Crystal structures of the cleavage complexes of topoisomerase IV from Gram-negative (K. pneumoniae) and Gram-positive (S. pneumoniae) bacterial pathogens stabilized by the clinically important antibacterial drug levofloxacin are presented, analysed and compared. For K. pneumoniae, this is the first high-resolution cleavage complex structure to be reported.
Keywords: Klebsiella pneumoniae, cleavage complex, quinolone, levofloxacin, topoisomerase IV, DNA binding, isomerase, isomerase–DNA complex, topoisomerases, Gram-negative complexes, X-ray crystallography, protein–DNA–drug complexes
Abstract
Klebsiella pneumoniae is a Gram-negative bacterium that is responsible for a range of common infections, including pulmonary pneumonia, bloodstream infections and meningitis. Certain strains of Klebsiella have become highly resistant to antibiotics. Despite the vast amount of research carried out on this class of bacteria, the molecular structure of its topoisomerase IV, a type II topoisomerase essential for catalysing chromosomal segregation, had remained unknown. In this paper, the structure of its DNA-cleavage complex is reported at 3.35 Å resolution. The complex is comprised of ParC breakage-reunion and ParE TOPRIM domains of K. pneumoniae topoisomerase IV with DNA stabilized by levofloxacin, a broad-spectrum fluoroquinolone antimicrobial agent. This complex is compared with a similar complex from Streptococcus pneumoniae, which has recently been solved.
1. Introduction
Klebsiella is a genus belonging to the Enterobacteriaceae family of Gram-negative bacilli, which is divided into seven species with demonstrated similarities in DNA homology: K. pneumoniae, K. ozaenae, K. rhinoscleromatis, K. oxytoca, K. planticola, K. terrigena and K. ornithinolytica. K. pneumoniae is the most medically important species of the genus owing to its high resistance to antibiotics. Significant morbidity and mortality has been associated with an emerging, highly drug-resistant strain of K. pneumoniae characterized as producing the carbapenemase enzyme (KPC-producing bacteria; Nordmann et al., 2009 ▸). The best therapeutic approach to KPC-producing organisms has yet to be defined. However, common treatments (based on in vitro susceptibility testing) are the polymyxins, tigecycline and, less frequently, aminoglycoside antibiotics (Arnold et al., 2011 ▸). Another effective strategy involves the limited use of certain antimicrobials, specifically fluoroquinolones and cephalosporins (Gasink et al., 2009 ▸). Several new antibiotics are under development for KPC producers. These include combinations of existing β-lactam antibiotics with new β-lactamase inhibitors able to circumvent KPC resistance. Neoglycosides are novel aminoglycosides that have activity against KPC-producing bacteria that are also being developed (Chen et al., 2012 ▸).
Type II topoisomerase enzymes play important roles in prokaryotic and eukaryotic DNA replication, recombination and transcription (Drlica et al., 2008 ▸; Laponogov et al., 2013 ▸; Lee et al., 2013 ▸; Nitiss, 2009a ▸,b ▸; Schoeffler & Berger, 2008 ▸; Sissi & Palumbo, 2009 ▸; Vos et al., 2011 ▸; Wendorff et al., 2012 ▸; Wu et al., 2011 ▸, 2013 ▸). In bacteria, topoisomerase IV, a tetramer of two ParC and two ParE subunits, unlinks daughter chromosomes prior to cell division, whereas the related enzyme gyrase, a GyrA2GyrB2 tetramer, supercoils DNA and helps unwind DNA at replication forks. Both enzymes act via a double-strand DNA break involving a cleavage complex and are targets for quinolone antimicrobials that act by trapping these enzymes at the DNA-cleavage stage and preventing strand re-joining (Drlica et al., 2008 ▸).
Levofloxacin is a broad-spectrum third-generation fluoroquinolone antibiotic. It is active against Gram-positive and Gram-negative bacteria and functions by inhibiting gyrase and topoisomerase IV (Drlica & Zhao, 1997 ▸; Laponogov et al., 2010 ▸). Acquiring a deep structural and functional understanding of the mode of action of fluoroquinolones (Tomašić & Mašič, 2014 ▸) and the development of new drugs targeted against topoisomerase IV and gyrase from a wide range of Gram-positive and Gram-negative pathogenic bacteria are highly active areas of current research directed at overcoming the vexed problem of drug resistance (Bax et al., 2010 ▸; Chan et al., 2015 ▸; Drlica et al., 2014 ▸; Mutsaev et al., 2014 ▸; Pommier, 2013 ▸; Srikannathasan et al., 2015 ▸).
Here, we report the first three-dimensional X-ray structure of a K. pneumoniae topoisomerase IV ParC/ParE cleavage complex with DNA stabilized by levofloxacin. The crystal structure provides structural information on topoisomerase IV from K. pneumoniae, a pathogen for which drug resistance is a serious concern. The structure of the ParC/ParE–DNA–levofloxacin binding site highlights the details of the cleavage-complex assembly that are essential for the rational design of Klebsiella topoisomerase inhibitors.
2. Materials and methods
2.1. Cloning, expression and purification of K. pneumoniae and Streptococcus pneumoniae ParC55/ParE30
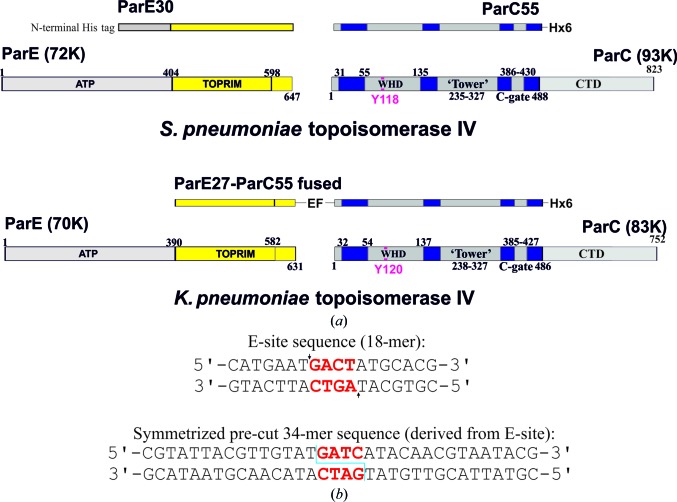
Cloning, expression and purification protocols are described in detail in the Supporting Information. Table 1 ▸ contains the sequence information for all of the components of the complexes. Fig. 1 ▸ provides information about the protein and DNA constructs used in the experimental work.
