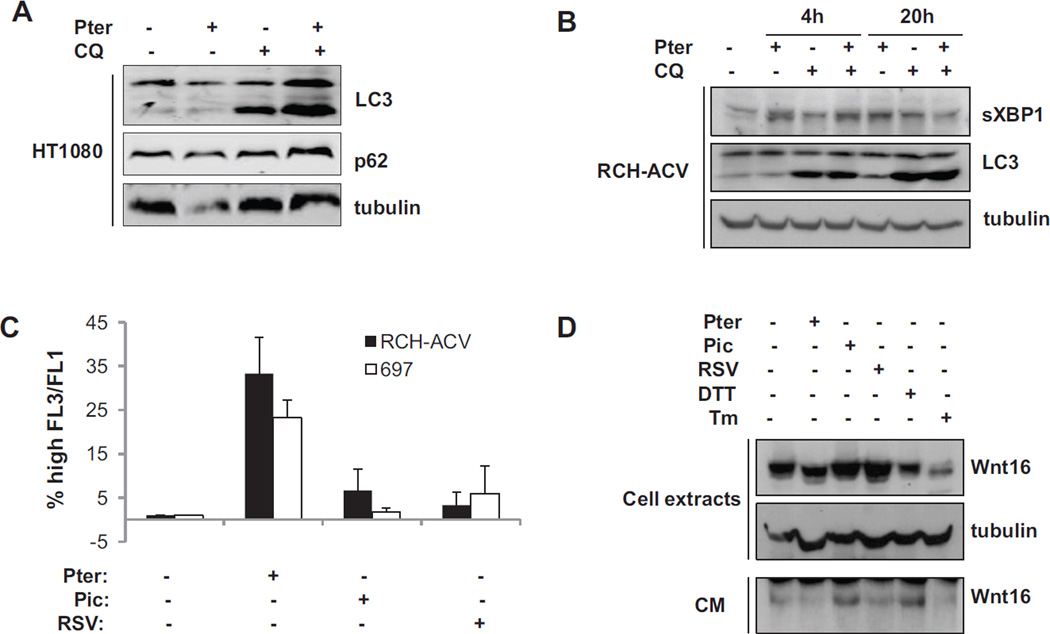
If chloroquine was adding to the ER stress by reducing the protein turnover in the autophagolysosome, we hypothesized that cells with a significant ER load would be particularly sensitive to this treatment. We have previously reported that myeloma cells are an example of cancer cells that are sensitive to inhibition of XBP1 activation [5]. Also, several mechanisms of toxicity of pterostilbene have been identified [11], so we carefully examined the effect of the interaction of pterostilbene with chloroquine on both LC3 processing and depletion of the adaptor protein p62/SQSTM1. Figure 4A confirms that interaction of the two drugs in stimulating autophagy because we find a modest decrease in p62 levels with pterostilbene treatment, and rescue of the protein expression with chloroquine. We also find a stimulation of LC3 processing with the combination of the pterostilbene and chloroquine, suggesting that pterostilbene has increased LC3 processing, and chloroquine has blocked the turnover.
Figure 4.
Molecular characterization of the effect of pterostilbene on autophagy in HT1080 and WNT16 expressing acute lymphoblastic leukemia (ALL) cells.
Panel A: Western blot analysis of autophagy markers in HT1080 cells exposed to 24 hours of pterostilbene, chloroquine, or the combination. Note decreased levels of adapter protein p62, and an enhanced induction of LC3 processing in the cells exposed to both drugs.
Panel B: Western blot analysis of XBP1 splicing and LC3 processing in RCH-ACV cells treated for 4 or 20 hours with 20 µM pterostilbene, chloroquine or the combination.
Panel C: Quantification of acidic vesicles in ALL cells after treatment for 20 hours with 20 µM of either resveratrol, piceatannol or pterostilbene as measured by acridine orange staining and fluorescent activated cell sorting (FACS).
