Significance
AcrB is one of the major multidrug resistance-conferring antibiotic efflux pumps from pathogenic bacteria. We have designed and produced the periplasmic, substrate binding domain of AcrB and solved its crystal structure in complex with multiple novel pyranopyridine inhibitors, as well as with drugs transported by AcrB. The structural data are corroborated by various cellular assays and molecular dynamics (MD) simulations, and allow us to propose a mechanism for AcrB efflux inhibition. Furthermore, the results provide a molecular platform for the development of combinational therapies against pathogenic Enterobacteriaceae.
Keywords: RND efflux transporters, multidrug resistance, efflux pump inhibitors, X-ray crystallography, molecular dynamics simulation
Abstract
The Escherichia coli AcrAB-TolC efflux pump is the archetype of the resistance nodulation cell division (RND) exporters from Gram-negative bacteria. Overexpression of RND-type efflux pumps is a major factor in multidrug resistance (MDR), which makes these pumps important antibacterial drug discovery targets. We have recently developed novel pyranopyridine-based inhibitors of AcrB, which are orders of magnitude more powerful than the previously known inhibitors. However, further development of such inhibitors has been hindered by the lack of structural information for rational drug design. Although only the soluble, periplasmic part of AcrB binds and exports the ligands, the presence of the membrane-embedded domain in AcrB and its polyspecific binding behavior have made cocrystallization with drugs challenging. To overcome this obstacle, we have engineered and produced a soluble version of AcrB [AcrB periplasmic domain (AcrBper)], which is highly congruent in structure with the periplasmic part of the full-length protein, and is capable of binding substrates and potent inhibitors. Here, we describe the molecular basis for pyranopyridine-based inhibition of AcrB using a combination of cellular, X-ray crystallographic, and molecular dynamics (MD) simulations studies. The pyranopyridines bind within a phenylalanine-rich cage that branches from the deep binding pocket of AcrB, where they form extensive hydrophobic interactions. Moreover, the increasing potency of improved inhibitors correlates with the formation of a delicate protein- and water-mediated hydrogen bond network. These detailed insights provide a molecular platform for the development of novel combinational therapies using efflux pump inhibitors for combating multidrug resistant Gram-negative pathogens.
Overexpression of resistance nodulation cell division (RND)-type efflux pumps is a major factor in multidrug resistance (MDR) in Gram-negative pathogens (1). These pumps recognize and extrude a chemically diverse compound range from the periplasm to the exterior of the cell (2). The major efflux pump of Escherichia coli, AcrAB-TolC, is the prototypical RND family pump system (3). It comprises a tripartite structure consisting of the integral membrane transporter AcrB; the outer membrane channel TolC; and the periplasmic protein adaptor AcrA, which stabilizes the interaction between AcrB and TolC (4). The AcrB transporter functions as an asymmetrical homotrimer (5–7) in which each protomer adopts a different conformation that represents a distinct step in the translocation pathway (8–10). The individual protomer conformations of the pump have been described as loose (L), tight (T), and open (O), which correspond to the initial interaction, poly-specific binding, and extrusion of substrates to the TolC channel, respectively (11). The conformational changes are driven by the proton motive force, which is transduced by the AcrB transmembrane domain (10, 12). Orthologs of AcrB are present in all of the emerging Gram-negative pathogens, including Pseudomonas aeruginosa, where the MexAB-OprM and MexXY-OprM pumps play an important role for MDR (13).
One class of efflux pump inhibitors (EPIs) that have been developed against RND pumps for clinical use within the past 15 y (14) are peptidomimetic compounds derived from the Phe-Arg β-naphthylamide (PAβN) (15). They inhibit RND efflux pumps from a broad spectrum of Gram-negative bacteria, but they are required at relatively high concentrations (e.g., 50 μM) and could not be developed for clinical use because of the issues of tissue accumulation (16). Other inhibitors that show broad activity are aryl-piperazines exemplified by 1-(1-naphthylmethyl)piperazine (NMP), but they show full activity only at high concentrations (≥50 μM) (17). For P. aeruginosa, tert-butylthiazolyl aminocarboxyl pyridopyrimidines (e.g., D13-9001) are effective (18), but their activity apparently relies on the utilization of specialized outer membrane channels for uptake, and they do not show much activity for Enterobacteriaceae. Further, the pyridopyrimidines do not inhibit MexY, and this property has hindered their further development (19). Crystal structures of D13-9001 bound to AcrB and MexB demonstrated binding to a unique site near the substrate deep binding pocket in the periplasmic domain known as the hydrophobic trap (20). The discovery of the hydrophobic trap is a significant advance, because it will enable the design of EPIs with increased potency.
