Abstract
The biological membrane is an efficient barrier against water-soluble substances. Solute transporters circumvent this membrane barrier by transporting water-soluble solutes across the membrane to the other sides. These transport proteins are thus required for all living organisms. Microorganisms, such as bacteria, effectively exploit solute transporters to acquire useful nutrients for growth or to expel substances that are inhibitory to their growth. Overall, there are distinct types of related solute transporters that are grouped into families or superfamilies. Of these various transporters, the major facilitator superfamily (MFS) represents a very large and constantly growing group and are driven by solute- and ion-gradients, making them passive and secondary active transporters, respectively. Members of the major facilitator superfamily transport an extreme variety of structurally different substrates such as antimicrobial agents, amino acids, sugars, intermediary metabolites, ions, and other small molecules. Importantly, bacteria, especially pathogenic ones, have evolved multidrug efflux pumps which belong to the major facilitator superfamily. Furthermore, members of this important superfamily share similar primary sequences in the form of highly conserved sequence motifs that confer useful functional properties during transport. The transporters of the superfamily also share similarities in secondary structures, such as possessing 12- or 14-membrane spanning α-helices and the more recently described 3-helix structure repeat element, known as the MFS fold. The three-dimensional structures of bacterial multidrug efflux pumps have been determined for only a few members of the superfamily, all drug pumps of which are surprisingly from Escherichia coli. This review briefly summarizes the structural properties of the bacterial multidrug efflux pumps of the major facilitator superfamily in a comparative manner and provides future directions for study.
Keywords: bacteria, multidrug efflux, antimicrobial resistance, major facilitator superfamily
Importance of solute transport
All living prokaryotic and eukaryotic cells are enclosed by a phospholipid bilayer membrane. This biological membrane is a barrier that prevents the movement of water-soluble solutes and ions, most of which are necessary for the life of the cell and must be contained intracellularly and in appropriate concentrations. These living cells, therefore, must have the ability to acquire and keep solutes that are necessary for life while simultaneously preventing the entry of and exporting solutes that are harmful. The biological membrane resolves both of these barrier and homeostatic challenges by using membrane transporter proteins which catalyze the entry and efflux of solutes across the membrane. All living cells harbor these membrane-bound solute transporters which are, thus, critical for all life [1–3].
Types of solute transporters
In terms of bioenergetics, solute transporters in living cells can be generally divided into two main categories. In the first category, solute transporters, called passive transporters, do not use biological energy to accomplish solute transport across the membrane. In these passive transport systems, the solutes are driven across the membrane by the solute gradient from a high solute concentration towards a low solute concentration. Examples of this system include carriers and porins that mediate facilitated diffusion. In the second category, solute transporters called active transporters use biological energy to transport solutes across the membrane in which solutes accumulate on one side of the membrane. Within these active solute transporters, there are two groups of solute transport systems. Primary active transport represents the first system, and it uses energy contained within the hydrolysis of adenosine triphosphate (ATP) to mediate passage of solutes across the membrane from a low solute concentration on one side of the membrane to a high solute concentration on the other side of the membrane [4]. Secondary active transport represents the second system, and it uses the energy contained within ion gradients by moving these ions across the membrane down their concentration gradients in order to transport solutes across the membrane against the solute gradient to mediate solute accumulation [5]. A third group of active transport consists of group translocation in which solute is enzymatically modified during transport, such as those seen in the phosphotransferase systems (PTS) for carbohydrates [6].
Superfamilies of solute transporters
In terms of phylogenetic relationships, solute transporters can be categorized into superfamilies consisting of families of related and homologous solute transporters. The individual solute transporters contained within these superfamilies share related primary amino acid sequences and protein structures. With respect to primary active transporters, a well-known and highly studied superfamily is that of the ATP-binding cassette (ABC) transporter superfamily [7]. On the other hand, secondary active transporters for antimicrobial agents are exemplified by several distinctive major superfamilies. These particular superfamilies include the multidrug and toxic compound extrusion (MATE) superfamily [8], the resistance nodulation cell division (RND) superfamily [9], the small multidrug resistance (SMR) superfamily [10], and the major facilitator superfamily (MFS) [11, 12].
Major facilitator superfamily
Due to their inherent hydrophobic and flexible nature, solute transporters are recalcitrant to protein purification with biochemical techniques. Therefore, molecular biological techniques have fostered the cloning of the genes encoding solute transporters, such as that seen for a key solute transporter, LacY, the lactose permease of Escherichia coli [13]. As a consequence of these early developments in gene cloning, nucleotide sequence elucidation became possible for genes encoding solute transporters [14]. Shortly after the DNA sequences became available for additional genes encoding solute transporters, Henderson and colleagues made the important and groundbreaking discovery that distinctive sugar transporters were homologous to each other [15], indicating that these various sugar transporters shared a common evolutionary origin despite that fact that these proteins were from different prokaryotic and eukaryotic organisms. As additional DNA sequences of genes coding for solute transporters became available and their deduced primary sequences were compared to each other, investigators began to group these seemingly unrelated transporters in families and superfamilies based on their sequence relatedness. These transporter groups were initially called the transporter superfamily (TSF) [16] or the uniporter, symporter, antiporter family (USA) [17], and the generally accepted term is major facilitator superfamily (MFS) [18].
Presently, the MFS harbors thousands of transporters conveniently organized in the well maintained Transporter Classification Database (TCD) www.tcdb.org [19]. Currently, the MFS contains over 15,000 individual solute transporters [19] and is a well-studied constellation of solute transporters from all known taxa [11, 12, 20–22]. The substrates of the transporters in the MFS are structurally diverse and include distinctive low molecular weight molecules such as sugars, antimicrobial agents, amino acids, nucleic acids, and intermediary metabolites.