Panel D: Molecular characterization of WNT16 processing in RCH-ACV cells treated with 20 µM stilbenes or classical ER-stressing agents DTT (1 mM) or tunicamycin (2.5 µg/ml). Top panels are western blot of cellular lysates and bottom panel is Immunoblot of equal volumes conditioned media from dishes after 24 hours treatments as indicated.
We have also found that WNT proteins are highly processed secreted glycoproteins that can be blocked from secretion by ER stress [7, 13]. We therefore chose to investigate the effects of stilbenes on cells that express this one protein as a means of stressing the ER. Acute lymphoblastic leukemia cells can express WNT16 due to the presence of the E2A-PBX1 fusion protein [14, 15]. We therefore examined the effect of pterostilbene and chloroquine treatment on RCH-ACV acute lymphoblastic leukemia (ALL) cells. Figure 4B shows by immunoblot analysis that treatment of RCH-ACV cells with pterostilbene alone, or in combination with chloroquine resulted in significant increase in spliced XBP1, and increased LC3 processing. Furthermore, figure 4C shows by FACS analysis that there is an increase in acridine orange staining of acidic vesicles in both RCH-ACV and 697 ALL cells after treatment with pterostilbene, supporting the concept the pterostilbene can induce autophagy[16, 17], especially in WNT expressing cells (figures 2 and 4).
We next examined the specific processing and secretion of the Wnt16 protein in ER-stressed RCH-ACV cells. Cells were treated with stilbenes or the classical ER-stressing agents dithiothreitol or tunicamycin as indicated. Cell extracts and conditioned media were collected 24 hours later. Comparison of intracellular levels of Wnt16 in the treated cells shows that ER stress can inhibit intracellular Wnt16 protein levels as well as Wnt16 secretion (Figure 4D). The effect of pterostilbene appears to be comparable to that of DTT and tunicamycin on intracellular Wnt16, and tunicamycin on secreted Wnt in the conditioned media. Therefore, pterostilbene could have a negative effect on Wnt signaling as well as inducing ER stress.
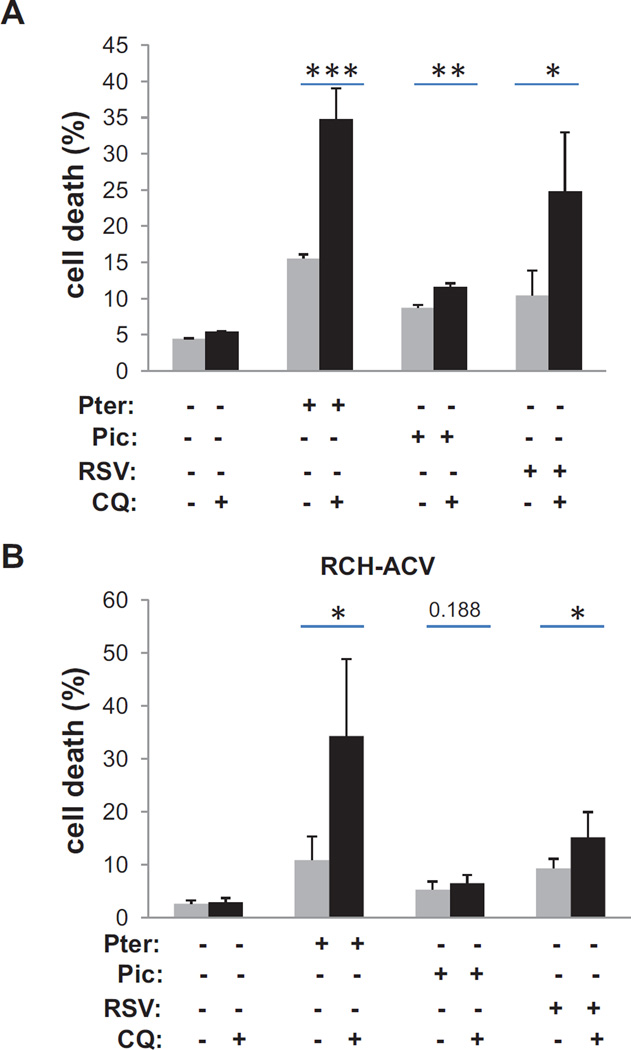
In order to determine the cellular effect of stilbenes on ALL cells, we treated RCH-ACV and 697 cells with the various stilbenes alone, and in combination with chloroquine to block autophagic decompression of the ER. In figure 5, we report that pterostilbene appears to have modest toxicity to either cell line alone, while chloroquine alone was not toxic at all. However, combining the two drugs together resulted in significant toxicity. Combination of resveratrol with chloroquine also showed some increased toxicity relative to resveratrol alone in both cell lines.