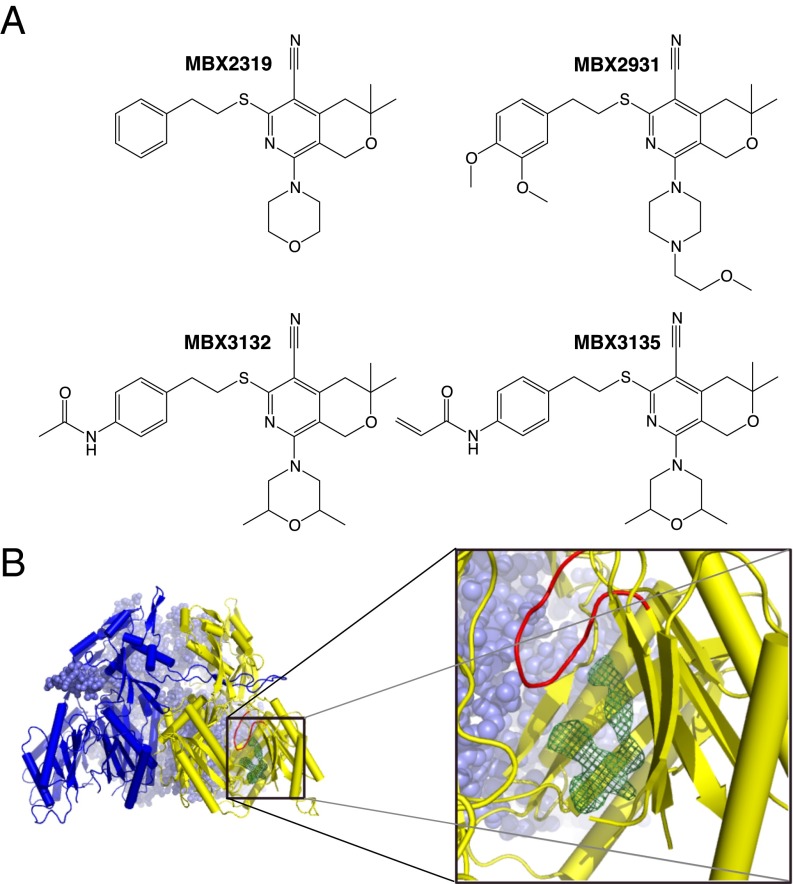
We previously described a novel EPI, MBX2319, comprising a pyranopyridine core with five substituents around the core (Fig. 1A). MBX2319 increases the potency of a broad range of antibiotics against E. coli and other Enterobacteriaceae and does not exhibit membrane-disrupting or antibacterial activity (21). Importantly, MBX2319 fully potentiated the activity of levofloxacin and piperacillin at concentrations as low as 3 μM, which is about an order of magnitude lower than the concentrations required for full activity of the earlier inhibitors mentioned above. Mechanism of action studies in E. coli indicated that the most likely target of MBX2319 is AcrB. More recently, we described new derivatives of MBX2319 with increased activity. Notably MBX3132 and MBX3135 showed full activity even at 0.1 μM, that is, at concentrations 500-fold lower than the classical inhibitors like PAβN (22). Here, we report on microbiological, crystallographic, and computational studies on the interaction of AcrB with these novel and powerful inhibitors.
Fig. 1.
Structure of inhibitors and the inhibitor-bound AcrBper. (A) Chemical structures of the studied MBX compounds. (B) Inhibitor binding is observed to the hydrophobic trap and associated deep binding pocket distal from the switch-loop (red) of the T monomer (yellow-colored cartoon) as represented by the MBX3132 Fo-Fc omit map (green mesh, contoured at 4.0 σ). The L protomers of AcrBper are shown in a blue surface sphere and cartoon representation.
Results and Discussion
Crystallization of the AcrB Periplasmic Domain.
To facilitate structural studies of inhibitors and substrates bound to AcrB, we designed a soluble form of the AcrB periplasmic domain (AcrBper) that is composed of the two substrate binding periplasmic AcrB loops (between transmembrane helices 1–2 and 7–8), which are connected by a nine-residue linker (GGSGGSGGS). The asymmetrical apo-AcrBper trimer was crystallized (diffracting up to a resolution of 1.8 Å) in complex with three DARPin (designed ankyrin repeat proteins) molecules (7) in space group P212121 (Experimental Procedures).