Members of the MFS include uniporters, symporters and antiporters [23]. A uniporter catalyzes the facilitated diffusion of a single substrate across the membrane down its substrate concentration gradient [18]. Symporters catalyze ion-gradient driven secondary active transport of solute and ion in the same direction across the membrane, and antiporters catalyze ion-driven secondary active transport of substrate and ion across the membrane but in opposite directions [11, 20]. Both symporters and antiporters accumulate their substrates on one side of the membrane against their concentration gradients.
Conserved amino acid sequence motifs of the MFS
Early studies showing high degrees of relatedness among members of the MFS also definitively demonstrated that highly conserved amino acid sequence motifs were shared [24–28]. One of these motifs, now called Motif A, was discovered by Henderson and colleagues, has residues “G (X)3 D R/K X G R R/K” and is found in the loop between helices 2 and 3 of virtually all of the MFS transporters [15, 29]. One of the first structure-function studies of Motif A in an MFS transporter showed that the Ser-65 – Asp-66 dipeptide within the motif of the Tn10 TetA(B) tetracycline efflux pump [30], required a negative charge and the inter-helical loop for gating but not for substrate binding [31]. The lack of requirement for a negative charge in the loop during substrate binding was in contrast to previous work [32] implicating residues in the loop between helices 2 and 3 as participating in substrate binding. Further studies showed that Asp-84 of helix 3 and Gly-62, Asp-66, Ser-77 and Arg-70 (residues of Motif A) formed a tetracycline channel structure and possibly mediated conformational changes during tetracycline efflux [33, 34].
Another important conserved sequence motif, now called Motif C, has residues “G (X)8 G (X)3 G P (X)2 G G” discovered by Rouch et al. to reside within the fifth transmembrane domain of transporters of the MFS [28]. Initially thought to be present only in antiporters of the MFS but not in symporters or uniporters, Motif C had been referred to as the “antiporter motif” [35, 36]. Recently, however, manual adjustments made to an extensive multiple sequence comparison surprisingly showed that elements of the “antiporter motif” were indeed present in both symporters and uniporters of the MFS [37]. Varela et al. conducted the first structure-function analysis to examine the functional importance of the most highly conserved residue of Motif C, Gly-147 of TetA(C), a pBR322 encoded tetracycline efflux pump [35]. Molecular modeling mechanics further suggested that helix 5 formed a slight bend or kink in the wild-type TetA(C) [35].
Structures of MFS transporters
In general terms, the transporters of the MFS have 12 or 14 transmembrane-spanning segments (TMS) [38–40]. As of this writing, crystal structures have been elucidated for over a dozen MFS transporters. These MFS protein structures include the multidrug efflux pump, EmrD, from E. coli [41], the fucose transporter, FucP, from E. coli [42], the glucose-H+ symporter, GlcPSc, from Staphylococcus epidermidis [43], the glycerol-3-phosphate transporter, GlpT, from E. coli [44], the glucose transporter, GLUT1, from Homo sapiens [45], the lactose symporter, LacY, from E. coli [46], the multidrug efflux pump, MdfA, from E. coli [47], the nitrate/nitrite antiporter, NarK, from E. coli [48], the nitrate/nitrIte exchanger, NarU, from E. coli [49], the oligopeptide-H+ symporter, PepTSo, from Shewanella oneidensis [50], the phosphate transporter, PipT, from Piriformospora indica [51], the xylose transporter, XylE, from E. coil [45], the multidrug transporter, YajR, from E. coli [52], the peptide transporter, YbgH, from E. coli [53], and most recently, the mammalian fructose transporter, GLUT5, from Rattus norvegicus and Bos taurus [54].
Thus far, the general properties of the elucidated protein structures show that the MFS transporters consist of two structurally symmetrical and functionally asymmetrical bundles, also called N-terminal domains (NTDs) and C-terminal domains (CTDs), and each of these bundles are in turn composed of the N-terminal TMSs 1 through 6 and of the C-terminal TMSs 7 through 12, respectively [11, 21]. In essence, an ancestral transporter with 6-TMSs underwent an internal gene sequence duplication, and then a tandem connection of the duplicated segments gave rise to the modern 12-TMS MFS transporter with the two bundles [26]. Additionally, a common structural feature of the MFS transporters is a large central aqueous cavity formed by elements of the two bundles, supporting prior genetic analyses of the tetracycline efflux pump, TetA(C), in which it was proposed that the N- and C-termini bundles or domains interact functionally [55, 56] and supporting Mitchell’s notion in which he proposed that a proton gradient is an energy source for driving solute transport across the membrane [23, 57]. When one considers how these structural bundles and the central aqueous cavity relate to substrate transport across the membrane, the so-called alternating access mechanism has been hypothesized [5, 58–60]. In this proposed mechanism, the substrate binding site of the resting MFS transporter faces one side of the membrane and then upon substrate binding orients itself via a conformational change such that the binding site faces the other side to facilitate transport across the membrane [20, 61]. The MFS transporters use flexible gating structures to form inward or outward facing states that are occluded in order to prevent leakage and dissipation of the ion gradients [62]. Inherent in the overall conserved protein structures of the MFS transporters is the “MFS fold” which is composed of an inverted triple helix motif that is repeated four times to form four 3-helix inverted-topology repeats; these repeats constitute the MFS fold in the conserved MFS transporters [63].