Figure 5.
Potentiation of stilbene toxicity by addition of the autophagy inhibitor chloroquine
Panel A: Quantitation of cell death in 697 cells after 72 hours treatment with either 20 µM stilbenes or chloroquine alone, or in combination. Dead cells were measured by loss of trypan blue exclusion. Statistical significance for duplicate experiments performed in triplicate (* = p< 0.05, ** .05<p<0.01)
Panel B: Similar analysis as in A for the RCH-ACV cells.
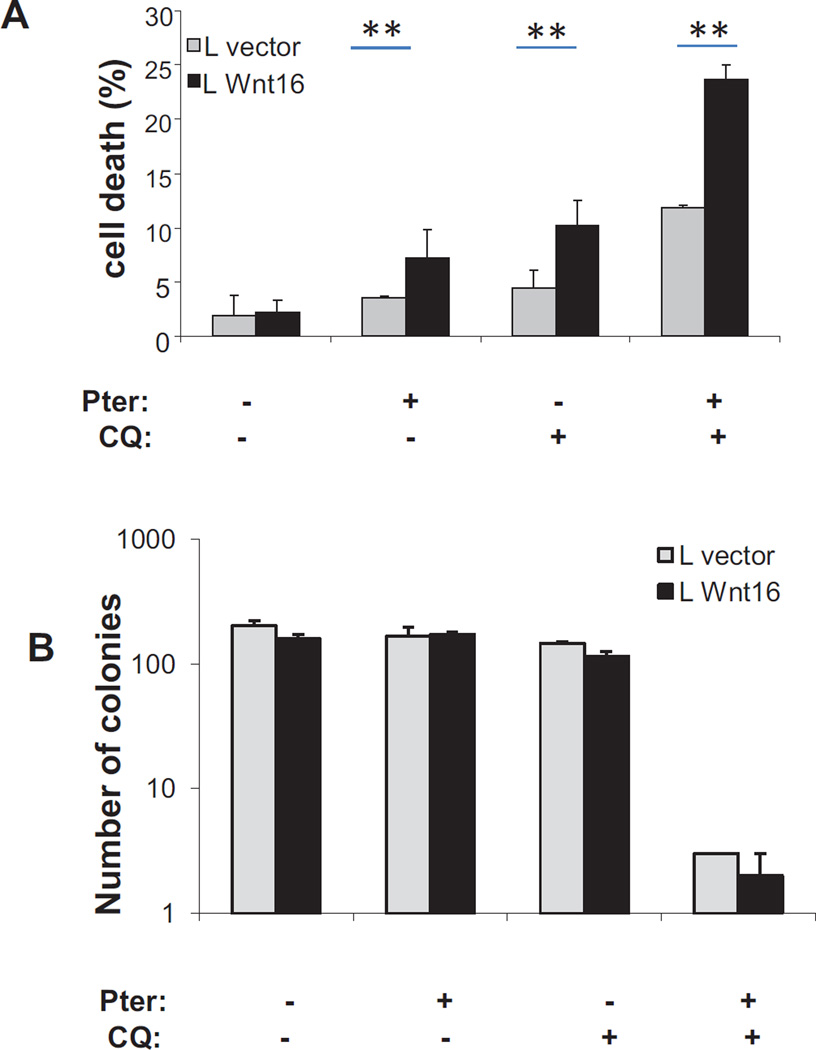
Mechanistically, it was not clear if the expression of Wnt16 was responsible for the sensitivity of these cells, and knockdown of WNT16 in these cells results in acute toxicity. We therefore generated a pool of mouse L cells that stably expressed WNT16. L cells are used in the study of WNT proteins because they produce and secrete high levels of WNT proteins without being dependent on WNT signals for growth [18]. We transduced L cells with either empty vector or retrovirus expressing WNT16 and selected drug resistant cells. WNT16 expression was confirmed by western blot (data not shown). We then challenged the two derived cell lines with pterostilbene in the presence or absence of chloroquine and measured acute toxicity at 72 hours by trypan blue exclusion. We find that the Wnt expressing cells are slightly more sensitive to either drug alone, but are further potentiated to killing by the combination. To determine if this effect was maintained over the long term, we performed colony formation assays on similarly treated cells and found that the combination of pterostilbene with chloroquine was very toxic, resulting in almost two logs of cell kill, although enhanced kill in the cells expressing Wnt16 did not retain statistical significance. In addition to cell toxicity, there may be additional anti-proliferative signals from pterostilbene as well that would inhibit colony formation.
Plant stilbenes are produced in times of stress and are thought to help the plant adapt to the insult [19]. Investigations into the effect of stilbenes on mammalian cells have been spurred by the realization that the stilbene resveratrol can activate SIRT proteins [20]. Potential applications of stilbenes in the areas of aging and cancer have been investigated [21]. However, to date, there is not a recognized, FDAapproved therapeutic application of stilbenes in humans, although toxicity studies have shown pterostilbene to be safe to humans up to a dose of 250 mg/day [22].
CONCLUSIONS