Structural alignments of the AcrBper monomers with the periplasmic domains of each protomer of the asymmetrical full-length AcrB crystal forms [Protein Data Bank (PDB) ID code 4DX5, P212121 space group; and PDB ID code 2GIF, C2 space group] indicate that the AcrBper conformers (chains A, B, and C) are L-T (access-access binding), respectively, hereafter referred to as LLT (6, 23) (Fig. 1B). The PC1 subdomains (residues 595–605) of the L conformers, but not the T conformer, are involved in crystal contact of ∼400 Å2 with symmetry-related protomers. The T conformer is involved in different and less substantial crystal contacts. These crystal contacts most likely stabilize the LLT conformation, resembling the stabilization by crystal contacts of the symmetrical (LLL) full-length AcrB in the R32 lattice (24). The 3D structures of the individual protomers of AcrBper are nearly identical to the periplasmic domains of the L and T conformers of full-length AcrB structures [PDB ID code 2GIF (6) and PDB ID code 4DX5 (23)], and the rmsd values after Cα-alignment between individual protomers of AcrBper and the periplasmic domains of the L and T protomers from full-length AcrB were below 0.7 Å (SI Appendix, Fig. S1 and Table S1). The two L monomers within AcrBper were nearly identical, with an rmsd of 0.24 Å. The (GGS)3 linker and residues between Thr329 and Thr569, the loop region between Pro669 and Thr676, and the residues C-terminal of Tyr864 appeared to be disordered, and electron densities for these regions were not visible in any of the structures presented in this study.
Crystal Structure of AcrBper Bound to Minocycline and Rhodamine 6G.
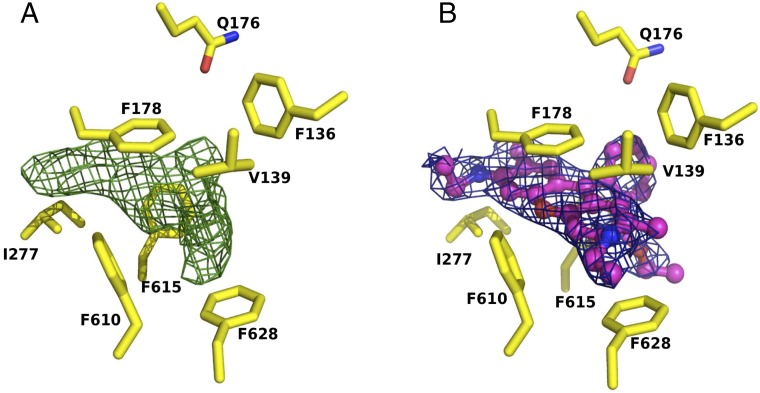
To confirm that the properties of the deep binding pocket of the AcrBper T protomer are equivalent to the properties of the deep binding pocket of full-length AcrB, we solved the crystal structure of AcrBper with bound minocycline (MIN). We found that MIN (SI Appendix, Fig. S2) binds to the same site on AcrBper as has been previously reported for full-length AcrB (5, 10, 23). To demonstrate that AcrBper retains broad substrate specificity (25, 26), we determined its structure in complex with Rhodamine 6G (R6G) and found that R6G binds to a position in the deep binding pocket of the T protomer that is similar to the position in doxorubicin (23), where it interacts mainly with the F178 and F628 side chains (Fig. 2). These results confirm that the substrate binding properties of the T protomers of AcrBper are essentially identical to the substrate binding properties of the full-length AcrB.
Fig. 2.
R6G binding to AcrBper is observed in the deep binding pocket of the T monomer. (A) Fo-Fc omit map of the R6G ligand is shown as green mesh, contoured at 3.0 σ. (B) Blue mesh (contoured at 1.0 σ) represents the R6G 2Fo-Fc electron density map after refinement of the complex structure. R6G is shown in a ball-and-stick representation (carbon, magenta; oxygen, red; nitrogen, blue). Side-chain residues of the AcrB deep binding pocket are shown as sticks (carbon, yellow; oxygen, red; nitrogen, blue).
Structure of AcrBper in Complex with MBX2319.
