Abstract
Plasma homocysteine, a metabolite involved in key cellular methylation processes seems to be implicated in cognitive functions and cardiovascular health with its high levels representing a potential modifiable risk factor for Alzheimer’s disease (AD) and other dementias. A better understanding of the genetic factors regulating homocysteine levels, particularly in non-white populations, may help in risk stratification analyses of existing clinical trials and may point to novel targets for homocysteine-lowering therapy. To identify genetic influences on plasma homocysteine levels in individuals with African ancestry, we performed a targeted gene and pathway-based analysis using a priori biological information and then to identify new association performed a genome-wide association study. All analyses used combined data from the African American and Yoruba cohorts from the Indianapolis-Ibadan Dementia Project. Targeted analyses demonstrated significant associations of homocysteine and variants within the CBS (Cystathionine beta-Synthase) gene. We identified a novel genome-wide significant association of the AD risk gene CD2AP (CD2-associated protein) with plasma homocysteine levels in both cohorts. Minor allele (T) carriers of identified CD2AP variant (rs6940729) exhibited decreased homocysteine level. Pathway enrichment analysis identified several interesting pathways including the GABA receptor activation pathway. This is noteworthy given the known antagonistic effect of homocysteine on GABA receptors. These findings identify several new targets warranting further investigation in relation to the role of homocysteine in neurodegeneration.
Keywords: African Continental Ancestry Group, CD2-associated protein, cystathionine beta-synthase, genome-wide association study, homocysteine, metabolic networks and pathways, metabolomics
INTRODUCTION
Homocysteine (HCY) is a sulfur-containing amino acid produced in the metabolism of the essential amino acid methionine. It exists at a critical biochemical juncture between methionine metabolism and the biosynthesis of the amino acids cysteine and taurine. HCY (Fig. 1) is normally metabolized via two biochemical pathways: re-methylation, which converts homocysteine back to methionine, and trans-sulfuration, which converts homocysteine to cysteine and taurine. Abnormally high blood levels of HCY signal a breakdown in this biochemical process, resulting in far-reaching biochemical and life consequences such as increased cardiovascular risks and cognitive decline in Alzheimer’s disease (AD). Increased HCY levels have been associated with cerebral atrophy [1-3] and cognitive impairment [4-7]. Meta-analyses have demonstrated a positive association between increased HCY levels and dementia risk and cognitive function in AD patients [8, 9] although the results have been inconsistent [10-15]. Recent metabolomics studies have revealed that methionine and the pathway leading to cysteine and glutathione production may be dysregulated in AD patients [16] suggesting aberrant methylation processes that can contribute to disease pathogenesis (see the study by Fuso and Scarpa [17] for short review). Most studies with homocysteine have been conducted using participants with European ancestry. Population differences in the role of HCY regulation remains under-investigated. Two studies have employed participants of South Korean [14] and African ancestry [11] and showed positive associations between elevated HCY and increased dementia risk.
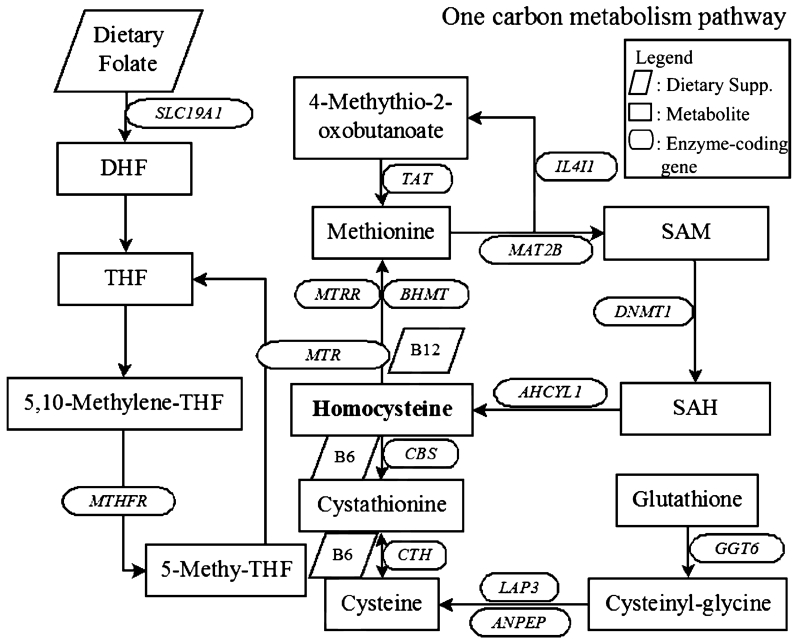
Fig. 1.
Schema of one-carbon metabolism pathways. DHF, dihydrofolate; THF, tetrahydrofolate; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; SLC19A1, solute carrier family 19 (folate transporter), member 1; MTHFR, methylenetetrahydrofolate reductase (NAD(P)H); TAT, tyrosine aminotransferase; MTRR, 5-methyltetrahydrofolate-homocysteine methyltransferase reductase; BHMT, betaine–homocysteine S-methyltransferase; MTR, 5-methyltetrahydrofolate-homocysteine methyltransferase; CBS, cystathionine-beta-synthase; CTH, cystathionase (cystathionine gamma-lyase); IL4I1, interleukin 4 induced 1; MAT2B, methionine adenosyltransferase II, beta; AHCYL1, adenosylhomocysteinase-like 1; DNMT1, DNA (cytosine-5-)-methyltransferase 1; LAP3, leucine aminopeptidase 3; ANPEP, alanyl (membrane) aminopeptidase; GGT6, gamma-glutamyltransferase 6.
Potential mechanisms of HCY effects contributing to cardiovascular diseases and dementias include upregulation of arterial smooth muscle cell collagen production [18], extracellular matrix remodeling [19], potassium channel inhibition [20], microvascular remodeling and increased permeability of the blood-brain barrier by reducing gamma-aminobutyric acid (GABA)-A receptor [21, 22], cytoskeletal remodeling [23, 24], increased cell adhesion [25], and induction of MMP-9 (matrix metalloproteinase-9) activation that can lead to blood-brain barrier dysfunction [26]. HCY can also increase neurotoxicity through overstimulation of N-methyl-D-aspartate receptors [27] and promote apoptosis by increasing DNA damage in neurons [28].
Considering the high estimated heritability (57%) of HCY [29], it is likely that there is substantial genetic predisposition regulating blood levels of HCY. There are several genome-wide association studies (GWAS) of HCY in clinical samples [30, 31], normal older controls [32] and healthy women [33]. A case-control GWAS in a dementia cohort [34] found the gene MTHFD1L (methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 1-like) that was involved in the folate-pathway. Most genetic studies with HCY levels including these GWAS except one GWAS with Filipino women [33] have investigated genetic factors in participants of European ancestry. Even though HCY increased dementia risk in African ancestry population [11], no studies to date were carried out to identify genetic factors influencing HCY in populations of African ancestry. Here we performed a genetic association analysis to investigate genetic influences on HCY by analyzing data from African American and Yoruba Ibadan Nigerian participants who participated in the Indianapolis-Ibadan Dementia Project.
MATERIALS AND METHODS
Indianapolis-Ibadan Dementia Project (IIDP)
The IIDP is a longitudinal prospective community-based study, started in 1992, of the prevalence, incidence, and risk factors for AD and dementia in two populations of African origin: older African Americans living in Indianapolis, Indiana, USA and Yoruba living in Ibadan, Nigeria. The study was approved by the institutional review boards of Indiana University School of Medicine and University of Ibadan. Informed consent was obtained from all participants. Details of the study design and participants have been described elsewhere [11, 35-38] and are briefly summarized here. In the IIDP, all participants were regularly followed-up for cognitive and functional evaluation every two to three years after baseline evaluation. Each evaluation was conducted in a two-stage design: 1) in-home cognitive and functional evaluation for all participants and 2) a full diagnostic evaluation of selected participants based on their cognitive test performance. The Community Screening Interview for Dementia (CSID) for a cognitive assessment [39] and an interview with a close relative for evaluation of daily function were used in the in-home evaluation. Diagnostic evaluation used 1) a neuropsychological battery, 2) a standardized neurologic and physical exam, and 3) a structured interview with a close relative, and diagnosis was made in a consensus conference of clinicians based on these assessments. Since the beginning of the IIDP, 1,893 African Americans and 1,939 Yoruba have been added to 1,649 participants of the original cohort in 2001. The original participants and additional participants were similar in basic demographics. From 2,764 out of these participants, blood samples were collected in 2001.
Participants
Included in this study are the IIDP participants, African Americans and Yoruba. Of 2,764 participants whose blood samples were obtained in 2001, 1,858 participants had genome-wide genotype data, blood biomarkers (levels of homocysteine, folate, vitamin B12), and cognitive performance measures administered in 2001 and were included in this study. Participant characteristics are shown in Table 1.
Table 1.