These studies identified stilbenes as potent agents that induce ER stress using an unbiased small molecule screen. However, inhibitors of many essential ER functions such as glycosylation or protein folding can result in ER stress. Investigations are ongoing to identify precise molecular pathway(s) that are specifically inhibited by stilbenes that could result in the accumulation of unfolded proteins. Many molecular activities have been attributed to pterostilbene, such as cell cycle arrest, apoptosis, autophagy, oxidative stress, or stem cell depletion [17, 23–31]. Pterostilbene has even been shown to have enhanced toxicity with autophagy inhibitors through somewhat empirical analysis [16, 32]. However, this work shows that it is possible that these effects could be a secondary consequence of a block to ER function, especially in cells with elevated ER load. As with other agents that induced ER stress, prolonged treatment with stilbenes has some cellular toxicity [24, 27, 33, 34].
The unfolded protein response/integrated stress response has inherent redundancy in the three arms (XBP1/ATF4/ATF6). However, our data supports the concept that there are differences in the downstream effects, and complementary roles for the autophagic machinery. We are able to effectively block stilbene-induced activation of the XBP1 transcription factor with STF-083010 (figure 2C) and this results in a compensatory increase in the ATF4/CHOP response. Interestingly, the combination of pterostilbene and STF-083010 does not result in increased toxicity in HT1080 cells in vitro; the compensation for lack of XBP1 is sufficient. On the other hand, the cellular induction of autophagy in response to pterostilbene appears to be related to clearance of proteins from the ER, and does not have a redundant pathway because addition of chloroquine adds significant toxicity to pterostilbene treatment.
This work identifies some of the pathways that are affected by stilbenes in order to exploit or potentiate their toxicity as anti-cancer agents. We present data supporting a role for ER-stress as a primary generator of toxicity by pterostilbene. However, this may not be the only cellular process that is affected, and additional mechanisms may be engaged as well. These alternative pathways may also help to explain some of the differences in the effects of the tested stilbenes. Reports have identified effects of stilbenes on stem cells, apoptosis, inflammation, oxidative stress, and cell cycle. It is possible that many of these effects are mediated through an inherent imbalance in the function of the ER and the secretome. In fact, a recent report of piceatannol identifies an atypical sensitivity of myeloma cells to the agent [35]. Such sensitivity would be consistent with an induction of ER stress because myeloma cells have been shown to be sensitive to ER-stressing agents [36].