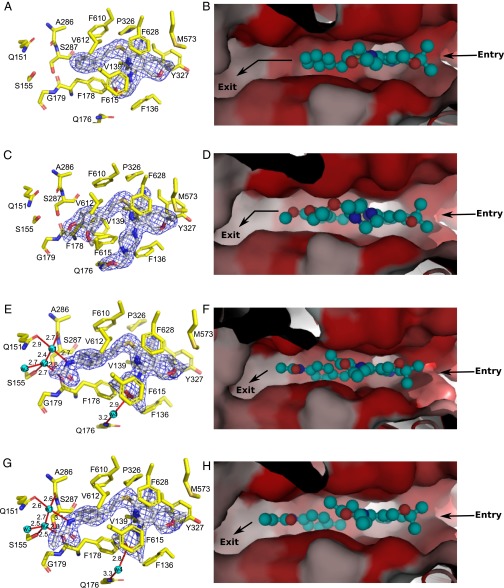
To determine the structural basis of AcrB inhibition by MBX2319, we solved the structure of MBX2319 bound to AcrBper. MBX2319 binds to the T protomer, and is engaged in multiple hydrophobic interactions with various side chains lining the deep binding pocket and hydrophobic trap (Fig. 3 A and B and SI Appendix, Fig. S3) as previously predicted by molecular dynamics (MD) simulation (27). The central, aromatic pyridine ring of MBX2319 is oriented parallel to the F628 aromatic side chain with a distance of ∼3.5 Å, resulting in an extensive π–π stacking interaction. Similarly, the phenyl and morpholinyl groups interact with F178 and F615 at a distance of ∼4 Å. The F610 side chain is orthogonally packed against the dimethylenesulfide moiety of MBX2319 that connects the pyranopyridine core to the phenyl group. The side chains of Y327 and M573 bind to the gem-dimethyl group (distance of ∼3.9 Å). An analysis by MD simulations indicates that the F178 and F628 side chains are the major contributors to the inferred tight binding (Table 1). The binding location of MBX2319 on the T protomer of AcrB appears to be similar to the binding location of the published pyridopyrimidine inhibitor D13-9001 (20) (SI Appendix, Fig. S4), substantiating that the hydrophobic trap is a promising target for EPIs. Furthermore, MBX2319 appears to hinder the binding of other substrates (Fig. 3B).
Fig. 3.
Details of the binding of inhibitors with AcrBper. MBX2319 (A and B) and MBX2931 (C and D) mainly interact via hydrophobic stacking interactions with residues comprising the deep binding pocket and the hydrophobic trap. MBX3132 (E and F) and MBX3135 (G and H) are additionally engaged in a water-mediated hydrogen bond network, extending from the acetamide and acrylamide groups, respectively. Hydrogen bonds and water molecules are shown as red lines (with distances in angstroms) and as cyan-colored spheres, respectively. MBX compounds are shown as sticks (carbon, gray; oxygen, red; nitrogen, blue; sulfur, yellow). (Left) AcrB residues involved in inhibitor binding are shown as sticks (carbon, yellow; oxygen, red; nitrogen, blue; sulfur, gold), and the 2Fo-Fc electron density maps (blue-colored mesh) are contoured at 1.0 σ (MBX2319 and MBX2931) and at 1.5 σ (MBX3132 and MBX3135), respectively. (Right) AcrB deep binding pocket surface is colored according to its hydrophobicity (red, hydrophobic; gray, hydrophilic), and the substrate pathway is indicated with arrows. MBX compounds are shown in a ball-and-stick representation (cyan-colored carbon atoms).
Table 1.
In silico thermodynamics of MBX binding to the AcrB deep binding pocket and hydrophobic trap
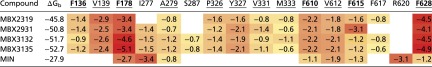 |
The free energies of binding (ΔGb) and the relative per-residue contributions (in kilocalories per mole) were calculated for the inhibitors MBX2319, MBX2931, MBX3132, and MBX3135 bound to AcrB. The values are compared with estimates calculated for MIN (23). Cells reporting per-residue free energies are colored according to the value of the contribution, from red (largest) to light yellow (lowest). Only residues contributing more than ∼0.6 kcal/mol (kBT at room temperature, where kB is the Boltzmann constant and T is the absolute temperature) are reported. Residues contributing to the binding of all four MBX inhibitors are underlined, and residues comprising the hydrophobic trap (19) are shown in bold.
Development and Characterization of Improved MBX Inhibitors.