Table 1. Macromolecule-production information.
(a).
K. pneumoniae topoisomerase IV.
| Source organism | K. pneumoniae (strain ATCC 35657) |
| Expression vector | pET-29a |
| Expression host | E. coli BL21(λDE3) pLysS |
| Complete amino-acid sequence of the construct produced | |
| Topoisomerase IV (ParE CTD 390–631 and ParC NTD 1–490 fused) | MKKLTSGPALPGKLADCTAQDLNRTELFLVEGDSAGGSAKQARDREYQAIMPLKGKILNTWEVSSDEVLASQEVHDISVAIGIDPDSDDLSQLRYGKICILADADSDGLHIATLLCALFVRHFRTLVKEGHVYVALPPLYRIDLGKEVYYALTEEEKTGVLEQLKRKKGKPNVQRFKGLGEMNPMQLRETTLDPNTRRLVQLVISDEDEQQTTAIMDMLLAKKRSEDRRNWLQEKGDMADLEVEFMSDMAERLALHEFTENAYLNYSMYVIMDRALPFIGDGLKPVQRRIVYAMSELGLNASAKFKKSARTVGDVLGKYHPHGDSACYEAMVLMAQPFSYRYPLGDGQGNWGAPDDPKSFAAMRYTESRLSKYAELLLSELGQGTVDWVPNFDGTLQEPKMLPARLPNILLNGTTGIAVGMATDIPPHNLREVAKAAITLIEQPKTTLDELLDIVQGPDFPTEAEIITSRAEIRKIYQNGRGSVRMRAVWSKEDGAVVITALPHQVSGAKVLEQIAAQMRNKKLPMVDDLRDESDHENPTRLVIVPRSNRVDMEQVMNHLFATTDLEKSYRINLNMIGLDGRPAVKNLLEILSEWLVFRRDTVRRRLNHRLEKVLKRLHILEGLLVAFLNIDEVIEIIRTEDEPKPALMSRFGISETQAEAILELKLRHLAKLEEMKIRGEQSELEKERDQLQAILASERKMNNLLKKELQADADAFGDDRRSPLHEREEAKAMSHHHHHH |
| Symmetrized E-site (pre-cut) DNA1 | 5′-CGTATTACGTTGTAT-3′ |
| Symmetrized E-site (pre-cut) DNA2 | 5′-GATCATACAACGTAATACG-3′ |
(b).
S. pneumoniae topoisomerase IV.
| Source organism | S. pneumoniae (isolate 7785 St George’s Hospital; Pan & Fisher, 1996 ▸) |
| Expression vector | pET-19b (N-terminal His10), pET-29a (C-terminal His6) |
| Expression host | E. coli BL21(λDE3) pLysS |
| Complete amino-acid sequence of the construct produced | |
| ParC55 | MSNIQNMSLEDIMGERFGRYSKYIIQDRALPDIRDGLKPVQRRILYSMNKDSNTFDKSYRKSAKSVGNIMGNFHPHGDSSIYDAMVRMSQNWKNREILVEMHGNNGSMDGDPPAAMRYTEARLSEIAGYLLQDIEKKTVPFAWNFDDTEKEPTVLPAAFPNLLVNGSTGISAGYATDIPPHNLAEVIDAAVYMIDHPTAKIDKLMEFLPGPDFPTGAIIQGRDEIKKAYETGKGRVVVRSKTEIEKLKGGKEQIVITEIPYEINKANLVKKIDDVRVNNKVAGIAEVRDESDRDGLRIAIELKKDANTELVLNYLFKYTDLQINYNFNMVAIDNFTPRQVGIVPILSSYIAHRREVILARSRFDKEKAEKRLHIVEGLIRVISILDEVIALIRASENKADAKENLKVSYDFTEEQAEAIVTLQLYRLTNTDVVVLQEEEAELREKIAMLAAIIGDERTMYNLMKKELREVKKKFATPRLSSLEDTAKALEHHHHHH |
| ParE30 | MGHHHHHHHHHHSSGHIDDDDKHMKNKKDKGLLSGKLTPAQSKNPAKNELYLVEGDSAGGSAKQGRDRKFQAILPLRGKVINTAKAKMADILKNEEINTMIYTIGAGVGADFSIEDANYDKIIIMTDADTDGAHIQTLLLTFFYRYMRPLVEAGHVYIALPPLYKMSKGKGKKEEVAYAWTDGELEELRKQFGKGATLQRYKGLGEMNADQLWETTMNPETRTLIRVTIEDLARAERRVNVLMGDKVEPRRKWIEDNVKFTLEEATVF |
| E-site DNA1 | 5′-CATGAATGACTATGCACG-3′ |
| E-site DNA2 | 5′-CGTGCATAGTCATTCATG-3′ |
Figure 1.
Protein and DNA used in the co-crystallization experiment. (a) Coloured diagram of the protein constructs used in crystallization. (b) DNA sequences used in crystallization. The 4 bp overhang is shown in red. Cleavage points are indicated by arrows.
2.2. Preparation of the DNA oligomer
For the K. pneumoniae cleavage complex, two DNA oligomers (5′-CGTATTACGTTGTAT-3′ and 5′-GATCATACAACGTAATACG-3′) were synthesized by solid-phase phosphoramidite chemistry and doubly HPLC purified by Metabion, Munich, Germany. The DNA sequence was designed to make a complementary DNA 34-mer that contained the ‘pre-cut’ binding-site fragment: 5′-CGTATTACGTTGTAT↓GATCATACAACGTAATACG-3′ and 3′-GCATAATGCAACATACTAG↓TATGTTGCATTATGC-5′ (the cuts are shown by arrows; see Fig. 1 ▸ b).
For the S. pneumoniae cleavage complex, two DNA oligomers (5′-CATGAATGACTATGCACG-3′ and 5′-CGTGCATAGTCATTCATG-3′) were synthesized by solid-phase phosphoramidite chemistry and doubly HPLC purified by Metabion, Munich, Germany. The DNA sequence corresponds to the E-site 18-mer, which was found to be a better DNA length for crystallization of the S. pneumoniae topoisomerase IV cleavage complexes in order to give stable reproducible crystals (see Fig. 1 ▸ b).
