Significance
Many bacteria swim using helical propellers, flagella. Intriguingly, different bacteria show different swimming abilities, strikingly illustrated by the abilities of some to bore through viscous fluids (e.g., gastrointestinal mucus) in which others are completely immobilized. We used 3D electron microscopy to show that differences can be explained by the structures of the torque-generating motors: two diverse high-torque motors position additional torque-generating complexes at wider radii from the axial driveshaft than in the model enteric bacteria; this positioning is consistent with the exertion of greater leverage to rotate the flagellum and thus greater torque generation. Intriguingly, these torque-generating complexes are scaffolded at wider radii by a conserved but divergent family of structures, suggesting an ancient origin of reconfiguring torque output.
Keywords: bacterial flagellar motors, electron cryo-tomography, macromolecular evolution, torque, Campylobacter
Abstract
Although it is known that diverse bacterial flagellar motors produce different torques, the mechanism underlying torque variation is unknown. To understand this difference better, we combined genetic analyses with electron cryo-tomography subtomogram averaging to determine in situ structures of flagellar motors that produce different torques, from Campylobacter and Vibrio species. For the first time, to our knowledge, our results unambiguously locate the torque-generating stator complexes and show that diverse high-torque motors use variants of an ancestrally related family of structures to scaffold incorporation of additional stator complexes at wider radii from the axial driveshaft than in the model enteric motor. We identify the protein components of these additional scaffold structures and elucidate their sequential assembly, demonstrating that they are required for stator-complex incorporation. These proteins are widespread, suggesting that different bacteria have tailored torques to specific environments by scaffolding alternative stator placement and number. Our results quantitatively account for different motor torques, complete the assignment of the locations of the major flagellar components, and provide crucial constraints for understanding mechanisms of torque generation and the evolution of multiprotein complexes.
Flagellated bacteria have tailored their motility to diverse habitats. For example, the enteric model organisms Salmonella enterica serovar Typhimurium and Escherichia coli colonize animal digestive tracts and can reside outside a host, assembling flagella over their cell body to swim. However, a diverse spectrum of flagellar swimming ability is seen across the bacterial kingdom. Caulobacter crescentus inhabits low-nutrient freshwater environments where it swims using a high-efficiency flagellar motor (1, 2), whereas Vibrio species produce high-speed, sodium-driven polar flagella to capitalize on the high sodium gradient of their marine habitat (3). On the other hand, the ε-proteobacteria and spirochetes, many of which thrive exclusively in association with a host, have evolved characteristically rapid and powerful swimming capabilities that enable them to bore through mucous layers coating epithelial cells or between tissues. Indeed, the ε-proteobacteria Campylobacter jejuni and Helicobacter pylori are capable of continued swimming in high-viscosity media that immobilize E. coli or Vibrio cells (4–6), and similar behavior is observed for spirochetes (7, 8).
Despite differences in the organisms’ swimming ability, the flagellar motor is composed of a conserved core of ∼20 structural proteins (9). The mechanism of flagellar motility is conserved (10), with torque generated by rotor and stator components (9). Stator complexes, heterooligomers of four motility A (MotA) and two motility B (MotB) proteins, are thought to form a ring that surrounds the axial driveshaft. Transmembrane helices of MotA and MotB form an ion channel, and MotB features a large periplasmic domain that binds peptidoglycan (11, 12) and the flagellar structural component, the P-ring (13). The stator complex couples ion flux to exertion of force on the cytoplasmic rotor ring (the C-ring), which transmits torque to the axial driveshaft (the rod), universal joint (the hook), and helical propeller (the filament), culminating in propulsion of the bacterium. Biophysical (14) and freeze-fracture (15) studies together with modeling (16) have proposed that a tight ring of ∼11 stator complexes dynamically assembles around the rod above the outer lobe of the C-ring in closely related Salmonella and E. coli motors (which we collectively refer to as the “enteric motor”). However, despite these conclusions, and although the structures observed in subtomogram averages have been proposed to be the stator complexes (17–19), the locations and stoichiometries of the stator complexes remain to be confirmed.