We systematically varied the substituents around the pyranopyridine core (22). We found that the nitrile, dimethylenesulfide, and gem-dimethyl groups could not be varied without negatively affecting the activity, whereas substitutions at the phenyl and morpholinyl groups (MBX2931, MBX3132, and MBX3135) improved potency and stability (Fig. 1A). In particular, the acetamide- and acrylamide-containing compounds MBX3132 and MBX3135 exhibit a 10- to 20-fold increase in potency compared with MBX2319 against a panel of representative Enterobacteriaceae in checkerboard assays (SI Appendix, Table S2) and in time-kill assays (Fig. 4 A–C). Moreover, the increased potency of MBX3132 and MBX3135 against AcrAB-TolC is demonstrated by the increased levels of accumulation of Hoechst 33342 (H33342), a fluorescent substrate of AcrB, in E. coli AB1157 (Fig. 4 D–G), confirming efflux pump inhibition. The accumulation of H33342 in the presence of 12.5 μM MBX3132 or MBX3135 significantly exceeded the level of H33342 accumulation over the level of H33342 accumulation of the ΔacrB strain (Fig. 4 F and G), suggesting that MBX3132 and MBX3135 inhibit additional RND family efflux pumps, because they do not perturb the inner or outer E. coli membrane (22). In the presence of 10 nM MBX3132 or MBX3135, the kinetics of AcrAB-TolC–mediated nitrocefin efflux were severely affected, whereas the effect of 10 nM MBX2319 was negligible (Fig. 4 H–J). MBX2319 affects the nitrocefin efflux activity only at >200 nM (21).
Fig. 4.
Analogs of the pyranopyridine EPI MBX2319 exhibit improved efflux pump inhibition. The bactericidal activity of a minimally bactericidal dose of ciprofloxacin (0.01 μg/mL) is potentiated against E. coli by MBX2319 (A), MBX3132 (B), and MBX3135 (C). The concentrations of MBX3132 and MBX3135 used are 10-fold lower than the concentration of MBX2319. Inhibition of efflux pump activity is shown in a cell-based assay that measures accumulation of the fluorescent dye H33342 in the presence of MBX2319 (D), MBX2931 (E), MBX3132 (F), and MBX3135 (G). Effect of 10 nM MBX2319 (H), MBX3132 (I), and MBX3135 (J) on the Michaelis–Menten kinetics of AcrAB-TolC using the nitrocefin efflux assay (31). CIP, ciprofloxacin; Cmpd, MBX compound alone; Cp, periplasmic concentration of nitrocefin (micromolar); RFU, relative fluorescence unit; Ve, efflux pump velocity (in nanomoles per second per milligram).
Interaction Between AcrB and More Recent Analogs of MBX2319.
MBX2913, which shows no or only a slight improvement in potency over MBX2319 (Fig. 4 and SI Appendix, Table S2), bound similar to MBX2319 (Fig. 3 C and D), and the binding energy contributed by the pocket was also similar, except for the increased contribution by F615 (Table 1). In contrast, with MBX3132 and MBX3135, which exhibited much stronger activity (Fig. 4 and SI Appendix, Table S2), a stronger contribution by F178 in the binding was evident (Table 1). A better indication of stronger and more stable binding of these compounds can be found in the greatly decreased temperature B-factors for these compounds (Table 2).
Table 2.
Temperature factor B from crystallography and RMSF and B factor from MD trajectories
| Compound | B-factor, Å2 | RMSF, Å | B-factor, Å2 |
| Crystallography | MD simulation | ||
| MBX2319 | 52.4 | 1.31 | 45.2 |
| MBX2931 | 52.2 | 1.19 | 37.2 |
| MBX3132 | 29.7 | 0.88 | 20.3 |
| MBX3135 | 27.4 | 0.95 | 23.5 |
RMSF, rms fluctuation.
Detailed analysis of high-resolution AcrBper crystal structures containing these compounds was instructive (Fig. 3 E and G and SI Appendix, Fig. S3). The respective acetamide and acrylamide extensions are engaged in sophisticated hydrogen bond networks centered on a highly coordinated solvent water molecule (w1; Fig. 3 E and G and SI Appendix, Fig. S5), which serves as a hydrogen bond donor to the AcrB A286 carbonyl backbone oxygen and the Q151 side chain. In turn, w1 serves as a hydrogen bond acceptor for the acetamide and acrylamide groups of MBX3132 and MBX3135, respectively, and for another water molecule (w2). The latter hydrogen bonds to the S155 hydroxyl side chain, the F178 carbonyl oxygen, and water molecule w3. Moreover, MBX3132 and MBX3135 form a hydrogen bond between the oxygen atom of their morpholinyl group and another water molecule, which, in turn, hydrogen-bonds to the Q176 side chain (w4; Fig. 3 E and G and SI Appendix, Fig. S5). Presumably, these hydrogen bond networks are critical for accurately positioning MBX3132 and MBX3135 in the hydrophobic trap of AcrB, which is reflected in the significantly reduced flexibilities for these compounds compared with MBX2319 and MBX2931, as observed by the reduced temperature factors and MD simulations (Table 2 and SI Appendix, Table S3). Overall, MBX3132 and MBX3135 show a far more substantial free energy release upon binding to AcrB than the free energy release of a typical substrate, MIN (Table 1), or of the earlier inhibitors PAβN and NMP (ca. 20–30 kcal/mol from MD simulations) (26). It appears that the stabilized positioning of the acetamide- and acrylamide-containing inhibitors contributes to this tighter binding (Fig. 3 E and G, Table 2, and SI Appendix, Fig. S6 and Table S4). Thus, both structural and computational data provide a consistent molecular explanation for the observed increased potency of MBX3132 and MBX3135.