DNA stock solutions were made by mixing the required oligomers (at 1 mM in 20 mM Tris pH 7.5, 200 mM NaCl, 1 mM β-mercaptothanol, 0.05% NaN3) in equal volumes. For DNA annealing, the mixtures of complementary oligomers were heated to 98°C and then slowly cooled to 4°C over a 48 h period.
2.3. Crystallization and data collection
Crystallization information for both the S. pneumoniae and the K. pneumoniae topoisomerase IV cleavage complexes is summarized in Table 2 ▸. Data-collection statistics and details are provided in Table 3 ▸. Structure-solution and refinement details are provided in Table 4 ▸.
Table 2. Crystallization.
| K. pneumoniae topoisomerase IV | S. pneumoniae topoisomerase IV | |
|---|---|---|
| Method | Vapour diffusion | Capillary counter-diffusion |
| Plate type | 24-well Limbro | N/A |
| Temperature (K) | 298 | 304 |
| Protein concentration (mg ml−1) | 4.5 | 4 |
| Buffer composition of protein solution | 20 mM Tris pH 7.5, 100 mM NaCl, 1 mM β-mercaptoethanol, 0.05% NaN3 | |
| Composition of reservoir solution | 0.1 M Tris pH 7.5–8.0, 0–50 mM NaCl, 4–8% PEG 4000, 12–15% glycerol | 50 mM sodium cacodylate pH 6.5, 2.5% Tacsimate, 7% 2-propanol, 62.5 mM KCl, 7.5 mM MgCl2 |
| Volume and ratio of drop | 4 + 2 µl | N/A |
| Volume of reservoir (ml) | 0.5 | N/A |
Table 3. Data collection and processing.
Values in parentheses are for the outer shell.
| K. pneumoniae topoisomerase IV | S. pneumoniae topoisomerase IV | |
|---|---|---|
| Diffraction source | Beamline I03, Diamond Light Source | |
| Wavelength (Å) | 0.97620 | 0.97630 |
| Temperature (K) | 100.0 | 100.0 |
| Detector | Pilatus 6M-F | ADSC Quantum 315 |
| Crystal-to-detector distance (mm) | 502.22 | 377.629 |
| Rotation range per image (°) | 0.2 | 0.25 |
| Total rotation range (°) | 180 | 75 |
| Exposure time per image (s) | 0.2 | 1.0 |
| Space group | P21 | P3121 |
| a, b, c (Å) | 102.07, 161.53, 138.60 | 157.83, 157.83, 211.15 |
| α, β, γ (°) | 90, 94.22, 90 | |
| Mosaicity (°) | 0.237 | 0.466 |
| Resolution range (Å) | 86.12–3.35 (3.53–3.35) | 50–2.90 (3.00–2.90) |
| Total No. of reflections | 160764 | 311576 |
| No. of unique reflections | 63406 | 67471 |
| Completeness (%) | 98.5 (98.4) | 99.4 (99.9) |
| Multiplicity | 2.53 (2.59) | 4.6 (4.7) |
| 〈I/σ(I)〉 | 3.48 (1.95) | 16.14 (3.48) |
| R r.i.m. † | 0.116 (0.434) | 0.08 (0.515) |
| Overall B factor from Wilson plot (Å2) | 53.29 | 73.37 |
Estimated R r.i.m. = R merge[N/(N − 1)]1/2, where N is the data multiplicity.
Table 4. Structure solution and refinement.
Values in parentheses are for the outer shell.
| K. pneumoniae topoisomerase IV | S. pneumoniae topoisomerase IV | |
|---|---|---|
| Resolution range (Å) | 85.01–3.35 (3.40–3.35) | 41.83–2.90 (2.93–2.90) |
| Completeness (%) | 98.3 | 99.5 |
| σ Cutoff | F > 1.350σ(F) | F > 1.34σ(F) |
| No. of reflections, working set | 60158 (2615) | 67471 (1992) |
| No. of reflections, test set | 3208 (142) | 6838 (218) |
| Final R cryst | 0.224 (0.2990) | 0.186 (0.2806) |
| Final R free | 0.259 (0.3537) | 0.226 (0.3562) |
| No. of non-H atoms | ||
| Protein | 18741 | 10338 |
| Nucleic acid | 1608 | 730 |
| Ligand | 104 | 52 |
| Ion | 8 | 6 |
| Water | — | 54 |
| Total | 20461 | 11180 |
| R.m.s. deviations | ||
| Bonds (Å) | 0.002 | 0.008 |
| Angles (°) | 0.611 | 1.221 |
| Average B factors (Å2) | ||
| Protein | 58.05 | 76.7 |
| Nucleic acid | 64.85 | 90.7 |
| Ligand | 60.14 | 95.7 |
| Ion | 42.62 | 84.5 |
| Water | — | 64.2 |
| Ramachandran plot | ||
| Most favoured (%) | 93 | 94 |
| Allowed (%) | 6 | 6 |
2.3.1. S. pneumoniae topoisomerase IV
Protein was mixed with DNA in a 1:1:1.2 molar ratio (ParC55:ParE30:18-mer E-site DNA) with an overall concentration of 4 mg ml−1. Levofloxacin and magnesium chloride were added to final concentrations of 2 and 10 mM, respectively. The mixture was pre-incubated at room temperature overnight. Initial crystallization screening was performed by sitting-drop vapour diffusion in a 96-well MRC crystallization plate (600 nl protein mixture + 400 nl reservoir solution) using a Mosquito robot (TTP Labtech; http://www.ttplabtech.com). The best crystals were obtained using capillary counter-diffusion against 50 mM sodium cacodylate pH 6.5, 2.5% Tacsimate (Hampton Research; McPherson & Cudney, 2006 ▸), 7% 2-propanol, 62.5 mM KCl, 7.5 mM MgCl2 at 304 K. The crystals were flash-cooled at 100 K in cryoprotectant buffer C [50 mM sodium cacodylate pH 6.5, 2.5% Tacsimate, 62.5 mM KCl, 7.5 mM MgCl2, 1 mM β-mercaptoethanol, 30%(v/v) MPD]. The best data set was collected on beamline I03 at Diamond Light Source at a wavelength of 0.9763 Å using an ADSC Quantum 315 detector. The data extended to 2.6 Å resolution anisotropically and were used in refinement with a maximum-likelihood target in the initial refinement cycles; they were deposited in the PDB without introducing a resolution cutoff. However, owing to the high R merge values in the outer shells, the final resolution is given as 2.9 Å and the statistics are reported according to this ‘trimmed’ resolution. The resolution cutoff was based on the rejection criteria R merge < 50% and I/σ(I) > 1.5 in the highest resolution shell. The data were integrated using HKL-2000 (Otwinowski & Minor, 1997 ▸). The space group was determined to be P3121, with unit-cell parameters a = b = 157.83, c = 211.15 Å.