Sample characteristics
| Characteristics | ALL | CN | Dementia | p * |
|---|---|---|---|---|
| Indianapolis + Ibadan | ||||
| N | 1,858 | 1,787 | 71 | |
| Age (years; mean ± SD) | 77.2 ± 5.46 | 77.0 ± 5.31 | 82.2 ± 6.77 | 3.22E-15 |
| Gender (male/female) | 644/1214 | 615/1172 | 29/42 | 2.64E-01 |
| APOE (ε4−/ε4+) | 1159/699 | 1130/657 | 29/42 | 1.34E-04 |
| Vitamin B12 (pg/mL; mean ± SD) | 699.4 ± 332.3 | 704.8 ± 333.7 | 562.0 ± 262.3 | 3.78E-04 |
| HCY_LOG10 (umol/L; mean ± SD) | 1.210 ± 0.16 | 1.208 ± 0.16 | 1.261 ± 0.18 | 5.23E-03 |
| Indianapolis | ||||
| N | 898 | 853 | 45 | |
| Age (years; mean ± SD) | 77.6 ± 5.47 | 77.3 ± 5.31 | 82.5 ± 6.25 | 3.00E-10 |
| Gender (male/female) | 308/590 | 284/569 | 24/21 | 5.78E-03 |
| APOE (ε4−/ε4+) | 572/326 | 556/297 | 16/29 | 5.63E-05 |
| Vitamin B12 (pg/mL; mean ± SD) | 612.4 ± 342.6 | 618.6 ± 345.8 | 496.0 ± 250.0 | 1.93E-02 |
| HCY_LOG10 (umol/L; mean ± SD) | 1.201 ± 0.17 | 1.198 ± 0.16 | 1.257 ± 0.17 | 2.03E-02 |
| Ibadan | ||||
| N | 960 | 934 | 26 | |
| Age (years; mean ± SD) | 76.8 ± 5.42 | 76.7 ± 5.29 | 81.5 ± 7.66 | 7.93E-06 |
| Gender (male/female) | 336/624 | 331/603 | 5/21 | 8.74E-02 |
| APOE (ε4−/ε4+) | 587/373 | 574/360 | 13/13 | 2.37E-01 |
| Vitamin B12 (pg/mL; mean ± SD) | 780.7 ± 300.6 | 783.6 ± 301.5 | 676.3 ± 247.3 | 7.26E-02 |
| HCY_LOG10 (umol/L; mean ± SD) | 1.219 ± 0.15 | 1.217 ± 0.15 | 1.268 ± 0.19 | 8.28E-02 |
CN, cognitively normal; APOE, apolipoprotein E; HCY, homocysteine.
p-values are computed using chi-square test for categorical variables and one-way analysis of variance for continuous variables.
Cognitive assessment
Cognitive function of study participants was assessed by the Community Screening Interview for Dementia (CSID), a widely used screening tool for dementia that evaluates multiple cognitive domains including language, attention, memory, orientation, praxis, comprehension, and motor response [39]. The CSID was administered for all participants every two or three years. The CSID total score is the sum of all domain scores with a score range from 0 to 80 with higher score indicating better cognitive function [38].
Biomarkers and quality control procedures
Peripheral blood samples were collected in 2001. They were drawn in 10-mL EDTA Vacutainer tubes and frozen plasma and buffy coat biosamples were shipped to and processed at Indiana University. Levels of plasma homocysteine (HCY), folate, and vitamin B12 were measured by using commercial kits from BioRad, Hercules, CA, and Diasorin, Stillwater, MN, USA [11]. HCY underwent further quality control (QC) procedures including log (base 10) transformation due to skewed distribution and removal of outliers (samples more than ±4 standard deviations from mean).
Genetic data and quality control procedures
Genome-wide genotype data were collected by using the Illumina HumanOmni1-Quad for African Americans and HumanOmni2.5-8v1 BeadChips (San Diego, CA, USA) for Yoruba. Collected genotype data underwent standard QC procedures using PLINK v1.07 [40] (http://pngu.mgh.harvard.edu/purcell/plink/) in the two samples independently. Sample and genotype markers were excluded based on the following criteria: call rate per sample <95%, gender ambiguity, groups of genetically related individuals by identity-by-descent (IBD) check, call rate per marker <95%, minor allele frequency (MAF) <1%, and Hardy-Weinberg Equilibrium (HWE) test p < 1 × 10−6 in cognitively normal participants only. For each sibling pair identified by IBD analysis, only one sample was randomly selected and after QC steps, population stratification analysis was performed to make sure that all samples in this study were grouped with Africans in HapMap3 samples by using a procedure described elsewhere [41]. After standard QC, genotype data were imputed to the 1000 Genome reference panel (http://www.1000genomes.org/) following the Enhancing Neuroimaging Genetics through Meta-Analysis 2 (ENIGMA 2) imputation protocol (http://enigma.ini.usc.edu/wp-content/uploads/2012/07/ENIGMA2_1KGP_cookbook_v3.pdf) as described previously [41-43]. Some imputed single nucleotide polymorphisms (SNPs) were removed based on the following criteria: r2 < 0.5 between imputed and the nearest genotyped SNPs, MAF <5%, and HWE p < 1 × 10−6. Apolipoprotein E (APOE) ε4 allele was a risk factor for AD in these cohorts [36] and APOE genotype was separately collected from genomic DNA derived from peripheral blood [37]. APOE genotype data were merged with the imputed genotype dataset. After all QC steps for HCY and genotype data, there were a total of 1858 samples with HCY and genotype data available for analysis (Table 1).
Statistical analysis
All genetic association testing utilized linear regression based on an additive model of SNP effect. We performed combined analyses including all available African Americans and Yoruba. Principal components (PC) were computed by using commonly genotyped SNPs in the two genotype platforms and the 1st PC was selected for inclusion as a covariate based on scree plot analysis. Because levels of folate and vitamin B12 can affect HCY metabolism, a multiple linear regression model was applied to determine if levels of folate and B12 were associated with HCY. Vitamin B12 level was significantly (p < 0.05) associated with HCY. Therefore, multiple regression model included age at the time of HCY measure, gender, the 1st PC, and level of vitamin B12 as covariates. Dichotomous diagnosis (cognitively normal control (CN) versus demented) made in 2001 was also included in the model.
Candidate gene- and pathway-based analysis
As shown in Fig. 1, one-carbon metabolism pathway is involved in production and degradation of HCY and there are multiple genes involved in one-carbon metabolism that can affect HCY level. Given this extensive a priori biological information, as a first step we investigated effect of enzyme-coding genes in this pathway on HCY. An analysis was performed with 15 genes (shown in oval circles in Fig. 1). Chromosomal position of each gene was determined based on hg19. 2,222 SNPs within ±10 kb from the 15 gene boundaries existed in the data and were analyzed. Association with p < 2.25 × 10−5 (Bonferroni correction threshold: 0.05/2,222 SNPs) was considered significant in the targeted genetic association analysis.
Genome-wide association analysis
Considering the high estimated heritability (57%) of HCY, there may exist other genes beyond ones in one-carbon metabolism pathway affecting HCY level. Therefore, GWAS was performed as an unbiased approach. Association with p < 5 × 10−8 in the GWAS results was considered genome-wide significant based on the Bonferroni correction of one million independent SNP tests [44]. A Manhattan plot of GWAS results was created with Haploview [45].
Pathway enrichment analysis
As a complementary approach to extend the GWAS findings, a pathway enrichment analysis was performed to identify biological pathways enriched in the GWAS results. GSA-SNP [46] was used with three curated pathway sets (BioCarta, KEGG, and Reactome) downloaded from the Molecular Signature Database, version 5.0 (http://www.broadinstitute.org/gsea/msigdb/index.jsp). In order to reduce the potential bias due to gene-set size, analysis was restricted to pathways with 5–100 genes [47]. All SNPs within ±10 kb from each gene boundary were included in the analysis and the false discovery rate (FDR) was used to correct p-values for pathway-level multiple comparisons [48]. All pathways with corrected p-value <0.01 were considered significant.
RESULTS
Candidate gene- and pathway-based analysis
In the candidate gene- and pathway-based analysis, four SNPs in and near cystathionine-beta-synthase (CBS) gene were significantly associated with HCY after correction for multiple testing (p < 2.25 × 10−5). The most significant SNP (rs28635199; p = 5.68 × 10−06) is located within 5 kb downstream of the gene. This SNP (rs28635199) accounted for 1% additional variation while 7.2% of the total variation of HCY was accounted for by a statistical model without this SNP. The regulome DB (http://regulome.stanford.edu/index) indicates its function as transcription factor binding or DNase peak. The association of this SNP with HCY was strong in each cohort separately. All four SNPs are in strong linkage disequilibrium (LD) (pairwise r2 > 0.95).