Conclusion
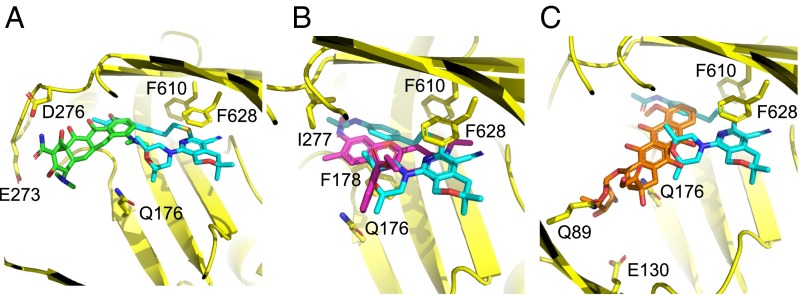
The high-resolution crystal structures described above show clearly how these potent pyranopyridine inhibitors prevent the binding of substrates to the deep binding pocket of AcrB substrates through steric hindrance (Figs. 3 B, D, F, and H and 5). Because these compounds bind more tightly than the usual substrates (Table 1), effective inhibition can be achieved at low inhibitor concentrations. It appears that all known potent inhibitors, including the MBX compounds, bind to the hydrophobic trap. In this way, they may prevent the T-to-O conformer transition, effectively preventing the functional rotation of the AcrB trimer, which is similar to a mechanism that has been proposed for the defect in efflux activity of the AcrB F610A substitution (2, 28). Furthermore, the structures provide a molecular rationale for further optimization of the pyranopyridine EPIs, which can be used to improve the drug-like properties of these compounds while maintaining or improving potency (a possible site of extension is shown in Fig. 6). The high-resolution structures of the MBX–AcrBper complexes allow the unambiguous assignment of inhibitor and side-chain positions, coordinated water molecules and the resulting interactions. This knowledge, combined with atomistic simulations, enables a more reliable assignment of free energy contributions between ligands, protein, and solvent, which is a necessary prerequisite for structure-based drug design (29). The successful production of well-diffracting AcrBper crystals will also enable convenient structural analysis of substrate binding to AcrB in a detergent-free background. This development should guide the future exploitation of additional substrate and inhibitor binding to AcrB and other RND family homologs for which structural data are sparse.
Fig. 5.
MBX compound binding site overlaps with substrate binding sites. The superimposition of MBX3132 coordinates (carbon, cyan; oxygen, red; nitrogen, blue; sulfur, yellow) with MIN [A; carbon, green; PDB ID code 4DX5 (21)], R6G (B; carbon, magenta; this study), and doxorubicin [C; carbon, orange; PDB ID code 4DX7 (21)] is shown, indicating that the EPI sterically prevents substrate binding to the AcrB deep binding pocket. AcrB side chains involved in the binding of substrates or EPIs are indicated and shown as sticks (carbon, yellow).
Fig. 6.