Importance of drug and multidrug resistance
Of the many solute transporters that are members of the major facilitator superfamily, the multidrug efflux pumps are of tremendous interest as these integral membrane transport proteins confer multidrug resistance in serious bacterial pathogens and confound the efficacy and efficiency of chemotherapy against infectious disease [64]. Thus, it is of critical importance that these multidrug efflux pumps be studied at the mechanistic and physiological levels [64–66]. The multidrug efflux pumps of pathogenic bacteria make excellent candidate targets for modulation [67] in order to eventually restore clinical efficacy of chemotherapy of bacterial infections.
Structures of multidrug efflux pumps from the major facilitator superfamily
As previously mentioned, high resolution structures for more than a dozen MFS transporters have been determined. Of these, only three of the transporters are believed to be bacterial multidrug efflux pumps. Below we discuss the structures of these known drug efflux systems.
EmrD from E. coli
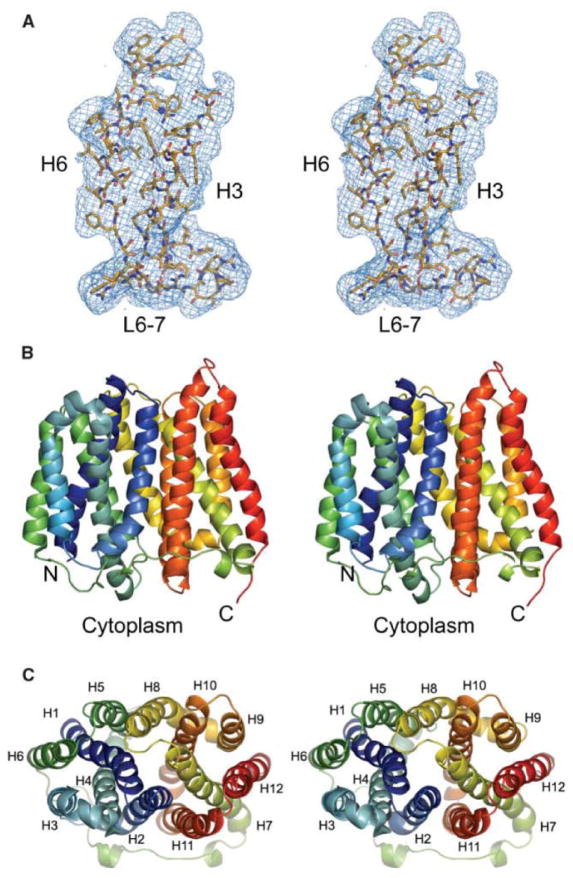
The E. coli EmrD transporter was the first multidrug efflux pump of the MFS to be crystallized and its three-dimensional protein structure to be determined at high resolution, Figure 1 [41]. As had been predicted [12], the EmrD protein structure consists of 12 transmembrane α-helices that cross the inner membrane of E. coli in a so-called zig-zag conformation with the N- and C-termini located in the cytoplasm [41]. The overall EmrD structure consists of two 6-helix pseudo-symmetrical perpendicular bundles, composed of helices 1–6 and helices 7–12, and referred to as the MFS bundle structure [41]. Interestingly, the EmrD structure has two intra-helical loops, one loop formed between helices 4 and 5 and the other formed between helices 10 and 11 [41], and both of these loops protrude into the inner leaflet of the bacterial cytoplasmic membrane [41]. This structural element formed by the loop between helices 10 and 11 is proposed to play a role in the lateral diffusion of hydrophobic drugs that are embedded in the membrane, achieving efflux of these types of drugs into the periplasm [68]. Inherent in the overall 3-dimensional structure are four inverted triple-helix topology repeat elements (see Figure 2), composed of helices 1–3, helices 4–6, helices 7–9 and helices 10–12 of the transporter, and known as the MFS fold [37, 63]. The fourth helix of EmrD has charged amino acids Arg-118, Arg-122, Asp-123, Glu-126, Arg-127, and Arg-131, all or most of which are believed to constitute a selectivity filter and a substrate recognition site [41]. Additionally, during the transport cycle, EmrD has an intermediate occluded conformational state that forms between the open and closed conformational states in which the large flexible central internal cavity is paramount in the cycle and is primarily composed of aromatic and bulky amino acids, such as Ile, Tyr, Trp and Phe, which may accommodate binding and transport of drug substrates [41]. Molecular dynamics simulations, using predicted protein models based on the known EmrD protein structure, implicated Val-45 and Leu-233 as mediating global conformational changes during carbonyl cyanide m-chlorophenylhydrazone transport and thus identifying residues that potentially dictate interactions with substrates and in mediating conformational changes that occur during the release of drug from the transporter into the periplasm at the end of the transport cycle [69]. Using structure-functional approaches it remains to be determined whether these residues play such functional roles at the physiological level in EmrD and whether closely and distantly related MFS multidrug efflux pumps work in the same way. Also unclear is whether EmrD and other MFS drug transporters bind and transport structurally different substrates using one conserved but structurally flexible site, e.g., the central aqueous cavity adjusts to accommodate its multiple substrates (i.e., one binding site) during solute transport. Alternatively, it may be possible that EmrD and MFS drug transporters have multiple substrate binding sites. Thus, much work remains to be completed in order to clearly understand multiple drug efflux.
Figure 1. Structure of EmrD from E. coli.