GWAS results
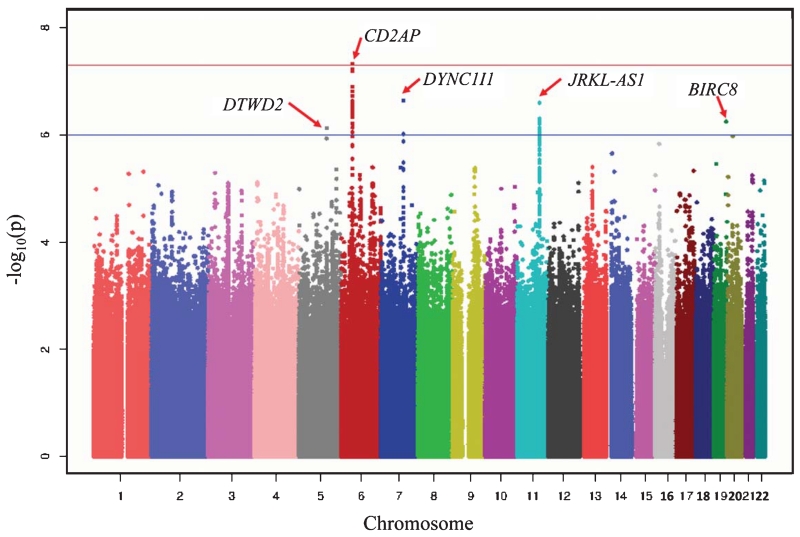
GWAS identified one novel finding, a significant association of SNP (rs6940729; p = 4.71 × 10−08) in the intronic region of CD2-associated protein (CD2AP) gene (Table 2). This SNP explains 1.5% additional variation while 7.2% of total variation of HCY was explained by a statistical model without the SNP in these cohorts, independent of the effect of CBS SNP (rs28635199). To date, there is no study reporting the direct association of CD2AP gene with plasma homocysteine. Of note, rs6940729 is not in strong LD with AD candidate SNPs in CD2AP (all pairwise r2 < 0.2) in European ancestry population [49-51]. The association of rs6940729 with HCY was tested separately for the African Americans and Yoruba to check whether this association was driven only by one cohort and association in both cohorts was strong, resulting in the genome-wide significance in the combined samples. In both cohorts, minor allele (T) of rs6940729 was associated with decreased level of HCY. Genomic inflation factor (λ = 1) indicated that the GWAS results did not seem to be inflated by other confounding factors. Several nearby SNPs showed similar significance although they did not reach genome-wide significance. Beyond CD2AP, there are several SNPs near DTW domain containing 2 (DTWD2), dynein, cytoplasmic 1, intermediate chain 1 (DYNC1I1), JRKL antisense RNA 1 (JRKL-AS1), baculoviral IAP repeat containing 8 (BIRC8) genes at suggestive association level (p < 1 × 10−6). Figure 2 shows the Manhattan plot of GWAS results displaying the genome-wide significant and suggestive loci.
Table 2.
Significant genetic association results. Significant SNPs associated with homocysteine in the GWAS and targeted approach are shown. For the most significant SNPs from the GWAS and targeted approach, association results in each cohort are presented. Significant association p-values are highlighted in bold face
| Cohorts | CHR | SNP | BP | Minor Allele | MAF | Gene | BETA | p |
|---|---|---|---|---|---|---|---|---|
| GWAS | ||||||||
| Indianapolis + Ibadan | 6 | rs6940729 | 47552920 | T | 0.4775 | CD2AP | −0.027 | 4.71E-08 |
| Indianapolis | 0.4773 | −0.032 | 2.65E-05 | |||||
| Ibadan | 0.4777 | −0.020 | 1.44E-03 | |||||
| Targeted (One carbon metabolism pathway) | ||||||||
| Indianapolis + Ibadan | 21 | rs28635199 | 44469734 | C | 0.1717 | CBS | −0.030 | 5.68E-06 |
| Indianapolis | 0.1635 | −0.030 | 2.99E-03 | |||||
| Ibadan | 0.1793 | −0.028 | 6.78E-04 | |||||
| Indianapolis + Ibadan | 21 | rs8127973 | 44474949 | T | 0.177 | CBS | −0.029 | 6.74E-06 |
| Indianapolis + Ibadan | 21 | rs28825153 | 44471639 | T | 0.1711 | CBS | −0.029 | 7.03E-06 |
| Indianapolis + Ibadan | 21 | rs12613 | 44473691 | T | 0.1711 | CBS | −0.029 | 7.38E-06 |
CHR, chromosome; BP, base position; SNP, single nucleotide polymorphism; MAF, minor allele frequency; BETA, regression coefficient of SNP; GWAS, genome-wide association study.
Fig. 2.
Manhattan plot of GWAS results. Red solid line indicates the genome-wide significant level (p = 5 × 10−8) and blue line shows p = 1 × 10−6.
Pathway enrichment analysis results
Pathway enrichment analysis found nine significant biological pathways (FDR-corrected p < 0.01, Table 3), including pathways related to blood vessels and cardiovascular risk factors and one pathway related to GABA receptors in which function has been reported as dysregulated in the AD brain tissue [52].
Table 3.
List of significant canonical pathways
| Pathways (Source database) | Set size* | uncorrected p-value | corrected p-value |
|---|---|---|---|
| COLLAGEN_FORMATION (Reactome) | 54 (58) | 4.12E-07 | 4.00E-04 |
| POTASSIUM_CHANNELS (Reactome) | 96 (98) | 4.75E-06 | 2.31E-03 |
| EXTRACELLULAR_MATRIX_ORGANIZATION (Reactome) | 82 (87) | 9.98E-06 | 3.23E-03 |
| SIGNALING_BY_RHO_GTPASES (Reactome) | 100 (113) | 2.27E-05 | 5.51E-03 |
| INSULIN_SYNTHESIS_AND_PROCESSING (Reactome) | 20 (21) | 4.47E-05 | 8.68E-03 |
| GABA_RECEPTOR_ACTIVATION (Reactome) | 50 (52) | 5.58E-05 | 9.02E-03 |
| HEPARAN_SULFATE_HEPARIN_HS_GAG_METABOLISM (Reactome) | 46 (52) | 5.79E-05 | 9.02E-03 |
| ETHANOL_OXIDATION (Reactome) | 10 (10) | 7.18E-05 | 9.02E-03 |
| ION_CHANNEL_TRANSPORT (Reactome) | 48 (55) | 7.41E-05 | 9.02E-03 |
Number of genes from study data (number of genes in the pathway).
DISCUSSION
In order to assess the influence of enzyme-coding genes involved in one-carbon metabolism pathway where HCY is produced, targeted gene association analysis was employed. Main effects of rs28635199 in CBS on HCY may indicate that this gene is a potential genetic risk factor for AD and/or dementia considering that HCY is a potential risk factor for AD and/or dementia. CBS is located at chromosome 21q22.3 and encodes an enzyme catalyzing the conversion of homocysteine to cystathionine as the first step of trans-sulfuration pathway that subsequently produces glutathione, taurine, and alpha-ketobutyrate (αKB). Careful regulation of CBS activity is very important to prevent subsequent disordered conditions. Hyperhomocysteinemia and homocystinuria, characterized by an abnormally upregulated homocysteine, can be caused by CBS deficiency [53, 54]. Upregulation of CBS can cause increased ammonia level, decreased glutathione synthesis and high loss of methyl groups due to increased process of homocysteine through trans-sulfuration pathway instead of re-methylation cycle although clinical outcomes of CBS upregulation can vary individually. In addition, one large-scale GWAS of human blood metabolites [55] identified the minor allele (T) of another SNP (rs2851391) in CBS gene significantly associated with decreased plasma betaine levels, which can reduce AD-like pathological changes and memory impairment induced by HCY [56] while high HCY is known to deplete betaine [57].
Considering that HCY has been shown to be elevated in patients with AD and the minor allele (C) of CSB SNP (rs28635199) was associated with decreased level of HCY in this study, this SNP appears to be a protective variant against elevation of HCY. However, relationship between CBS enzyme and HCY may not be simple and need to be thought from a more complex systems biology perspective [58] considering many other genes and environmental factors that can affect the level of HCY and CBS enzyme. One study found that the CBS 844 in 68 polymorphism (a 68 bp insertion at 844 in the exon 8) was associated with mild hyperhomocysteinemia and could be a risk factor for AD [59], while another study showed that CBS enzyme level was increased in postmortem brains of Down’s syndrome patients who often develop AD compared to levels in brains of normal individuals and CBS enzyme was located to astrocytes and those surrounding senile plaques in the brains of Down’s syndrome patients [60].