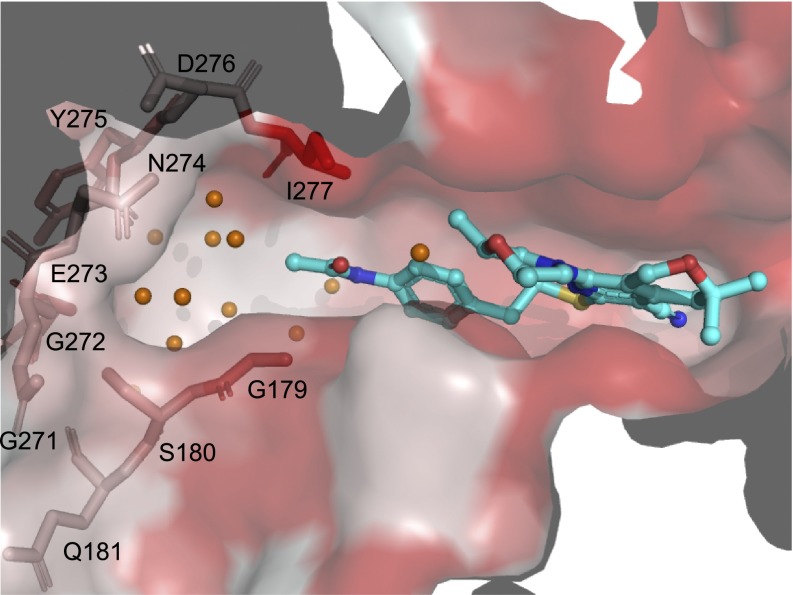
Hydrophilic, water-filled cavity provides potential for MBX inhibitor improvement. The AcrB deep binding pocket surface is colored as in Fig. 1. MBX3132 is shown in a ball-and-stick representation (carbon, cyan; oxygen, red; nitrogen, blue; sulfur, yellow). Coordinated water molecules in the extended hydrophilic pocket between the acetamide moiety of MBX3132 and residues 179–181 and 271–277 (shown as sticks) are shown as orange spheres. Augmented inhibitors could potentially protrude into this solvent-filled pocket.
Experimental Procedures
Most of the microbiological methods were described earlier (21). The gene coding for AcrBper was created by amplifying the acrB segments coding for A39-T329 and S561-S869 with primers that connect the two regions via a GGSGGSGGS linker upon BamHI digestion and ligation (30). AcrBper was expressed in E. coli MC1061 (30) and was purified by using its C-terminal His tag sequence. A vapor diffusion technique was used for crystallization, and ligands were introduced by soaking. Details are described in SI Appendix.
Supplementary Material
Acknowledgments
We thank the Swiss Light Source, SOLEIL Synchrotron, and Deutsches Elektronen Synchrotron for access to their facilities and the beamline scientists for support during data collection. This work was supported by the National Institute of Allergy and Infectious Diseases (Grant R44 AI100332). H.S., A.R.O., and K.M.P. are supported by the Deutche Forschungsgemeinschaft (Sonderforschungsbereich 807, Transport and Communication Across Biological Membranes), Deutsche Forschungsgemeinschaft EXC115 (Cluster of Excellence Frankfurt-Macromolecular Complexes), German-Israeli Foundation, and Human Frontiers Science Program. The research of K.M.P., H.S., A.R.O., A.V.V., and P.R. was conducted as part of the Translocation Consortium (www.translocation.eu) and has received support from the Innovative Medicines Initiative Joint Undertaking under Grant 115525, resources that are composed of financial contributions from the European Union’s Seventh Framework Programme (FP7/2007-2013) and European Federation of Pharmaceutical Industries and Associations (EFPIA) companies.
Footnotes
Conflict of interest statement: K.M.P. and reviewer B.L. coauthored a review article in 2015.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [5EN5 (apo form), 5ENO (MBX2319-bound), 5ENP (MBX2931-bound), 5ENQ (MBX3132-bound), 5ENR (MBX3135-bound), 5ENS (rhodamine 6G-bound), and 5ENT (minocycline-bound)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1602472113/-/DCSupplemental.