METHODS
Small-molecule library screening
High Throughput Screen
Two libraries of known modulators/FDA approved drugs were screened at the Stanford High- Throughput Bioscience Center (HTBC) including the Library of Pharmacologically Active Compounds (LOPAC1280) and the NIH Clinical Collection (NIHCC446). HT1080 reporter cells were plated at 104 cells per well in 384-well plates. The next day, duplicate wells were tested at 10 µM for each compound in both the XBP1-luciferase and the CMV luciferase reporter cell lines. 24 hours later luciferase signal was detected and relative change in the XBP1 reporter cells was normalized to changes in the CMV reporter cells.
Secondary Reporter assays
HT1080 cells expressing the XBP1-luciferase reporter were plated and 24 hours later were treated with the indicated compound at 10 µM. 24 hours later, cells were lysed, and reporter activity was measured using a Monolight 2010 luminometer (Analytical Luminescence Laboratory, San Diego, CA) and a luciferase reporter assay kit (Roche, Indianapolis, IN). The data are normalized for expression of the CMV-luciferase expressing HT1080 cells. Values are reported as mean +S.D. for three independent experiments.
Western Blot Analysis
In brief, treated cells were harvested in RIPA buffer (1% Triton X-100, 150mM NaCl, 20mM Hepes (pH 7.5), 10% glycerol, 1mM EDTA, 100mM NaF, 17.5mM β-glycerophosphate, 1mM PMSF, 4µg/ml aprotinin, 2µg/ml pepstatin A), lysates were sonicated and cleared by centrifugation, protein concentrations were quantitated. 25–50 µg were electrophoresed on a reducing Tris-Tricine gel, and electroblotted to PVDF membrane. Antibodies used were anti-β-catenin and anti-Wnt16 (BD Biosciences Pharmingen), anti-p-eIF2#, anti-CHOP (Cell Signaling Technology), anti-LC3 (MBL International), anti-sXbp1 (Biolegend), anti-β-actin (Abcam) and anti-tubulin (Santa Cruz Biotechnology). Primary antibodies were detected with species-specific HRP-secondary antibodies (Invitrogen) and visualized with ECL (Amersham).
Cell number assays
For trypan blue exclusion death assay, cells were plated in 12-well plates (2×104 cells/well) and treated. All cells were harvested, and resuspended in PBS. An equal volume of cell suspension was mixed with 0.4% Trypan Blue solution (Sigma-Aldrich, Saint Louis, MO) and the number of blue cells determined on a hemocytometer with the percent of total cells reported. Assays were performed in triplicate. For cell proliferation, the Vybrant® MTT proliferation kit (Life Technologies) was used. Briefly, 6,000 cells were plated in 96 well plates in phenol red-free media and treated with increasing concentrations of chloroquine and/or pterostilbene in triplicate wells for 72h as indicated. 120 µM of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) was added and the formazan precipitate produced over 4 hours. Precipitate was solubilized in 70% DMSO, and absorbance was measured at 540nm.
Acridine orange (AO) staining
Changes in intracellular acidic compartments were determined according to Klionsky with minor modifications [37]. Briefly, following treatment, cells were stained with 2.5 µg/ml AO for 15 min, resuspended in PBS, and immediately analyzed in a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA, USA). Green and red fluorescence were measured on a linear scale using the FL1 and FL3 channels, respectively.
Statistical Analysis
Pairwise comparisons were analyzed by two tailed students T test to determine statistical significance. Significance is indicated on figures with * P<0.05, ** 0.05<P<0.01, *** 0.01<P<0.001
Figure 6.