The structure was solved by molecular replacement using Phaser (McCoy et al., 2007 ▸) as implemented within the CCP4 suite (Winn et al., 2011 ▸) and our previously published topoisomerase IV–levofloxacin structure (PDB entry 3k9f; Laponogov et al., 2010 ▸). Refinement was performed in PHENIX (Adams et al., 2002 ▸, 2010 ▸) with manual inspection and corrections performed in WinCoot (Emsley & Cowtan, 2004 ▸; Emsley et al., 2010 ▸).The structure was verified using WinCoot and PROCHECK (Laskowski et al., 1993 ▸).
2.3.2. K. pneumoniae topoisomerase IV
ParC55/ParE30 protein stock in incubation buffer (at 4.5 mg ml−1) was mixed with the ‘pre-cut’ 34-mer DNA stock in a 1:1.2 protein:DNA molar ratio. High-concentration stocks of levofloxacin and MgCl2 were added to give final concentrations of 2 and 10 mM, respectively. The mixture was incubated overnight at room temperature. Initial crystallization screening was performed by sitting-drop vapour diffusion in 96-well MRC crystallization plates (600 nl protein mixture + 300 nl reservoir solution) using a Mosquito robot. When the optimal crystallization conditions had been established, conventional hanging-drop vapour diffusion in 24-well Linbro plates (4 µl protein mixture + 2 µl reservoir solution) was used to increase the crystal size.
Crystals formed after ∼7–10 d at room temperature. The crystallization conditions varied slightly from batch to batch in the range 0.1 M Tris pH 7.5–8.0, 0–50 mM NaCl, 4–8% PEG 4000, 12–15% glycerol.
It should be mentioned that several other DNA oligomers with the same binding-site sequence were tried for crystallization (i.e. 20-mer, ‘pre-cut’ 20-mer and 34-mer DNA sequences). However, these protein–DNA–drug complexes did not produce good-quality crystals for data collection.
Crystals were tested in-house for diffraction quality using an Oxford Xcalibur Nova CCD diffractometer and were then transported for high-resolution data collection at Diamond Light Source (Harwell Science and Innovation Campus, Oxfordshire, England). The data were collected on beamline I03 (wavelength 0.9762 Å) using a Pilatus 6M-F detector (0.2° oscillation per image, 100 K nitrogen stream). The best crystals diffracted to ∼3.2 Å resolution.
All data sets were integrated with MOSFLM (Leslie & Powell, 2007 ▸) and merged with SCALA (Evans, 2006 ▸) as implemented in CCP4 (Winn et al., 2011 ▸). The ParC55/ParE30–DNA–levofloxacin crystals belonged to space group P21, with unit-cell parameters a = 102.07, b = 161.53, c = 138.60 Å, α = 90.00, β = 94.22, γ = 90.00°. They contained two ParC/ParE–DNA heterodimers in the asymmetric unit.
Several data sets were collected, some of which contained visible diffraction to 3.2 Å resolution, but owing to potential internal twinning and space-group ambiguity (most data sets could be integrated in space groups P21 and P212121) and the fact that the structure solution could be obtained in both space groups, careful selection of the integration ranges as well as appropriate data truncation were necessary. The best region of data was integrated to 3.35 Å (see Table 3 ▸ for statistics). The resolution cutoff was based on the rejection criteria R merge < 50% and I/σ(I) > 1.5 in the highest resolution shell.
The structure was solved by the molecular-replacement method in Phaser (McCoy et al., 2007 ▸) using the levofloxacin–DNA cleavage complex of topoisomerase IV from S. pneumoniae as a search model (PDB entry 3rae; ∼41.8% sequence identity). Refinement was performed in PHENIX (Adams et al., 2002 ▸, 2010 ▸) using secondary-structure restraints derived by superposition of the K. pneumoniae ParC/ParE model with the previously solved complex of S. pneumoniae ParC/ParE. Rigid-body, positional and TLS refinements were performed. Levofloxacin molecules and magnesium ions were placed during the final stages of refinement based on missing electron density in the σA-weighted 2F obs − F calc and F obs − F calc maps. WinCoot (Emsley & Cowtan, 2004 ▸) was used for interactive model fitting. The structure was verified using WinCoot and PROCHECK (Laskowski et al., 1993 ▸). The resulting model had good geometry, with 87.8, 9.9 and 1.3% of residues in the favoured, allowed and generously allowed regions of the Ramachandran plot, respectively, and no more than 1% of residues in disallowed regions. The data-collection and final refinement statistics are given in Tables 3 ▸ and 4 ▸. Sequence alignment was performed in ClustalW (Larkin et al., 2007 ▸, McWilliam et al., 2013 ▸). Figures were prepared using PyMOL (DeLano, 2008 ▸), CHEMDRAW (Evans, 2014 ▸) and CorelDRAW (http://www.coreldraw.com).