The crystal structure of the multidrug efflux pump EmrD from E. coli is shown; stereo views. (A) The electron density profile for helices 3 and 6 and the loop between helices 6 and 7 are shown. (B) The ribbons in color show the α-helices, side view; the N- and C-termini are facing the cytoplasm; the membrane is not shown. (C) Top view of EmrD; the α-helices are numbered (Reprinted from Yin, Y., He, X., Szewczyk, P., Nguyen, T. and Chang, G. 2006, Science, 312, 741–744 with permission from AAAS).
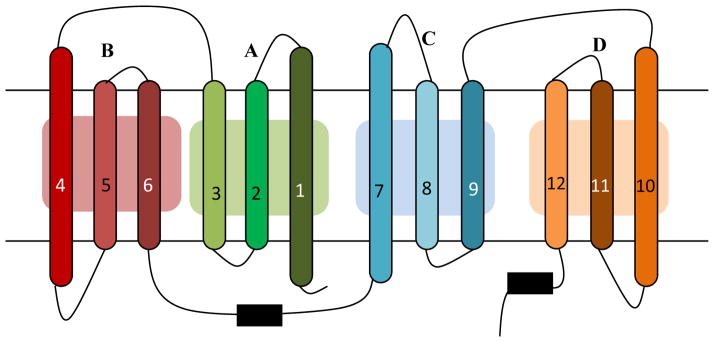
Figure 2. The MFS fold.
The MFS fold consists of four inverted triple-helix topology repeat elements. The transmembrane α-helices 1 through 12 are shown as numbered long horizontal bars. The bold letters A through D indicate each of the four triple-helical inverted repeats. Adapted from Yaffe et al. [37] and Radestock et al. [63].
YajR from E. coli
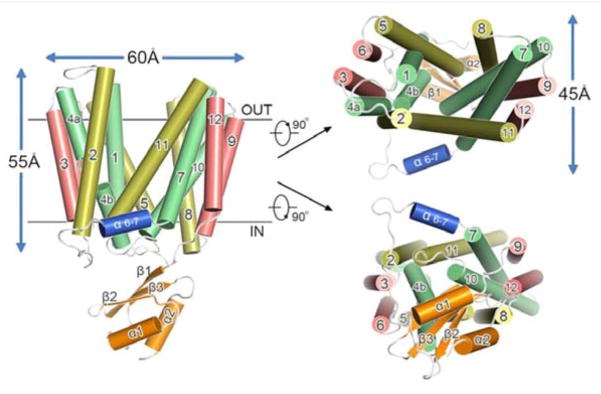
The crystal structure of YajR, a multidrug efflux pump from E. coli, was determined [52], and is shown in Figure 3. The YajR structure was present in an outward facing conformation, and interestingly, functional roles for residues of Motif A were postulated in the same study [52]. In particular, Gly-69 interacted closely with Gly-337 and Gly-341 of helix 11, forming part of an interface between the two N- and C-terminal domains (i.e., bundles) in order to stabilize the outward facing conformation of YajR [52]. In the YajR structure, Asp-73 is adjacent to helix 11 and buried deep within the bundle interface, possibly stabilizing both the structure of helix 11 and the bundle interface conformation by using a postulated dipole-helix interaction; and when Asp-73 was changed to Arg the melting temperature decreased, suggesting that Asp-73 becomes accessible to solvent (i.e., unburied) when the inward-facing conformation forms [52]. Arg-74 of YajR is further thought to interact closely with phospholipid molecules, thus providing structural stability while in the membrane [52]. With respect to Gly-76 of Motif A in YajR, this conserved residue is believed to stabilize molecular interactions within N-terminal bundle, and thus dictate intradomain stability [52]. Conversely, Arg-77 may form salt bridges with Asp-73 of Motif A and Asp-126, which is near the C-terminal part of helix 4 [52]. It is interesting to note that a similar type of salt-bridge formation occurs in the lactose permease of E. coli, in which Lys-319 forms alternating ion-pairs with Asp-240 and Glu-269 of LacY [70, 71]. Lastly, in Motif A, Lys-73 of YajR possibly interacts with the C-terminal end of helix 6 [52]. In essence, Motif A of YajR stabilizes an outward facing conformation and may further mediate the conformational changes between the outward and inward facing structures formed during multidrug transport [52]. Strikingly, sequence elements of Motif A of loop 2–3 (called L2–3) are present to a certain extent in loops between helices 5 and 6 (L5–6), 8 and 9 (L8–9) and 11 and 12 (L11–12) as well, suggesting a universal importance of structures formed by Motif A and Motif A-like sequences not only throughout a given MFS solute transporter, but in all transporters of the MFS [52].
Figure 3. Structure of the multidrug efflux pump YajR from E. coli.
The figure shows the α-helices of YajR as numbered cylinders, and the outside facing aqueous cavity is shown in the structure on the left. The views of the pump from the top and bottom are shown on the right (Reprinted from Jiang, D., Zhao, Y., Wang, X., Fan, J., Heng, J., Liu, X., Feng, W., Kang, X., Huang, B., Liu, J. and Zhang X. C. 2013, Proc. Natl. Acad. Sci. USA, 110, 14664–14669).