Expression of WNT16 in mouse L cells can sensitize them to treatment with stilbenes and autophagy inhibitors
Panel A: Quantification of cell death in empty vector and WNT16 expressing L cells after 72 hours treatment with 20 µM stilbene, chloroquine or the combination as indicated. Death determined by loss of trypan blue exclusion in duplicate experiments performed in triplicate (* = p< 0.05, ** .05<p<0.01).
Panel B: Quantitation of colony formation in empty vector and WNT16 expressing L cells treated with 20 µM stilbenes, chloroquine or the combination and allowed to grow for 10 days. Colonies were fixed, stained with crystal violet, and counted if they were over 50 cells.
Highlights.
Plant stilbenes reveratrol, piceatannol and pterostilbene were identified in a high throughput screen as inducers of ER stress.
Autophagy is a component of the cellular adaptation to stilbene exposure.
Combined treatment with stilbenes and autophagy inhibitors yields more than additive cell kill.
Acknowledgments
We thank David Solow-Cordero (Stanford Bioscience Screening Facility) for assistance with the high throughput small molecule screen. This study was funded by NCI P01 CA67166 (NCD and ACK).
ABBREVIATIONS
- ER
endoplasmic reticulum
- UPR
unfolded protein response
- AO
acridine orange
- LC3
microtubule-associated protein 1 light chain 3
- ERAD
ER associated degradation
- XBP1
X-box binding protein 1
- IRE1
Inositol-requiring enzyme-1
- PERK
pancreatic eIF2 kinase
- CHOP
C/EBP homologous protein 10
- ATF4
activating transcription factor 4
- LOPAC
library of pharmacologically active compounds
- NIHCC
NIH clinical collection
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors report no conflicts of interest.
AUTHOR CONTRIBUTIONS
IP and MV performed the small molecule screen, the molecular characterization experiments, statistical analysis and helped interpreted the results. BMC performed protein expression experiments. ND and AK funded and designed the experiments and helped in data interpretation. ND drafted the manuscript and IP, MV and AK edited the manuscript.
Contributor Information
Ioanna Papandreou, Email: Ioanna.papandreou@osumc.edu.
Meletios Verras, Email: mverras@upatras.gr.
Betina McNeil, Email: betina.mcneil@osumc.edu.
Albert C Koong, Email: albert.koong@stanford.edu.
Nicholas C Denko, Email: Nicholas.denko@osumc.edu.
Literature Cited
- 1.Hampton RY. ER stress response: getting the UPR hand on misfolded proteins. Curr Biol. 2000;10(14):R518–R521. doi: 10.1016/s0960-9822(00)00583-2. [DOI] [PubMed] [Google Scholar]
- 2.Koumenis C. ER stress, hypoxia tolerance and tumor progression. Curr Mol Med. 2006;6(1):55–69. doi: 10.2174/156652406775574604. [DOI] [PubMed] [Google Scholar]
- 3.Merulla J, et al. Specificity and regulation of the endoplasmic reticulum-associated degradation machinery. Traffic. 2013;14(7):767–777. doi: 10.1111/tra.12068. [DOI] [PubMed] [Google Scholar]
- 4.Sano R, Reed JC. ER stress-induced cell death mechanisms. Biochim Biophys Acta. 2013;1833(12):3460–3470. doi: 10.1016/j.bbamcr.2013.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Papandreou I, et al. Identification of an Ire1alpha endonuclease specific inhibitor with cytotoxic activity against human multiple myeloma. Blood. 