This study is the first GWAS of plasma HCY in samples of African ancestry at risk for AD or other dementia. By analyzing 1,858 plasma homocysteine samples collected from older African Americans and Yoruba Nigerians, we identified one novel genomewide significant association of SNP (rs6940729) in CD2AP gene. The CD2AP gene is also one of the AD candidate genes for the white European ancestry population and three SNPs (rs9349407, rs9296559, rs10948363) in CD2AP have been identified as susceptibility loci for AD [49-51] and one AD candidate SNP (rs9349407) in this gene was associated with neuritic plaque burden [61]. These SNPs failed replication attempts in other African American cohorts [62, 63]. However, the GWAS SNP in this study is not in strong LD with these SNPs (all pairwise r2 < 0.2). The effect of this SNP (rs6940729) as a genetic risk factor for AD and/or dementia needs to be further investigated in larger Africa-originated cohorts.
CD2AP is a protein-coding gene with 18 exons located at chromosome 6p12 and its product is proposed to play important roles in the immune system, endocytosis, cytoskeletal reorganization, cell adhesion, and vesicle trafficking [64-70]. Suppression of CD2AP in an APP transgenic mouse model resulted in decreased Aβ release and lower Aβ42/Aβ40 ratio in brain [71] and a drosophila AD model with knockdown of CD2AP fly ortholog showed enhanced tau toxicity [70]. However, CD2AP mRNA expression was not altered in AD brains [72] although the gene was expressed in brain [71]. These inconsistent findings of CD2AP in relation to gene expression in AD brains and AD relevant biomarkers may imply an indirect influence of CD2AP variation on AD through interaction with other genes/pathways and/or genetic influences on other AD mechanisms including cardiovascular risk factors.
To date, it is unknown how CD2AP affects the level of plasma homocysteine as no study has investigated the relationship between CD2AP and plasma homocysteine. Considering that CD2AP is a regulator of cytoskeletal structure and remodeling, while HCY potentially exerts its effects on cells through effects on cytoskeletal structure and production of reactive oxygen species (ROS), genetic variation in CD2AP may influence the relative sensitivity (resistance) of neuronal cells to the cytoskeletal and ROS effects of HCY, thus impacting the balance of cell life and plasticity and cell death [23, 24]. Another potential mechanism linking CD2AP and HCY may be cell adhesion in which both CD2AP and HCY are involved [70]. Due to lack of studies, potential mechanisms to connect CD2AP and HCY deserve investigation.
Suggestive associations in the GWAS results (Fig. 2) included SNPs near DTWD2 that was associated with subcutaneous adipose tissue (SAT) in European ancestry men [73]. Although visceral adipose tissue is more strongly correlated with metabolic risk factors than SAT [74], SAT can increase cardiometabolic risk which may affect AD risk subsequently [75]. Two variants in DYNC1I1 were associated with fat oxidation and total energy expenditure in the Hispanic population [76], implying genetic influence on cardiovascular risk. JRKL-AS1 was associated with formal thought disorder in schizophrenia in European ancestry cohort [77], but no studies to date reported the association with HCY.
Our enriched pathway analysis indicates HCY as a cardiovascular risk by upregulating arterial smooth muscle cell collagen production [18], remodeling extracellular matrix [19], inhibiting potassium channels [20], and antagonizing GABA-A receptor inducing microvascular remodeling [21] and increasing permeability of blood-brain barrier [22], all of which are related to vascular dysfunction. Among enriched pathways, GABA receptor activation appears particularly interesting in light of data on GABA from reactive astrocyte impairing memory in an AD mouse model [78] and its disruption in AD patients (see the study by Lanctot et al. [79] for review). Subunits of GABA-A receptors were dysregulated in AD brain compared to normal brains [52]. In addition, enhanced GABA-A receptor activity was shown to reduce HCY-induced MMP-9 activation by ERK (extracellular signal-regulated kinase) pathway [26] and plasma level of MMP-9 was shown to be increased in AD patients [80].
This study has some limitations, including unbalanced sample size between cognitively normal controls and demented participants and mixed dementia patients. Due to the small number of demented patients (n = 71), it is difficult to exactly assess diagnostic potential of genetic findings in this study and all finding should be replicated in larger well-balanced cohorts in the future study. The dementia group contained some mixed dementia patients although 65 of 71 patients were diagnosed with AD perhaps limiting this influence. The issue of whether HCY is an AD risk factor and the role of identified SNPs on AD pathophysiology warrants further investigation from a systems biology perspective in a carefully controlled environment considering that biological networks are complicatedly interconnected affecting one another to maintain its homeostasis via potential regulatory feedback networks. This study did not consider B vitamin metabolism in relation to HCY metabolism because B vitamin metabolism is beyond the scope of the present manuscript although this topic is interesting and potentially important. We hope to include this in future studies.
In conclusion, we discovered a novel association of CD2AP with plasma homocysteine in participants with African ancestry and found a new variant in the candidate gene CBS associated with HCY. Many clinical trials investigating HCY reduction as a potential modifiable risk factor employed vitamin B6, B12, and folic acid (B9) to reduce HCY although the clinical benefit is still under debate [81-84]. The inconsistent results of clinical trials may partly be due to unexplained genetic factors influencing HCY. Therefore, the findings in this study merit further investigation regarding their potential role as modifiable AD risk factors.
ACKNOWLEDGMENTS
This research was supported by NIH R01 AG09956 and P30 AG10133. Additional support for analyses included in the present report was provided by the following sources: NIA R01 P30 AG10133, NIA AG19771, NIA R03 AG050856, the Biomarkers Across Neurodegenerative Diseases (BAND) Award (Alzheimer’s Association, Alzheimer’s Research UK, Michael J. Fox Foundation, Weston Brain Institute), NLM R01 LM011360, and NLM R00 LM011384. PMD has received research grants and fees from pharmaceutical and health companies for other projects, and owns stock in AEI, Anthrotronix, Maxwell Health, Muses Labs and Evidation whose products are not discussed in this article. The authors acknowledge the collaboration with recently initiated Alzheimer’s Disease Metabolomics Consortium (http://sites.duke.edu/adnimetab/about-us/metabolomics-and-alzheimers-disease/).
Footnotes
Authors’ disclosures available online (http://j-alz.com/manuscript-disclosures/15-0651r1).
REFERENCES
- [1].Rajagopalan P, Hua X, Toga AW, Jack CR, Jr, Weiner MW, Thompson PM. Homocysteine effects on brain volumes mapped in 732 elderly individuals. Neuroreport. 2011;22:391–395. doi: 10.1097/WNR.0b013e328346bf85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Madsen SK, Rajagopalan P, Joshi SH, Toga AW, Thompson PM. Higher homocysteine associated with thinner cortical gray matter in 803 participants from the Alzheimer’s Disease Neuroimaging Initiative. Neurobiol Aging. 2015;36(Suppl 1):S203–S210. doi: 10.1016/j.neurobiolaging.2014.01.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].den Heijer T, Vermeer SE, Clarke R, Oudkerk M, Koudstaal PJ, Hofman A, Breteler MM. Homocysteine and brain atrophy on MRI of non-demented elderly. Brain. 2003;126:170–175. doi: 10.1093/brain/awg006. [DOI] [PubMed] [Google Scholar]