How can we explain the wide diversity in flagellar swimming abilities in the context of a conserved core flagellar motor? Biophysical studies suggest that the source of the difference lies, at least in part, in variations in the mechanical output of the motors themselves. Torques of motors from different bacteria have been shown to range over an order of magnitude, and torque correlates with swimming speed and the ability of bacteria to propel themselves through different viscosities, indicating that adaptations are likely to be at the level of the motor itself. [Torque also varies within a single species, up to a maximum value, as a function of the number of stator complexes incorporated into the motor (14)]. For example, C. crescentus motors have been measured to produce torques of 350 pN⋅nm (2). Estimates for the torque of the enteric motor ranges from ∼1,300 to ∼2,000 pN⋅nm (20, 21). The ε-proteobacterium H. pylori has been estimated to swim with torque of 3,600 pN⋅nm (22), and spirochetes are capable of swimming with 4,000 pN⋅nm of torque (21, 23). Sodium-driven motor torques in Vibrio spp. have been measured between ∼2,000 and 4,000 pN⋅nm (24), depending on the magnitude of the sodium gradient. It is noteworthy, however, that an estimated sodium motive force in Vibrio spp. that is lower than the standard E. coli proton motive force nevertheless drives the Vibrio motor with higher torque than the E. coli motor (24, 25), further suggesting that torque differences likely exist at the level of the motor. However, the molecular mechanism by which different motors might produce different torques has not been investigated.
The simplest scenario for tuning motor torque would be evolved adaptation of motor architecture. In support of this scenario, we recently showed that many motors have evolved additional structures not found in the well-studied enteric motors (18), and we observed that the C-ring radius varies among species (17, 18). One of the most widespread novel structures is a periplasmic basal disk directly beneath the outer membrane, often co-occurring with varied uncharacterized additional structures, which we collectively term “disk complexes.” Consistently, disk complexes have been seen only in motors that produce torque higher than that in E. coli or Salmonella. For example, the sodium-driven ∼2,000+ pN⋅nm torque motors of Vibrio species assemble a disk complex featuring a basal disk beneath the outer membrane (18) in addition to smaller H- and T-rings composed of FlgOT (flagella O, T) and MotXY (motility X, Y), respectively (26, 27). It has been shown that the T-ring interacts with stator complexes in Vibrio spp. (28), although the exact location and number of stator complexes in Vibrio spp. remains unclear. ε-Proteobacteria such as Helicobacter species, C. jejuni, and Wolinella succinogenes also assemble disk complexes composed of large basal disks beneath the outer membrane together with additional smaller disks (18, 29). Although these and other cases of additional disks have been reported (18, 30), their relation to flagellar function remains enigmatic, and it is unclear if these widespread disk complexes are homologous or analogous.
In this study, we hypothesized that bacteria have tuned their swimming abilities by evolving structural adaptations to their flagellar motors that would result in altered torque generation. Using electron cryo-tomography and subtomogram averaging, we found that Vibrio polar γ-proteobacterial and Campylobacter ε-proteobacterial flagellar motors incorporate 13 and 17 stator complexes, respectively, compared with the ∼11 in enteric bacteria. In both cases, these stator complexes are scaffolded into wider stator rings relative to the enteric motor by components of their respective disk complexes. The wider C. jejuni stator ring is further reflected in a considerably wider rotor C-ring. Further analysis of the components of the Vibrio and C. jejuni disk complexes reveals that they share a core protein, FlgP, but each has acquired diverse additional components to form divergent disk-complex architectures. We conclude by showing that our structural data of wider stator rings featuring additional stator complexes can quantitatively account for the differences in torque between different flagellar motors.
Results
Three Flagellar Motors That Produce Different Torques Are Structurally Diverse.
We hypothesized that disk complexes might offer a structural rationale for differences in motor torque. To explore this hypothesis, we performed a comparative electron cryo-tomography study of high-torque motor structures in situ, using Vibrio fischeri and C. jejuni as models for Vibrio spp. and ε-proteobacteria, respectively, and compared them with the Salmonella motor as a well-studied model enteric motor. For data collection we developed a high-throughput electron cryo-tomography pipeline on a 200-kV field emission gun (FEG) electron microscope with a side-entry cryo-holder and direct electron detector camera and imaged motors in situ in Salmonella, V. fischeri, and C. jejuni. For Salmonella, we used a mutant that produced minicells (Fig. 1A) to circumvent problems caused by excessive inelastic and multiple electron scattering (31), but we were able to image the motors in WT V. fischeri and C. jejuni (Fig. 1 B and C).