2011;117(4):1311–1314. doi: 10.1182/blood-2010-08-303099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Spiotto MT, et al. Imaging the unfolded protein response in primary tumors reveals microenvironments with metabolic variations that predict tumor growth. Cancer Res. 2010;70(1):78–88. doi: 10.1158/0008-5472.CAN-09-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Verras M, et al. Tumor hypoxia blocks Wnt processing and secretion through the induction of endoplasmic reticulum stress. Mol Cell Biol. 2008;28(23):7212–7224. doi: 10.1128/MCB.00947-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cho SJ, et al. Evolutionary dynamics of the wnt gene family: a lophotrochozoan perspective. Mol Biol Evol. 2010;27(7):1645–1658. doi: 10.1093/molbev/msq052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Calfon M, et al. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415(6867):92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- 10.Sayers CM, et al. Identification and characterization of a potent activator of p53-independent cellular senescence via a small-molecule screen for modifiers of the integrated stress response. Mol Pharmacol. 2013;83(3):594–604. doi: 10.1124/mol.112.081810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mena S, et al. Pterostilbene-induced tumor cytotoxicity: a lysosomal membrane permeabilization-dependent mechanism. PLoS One. 2012;7(9):e44524. doi: 10.1371/journal.pone.0044524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCormack D, McFadden D. Pterostilbene and cancer: current review. J Surg Res. 2012;173(2):e53–e61. doi: 10.1016/j.jss.2011.09.054. [DOI] [PubMed] [Google Scholar]
- 13.Verras M, Papandreou I, Denko NC. WNT16-expressing Acute Lymphoblastic Leukemia Cells are Sensitive to Autophagy Inhibitors after ER Stress Induction. Anticancer Res. 2015;35(9):4625–4631. [PMC free article] [PubMed] [Google Scholar]
- 14.Casagrande G, te Kronnie G, Basso G. The effects of siRNA-mediated inhibition of E2A-PBX1 on EB-1 and Wnt16b expression in the 697 pre-B leukemia cell line. Haematologica. 2006;91(6):765–771. [PubMed] [Google Scholar]
- 15.Mazieres J, et al. Inhibition of Wnt16 in human acute lymphoblastoid leukemia cells containing the t(1;19) translocation induces apoptosis. Oncogene. 2005;24(34):5396–5400. doi: 10.1038/sj.onc.1208568. [DOI] [PubMed] [Google Scholar]
- 16.Chen WC, et al. The anti-tumor efficiency of pterostilbene is promoted with a combined treatment of Fas signaling or autophagy inhibitors in triple negative breast cancer cells. Food Funct. 2014;5(8):1856–1865. doi: 10.1039/c4fo00145a. [DOI] [PubMed] [Google Scholar]
- 17.Siedlecka-Kroplewska K, et al. Pterostilbene induces accumulation of autophagic vacuoles followed by cell death in HL60 human leukemia cells. J Physiol Pharmacol. 2013;64(5):545–556. [PubMed] [Google Scholar]
- 18.Shibamoto S, et al. Cytoskeletal reorganization by soluble Wnt-3a protein signalling. Genes Cells. 1998;3(10):659–670. doi: 10.1046/j.1365-2443.1998.00221.x. [DOI] [PubMed] [Google Scholar]
- 19.Schmidlin L, et al. A stress-inducible resveratrol O-methyltransferase involved in the biosynthesis of pterostilbene in grapevine. Plant Physiol. 2008;148(3):1630–1639. doi: 10.1104/pp.108.126003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baur JA, Sinclair DA. Therapeutic potential of resveratrol: the in vivo evidence. Nat Rev Drug Discov. 2006;5(6):493–506. doi: 10.1038/nrd2060. [DOI] [PubMed] [Google Scholar]
- 21.Balcerczyk A, Pirola L. Therapeutic potential of activators and inhibitors of sirtuins. Biofactors. 2010;36(5):383–393. doi: 10.1002/biof.112. [DOI] [PubMed] [Google Scholar]