- [4].Lehmann M, Gottfries CG, Regland B. Identification of cognitive impairment in the elderly: Homocysteine is an early marker. Dement Geriatr Cogn Disord. 1999;10:12–20. doi: 10.1159/000017092. [DOI] [PubMed] [Google Scholar]
- [5].Morris MS, Jacques PF, Rosenberg IH, Selhub J. Hyperhomocysteinemia associated with poor recall in the third National Health and Nutrition Examination Survey. Am J Clin Nutr. 2001;73:927–933. doi: 10.1093/ajcn/73.5.927. [DOI] [PubMed] [Google Scholar]
- [6].Riggs KM, Spiro A, 3rd, Tucker K, Rush D. Relations of vitamin B-12, vitamin B-6, folate, and homocysteine to cognitive performance in the Normative Aging Study. Am J Clin Nutr. 1996;63:306–314. doi: 10.1093/ajcn/63.3.306. [DOI] [PubMed] [Google Scholar]
- [7].Kim JM, Kim SW, Shin IS, Yang SJ, Park WY, Kim SJ, Shin HY, Yoon JS. Folate, vitamin B(12), and homocysteine as risk factors for cognitive decline in the elderly. Psychiatry Investig. 2008;5:36–40. doi: 10.4306/pi.2008.5.1.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Wald DS, Kasturiratne A, Simmonds M. Serum homocysteine and dementia: Meta-analysis of eight cohort studies including 8669 participants. Alzheimers Dement. 2011;7:412–417. doi: 10.1016/j.jalz.2010.08.234. [DOI] [PubMed] [Google Scholar]
- [9].Wang B, Zhong Y, Yan H, Cui L. Meta-analysis of plasma homocysteine content and cognitive function in elderly patients with Alzheimer’s disease and vascular dementia. Int J Clin Exp Med. 2014;7:5118–5123. [PMC free article] [PubMed] [Google Scholar]
- [10].Seshadri S, Beiser A, Selhub J, Jacques PF, Rosenberg IH, D’Agostino RB, Wilson PW, Wolf PA. Plasma homocysteine as a risk factor for dementia and Alzheimer’s disease. N Engl J Med. 2002;346:476–483. doi: 10.1056/NEJMoa011613. [DOI] [PubMed] [Google Scholar]
- [11].Hendrie HC, Baiyewu O, Lane KA, Purnell C, Gao S, Hake A, Ogunniyi A, Gureje O, Unverzagt FW, Murrell J, Deeg MA, Hall K. Homocysteine levels and dementia risk in Yoruba and African Americans. Int Psychogeriatr. 2013;25:1859–1866. doi: 10.1017/S1041610213001294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Luchsinger JA, Tang MX, Shea S, Miller J, Green R, Mayeux R. Plasma homocysteine levels and risk of Alzheimer disease. Neurology. 2004;62:1972–1976. doi: 10.1212/01.wnl.0000129504.60409.88. [DOI] [PubMed] [Google Scholar]
- [13].Haan MN, Miller JW, Aiello AE, Whitmer RA, Jagust WJ, Mungas DM, Allen LH, Green R. Homocysteine, B vitamins, and the incidence of dementia and cognitive impairment: Results from the Sacramento Area Latino Study on Aging. Am J Clin Nutr. 2007;85:511–517. doi: 10.1093/ajcn/85.2.511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Kim JM, Stewart R, Kim SW, Shin IS, Yang SJ, Shin HY, Yoon JS. Changes in folate, vitamin B12 and homocysteine associated with incident dementia. J Neurol Neurosurg Psychiatry. 2008;79:864–868. doi: 10.1136/jnnp.2007.131482. [DOI] [PubMed] [Google Scholar]
- [15].Kivipelto M, Annerbo S, Hultdin J, Backman L, Viitanen M, Fratiglioni L, Lokk J. Homocysteine and holotranscobalamin and the risk of dementia and Alzheimers disease: A prospective study. Eur J Neurol. 2009;16:808–813. doi: 10.1111/j.1468-1331.2009.02590.x. [DOI] [PubMed] [Google Scholar]
- [16].Kaddurah-Daouk R, Zhu H, Sharma S, Bogdanov M, Rozen SG, Matson W, Oki NO, Motsinger-Reif AA, Churchill E, Lei Z, Appleby D, Kling MA, Trojanowski JQ, Doraiswamy PM, Arnold SE. Alterations in metabolic pathways and networks in Alzheimer’s disease. Transl Psychiatry. 2013;3:e244. doi: 10.1038/tp.2013.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Fuso A, Scarpa S. One-carbon metabolism and Alzheimer’s disease: Is it all a methylation matter? Neurobiol Aging. 2011;32:1192–1195. doi: 10.1016/j.neurobiolaging.2011.01.012. [DOI] [PubMed] [Google Scholar]
- [18].Majors AK, Sengupta S, Jacobsen DW, Pyeritz RE. Upregulation of smooth muscle cell collagen production by homocysteine-insight into the pathogenesis of homocystinuria. Mol Genet Metab. 2002;76:92–99. doi: 10.1016/s1096-7192(02)00030-6. [DOI] [PubMed] [Google Scholar]
- [19].Carroll JF, Tyagi SC. Extracellular matrix remodeling in the heart of the homocysteinemic obese rabbit. Am J Hypertens. 2005;18:692–698. doi: 10.1016/j.amjhyper.2004.11.035. [DOI] [PubMed] [Google Scholar]
- [20].Cai BZ, Gong DM, Liu Y, Pan ZW, Xu CQ, Bai YL, Qiao GF, Lu YJ, Yang BF. Homocysteine inhibits potassium channels in human atrial myocytes. Clin Exp Pharmacol Physiol. 2007;34:851–855. doi: 10.1111/j.1440-1681.2007.04671.x. [DOI] [PubMed] [Google Scholar]
- [21].Lominadze D, Tyagi N, Sen U, Ovechkin A, Tyagi SC. Homocysteine alters cerebral microvascular integrity and causes remodeling by antagonizing GABA-A receptor. Mol Cell Biochem. 2012;371:89–96. doi: 10.1007/s11010-012-1425-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Tyagi SC, Lominadze D, Roberts AM. Homocysteine in microvascular endothelial cell barrier permeability. Cell Biochem Biophys. 2005;43:37–44. doi: 10.1385/CBB:43:1:037. [DOI] [PubMed] [Google Scholar]
- [23].Loureiro SO, Heimfarth L, Pelaez Pde L, Vanzin CS, Viana L, Wyse AT, Pessoa-Pureur R. Homocysteine activates calcium-mediated cell signaling mechanisms targeting the cytoskeleton in rat hippocampus. Int J Dev Neurosci. 2008;26:447–455. doi: 10.1016/j.ijdevneu.2008.03.001. [DOI] [PubMed] [Google Scholar]
- [24].Loureiro SO, Romao L, Alves T, Fonseca A, Heimfarth L, Moura Neto V, Wyse AT, Pessoa-Pureur R. Homocysteine induces cytoskeletal remodeling and production of reactive oxygen species in cultured cortical astrocytes. Brain Res. 2010;1355:151–164. doi: 10.1016/j.brainres.2010.07.071. [DOI] [PubMed] [Google Scholar]
- [25].Cacciapuoti G, Manna C, Napoli D, Zappia V, Porcelli M. Homocysteine-induced endothelial cell adhesion is related to adenosine lowering and is not mediated by S-adenosylhomocysteine. FEBS Lett. 2007;581:4567–4570. doi: 10.1016/j.febslet.2007.08.042. [DOI] [PubMed] [Google Scholar]
- [26].Tyagi N, Gillespie W, Vacek JC, Sen U, Tyagi SC, Lominadze D. Activation of GABA-A receptor ameliorates homocysteine-induced MMP-9 activation by ERK pathway. J Cell Physiol. 2009;220:257–266. doi: 10.1002/jcp.21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lipton SA, Kim WK, Choi YB, Kumar S, D’Emilia DM, Rayudu PV, Arnelle DR, Stamler JS. Neurotoxicity associated with dual actions of homocysteine at the N-methyl-D-aspartate receptor. Proc Natl Acad Sci U S A. 1997;94:5923–5928. doi: 10.1073/pnas.94.11.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kruman II, Culmsee C, Chan SL, Kruman Y, Guo Z, Penix L, Mattson MP. Homocysteine elicits a DNA damage response in neurons that promotes apoptosis and hypersensitivity to excitotoxicity. J Neurosci. 2000;20:6920–6926. doi: 10.1523/JNEUROSCI.20-18-06920.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Siva A, De Lange M, Clayton D, Monteith S, Spector T, Brown MJ. The heritability of plasma homocysteine, and the influence of genetic variation in the homocysteine methylation pathway. QJM. 2007;100:495–499. doi: 10.1093/qjmed/hcm054. [DOI] [PubMed] [Google Scholar]