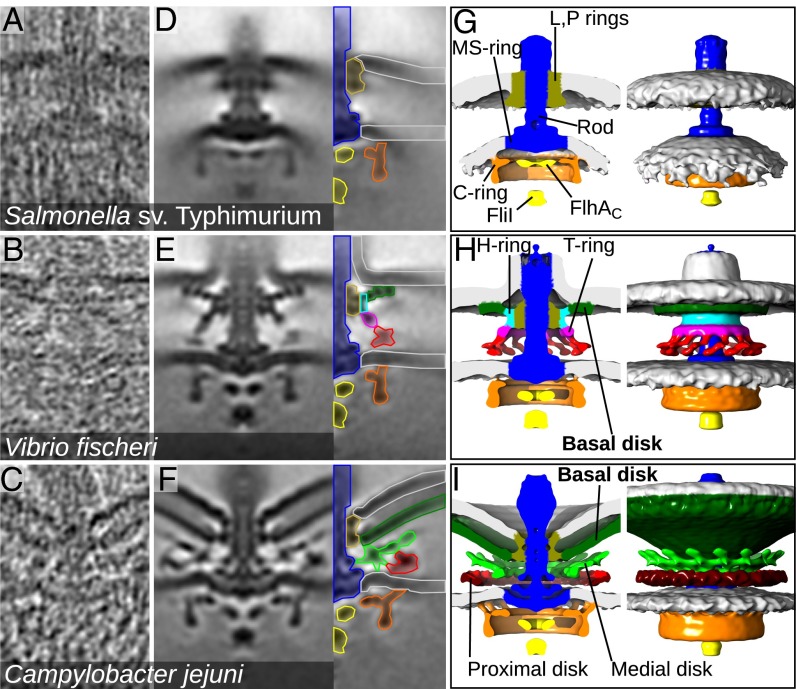
Fig. 1.
High-torque bacterial flagellar motors assemble large periplasmic disk complexes. (A–C) Tomographic slices through intact cells of Salmonella (A), V. fischeri (B), and C. jejuni (C) showing individual flagellar motors. Height of all image panels is 100 nm. (D–F) Slices (100 × 100 × 0.81 nm) through subtomogram averages of hundreds of motors. Color keys indicate the regions of the motor (named in G–I), of Salmonella (average of 286 motors) (D), V. fischeri (average of 302 motors) (E), and C. jejuni (average of 156 motors) (F). (G–I) Isosurface renderings of motors shown in D–F. FliI and FlhAC are components of the flagellar type III secretion system.
Subtomogram averaging of hundreds of motors enabled us to determine structures (Fig. 1) at resolutions sufficient for discerning individual protein domains (Fig. S1). Parts of the V. fischeri and C. jejuni motors showed clear 13-fold and 17-fold symmetry around their axes of rotation, respectively (Fig. 2 and Fig. S2); this symmetry was imposed to improve contrast. The Salmonella motor exhibited minor flexion in the rod between the MS- and P-rings, necessitating separate averaging of the cytoplasmic and periplasmic sections and merging the two structures at this flexion point. Because no symmetry was evident in the Salmonella motor, we applied 11-fold symmetry based on the consensus of stator-complex symmetry in Salmonella and E. coli (14, 32). Neither this 11-fold symmetry nor the other symmetries clarified any symmetric features in our reconstructions. Our Salmonella structure reproduced the expected features seen in previous studies (Fig. 1 D and G) (31, 33).
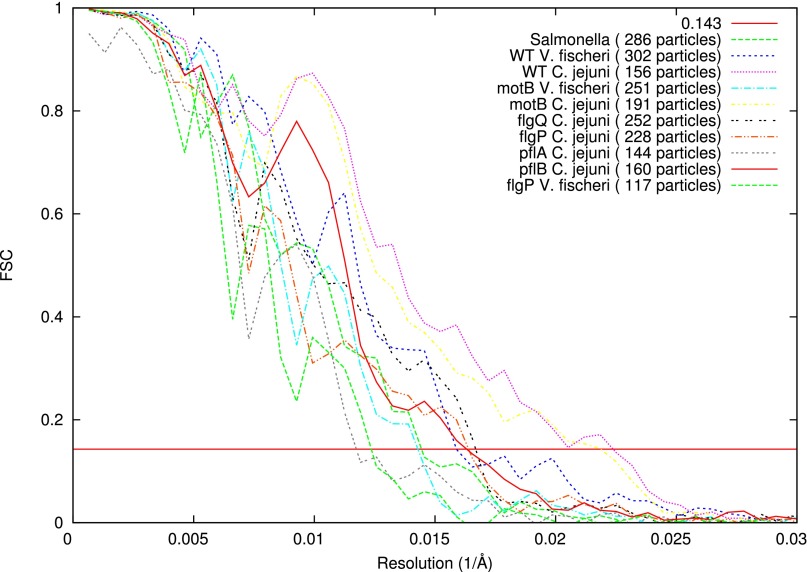
Fig. S1.