- 22.Riche DM, et al. Analysis of safety from a human clinical trial with pterostilbene. J Toxicol. 2013;2013:463595. doi: 10.1155/2013/463595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Acharya JD, Ghaskadbi SS. Protective effect of Pterostilbene against free radical mediated oxidative damage. BMC Complement Altern Med. 2013;13:238. doi: 10.1186/1472-6882-13-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen RJ, Ho CT, Wang YJ. Pterostilbene induces autophagy and apoptosis in sensitive and chemoresistant human bladder cancer cells. Mol Nutr Food Res. 2010;54(12):1819–1832. doi: 10.1002/mnfr.201000067. [DOI] [PubMed] [Google Scholar]
- 25.Dewi NI, Yagasaki K, Miura Y. Anti-proliferative effect of pterostilbene on rat hepatoma cells in culture. Cytotechnology. 2014 doi: 10.1007/s10616-014-9758-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsiao PC, et al. Pterostilbene simultaneously induced G0/G1-phase arrest and MAPK-mediated mitochondrial-derived apoptosis in human acute myeloid leukemia cell lines. PLoS One. 2014;9(8):e105342. doi: 10.1371/journal.pone.0105342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsieh MT, et al. The novel pterostilbene derivative ANK-199 induces autophagic cell death through regulating PI3 kinase class III/beclin 1/Atgrelated proteins in cisplatinresistant CAR human oral cancer cells. Int J Oncol. 2014;45(2):782–794. doi: 10.3892/ijo.2014.2478. [DOI] [PubMed] [Google Scholar]
- 28.Lin VC, et al. Activation of AMPK by pterostilbene suppresses lipogenesis and cell-cycle progression in p53 positive and negative human prostate cancer cells. J Agric Food Chem. 2012;60(25):6399–6407. doi: 10.1021/jf301499e. [DOI] [PubMed] [Google Scholar]
- 29.Lv M, et al. Pterostilbene attenuates the inflammatory reaction induced by ischemia/reperfusion in rat heart. Mol Med Rep. 2015;11(1):724–728. doi: 10.3892/mmr.2014.2719. [DOI] [PubMed] [Google Scholar]
- 30.Siedlecka-Kroplewska K, et al. Pterostilbene induces cell cycle arrest and apoptosis in MOLT4 human leukemia cells. Folia Histochem Cytobiol. 2012;50(4):574–580. doi: 10.5603/20257. [DOI] [PubMed] [Google Scholar]
- 31.Yang Y, et al. Pterostilbene exerts antitumor activity via the Notch1 signaling pathway in human lung adenocarcinoma cells. PLoS One. 2013;8(5):e62652. doi: 10.1371/journal.pone.0062652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hsieh MJ, et al. A combination of pterostilbene with autophagy inhibitors exerts efficient apoptotic characteristics in both chemosensitive and chemoresistant lung cancer cells. Toxicol Sci. 2014;137(1):65–75. doi: 10.1093/toxsci/kft238. [DOI] [PubMed] [Google Scholar]
- 33.Ferrer P, et al. Association between pterostilbene and quercetin inhibits metastatic activity of B16 melanoma. Neoplasia. 2005;7(1):37–47. doi: 10.1593/neo.04337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Harun Z, Ghazali AR. Potential chemoprevention activity of pterostilbene by enhancing the detoxifying enzymes in the HT-29 cell line. Asian Pac J Cancer Prev. 2012;13(12):6403–6407. doi: 10.7314/apjcp.2012.13.12.6403. [DOI] [PubMed] [Google Scholar]
- 35.Schmeel FC, et al. Piceatannol exhibits selective toxicity to multiple myeloma cells and influences the Wnt/ beta-catenin pathway. Hematol Oncol. 2014;32(4):197–204. doi: 10.1002/hon.2122. [DOI] [PubMed] [Google Scholar]
- 36.Mimura N, et al. Blockade of XBP1 splicing by inhibition of IRE1alpha is a promising therapeutic option in multiple myeloma. Blood. 2012;119(24):5772–5781. doi: 10.1182/blood-2011-07-366633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Klionsky DJ. Autophagy: from phenomenology to molecular understanding in less than a decade. Nat Rev Mol Cell Biol. 2007;8(11):931–937. doi: 10.1038/nrm2245. [DOI] [PubMed] [Google Scholar]