- [30].Keene KL, Chen WM, Chen F, Williams SR, Elkhatib SD, Hsu FC, Mychaleckyj JC, Doheny KF, Pugh EW, Ling H, Laurie CC, Gogarten SM, Madden EB, Worrall BB, Sale MM. Genetic associations with plasma B12, B6, and folate levels in an ischemic stroke population from the Vitamin Intervention for Stroke Prevention (VISP) trial. Front Public Health. 2014;2:112. doi: 10.3389/fpubh.2014.00112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].van Meurs JB, Pare G, Schwartz SM, Hazra A, Tanaka T, Vermeulen SH, Cotlarciuc I, Yuan X, Malarstig A, Bandinelli S, Bis JC, Blom H, Brown MJ, Chen C, Chen YD, Clarke RJ, Dehghan A, Erdmann J, Ferrucci L, Hamsten A, Hofman A, Hunter DJ, Goel A, Johnson AD, Kathiresan S, Kampman E, Kiel DP, Kiemeney LA, Chambers JC, Kraft P, Lindemans J, McKnight B, Nelson CP, O’Donnell CJ, Psaty BM, Ridker PM, Rivadeneira F, Rose LM, Seedorf U, Siscovick DS, Schunkert H, Selhub J, Ueland PM, Vollenweider P, Waeber G, Waterworth DM, Watkins H, Witteman JC, den Heijer M, Jacques P, Uitterlinden AG, Kooner JS, Rader DJ, Reilly MP, Mooser V, Chasman DI, Samani NJ, Ahmadi KR. Common genetic loci influencing plasma homocysteine concentrations and their effect on risk of coronary artery disease. Am J Clin Nutr. 2013;98:668–676. doi: 10.3945/ajcn.112.044545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Tanaka T, Scheet P, Giusti B, Bandinelli S, Piras MG, Usala G, Lai S, Mulas A, Corsi AM, Vestrini A, Sofi F, Gori AM, Abbate R, Guralnik J, Singleton A, Abecasis GR, Schlessinger D, Uda M, Ferrucci L. Genome-wide association study of vitamin B6, vitamin B12, folate, and homocysteine blood concentrations. Am J Hum Genet. 2009;84:477–482. doi: 10.1016/j.ajhg.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lange LA, Croteau-Chonka DC, Marvelle AF, Qin L, Gaulton KJ, Kuzawa CW, McDade TW, Wang Y, Li Y, Levy S, Borja JB, Lange EM, Adair LS, Mohlke KL. Genome-wide association study of homocysteine levels in Filipinos provides evidence for CPS1 in women and a stronger MTHFR effect in young adults. Hum Mol Genet. 2010;19:2050–2058. doi: 10.1093/hmg/ddq062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Naj AC, Beecham GW, Martin ER, Gallins PJ, Powell EH, Konidari I, Whitehead PL, Cai G, Haroutunian V, Scott WK, Vance JM, Slifer MA, Gwirtsman HE, Gilbert JR, Haines JL, Buxbaum JD, Pericak-Vance MA. Dementia revealed: Novel chromosome 6 locus for late-onset Alzheimer disease provides genetic evidence for folate-pathway abnormalities. PLoS Genet. 2010;6:e1001130. doi: 10.1371/journal.pgen.1001130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Deeg M, Baiyewu O, Gao S, Ogunniyi A, Shen J, Gureje O, Taylor S, Murrell J, Unverzagt F, Smith-Gamble V, Evans R, Dickens J, Hendrie H, Hall K. A comparison of cardiovascular disease risk factor biomarkers in African Americans and Yoruba Nigerians. Ethn Dis. 2008;18:427–433. [PMC free article] [PubMed] [Google Scholar]
- [36].Hendrie HC, Murrell J, Baiyewu O, Lane KA, Purnell C, Ogunniyi A, Unverzagt FW, Hall K, Callahan CM, Saykin AJ, Gureje O, Hake A, Foroud T, Gao S. APOE epsilon4 and the risk for Alzheimer disease and cognitive decline in African Americans and Yoruba. Int Psychogeriatr. 2014;26:977–985. doi: 10.1017/S1041610214000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Murrell JR, Price BM, Baiyewu O, Gureje O, Deeg M, Hendrie H, Ogunniyi A, Hall K. The fourth apolipoprotein E haplotype found in the Yoruba of Ibadan. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:426–427. doi: 10.1002/ajmg.b.30295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Liu H, Gao S, Hall KS, Unverzagt FW, Lane KA, Callahan CM, Hendrie HC. Optimal blood pressure for cognitive function: Findings from an elderly African-American cohort study. J Am Geriatr Soc. 2013;61:875–881. doi: 10.1111/jgs.12259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hall KS, Ogunniyi AO, Hendrie HC, Osuntokun BO, Hui SL, Musick BS, Rodenberg CA, Unverzagt FW, Guerje O, Baiyewu O. A cross-cultural community based study of dementias: Methods and performance of the survey instrument Indianapolis, USA, and Ibadan, Nigeria. Int J Methods Psychiatr Res. 1996;6:129–142. [Google Scholar]
- [40].Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kim S, Swaminathan S, Inlow M, Risacher SL, Nho K, Shen L, Foroud TM, Petersen RC, Aisen PS, Soares H, Toledo JB, Shaw LM, Trojanowski JQ, Weiner MW, McDonald BC, Farlow MR, Ghetti B, Saykin AJ. Influence of genetic variation on plasma protein levels in older adults using a multianalyte panel. PLoS One. 2013;8:e70269. doi: 10.1371/journal.pone.0070269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Nho K, Corneveaux JJ, Kim S, Lin H, Risacher SL, Shen L, Swaminathan S, Ramanan VK, Liu Y, Foroud T, Inlow MH, Siniard AL, Reiman RA, Aisen PS, Petersen RC, Green RC, Jack CR, Weiner MW, Baldwin CT, Lunetta K, Farrer LA, Furney SJ, Lovestone S, Simmons A, Mecocci P, Vellas B, Tsolaki M, Kloszewska I, Soininen H, McDonald BC, Farlow MR, Ghetti B, Huentelman MJ, Saykin AJ. Whole-exome sequencing and imaging genetics identify functional variants for rate of change in hippocampal volume in mild cognitive impairment. Mol Psychiatry. 2013;18:781–787. doi: 10.1038/mp.2013.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ramanan VK, Risacher SL, Nho K, Kim S, Swaminathan S, Shen L, Foroud TM, Hakonarson H, Huentelman MJ, Aisen PS, Petersen RC, Green RC, Jack CR, Koeppe RA, Jagust WJ, Weiner MW, Saykin AJ. APOE and BCHE as modulators of cerebral amyloid deposition: A florbetapir PET genome-wide association study. Mol Psychiatry. 2014;19:351–357. doi: 10.1038/mp.2013.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- [45].Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- [46].Nam D, Kim J, Kim SY, Kim S. GSA-SNP: A general approach for gene set analysis of polymorphisms. Nucleic Acids Res. 2010;38:W749–W754. doi: 10.1093/nar/gkq428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Ramanan VK, Shen L, Moore JH, Saykin AJ. Pathway analysis of genomic data: Concepts, methods, and prospects for future development. Trends Genet. 2012;28:323–332. doi: 10.1016/j.tig.2012.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- [49].Hollingworth P, Harold D, Sims R, Gerrish A, Lambert JC, Carrasquillo MM, Abraham R, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Jones N, Stretton A, Thomas C, Richards A, Ivanov D, Widdowson C, Chapman J, Lovestone S, Powell J, Proitsi P, Lupton MK, Brayne C, Rubinsztein DC, Gill M, Lawlor B, Lynch A, Brown KS, Passmore PA, Craig D, McGuinness B, Todd S, Holmes C, Mann D, Smith AD, Beaumont H, Warden D, Wilcock G, Love S, Kehoe PG, Hooper NM, Vardy ER, Hardy J, Mead S, Fox NC, Rossor M, Collinge J, Maier W, Jessen F, Ruther E, Schurmann B, Heun R, Kolsch H, van den Bussche H, Heuser I, Kornhuber J, Wiltfang J, Dichgans M, Frolich L, Hampel H, Gallacher J, Hull M, Rujescu D, Giegling I, Goate AM, Kauwe JS, Cruchaga C, Nowotny P, Morris JC, Mayo K, Sleegers K, Bettens K, Engelborghs S, De Deyn PP, Van Broeckhoven C, Livingston G, Bass NJ, Gurling H, McQuillin A, Gwilliam R, Deloukas P, Al-Chalabi A, Shaw CE, Tsolaki M, Singleton AB, Guerreiro R, Muhleisen TW, Nothen MM, Moebus S, Jockel KH, Klopp N, Wichmann HE, Pankratz VS, Sando SB, Aasly JO, Barcikowska M, Wszolek ZK, Dickson DW, Graff-Radford NR, Petersen RC, van Duijn CM, Breteler MM, Ikram MA, DeStefano AL, Fitzpatrick AL, Lopez O, Launer LJ, Seshadri S, Berr C, Campion D, Epelbaum J, Dartigues JF, Tzourio C, Alperovitch A, Lathrop M, Feulner TM, Friedrich P, Riehle C, Krawczak M, Schreiber S, Mayhaus M, Nicolhaus S, Wagenpfeil S, Steinberg S, Stefansson H, Stefansson K, Snaedal J, Bjornsson S, Jonsson PV, Chouraki V, Genier-Boley B, Hiltunen M, Soininen H, Combarros O, Zelenika D, Delepine M, Bullido MJ, Pasquier F, Mateo I, Frank-Garcia A, Porcellini E, Hanon O, Coto E, Alvarez V, Bosco P, Siciliano G, Mancuso M, Panza F, Solfrizzi V, Nacmias B, Sorbi S, Bossu P, Piccardi P, Arosio B, Annoni G, Seripa D, Pilotto A, Scarpini E, Galimberti D, Brice A, Hannequin D, Licastro F, Jones L, Holmans PA, Jonsson T, Riemenschneider M, Morgan K, Younkin SG, Owen MJ, O’Donovan M, Amouyel P, Williams J. Common variants at ABCA7, MS4A6A/MS4A4E, EPHA1, CD33 and CD2AP are associated with Alzheimer’s disease. Nat Genet. 