MdfA from E. coli
An important multidrug efflux pump from E. coli is MdfA [72], also known as Cmr [73] and CmlA [74–76], which confers chloramphenicol resistance and is distinct from CmlA on Tn1696 [77]. Studies of cationic drug and proton transport indicate that Asp-34 [78] and Glu-26 [79] play functional roles in transport. A study of transport involving MdfA indicated that the substrate binding pocket may be composed of two distinct structural motifs. The first such motif involves residues Val-125, Tyr-127, and Ala-128 located in helix 4, Ser-133 located in the cytoplasmic loop between helices 2 and 3, plus Met-146, Ala-147, and Ala-150, which reside in helix 6 [80]; whereas the second structural motif is thought to be composed of Cys-21, Val-23 in helix 1, Gly-39 located in the periplasmic loop between helices 1 and 2, and Val-54 and Thr-56 in helix 2 [80]. In an interesting study of MdfA conformational changes associated with the transport of multiple drugs, Fluman et al. [81] postulated a so-called “promiscuous conformation switch” mechanism in which substrate binding triggers conformational switching to a periplasm-facing orientation, and the proton binding to MdfA induces another conformational switch to reorient the substrate binding site back to the cytoplasmic side of the membrane [81, 82]. After having replaced a wild-type glycine residue at position 354 with a new negatively charged residue glutamate in the middle of helix 11 of MdfA, the proton stoichiometry was altered to transport additional protons during drug transport [83]; this finding suggests that residues at positions 34, 26 and 354 are in close proximity in MdfA. Subsequently, it was demonstrated that MdfA transports dequalinium, chlorhexidine and pentamidine, all of which are dicationic, in a process that involves binding and transport of one of the two cationic moieties of the substrate across the membrane before mediating binding and transport of the second charged moiety of the same molecule through the transporter, thus accomplishing dicationic drug transport in a successive fashion [84].
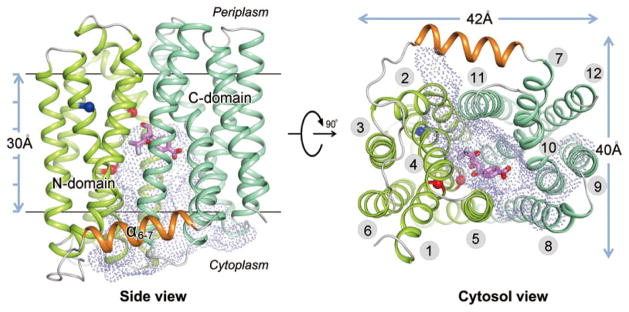
Recently, three structures of MdfA from E. coli were determined (Figure 4) all of which were in a cytoplasmic facing conformation and had the central aqueous cavity [47]. Furthermore, each of the three protein structures were bound to a different solute, i.e., chloramphenicol (an established substrate for MdfA) [72], plus deoxycholate and n-dodecyl-N,N-dimethylamide-N-oxide (two known substrate analogs) [47]. In this study, elements of the substrate binding site were found in helix 1 of MdfA, i.e., Glu-26 and Asp-34, which are conserved, and are surrounded by the highly conserved residues of Motif C in helix 5, i.e., Val-149, Ala-150, Ala-153, and Pro-154 [28]. Thus, Motif C is currently believed to lie in the interface between the two N- and C-terminal bundles, now referred to as the “inter-domain interface” [47]. As such, it was proposed that Motif C has functional roles during drug transport by participating with other helices to prevent proton leakage (and thus prevent proton gradient dissipation) and, importantly by stabilizing the interactions that occur between the helices that constitute the inter-domain interface in order to maintain the cytoplasmic-facing conformation [47]. Lastly, because the inter-helical loops between helices 2 and 3 of MdfA were not found to interact with residues of other loops, it was postulated that Motif A serves to stabilize the external-facing conformation of the multidrug efflux pump. Clearly, these and other highly conserved sequence motifs may play critical roles during the transport cycles of other multidrug efflux pumps of the MFS. Such structures formed by these sequence motifs may be good targets for efflux pump inhibitors [67].
Figure 4. Structure of MdfA from E. coli.
(Reprinted by permission from Macmillan Publishers Ltd: [Cell Research] (Heng, J., Zhao, Y., Liu, M., Liu, Y., Fan, J., Wang, X., Zhao, Y. and Zhang, X. C. 2015, Cell. Res., 25, 1060–1073), copyright (2015)).
Future studies
Presently, it remains unclear whether the various structures and functions elucidated already have universal application to all members of the MFS. Additionally, it is not known how these MFS transporters with their diverse substrates manage to maintain their individual substrate specificities while accomplishing their shared universal function, i.e., transport. It is especially unclear at the molecular level how these MFS transporters dictate transport of single versus multiple solutes as their substrates. Interestingly, very little work has been performed that addresses how these MFS transporters sort out passive versus secondary active transport. With respect to the energy mode, it is poorly understood how these transporters mediate H+ versus Na+ ion selectivity. One is curious whether a simple point mutation in a given MFS transporter would be enough to convert a passive carrier into an active transporter, or vice versa, as seen in LacY [71]. Further physiological, structure-function and structural studies will be necessary to ascertain whether all or sub-sets of functionally related transporters of the MFS work by a common transport mechanism, as has been previously proposed [26]. Whether the transporters of the MFS catalyze solute transport across the membrane by similar or distinctive means, such mechanisms will no doubt, for instance, be good targets for modulation in order to effectively restore the clinical efficacy of antimicrobial agents whose utilities are compromised by multidrug resistance observed in microbial pathogens. Along these lines, the MFS transporters may be altered to accommodate enhanced transport of desirable medically- or industrially-based substrates.
Acknowledgments
This publication was supported in part by a grant from the National Institute of General Medical Sciences (P20GM103451) of the National Institutes of Health and by an internal research grant, ENMU.
Footnotes
CONFLICT OF INTEREST STATEMENT
The authors declare no conflicts of interest.