References
- 1.Li XZ, Plésiat P, Nikaido H. The challenge of efflux-mediated antibiotic resistance in Gram-negative bacteria. Clin Microbiol Rev. 2015;28(2):337–418. doi: 10.1128/CMR.00117-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruggerone P, Murakami S, Pos KM, Vargiu AV. RND efflux pumps: Structural information translated into function and inhibition mechanisms. Curr Top Med Chem. 2013;13(24):3079–3100. doi: 10.2174/15680266113136660220. [DOI] [PubMed] [Google Scholar]
- 3.Du D, van Veen HW, Murakami S, Pos KM, Luisi BF. Structure, mechanism and cooperation of bacterial multidrug transporters. Curr Opin Struct Biol. 2015;33:76–91. doi: 10.1016/j.sbi.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 4.Du D, et al. Structure of the AcrAB-TolC multidrug efflux pump. Nature. 2014;509(7501):512–515. doi: 10.1038/nature13205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443(7108):173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 6.Seeger MA, et al. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science. 2006;313(5791):1295–1298. doi: 10.1126/science.1131542. [DOI] [PubMed] [Google Scholar]
- 7.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grütter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5(1):e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seeger MA, et al. Engineered disulfide bonds support the functional rotation mechanism of multidrug efflux pump AcrB. Nat Struct Mol Biol. 2008;15(2):199–205. doi: 10.1038/nsmb.1379. [DOI] [PubMed] [Google Scholar]
- 9.Nakashima R, Sakurai K, Yamasaki S, Nishino K, Yamaguchi A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature. 2011;480(7378):565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 10.Eicher T, et al. Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. eLife. 2014;3:1–26. doi: 10.7554/eLife.03145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pos KM. Drug transport mechanism of the AcrB efflux pump. Biochim Biophys Acta. 2009;1794(5):782–793. doi: 10.1016/j.bbapap.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 12.Su C-C, et al. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J Bacteriol. 2006;188(20):7290–7296. doi: 10.1128/JB.00684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dreier J, Ruggerone P. Interaction of antibacterial compounds with RND efflux pumps in Pseudomonas aeruginosa. Front Microbiol. 2015;6:660. doi: 10.3389/fmicb.2015.00660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Opperman TJ, Nguyen ST. Recent advances toward a molecular mechanism of efflux pump inhibition. Front Microbiol. 2015;6:421. doi: 10.3389/fmicb.2015.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lomovskaya O, et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob Agents Chemother. 2001;45(1):105–116. doi: 10.1128/AAC.45.1.105-116.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lomovskaya O, Bostian KA. Practical applications and feasibility of efflux pump inhibitors in the clinic--a vision for applied use. Biochem Pharmacol. 2006;71(7):910–918. doi: 10.1016/j.bcp.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 17.Bohnert JA, Kern WV. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob Agents Chemother. 2005;49(2):849–852. doi: 10.1128/AAC.49.2.849-852.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yoshida K, et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 7: Highly soluble and in vivo active quaternary ammonium analogue D13-9001, a potential preclinical candidate. Bioorg Med Chem. 2007;15(22):7087–7097. doi: 10.1016/j.bmc.2007.07.039. [DOI] [PubMed] [Google Scholar]
- 19.Yamaguchi A, Nakashima R, Sakurai K. Structural basis of RND-type multidrug exporters. Front Microbiol. 2015;6:327. doi: 10.3389/fmicb.2015.00327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakashima R, et al. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500(7460):102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 21.Opperman TJ, et al. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob Agents Chemother. 2014;58(2):722–733. doi: 10.1128/AAC.01866-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen ST, et al. Structure-activity relationships of a novel pyranopyridine series of Gram-negative bacterial efflux pump inhibitors. Bioorg Med Chem. 2015;23(9):2024–2034. doi: 10.1016/j.bmc.2015.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eicher T, et al. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc Natl Acad Sci USA. 2012;109(15):5687–5692. doi: 10.1073/pnas.1114944109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murakami S, Nakashima R, Yamashita E, Yamaguchi A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature. 2002;419(6907):587–593. doi: 10.1038/nature01050. [DOI] [PubMed] [Google Scholar]
- 25.Takatsuka Y, Chen C, Nikaido H. Mechanism of recognition of compounds of diverse structures by the multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci USA. 2010;107(15):6559–6565. doi: 10.1073/pnas.1001460107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vargiu AV, Nikaido H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc Natl Acad Sci USA. 2012;109(50):20637–20642. doi: 10.1073/pnas.1218348109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vargiu AV, Ruggerone P, Opperman TJ, Nguyen ST, Nikaido H. Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob Agents Chemother. 2014;58(10):6224–6234. doi: 10.1128/AAC.03283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vargiu AV, et al. Effect of the F610A mutation on substrate extrusion in the AcrB transporter: Explanation and rationale by molecular dynamics simulations. J Am Chem Soc. 2011;133(28):10704–10707. doi: 10.1021/ja202666x. [DOI] [PubMed] [Google Scholar]
- 29.Grey JL, Thompson DH. Challenges and opportunities for new protein crystallization strategies in structure-based drug design. Expert Opin Drug Discov. 2010;5(11):1039–1045. doi: 10.1517/17460441.2010.515583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geertsma ER, Dutzler R. A versatile and efficient high-throughput cloning tool for structural biology. Biochemistry. 2011;50(15):3272–3278. doi: 10.1021/bi200178z. [DOI] [PubMed] [Google Scholar]
- 31.Nagano K, Nikaido H. Kinetic behavior of the major multidrug efflux pump AcrB of Escherichia coli. Proc Natl Acad Sci USA. 2009;106(14):5854–5858. doi: 10.1073/pnas.0901695106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.