Gold-standard FSC curves of subtomogram averages. Datasets were fully separated into two half datasets, and two independent reconstructions were calculated using different template subtomograms from each dataset (59). FSC curves were calculated between the two aligned averages. Subsequently these two averages were averaged together and symmetrized to form the final averaged structure. Resolutions in Angstroms at a 0.143 threshold are Salmonella WT: 69.4; V. fischeri WT: 63.0; C. jejuni WT: 44.7; V. fischeri ΔmotB: 70.3; C. jejuni ΔmotB: 45.8; C. jejuni ΔflgQ: 60.1; C. jejuni ΔflgP: 61.2; C. jejuni ΔpflA: 85.3; C. jejuni ΔpflB: 61.9; and V. fischeri ΔflgP: 81.0.
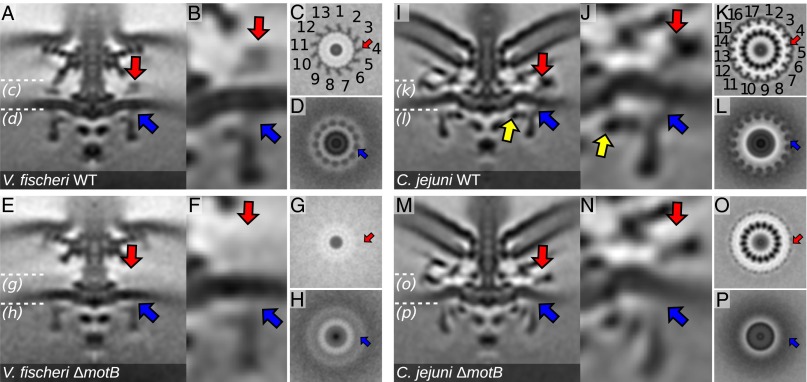
Fig. 2.
Disk complexes scaffold alternative stator-complex locations and numbers. (A) WT V. fischeri motor highlighting the putative MotB (red arrow) and the putative FliGC:MotA interface (blue arrow). (B) Close-up of putative MotB density and FliGC:MotA interface. (C and D) Thirteen-fold symmetry through the stator and C-ring planes shown by white dotted lines in A. (E–H) Images of V. fischeri ΔmotB equivalent to the images in A–D confirming the identity of proposed stator complexes. (I–P) Equivalent images for WT C. jejuni (I–L) and C. jejuni ΔmotB (M–P). Note 17 stator-complex densities observed in C. jejuni. Yellow arrows in I and J indicate an additional density on the inner lobe of the C. jejuni C-ring as compared with Salmonella and V. fischeri. Note that the odd-numbered stator counts result in an asymmetric appearance of motor cross-sections.
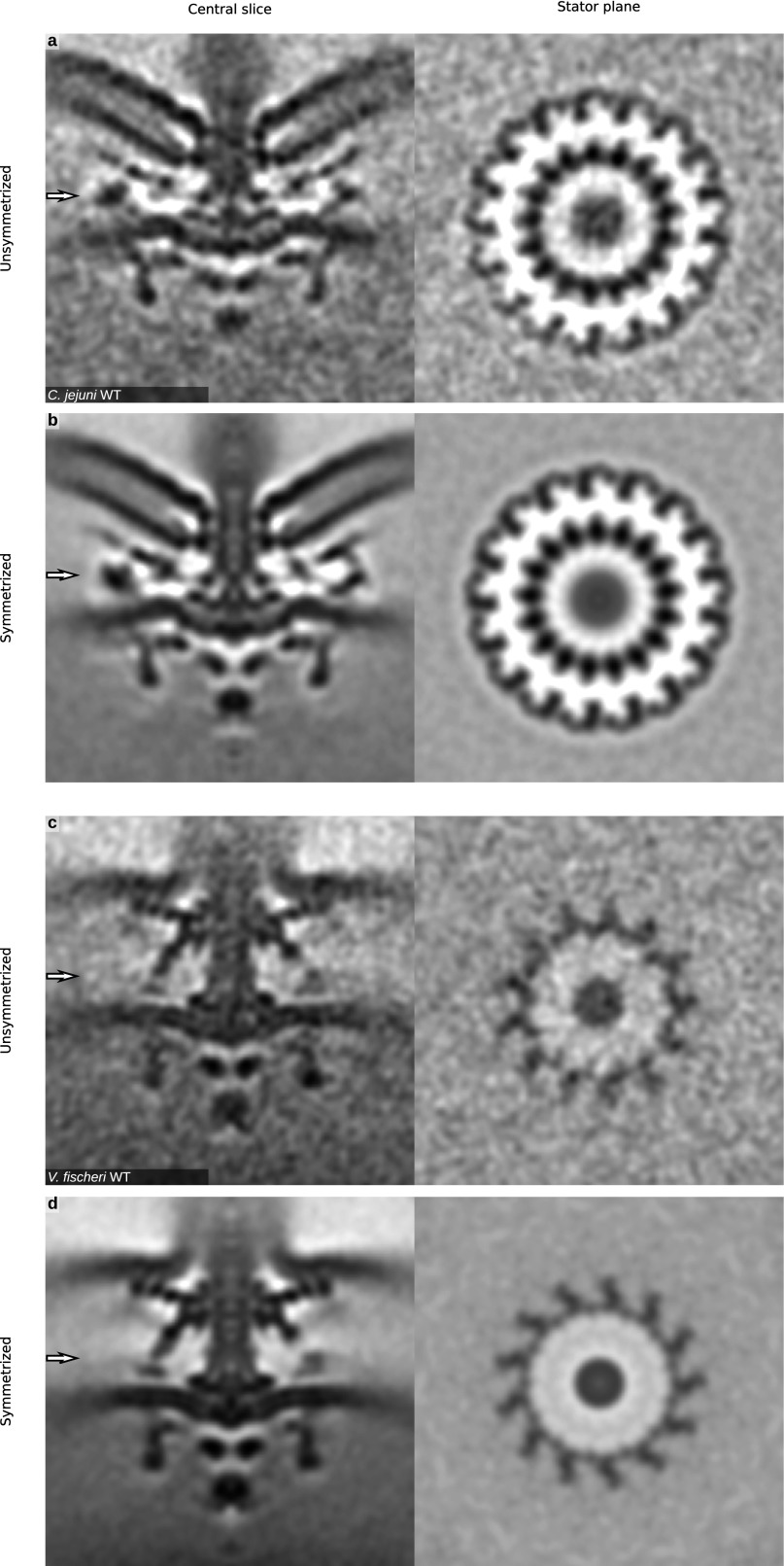
Fig. S2.
Unsymmetrized motors showing clear symmetries. C. jejuni motors (A, Left) exhibited clear 17-fold symmetry (A, Right), which was applied to improve the signal-to-noise ratio (B). V. fischeri exhibited 13-fold symmetry (C), which was applied to improve the signal-to-noise ratio (D).