References
- 1.Broome-Smith JK. Transport of Molecules Across Microbial Membranes. Cambridge: University Press; 1999. [Google Scholar]
- 2.Krämer R. Biochim Biophys Acta. 1994;1185:1–34. doi: 10.1016/0005-2728(94)90189-9. [DOI] [PubMed] [Google Scholar]
- 3.Poolman B, Konings WN. Biochim Biophys Acta. 1993;1183:5–39. doi: 10.1016/0005-2728(93)90003-x. [DOI] [PubMed] [Google Scholar]
- 4.Saidijam M, Bettaney KE, Szakonyi G, Psakis G, Shibayama K, Suzuki S, Clough JL, Blessie V, Abu-Bakr A, Baumberg S, Meuller J, Hoyle CK, Palmer SL, Butaye P, Walravens K, Patching SG, O’Reilly J, Rutherford NG, Bill RM, Roper DI, Phillips-Jones MK, Henderson PJ. Biochem Soc Trans. 2005;33:867–872. doi: 10.1042/BST0330867. [DOI] [PubMed] [Google Scholar]
- 5.West IC. Biochim Biophys Acta. 1980;604:91–126. doi: 10.1016/0005-2736(80)90586-6. [DOI] [PubMed] [Google Scholar]
- 6.Erni B. Int Rev Cytol. 1992;137:127–148. doi: 10.1016/s0074-7696(08)62675-3. [DOI] [PubMed] [Google Scholar]
- 7.Schneider E, Hunke SS. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 8.Kuroda T, Tsuchiya T. Biochim Biophys Acta. 2009;1794:763–768. doi: 10.1016/j.bbapap.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 9.Nikaido H, Takatsuka Y. Biochim Biophys Acta. 2009;1794:769–781. doi: 10.1016/j.bbapap.2008.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung YJ, Saier MH., Jr Curr Opin Drug Discov Devel. 2001;4:237–245. [PubMed] [Google Scholar]
- 11.Pao SS, Paulsen IT, Saier MH., Jr Microbiol Mol Biol Rev. 1998;62:1–34. doi: 10.1128/mmbr.62.1.1-34.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Paulsen IT, Brown MH, Skurray RA. Microbiol Rev. 1996;60:575–608. doi: 10.1128/mr.60.4.575-608.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Teather RM, Müller-Hill B, Abrutsch U, Aichele G, Overath P. Molecular & General Genetics: MGG. 1978;159:239–248. doi: 10.1007/BF00268260. [DOI] [PubMed] [Google Scholar]
- 14.Büchel DE, Gronenborn B, Müller-Hill B. Nature. 1980;283:541–545. doi: 10.1038/283541a0. [DOI] [PubMed] [Google Scholar]
- 15.Maiden MC, Davis EO, Baldwin SA, Moore DC, Henderson PJ. Nature. 1987;325:641–643. doi: 10.1038/325641a0. [DOI] [PubMed] [Google Scholar]
- 16.Henderson PJ. Curr Opin Cell Biol. 1993;5:708–721. doi: 10.1016/0955-0674(93)90144-f. [DOI] [PubMed] [Google Scholar]
- 17.Goswitz VC, Brooker RJ. Protein Sci. 1995;4:534–537. doi: 10.1002/pro.5560040319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marger MD, Saier MH., Jr Trends Biochem Sci. 1993;18:13–20. doi: 10.1016/0968-0004(93)90081-w. [DOI] [PubMed] [Google Scholar]
- 19.Saier MH, Jr, Reddy VS, Tamang DG, Vastermark A. Nucleic Acids Res. 2014;42:D251–258. doi: 10.1093/nar/gkt1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Law CJ, Maloney PC, Wang DN. Annu Rev Microbiol. 2008;62:289–305. doi: 10.1146/annurev.micro.61.080706.093329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saier MH, Jr, Beatty JR, Goffeau A, Harley KT, Heijne WH, Huang SC, Jack DL, Jahn PS, Lew K, Liu J, Pao SS, Paulsen IT, Tseng TT, Virk PS. J Mol Microbiol Biotechnol. 1999;1:257–279. [PubMed] [Google Scholar]
- 22.Maloney PC. Curr Opin Cell Biol. 1994;6:571–582. doi: 10.1016/0955-0674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 23.Mitchell P. Biosci Rep. 1991;11:297–344. doi: 10.1007/BF01130212. discussion 345–296. [DOI] [PubMed] [Google Scholar]
- 24.Henderson PJ. Journal of Bioenergetics and Biomembranes. 1990;22:525–569. doi: 10.1007/BF00762961. [DOI] [PubMed] [Google Scholar]
- 25.Henderson PJ. Research in Microbiology. 1990;141:316–328. doi: 10.1016/0923-2508(90)90005-b. [DOI] [PubMed] [Google Scholar]
- 26.Griffith JK, Baker ME, Rouch DA, Page MG, Skurray RA, Paulsen IT, Chater KF, Baldwin SA, Henderson PJ. Curr Opin Cell Biol. 1992;4:684–695. doi: 10.1016/0955-0674(92)90090-y. [DOI] [PubMed] [Google Scholar]
- 27.Henderson PJ, Roberts PE, Martin GE, Seamon KB, Walmsley AR, Rutherford NG, Varela MF, Griffith JK. Biochem Soc Trans. 1993;21:1002–1006. doi: 10.1042/bst0211002. [DOI] [PubMed] [Google Scholar]
- 28.Rouch DA, Cram DS, DiBerardino D, Littlejohn TG, Skurray RA. Mol Microbiol. 1990;4:2051–2062. doi: 10.1111/j.1365-2958.1990.tb00565.x. [DOI] [PubMed] [Google Scholar]
- 29.Henderson PJ, Maiden MC. Philos Trans R Soc Lond B Biol Sci. 1990;326:391–410. doi: 10.1098/rstb.1990.0020. [DOI] [PubMed] [Google Scholar]