2011;43:429–435. doi: 10.1038/ng.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, DeStafano AL, Bis JC, Beecham GW, Grenier-Boley B, Russo G, Thorton-Wells TA, Jones N, Smith AV, Chouraki V, Thomas C, Ikram MA, Zelenika D, Vardarajan BN, Kamatani Y, Lin CF, Gerrish A, Schmidt H, Kunkle B, Dunstan ML, Ruiz A, Bihoreau MT, Choi SH, Reitz C, Pasquier F, Cruchaga C, Craig D, Amin N, Berr C, Lopez OL, De Jager PL, Deramecourt V, Johnston JA, Evans D, Lovestone S, Letenneur L, Moron FJ, Rubinsztein DC, Eiriksdottir G, Sleegers K, Goate AM, Fievet N, Huentelman MW, Gill M, Brown K, Kamboh MI, Keller L, Barberger-Gateau P, McGuiness B, Larson EB, Green R, Myers AJ, Dufouil C, Todd S, Wallon D, Love S, Rogaeva E, Gallacher J, St George-Hyslop P, Clarimon J, Lleo A, Bayer A, Tsuang DW, Yu L, Tsolaki M, Bossu P, Spalletta G, Proitsi P, Collinge J, Sorbi S, Sanchez-Garcia F, Fox NC, Hardy J, Deniz Naranjo MC, Bosco P, Clarke R, Brayne C, Galimberti D, Mancuso M, Matthews F, Moebus S, Mecocci P, Del Zompo M, Maier W, Hampel H, Pilotto A, Bullido M, Panza F, Caffarra P, Nacmias B, Gilbert JR, Mayhaus M, Lannefelt L, Hakonarson H, Pichler S, Carrasquillo MM, Ingelsson M, Beekly D, Alvarez V, Zou F, Valladares O, Younkin SG, Coto E, Hamilton-Nelson KL, Gu W, Razquin C, Pastor P, Mateo I, Owen MJ, Faber KM, Jonsson PV, Combarros O, O’Donovan MC, Cantwell LB, Soininen H, Blacker D, Mead S, Mosley TH, Jr, Bennett DA, Harris TB, Fratiglioni L, Holmes C, de Bruijn RF, Passmore P, Montine TJ, Bettens K, Rotter JI, Brice A, Morgan K, Foroud TM, Kukull WA, Hannequin D, Powell JF, Nalls MA, Ritchie K, Lunetta KL, Kauwe JS, Boerwinkle E, Riemenschneider M, Boada M, Hiltuenen M, Martin ER, Schmidt R, Rujescu D, Wang LS, Dartigues JF, Mayeux R, Tzourio C, Hofman A, Nothen MM, Graff C, Psaty BM, Jones L, Haines JL, Holmans PA, Lathrop M, Pericak-Vance MA, Launer LJ, Farrer LA, van Duijn CM, Van Broeckhoven C, Moskvina V, Seshadri S, Williams J, Schellenberg GD, Amouyel P. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Naj AC, Jun G, Beecham GW, Wang LS, Vardarajan BN, Buros J, Gallins PJ, Buxbaum JD, Jarvik GP, Crane PK, Larson EB, Bird TD, Boeve BF, Graff-Radford NR, De Jager PL, Evans D, Schneider JA, Carrasquillo MM, Ertekin-Taner N, Younkin SG, Cruchaga C, Kauwe JS, Nowotny P, Kramer P, Hardy J, Huentelman MJ, Myers AJ, Barmada MM, Demirci FY, Baldwin CT, Green RC, Rogaeva E, St George-Hyslop P, Arnold SE, Barber R, Beach T, Bigio EH, Bowen JD, Boxer A, Burke JR, Cairns NJ, Carlson CS, Carney RM, Carroll SL, Chui HC, Clark DG, Corneveaux J, Cotman CW, Cummings JL, DeCarli C, DeKosky ST, Diaz-Arrastia R, Dick M, Dickson DW, Ellis WG, Faber KM, Fallon KB, Farlow MR, Ferris S, Frosch MP, Galasko DR, Ganguli M, Gearing M, Geschwind DH, Ghetti B, Gilbert JR, Gilman S, Giordani B, Glass JD, Growdon JH, Hamilton RL, Harrell LE, Head E, Honig LS, Hulette CM, Hyman BT, Jicha GA, Jin LW, Johnson N, Karlawish J, Karydas A, Kaye JA, Kim R, Koo EH, Kowall NW, Lah JJ, Levey AI, Lieberman AP, Lopez OL, Mack WJ, Marson DC, Martiniuk F, Mash DC, Masliah E, McCormick WC, McCurry SM, McDavid AN, McKee AC, Mesulam M, Miller BL, Miller CA, Miller JW, Parisi JE, Perl DP, Peskind E, Petersen RC, Poon WW, Quinn JF, Rajbhandary RA, Raskind M, Reisberg B, Ringman JM, Roberson ED, Rosenberg RN, Sano M, Schneider LS, Seeley W, Shelanski ML, Slifer MA, Smith CD, Sonnen JA, Spina S, Stern RA, Tanzi RE, Trojanowski JQ, Troncoso JC, Van Deerlin VM, Vinters HV, Vonsattel JP, Weintraub S, Welsh-Bohmer KA, Williamson J, Woltjer RL, Cantwell LB, Dombroski BA, Beekly D, Lunetta KL, Martin ER, Kamboh MI, Saykin AJ, Reiman EM, Bennett DA, Morris JC, Montine TJ, Goate AM, Blacker D, Tsuang DW, Hakonarson H, Kukull WA, Foroud TM, Haines JL, Mayeux R, Pericak-Vance MA, Farrer LA, Schellenberg GD. Common variants at MS4A4/MS4A6E, CD2AP, CD33 and EPHA1 are associated with late-onset Alzheimer’s disease. Nat Genet. 2011;43:436–441. doi: 10.1038/ng.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Limon A, Reyes-Ruiz JM, Miledi R. Loss of functional GABA(A) receptors in the Alzheimer diseased brain. Proc Natl Acad Sci U S A. 2012;109:10071–10076. doi: 10.1073/pnas.1204606109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ignoul S, Eggermont J. CBS domains: Structure, function, and pathology in human proteins. Am J Physiol Cell Physiol. 2005;289:C1369–C1378. doi: 10.1152/ajpcell.00282.2005. [DOI] [PubMed] [Google Scholar]
- [54].Hamelet J, Demuth K, Paul JL, Delabar JM, Janel N. Hyperhomocysteinemia due to cystathionine beta synthase deficiency induces dysregulation of genes involved in hepatic lipid homeostasis in mice. J Hepatol. 2007;46:151–159. doi: 10.1016/j.jhep.2006.07.028. [DOI] [PubMed] [Google Scholar]
- [55].Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, Walter K, Menni C, Chen L, Vasquez L, Valdes AM, Hyde CL, Wang V, Ziemek D, Roberts P, Xi L, Grundberg E, Waldenberger M, Richards JB, Mohney RP, Milburn MV, John SL, Trimmer J, Theis FJ, Overington JP, Suhre K, Brosnan MJ, Gieger C, Kastenmuller G, Spector TD, Soranzo N. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543–550. doi: 10.1038/ng.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Chai GS, Jiang X, Ni ZF, Ma ZW, Xie AJ, Cheng XS, Wang Q, Wang JZ, Liu GP. Betaine attenuates Alzheimer-like pathological changes and memory deficits induced by homocysteine. J Neurochem. 2013;124:388–396. doi: 10.1111/jnc.12094. [DOI] [PubMed] [Google Scholar]
- [57].Imbard A, Benoist JF, Esse R, Gupta S, Lebon S, De Vriese AS, Ogier De Baulny H, Kruger W, Schiff M, Blom HJ. High homocysteine induces betaine depletion. Biosci Rep. 2015;35:e00222. doi: 10.1042/BSR20150094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Saykin AJ, Shen L, Yao X, Kim S, Nho K, Risacher SL, Ramanan VK, Foroud TM, Faber KM, Sarwar N, Munsie LM, Hu X, Soares HD, Potkin SG, Thompson PM, Kauwe JSK, Kaddurah-Daouk R, Green RC, Toga AW, Weiner MW. Genetic studies of quantitative MCI and AD phenotypes in ADNI: Progress, opportunities, and plans. Alzheimers Dement. 2015;11:792–814. doi: 10.1016/j.jalz.2015.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Beyer K, Lao JI, Carrato C, Rodriguez-Vila A, Latorre P, Mataro M, Llopis MA, Mate JL, Ariza A. Cystathionine beta synthase as a risk factor for Alzheimer disease. Curr Alzheimer Res. 2004;1:127–133. doi: 10.2174/1567205043332243. [DOI] [PubMed] [Google Scholar]
- [60].Ichinohe A, Kanaumi T, Takashima S, Enokido Y, Nagai Y, Kimura H. Cystathionine beta-synthase is enriched in the brains of Down’s patients. Biochem Biophys Res Commun. 2005;338:1547–1550. doi: 10.1016/j.bbrc.2005.10.118. [DOI] [PubMed] [Google Scholar]