3. Results and discussion
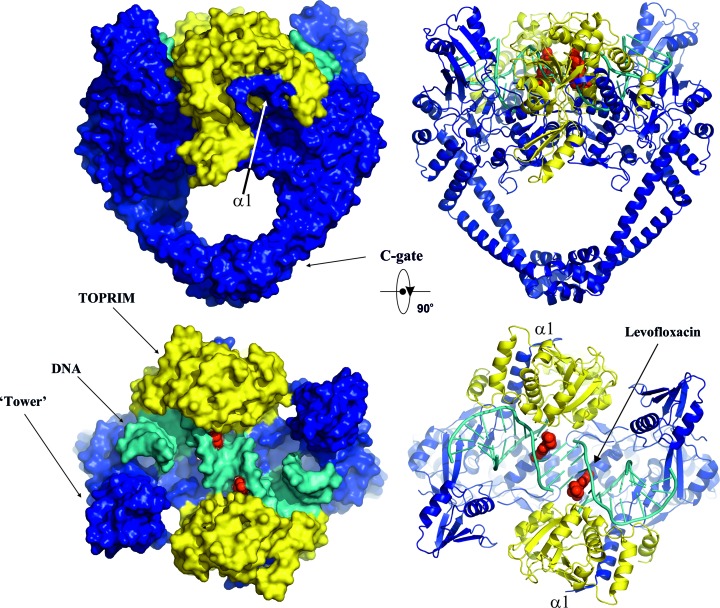
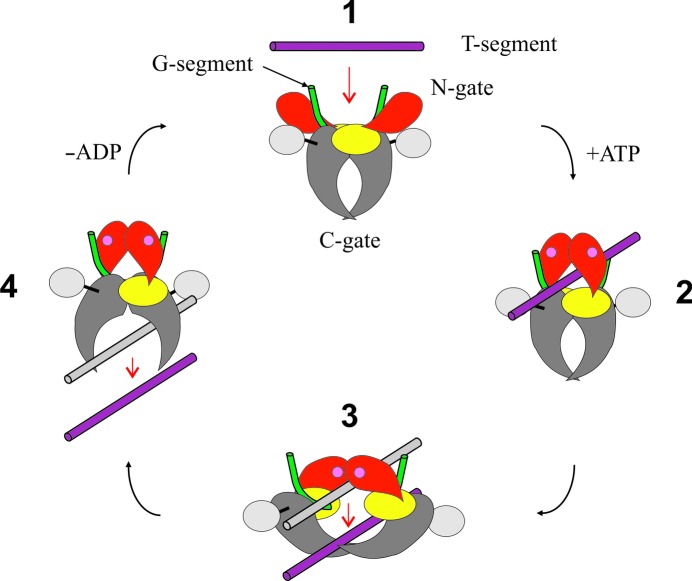
We have co-crystallized the K. pneumoniae topoisomerase IV ParC/ParE breakage-reunion domain (ParC55; residues 1–490) and ParE TOPRIM domain (ParE30; residues 390–631) with a precut 34 bp DNA duplex (the E-site), stabilized by levofloxacin. The X-ray crystal structure of the complex was determined to 3.35 Å resolution, revealing a closed ParC55 dimer flanked by two ParE30 monomers (Figs. 1 ▸, 2 ▸ and 3 ▸). The overall architecture of this complex is similar to that found for S. pneumoniae topoisomerase–DNA–drug complexes (Laponogov et al., 2009 ▸, 2010 ▸). Residues 6–30 of the N-terminal α-helix α1 of the ParC subunit again embrace the ParE subunit, ‘hugging’ the ParE subunits close to either side of the ParC dimer (Laponogov et al., 2010 ▸). Deletion of this ‘arm’ α1 results in loss of DNA-cleavage activity (Laponogov et al., 2007 ▸) and is clearly very important in complex stability (Fig. 3 ▸). This structural feature was absent in our original ParC55 structure (Laponogov et al., 2007 ▸; Sohi et al., 2008 ▸). The upper region of the topoisomerase complex consists of the E-subunit TOPRIM metal-binding domain formed of four parallel β-sheets and the surrounding α-helices. The C-subunit provides the WHD (winged-helix domain; a CAP-like structure; McKay & Steitz, 1981 ▸) and the ‘tower’ which form the U groove-shaped protein region into which the G-gate DNA binds with an induced U-shaped bend. The lower C-gate region (Fig. 3 ▸) consists of the same disposition of pairs of two long α-helices terminated by a spanning short α-helix forming a 30 Å wide DNA-accommodating cavity through which the T-gate DNA passes as found in the S. pneumoniae complex. Owing to the structural similarity, it appears that the topoisomerases IV from K. pneumoniae and S. pneumoniae are likely to follow a similar overall topoisomerase catalytic cycle as shown in Fig. 4 ▸; we have confirmation of one intermediate from our recent structure of the full complex (the holoenzyme less the CTD β-pinwheel domain) with the ATPase domain in the open conformation (Laponogov et al., 2013 ▸).
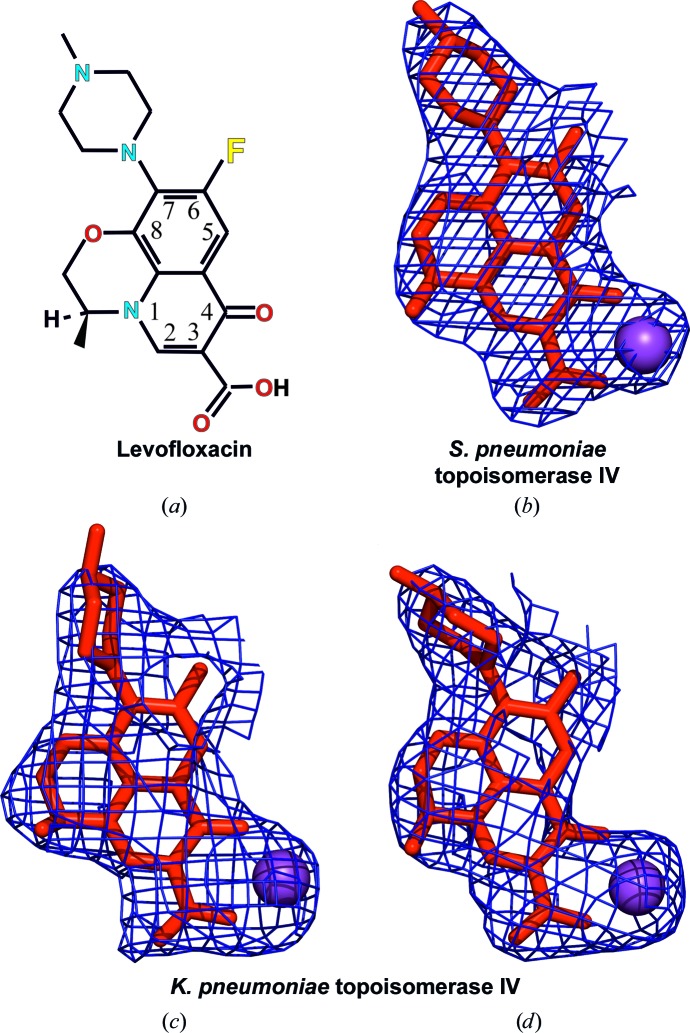
Figure 2.