- 30.McMurry L, Petrucci RE, Jr, Levy SB. Proc Natl Acad Sci USA. 1980;77:3974–3977. doi: 10.1073/pnas.77.7.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamaguchi A, Ono N, Akasaka T, Noumi T, Sawai TT. The J Biol Chem. 1990;265:15525–15530. [PubMed] [Google Scholar]
- 32.Chopra I. The Journal of Antimicrobial Chemotherapy. 1986;18(Suppl C):51–56. doi: 10.1093/jac/18.supplement_c.51. [DOI] [PubMed] [Google Scholar]
- 33.Kimura T, Shiina Y, Sawai T, Yamaguchi A. The J Biol Chem. 1998;273:5243–5247. doi: 10.1074/jbc.273.9.5243. [DOI] [PubMed] [Google Scholar]
- 34.Yamaguchi A, Akasaka T, Kimura T, Sakai T, Adachi Y, Sawai T. Biochemistry. 1993;32:5698–5704. doi: 10.1021/bi00072a027. [DOI] [PubMed] [Google Scholar]
- 35.Varela MF, Sansom CE, Griffith JK. Mol Membr Biol. 1995;12:313–319. doi: 10.3109/09687689509072433. [DOI] [PubMed] [Google Scholar]
- 36.Varela MF, Griffith JK. Antimicrob Agents Chemother. 1993;37:1253–1258. doi: 10.1128/aac.37.6.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yaffe D, Radestock S, Shuster Y, Forrest LR, Schuldiner S. Proc Natl Acad Sci USA. 2013;110:E1332–1341. doi: 10.1073/pnas.1220497110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moir JW, Wood NJ. Cell Mol Life Sci. 2001;58:215–224. doi: 10.1007/PL00000849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hirai T, Heymann JA, Maloney PC, Subramaniam S. J Bacteriol. 2003;185:1712–1718. doi: 10.1128/JB.185.5.1712-1718.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saidijam M, Benedetti G, Ren Q, Xu Z, Hoyle CJ, Palmer SL, Ward A, Bettaney KE, Szakonyi G, Meuller J, Morrison S, Pos MK, Butaye P, Walravens K, Langton K, Herbert RB, Skurray RA, Paulsen IT, O’Reilly J, Rutherford NG, Brown MH, Bill RM, Henderson PJ. Curr Drug Targets. 2006;7:793–811. doi: 10.2174/138945006777709575. [DOI] [PubMed] [Google Scholar]
- 41.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Science. 2006;312:741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dang S, Sun L, Huang Y, Lu F, Liu Y, Gong H, Wang J, Yan N. Nature. 2010;467:734–738. doi: 10.1038/nature09406. [DOI] [PubMed] [Google Scholar]
- 43.Iancu CV, Zamoon J, Woo SB, Aleshin A, Choe JY. Proc Natl Acad Sci USA. 2013;110:17862–17867. doi: 10.1073/pnas.1311485110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang Y, Lemieux MJ, Song J, Auer M, Wang DN. Science. 2003;301:616–620. doi: 10.1126/science.1087619. [DOI] [PubMed] [Google Scholar]
- 45.Sun L, Zeng X, Yan C, Sun X, Gong X, Rao Y, Yan N. Nature. 2012;490:361–366. doi: 10.1038/nature11524. [DOI] [PubMed] [Google Scholar]
- 46.Abramson J, Smirnova I, Kasho V, Verner G, Kaback HR, Iwata S. Science. 2003;301:610–615. doi: 10.1126/science.1088196. [DOI] [PubMed] [Google Scholar]
- 47.Heng J, Zhao Y, Liu M, Liu Y, Fan J, Wang X, Zhao Y, Zhang XC. Cell Res. 2015;25:1060–1073. doi: 10.1038/cr.2015.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zheng H, Wisedchaisri G, Gonen T. Nature. 2013;497:647–651. doi: 10.1038/nature12139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan H, Huang W, Yan C, Gong X, Jiang S, Zhao Y, Wang J, Shi Y. Cell Rep. 2013;3:716–723. doi: 10.1016/j.celrep.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 50.Newstead S, Drew D, Cameron AD, Postis VL, Xia X, Fowler PW, Ingram JC, Carpenter EP, Sansom MS, McPherson MJ, Baldwin SA, Iwata S. EMBO J. 2011;30:417–426. doi: 10.1038/emboj.2010.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pedersen BP, Kumar H, Waight AB, Risenmay AJ, Roe-Zurz Z, Chau BH, Schlessinger A, Bonomi M, Harries W, Sali A, Johri AK, Stroud RM. Nature. 2013;496:533–536. doi: 10.1038/nature12042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang D, Zhao Y, Wang X, Fan J, Heng J, Liu X, Feng W, Kang X, Huang B, Liu J, Zhang XC. Proc Natl Acad Sci USA. 2013;110:14664–14669. doi: 10.1073/pnas.1308127110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhao Y, Mao G, Liu M, Zhang L, Wang X, Zhang XC. Structure. 2014;22:1152–1160. doi: 10.1016/j.str.2014.06.008. [DOI] [PubMed] [Google Scholar]