- [61].Shulman JM, Chen K, Keenan BT, Chibnik LB, Fleisher A, Thiyyagura P, Roontiva A, McCabe C, Patsopoulos NA, Corneveaux JJ, Yu L, Huentelman MJ, Evans DA, Schneider JA, Reiman EM, De Jager PL, Bennett DA. Genetic susceptibility for Alzheimer disease neuritic plaque pathology. JAMA Neurol. 2013;70:1150–1157. doi: 10.1001/jamaneurol.2013.2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Logue MW, Schu M, Vardarajan BN, Buros J, Green RC, Go RC, Griffith P, Obisesan TO, Shatz R, Borenstein A, Cupples LA, Lunetta KL, Fallin MD, Baldwin CT, Farrer LA. A comprehensive genetic association study of Alzheimer disease in African Americans. Arch Neurol. 2011;68:1569–1579. doi: 10.1001/archneurol.2011.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Reitz C, Jun G, Naj A, Rajbhandary R, Vardarajan BN, Wang LS, Valladares O, Lin CF, Larson EB, Graff-Radford NR, Evans D, De Jager PL, Crane PK, Buxbaum JD, Murrell JR, Raj T, Ertekin-Taner N, Logue M, Baldwin CT, Green RC, Barnes LL, Cantwell LB, Fallin MD, Go RC, Griffith P, Obisesan TO, Manly JJ, Lunetta KL, Kamboh MI, Lopez OL, Bennett DA, Hendrie H, Hall KS, Goate AM, Byrd GS, Kukull WA, Foroud TM, Haines JL, Farrer LA, Pericak-Vance MA, Schellenberg GD, Mayeux R. Variants in the ATP-binding cassette transporter (ABCA7), apolipoprotein E4, and the risk of late-onset Alzheimer disease in African Americans. JAMA. 2013;309:1483–1492. doi: 10.1001/jama.2013.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Cormont M, Meton I, Mari M, Monzo P, Keslair F, Gaskin C, McGraw TE, Le Marchand-Brustel Y. CD2AP/CMS regulates endosome morphology and traffic to the degradative pathway through its interaction with Rab4 and c-Cbl. Traffic. 2003;4:97–112. doi: 10.1034/j.1600-0854.2003.40205.x. [DOI] [PubMed] [Google Scholar]
- [65].Dustin ML, Olszowy MW, Holdorf AD, Li J, Bromley S, Desai N, Widder P, Rosenberger F, van der Merwe PA, Allen PM, Shaw AS. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell. 1998;94:667–677. doi: 10.1016/s0092-8674(00)81608-6. [DOI] [PubMed] [Google Scholar]
- [66].Lehtonen S, Zhao F, Lehtonen E. CD2-associated protein directly interacts with the actin cytoskeleton. Am J Physiol Renal Physiol. 2002;283:F734–F743. doi: 10.1152/ajprenal.00312.2001. [DOI] [PubMed] [Google Scholar]
- [67].Lynch DK, Winata SC, Lyons RJ, Hughes WE, Lehrbach GM, Wasinger V, Corthals G, Cordwell S, Daly RJ. A Cortactin-CD2-associated protein (CD2AP) complex provides a novel link between epidermal growth factor receptor endocytosis and the actin cytoskeleton. J Biol Chem. 2003;278:21805–21813. doi: 10.1074/jbc.M211407200. [DOI] [PubMed] [Google Scholar]
- [68].Deckert M, Ticchioni M, Mari B, Mary D, Bernard A. The glycosylphosphatidylinositol-anchored CD59 protein stimulates both T cell receptor zeta/ZAP-70-dependent and -independent signaling pathways in T cells. Eur J Immunol. 1995;25:1815–1822. doi: 10.1002/eji.1830250704. [DOI] [PubMed] [Google Scholar]
- [69].Naderi S, Hofmann P, Seiter S, Tilgen W, Abken H, Reinhold U. CD2-mediated CD59 stimulation in keratinocytes results in secretion of IL-1alpha, IL-6, and GM-CSF: Implications for the interaction of keratinocytes with intraepidermal T lymphocytes. Int J Mol Med. 1999;3:609–614. doi: 10.3892/ijmm.3.6.609. [DOI] [PubMed] [Google Scholar]
- [70].Shulman JM, Imboywa S, Giagtzoglou N, Powers MP, Hu Y, Devenport D, Chipendo P, Chibnik LB, Diamond A, Perrimon N, Brown NH, De Jager PL, Feany MB. Functional screening in Drosophila identifies Alzheimer’s disease susceptibility genes and implicates Tau-mediated mechanisms. Hum Mol Genet. 2014;23:870–877. doi: 10.1093/hmg/ddt478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Liao F, Jiang H, Srivatsan S, Xiao Q, Lefton KB, Yamada K, Mahan TE, Lee JM, Shaw AS, Holtzman DM. Effects of CD2-associated protein deficiency on amyloid-beta in neuroblastoma cells and in an APP transgenic mouse model. Mol Neurodegener. 2015;10:12. doi: 10.1186/s13024-015-0006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Karch CM, Jeng AT, Nowotny P, Cady J, Cruchaga C, Goate AM. Expression of novel Alzheimer’s disease risk genes in control and Alzheimer’s disease brains. PLoS One. 2012;7:e50976. doi: 10.1371/journal.pone.0050976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Fox CS, Liu Y, White CC, Feitosa M, Smith AV, Heard-Costa N, Lohman K, Johnson AD, Foster MC, Greenawalt DM, Griffin P, Ding J, Newman AB, Tylavsky F, Miljkovic I, Kritchevsky SB, Launer L, Garcia M, Eiriksdottir G, Carr JJ, Gudnason V, Harris TB, Cupples LA, Borecki IB. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012;8:e1002695. doi: 10.1371/journal.pgen.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Fox CS, Massaro JM, Hoffmann U, Pou KM, Maurovich-Horvat P, Liu CY, Vasan RS, Murabito JM, Meigs JB, Cupples LA, D’Agostino RB, Sr, O’Donnell CJ. Abdominal visceral and subcutaneous adipose tissue compartments: Association with metabolic risk factors in the Framingham Heart Study. Circulation. 2007;116:39–48. doi: 10.1161/CIRCULATIONAHA.106.675355. [DOI] [PubMed] [Google Scholar]
- [75].Liu J, Fox CS, Hickson DA, May WD, Hairston KG, Carr JJ, Taylor HA. Impact of abdominal visceral and subcutaneous adipose tissue on cardiometabolic risk factors: The Jackson Heart Study. J Clin Endocrinol Metab. 2010;95:5419–5426. doi: 10.1210/jc.2010-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, Butte NF. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7:e51954. doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Wang KS, Zhang Q, Liu X, Wu L, Zeng M. PKNOX2 is associated with formal thought disorder in schizophrenia: A meta-analysis of two genome-wide association studies. J Mol Neurosci. 2012;48:265–272. doi: 10.1007/s12031-012-9787-4. [DOI] [PubMed] [Google Scholar]
- [78].Jo S, Yarishkin O, Hwang YJ, Chun YE, Park M, Woo DH, Bae JY, Kim T, Lee J, Chun H, Park HJ, Lee da Y, Hong J, Kim HY, Oh SJ, Park SJ, Lee H, Yoon BE, Kim Y, Jeong Y, Shim I, Bae YC, Cho J, Kowall NW, Ryu H, Hwang E, Kim D, Lee CJ. GABA from reactive astrocytes impairs memory in mouse models of Alzheimer’s disease. Nat Med. 2014;20:886–896. doi: 10.1038/nm.3639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Lanctot KL, Herrmann N, Mazzotta P, Khan LR, Ingber N. GABAergic function in Alzheimer’s disease: Evidence for dysfunction and potential as a therapeutic target for the treatment of behavioural and psychological symptoms of dementia. Can J Psychiatry. 2004;49:439–453. doi: 10.1177/070674370404900705. [DOI] [PubMed] [Google Scholar]
- [80].Lorenzl S, Albers DS, Relkin N, Ngyuen T, Hilgenberg SL, Chirichigno J, Cudkowicz ME, Beal MF. Increased plasma levels of matrix metalloproteinase-9 in patients with Alzheimer’s disease. Neurochem Int. 2003;43:191–196. doi: 10.1016/s0197-0186(03)00004-4. [DOI] [PubMed] [Google Scholar]
- [81].Malouf M, Grimley EJ, Areosa SA. Folic acid with or without vitamin B12 for cognition and dementia. Cochrane Database Syst Rev. 2003:CD004514. doi: 10.1002/14651858.CD004514. [DOI] [PubMed] [Google Scholar]
- [82].Malouf R, Grimley Evans J. Folic acid with or without vitamin B12 for the prevention and treatment of healthy elderly and demented people. Cochrane Database Syst Rev. 2008:CD004514. doi: 10.1002/14651858.CD004514.pub2. [DOI] [PubMed] [Google Scholar]
- [83].Malouf R, Areosa Sastre A. Vitamin B12 for cognition. Cochrane Database Syst Rev. 2003:CD004326. doi: 10.1002/14651858.CD004326. [DOI] [PubMed] [Google Scholar]
- [84].Agnew-Blais JC, Wassertheil-Smoller S, Kang JH, Hogan PE, Coker LH, Snetselaar LG, Smoller JW. Folate, vitamin B-6, and vitamin B-12 intake and mild cognitive impairment and probable dementia in the Women’s Health Initiative Memory Study. J Acad Nutr Diet. 2015;115:231–241. doi: 10.1016/j.jand.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]