Chemical structure of levofloxacin (a) and its conformations observed within the active sites of S. pneumoniae topoisomerase IV (b) and K. pneumoniae topoisomerase IV (c, d). Electron-density maps (2F obs − F calc) are shown as meshes for the drug molecules contoured at 1.5σ and are limited to a distance of 2.3 Å from the drug atoms.
Figure 3.
Overall orthogonal views of the cleavage complex of topoisomerase IV from K. pneumoniae in surface (left) and cartoon (right) representations. The ParC subunit is in blue, ParE is in yellow and DNA is in cyan. The bound quinolone molecules (levofloxacin) are in red and are shown using van der Waals representation.
Figure 4.
Schematic representation of the catalytic cycle of type II topoisomerases. The ParC N-terminal domain (ParC55) is in grey, the ParC C-terminal β-pinwheel domain is in silver, the ParE N-terminal ATPase domain is in red, the ParE C-terminal domain (ParE30) is in yellow, the G-gate DNA is in green and the T-segment DNA is in purple. Bound ATP is indicated by pink circles in the ATPase domains (reproduced with permission from Fig. 1 of Lapanogov et al., 2013).
The G-gate DNA for the S. pneumoniae complex consists of an 18-base-pair E-site sequence (our designation for a DNA site which we first found from DNA-mapping studies; Leo et al., 2005 ▸; Arnoldi et al., 2013 ▸; Fig. 1 ▸). The crystallized complex was formed by turning over the topoisomerase tetramer in the presence of DNA and levofloxacin and crystallizing the product. In contrast, the K. pneumoniae complex was formed by co-crystallizing the topoisomerase tetramer complex in the presence of a 34-base-pair pre-cleaved DNA in the presence of levofloxacin. In both cases the DNA is bent into a U-form and bound snugly against the protein of the G-gate. We have been able to unambiguously read off the DNA sequences in the electron-density maps.
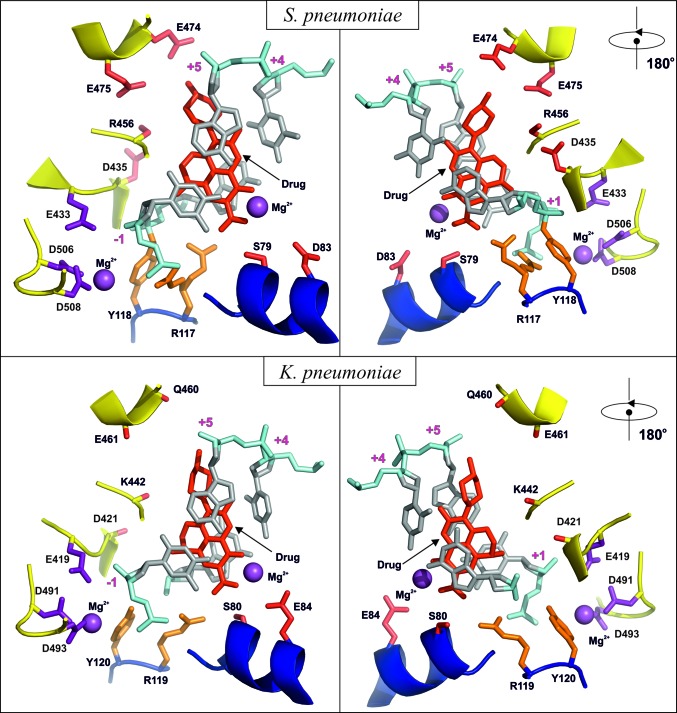
There is 41.6% sequence identity and 54.4% sequence homology between the ParE subunit of K. pneumoniae and that of S. pneumoniae. For the ParC subunits, the figures are 40.8 identity and 55.6% homology between the two organisms. The sequence alignment is given in Supplementary Fig. S1, with the key metal-binding residues and those which give rise to quinolone resistance highlighted. The binding of levofloxacin in the K. pneumoniae complex is shown in Figs. 2 ▸, 3 ▸ and 5 ▸ and is hemi-intercalated into the DNA and stacked against the DNA bases at the cleavage site (positions −1 and +1 of the four-base-pair staggered cut in the 34-mer DNA) which is similar to that found for the S. pneumoniae complex. Fig. 5 ▸ presents side-by-side views of the K. pneumoniae and S. pneumoniae active sites which shows that levofloxacin binds in a very similar manner in these two complexes with extensive π–π stacking interaction between the bases and the drug. The methylpiperazine at C7 (using the conventional quinolone numbering; C9 in the IUPAC numbering) on the drug extends towards residues Glu474 and Glu475 for S. pneumoniae and towards Gln460 and Glu461 for K. pneumoniae, where the glutamate at 474 is substituted by a glutamine at 460 in the Klebsiella strain. Interestingly, for S. pneumoniae we observe only one possible orientation of the C7 groups in both subunits, while for K. pneumoniae we can see two: one with the same orientation as in S. pneumoniae and other rotated 180° away. They both exist within the same crystal in the two different dimers in the asymmetric unit. The side chains surrounding them in ParE are quite disordered and are more defined in K. pneumoniae (even though this complex is at lower resolution) than in S. pneumoniae. There are no direct hydrogen bonds from the drug to these residues (although it is possible that some are formed through water, which cannot be observed at this resolution). Obviously, the drug–ParE interaction in this region is less strong compared with PD 0305970 binding to the S. pneumoniae DNA complex, where PD 0305970 forms a hydrogen bond to ParE residue Asp475 and can form one to Asp474 if the bond rotates (Laponogov et al., 2010 ▸). This may explain why drug-resistance mutations for levofloxacin are more likely to form in the ParC subunits rather than in the ParE subunits.
Figure 5.
Detailed views of the active sites of topoisomerase IV from S. pneumoniae and K. pneumoniae with quinolone molecules bound. The magnesium ions and their coordinating amino acids are shown in purple. The drug molecules and residues known to lead to drug resistance upon mutation are in red. The active-site tyrosine and arginine are in orange. The DNA is shown in silver/cyan. The ParC and ParE backbones are shown in blue and yellow, respectively.