- 54.Nomura N, Verdon G, Kang HJ, Shimamura T, Nomura Y, Sonoda Y, Hussien SA, Qureshi AA, Coincon M, Sato Y, Abe H, Nakada-Nakura Y, Hino T, Arakawa T, Kusano-Arai O, Iwanari H, Murata T, Kobayashi T, Hamakubo T, Kasahara M, Iwata S, Drew D. Nature. 2015;526:397–401. doi: 10.1038/nature14909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McNicholas P, McGlynn M, Guay GG, Rothstein DM. J Bacteriol. 1995;177:5355–5357. doi: 10.1128/jb.177.18.5355-5357.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McNicholas P, Chopra I, Rothstein DM. J Bacteriol. 1992;174:7926–7933. doi: 10.1128/jb.174.24.7926-7933.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mitchell P. Annual Review of Biochemistry. 1977;46:996–1005. doi: 10.1146/annurev.bi.46.070177.005024. [DOI] [PubMed] [Google Scholar]
- 58.Jencks WP. Advances in Enzymology and Related Areas of Molecular Biology. 1980;51:75–106. doi: 10.1002/9780470122969.ch2. [DOI] [PubMed] [Google Scholar]
- 59.Tanford C. Proc Natl Acad Sci USA. 1982;79:2882–2884. doi: 10.1073/pnas.79.9.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.West IC. Biochimica et Biophysica Acta. 1997;1331:213–234. doi: 10.1016/s0304-4157(97)00007-5. [DOI] [PubMed] [Google Scholar]
- 61.Henderson PJ. Biosci Rep. 1991;11:477–453. doi: 10.1007/BF01130216. discussion 534–478. [DOI] [PubMed] [Google Scholar]
- 62.Stelzl LS, Fowler PW, Sansom MS, Beckstein O. J Mol Biol. 2014;426:735–751. doi: 10.1016/j.jmb.2013.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Radestock S, Forrest LR. J Mol Biol. 2011;407:698–715. doi: 10.1016/j.jmb.2011.02.008. [DOI] [PubMed] [Google Scholar]
- 64.Andersen JL, He GX, Kakarla P, Ranjana KC, Kumar S, Lakra WS, Mukherjee MM, Ranaweera I, Shrestha U, Tran T, Varela MF. Int J Environ Res Public Health. 2015;12:1487–1547. doi: 10.3390/ijerph120201487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kumar S, Floyd JT, He G, Varela MF. Recent Research Developments in Antimicrobial Agents & Chemotherapy. Research Signpost. 2013:1–21. [Google Scholar]
- 66.Kumar S, Varela MF. Int J Mol Sci. 2012;13:4484–4495. doi: 10.3390/ijms13044484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kumar S, Mukherjee MM, Varela MF. Int J Bacteriol. 2013;2013:1–15. doi: 10.1155/2013/204141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Naroditskaya V, Schlosser MJ, Fang NY, Lewis K. Biochem Biophys Res Commun. 1993;196:803–809. doi: 10.1006/bbrc.1993.2320. [DOI] [PubMed] [Google Scholar]
- 69.Baker J, Wright SH, Tama F. Proteins. 2012;80:1620–1632. doi: 10.1002/prot.24056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee JI, Hwang PP, Wilson TH. J Biol Chem. 1993;268:20007–20015. [PubMed] [Google Scholar]
- 71.Varela MF, Wilson TH. Biochim Biophys Acta. 1996;1276:21–34. doi: 10.1016/0005-2728(96)00030-8. [DOI] [PubMed] [Google Scholar]
- 72.Edgar R, Bibi E. J Bacteriol. 1997;179:2274–2280. doi: 10.1128/jb.179.7.2274-2280.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nilsen IW, Bakke I, Vader A, Olsvik O, El-Gewely MR. J Bacteriol. 1996;178:3188–3193. doi: 10.1128/jb.178.11.3188-3193.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reeve EC. Genet Res. 1966;7:281–286. doi: 10.1017/s0016672300009708. [DOI] [PubMed] [Google Scholar]
- 75.Reeve EC, Suttie DR. Genet Res. 1968;11:97–104. doi: 10.1017/s0016672300011228. [DOI] [PubMed] [Google Scholar]
- 76.Bohn C, Bouloc P. J Bacteriol. 1998;180:6072–6075. doi: 10.1128/jb.180.22.6072-6075.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bissonnette L, Champetier S, Buisson JP, Roy PH. J Bacteriol. 1991;173:4493–4502. doi: 10.1128/jb.173.14.4493-4502.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sigal N, Molshanski-Mor S, Bibi E. J Bacteriol. 2006;188:5635–5639. doi: 10.1128/JB.00422-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Edgar R, Bibi E. EMBO J. 1999:18822–832. doi: 10.1093/emboj/18.4.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Adler J, Bibi E. J Biol Chem. 2004;279:8957–8965. doi: 10.1074/jbc.M313422200. [DOI] [PubMed] [Google Scholar]
- 81.Fluman N, Cohen-Karni D, Weiss T, Bibi E. J Biol Chem. 2009;284:32296–32304. doi: 10.1074/jbc.M109.050658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fluman N, Ryan CM, Whitelegge JP, Bibi E. Molecular Cell. 2012;47:777–787. doi: 10.1016/j.molcel.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tirosh O, Sigal N, Gelman A, Sahar N, Fluman N, Siemion S, Bibi E. Proc Natl Acad Sci USA. 2012;109:12473–12478. doi: 10.1073/pnas.1203632109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fluman N, Adler J, Rotenberg SA, Brown MH, Bibi E. Nature Communications. 2014;5:4615. doi: 10.1038/ncomms5615. [DOI] [PubMed] [Google Scholar]