For both complexes there is a Mg2+ ion bound to levofloxacin between the carbonyl group at position 4 of the quinolone and the carboxyl at position 6 (Figs. 2 ▸ and 5 ▸ and Supplementary Fig. 2 ▸). For S. pneumoniae topoisomerase IV, one of the O atoms of the carboxyl of Asp83 points towards the Mg2+ ion and is within hydrogen-bonding distance (5.04 Å) through an Mg2+-coordinated water. For K. pneumoniae both of the carboxyl O atoms are pointing towards the Mg2+ ion at distances of 4.86 and 4.23 Å. These residues are ordered in only one of the two dimers in the K. pneumoniae crystal (the one in which the C7 group is pointing towards the DNA away from ParE, although the conformations of these two groups on the drug are probably not correlated).
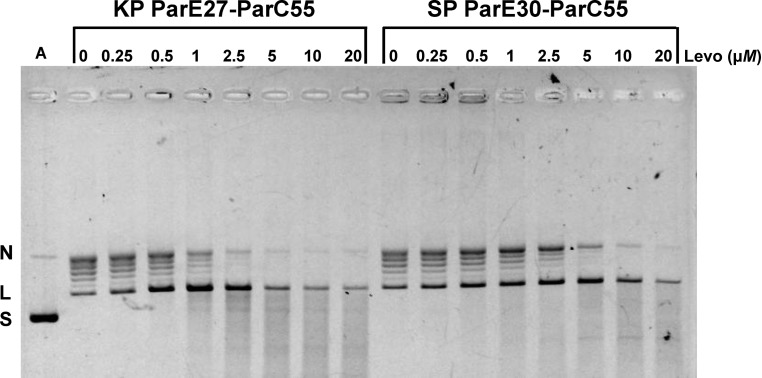
The topoisomerase IV ParE27-ParC55 fusion protein from K. pneumoniae was fully active in promoting levofloxacin-mediated cleavage of DNA (Fig. 6 ▸). In the absence of the drug and ATP, the protein converted supercoiled pBR322 into a ladder of bands corresponding to relaxed DNA. The inclusion of levofloxacin produced linear DNA in a dose-dependent and ATP-independent fashion. Similar behaviour was observed for the S. pneumoniae topoisomerase IV ParE30-ParC55 fusion protein. The CC25 (the drug concentration that converted 25% of the supercoiled DNA substrate to a linear form) was 0.5 µM for the Klebsiella enzyme and 1 µM for the pneumococcal enzyme. Interestingly, K. pneumoniae strains are much more susceptible to levofloxacin than S. pneumoniae, with typical MIC values of 0.016 and 1 mg l−1, respectively (Odenholt & Cars, 2006 ▸), reflecting differences in multiple factors (in addition to binding affinity) that influence drug responses, including membrane, peptidoglycan structure, drug-uptake and efflux mechanisms. Moreover, although topoisomerase IV is primarily the target of levofloxacin in S. pneumoniae, it is likely to be gyrase in the Gram-negative K. pneumoniae.
Figure 6.
Comparison of DNA cleavage by topoisomerase IV core ParE-ParC fusion proteins from K. pneumoniae (KP) and S. pneumoniae (SP) promoted by levofloxacin. Supercoiled plasmid pBR322 (400 ng) was incubated with topoisomerase IV proteins (400 ng) in the absence or presence of levofloxacin at the indicated concentrations. After 60 min incubation, samples were treated with SDS and proteinase K to remove proteins covalent bound to DNA, and the DNA products were examined by gel electrophoresis in 1% agarose. Lane A, supercoiled pBR322 DNA; N, L and S, nicked, linear and supercoiled pBR322, respectively.
In summary, we have determined the first structure of a quinolone–DNA cleavage complex involving a type II topoisomerase from K. pneumoniae. Given the current concerns about drug-resistant strains of Klebsiella, the structure reported here provides key information in understanding the action of currently used quinolones and should aid in the development of other topoisomerase-targeting therapeutics active against this major human pathogen.
Supplementary Material
PDB reference: topoisomerase IV cleavage complex from Klebsiella pneumoniae, 5eix
PDB reference: from Streptococcus pneumoniae, 3rae
Supporting Information.. DOI: 10.1107/S2059798316001212/mn5108sup1.pdf
Acknowledgments
We thank Diamond Light Source for access to beamlines I03, I02, I04, I04-1 and I24 (MX1220, MX7656 and MX9495) and thank the beamline staff for their help, which contributed to the results presented here. This research was supported by PTC Therapeutics Inc. and in part by funding from a Wellcome Trust Seeding Drug Discovery award (097753) to PTC Therapeutics. The research was also funded from Biotechnology and Biological Research Council project grants BB/H00405X/1 and BB/K10069/1. X-SP and IL were funded by the BBSRC. The open access charge was funded by the BBSRC. DAV, IL, X-SP, JS, AB, JN, JVNVP, LMF and MRS conceived and designed the experiments. DAV, IL, X-SP, JS, GBS and MRS performed the experiments. DAV, IL, X-SP, JS, GBS, LMF and MRS analysed and interpreted the data. X-SP, JS, AB, JN and LMF contributed reagents/materials/analysis tools. DAV, IL, X-SP, JS, AB, JN, LMF and MRS wrote the paper and critiqued the output for intellectual content prior to publication.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Arnold, R. S., Thom, K. A., Sharma, S., Phillips, M., Kristie Johnson, J. & Morgan, D. J. (2011). South. Med. J. 104, 40–45. [DOI] [PMC free article] [PubMed]
- Arnoldi, E., Pan, X.-S. & Fisher, L. M. (2013). Nucleic Acids Res. 41, 9411–9423. [DOI] [PMC free article] [PubMed]
- Bax, B. D. et al. (2010). Nature (London), 466, 935–940.
- Chan, P. F. et al. (2015). Nature Commun. 6, 10048. [DOI] [PMC free article] [PubMed]
- Chen, L. F., Anderson, D. J. & Paterson, D. L. (2012). Infect. Drug Resist. 5, 133–141. [DOI] [PMC free article] [PubMed]
- DeLano, W. L. (2008). PyMOL. http://www.pymol.org.
- Drlica, K., Malik, M., Kerns, R. J. & Zhao, X. (2008). Antimicrob. Agents Chemother. 52, 385–392. [DOI] [PMC free article] [PubMed]
- Drlica, K., Mustaev, A., Towle, T. R., Luan, G., Kerns, R. J. & Berger, J. M. (2014). ACS Chem. Biol. 19, 2895–2904. [DOI] [PMC free article] [PubMed]
- Drlica, K. & Zhao, X. (1997). Microbiol. Mol. Biol. Rev. 61, 377–392. [DOI] [PMC free article] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Evans, D. A. (2014). Angew. Chem. Int. Ed. 53, 11140–11145.
- Gasink, L. B., Edelstein, P., Lautenbach, E., Synnestvedt, M. & Fishman, N. (2009). Infect. Control Hosp. Epidemiol. 30, 1180–1185. [DOI] [PMC free article] [PubMed]
- Laponogov, I., Pan, X.-S., Veselkov, D. A., McAuley, K. E., Fisher, L. M. & Sanderson, M. R. (2010). PLoS One, 5, e11338. [DOI] [PMC free article] [PubMed]
- Laponogov, I., Sohi, M. K., Veselkov, D. A., Pan, X.-S., Sawhney, R., Thompson, A. W., McAuley, K. E., Fisher, L. M. & Sanderson, M. R. (2009). Nature Struct. Mol. Biol. 16, 667–669. [DOI] [PubMed]
- Laponogov, I., Veselkov, D. A., Crevel, I. M.-T., Pan, X.-S., Fisher, L. M. & Sanderson, M. R. (2013). Nucleic Acids Res. 41, 9911–9923. [DOI] [PMC free article] [PubMed]
- Laponogov, I., Veselkov, D. A., Sohi, M. K., Pan, X.-S., Achari, A., Yang, C., Ferrara, J. D., Fisher, L. M. & Sanderson, M. R. (2007). PLoS One, 2, e301. [DOI] [PMC free article] [PubMed]
- Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J. & Higgins, D. G. (2007). Bioinformatics, 23, 2947–2948. [DOI] [PubMed]
- Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993). J. Appl. Cryst. 26, 283–291.
- Lee, I., Dong, K. C. & Berger, J. M. (2013). Nucleic Acids Res. 41, 5444–5456. [DOI] [PMC free article] [PubMed]
- Leo, E., Gould, K. A., Pan, X.-S., Capranico, G., Sanderson, M. R., Palumbo, M. & Fisher, L. M. (2005). J. Biol. Chem. 280, 14252–14263. [DOI] [PubMed]
- Leslie, A. G. W. & Powell, H. R. (2007). Evolving Methods for Macromolecular Crystallography, edited by R. Read & J. Sussman, pp. 41–51. Dordrecht: Springer.
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- McKay, D. B. & Steitz, T. A. (1981). Nature (London), 290, 744–749. [DOI] [PubMed]
- McPherson, A. & Cudney, B. (2006). J. Struct. Biol. 156, 387–406. [DOI] [PubMed]
- McWilliam, H., Li, W., Uludag, M., Squizzato, S., Park, Y. M., Buso, N., Cowley, A. P. & Lopez, R. (2013). Nucleic Acids Res. 41, W597–W600. [DOI] [PMC free article] [PubMed]
- Mustaev, A., Malik, M., Zhao, X., Kurepina, N., Luan, G., Oppegard, L. M., Hiasa, H., Marks, K. R., Kerns, R. J., Berger, J. M. & Drlica, K. (2014). J. Biol. Chem. 289, 12300–12312. [DOI] [PMC free article] [PubMed]
- Nitiss, J. L. (2009a). Nature Rev. Cancer, 9, 327–337. [DOI] [PMC free article] [PubMed]
- Nitiss, J. L. (2009b). Nature Rev. Cancer, 9, 338–350. [DOI] [PMC free article] [PubMed]
- Nordmann, P., Cuzon, G. & Naas, T. (2009). Lancet Infect. Dis. 9, 228–236. [DOI] [PubMed]
- Odenholt, I. & Cars, O. J. (2006). J. Antimicrob. Chemother. 58, 960–965. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pan, X.-S. & Fisher, L. M. (1996). J. Bacteriol. 178, 4060–4069. [DOI] [PMC free article] [PubMed]
- Pommier, Y. (2013). ACS Chem. Biol. 8, 82–95. [DOI] [PMC free article] [PubMed]
- Schoeffler, A. J. & Berger, J. M. (2008). Q. Rev. Biophys. 41, 41–101. [DOI] [PubMed]
- Sissi, C. & Palumbo, M. (2009). Nucleic Acids Res. 37, 702–711. [DOI] [PMC free article] [PubMed]
- Sohi, M. K., Veselkov, D. A., Laponogov, I., Pan, X.-S., Fisher, L. M. & Sanderson, M. R. (2008). PLoS One, 3, e3201. [DOI] [PMC free article] [PubMed]
- Srikannathasan, V., Wohlkonig, A., Shillings, A., Singh, O., Chan, P. F., Huang, J., Gwynn, M. N., Fosberry, A. P., Homes, P., Hibbs, M., Theobald, A. J., Spitzfaden, C. & Bax, B. D. (2015). Acta Cryst. F71, 1242–1246. [DOI] [PMC free article] [PubMed]
- Tomašić, T. & Mašič, L. P. (2014). Curr. Top. Med. Chem. 14, 130–151. [DOI] [PubMed]
- Vos, S. M., Tretter, E. M., Schmidt, B. H. & Berger, J. M. (2011). Nature Rev. Mol. Cell Biol. 12, 827–841. [DOI] [PMC free article] [PubMed]
- Wendorff, T. J., Schmidt, B. H., Heslop, P., Austin, C. A. & Berger, J. M. (2012). J. Mol. Biol. 424, 109–124. [DOI] [PMC free article] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wu, C.-C., Li, T.-K., Farh, L., Lin, L.-Y., Lin, T.-S., Yu, Y.-J., Yen, T.-J., Chiang, C.-W. & Chan, N.-L. (2011). Science, 333, 459–462. [DOI] [PubMed]
- Wu, C.-C., Li, Y.-C., Wang, Y.-R., Li, T.-K. & Chan, N.-L. (2013). Nucleic Acids Res. 41, 10630–10640. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: topoisomerase IV cleavage complex from Klebsiella pneumoniae, 5eix
PDB reference: from Streptococcus pneumoniae, 3rae
Supporting Information.. DOI: 10.1107/S2059798316001212/mn5108sup1.pdf