The V. fischeri and C. jejuni motors featured disk complexes with prominent basal disks beneath the outer membrane (Fig. 1 H and I). We also could visualize the previously described H- and T-rings (26, 27) in the V. fischeri motor, although they were positioned 12 nm from the inner membrane, further than the 5 nm recently proposed in closely related Vibrio alginolyticus (34). In C. jejuni, two additional disks were present between the basal disk and the inner membrane, which we annotated as a periplasmic “medial disk” and an inner membrane-associated “proximal disk” (Fig. 1 F and I). Although the C. jejuni basal disk did not exhibit any clear symmetry, the periplasmic medial disk and inner membrane-associated proximal disk formed rings with clear 17-fold symmetry (Fig. 2 I–K and Fig. S2). Curiously, the cup-shaped C. jejuni basal disk appeared to push the outer membrane away from the cell body at an increased radius from the motor axis, producing a distortion—a concave indentation— in the outer membrane around the motor. The significance of this distortion is unclear, particularly in light of the observation that the closely related H. pylori motor has a flat basal disk and does not feature a corresponding indentation (18).
High-Torque Motors Incorporate Additional Stator Complexes.
We investigated our speculation that disk complexes might contribute to the ability of these motors to generate high torque by examining the torque-generating stator complexes. The enteric flagellar motor has been suggested to incorporate ∼11 stator complexes that interact with the outer lobe of the C-ring in the cytoplasm and with the peptidoglycan and the P-ring (13). The number of stator complexes incorporated into the enteric motor is proportional to the load on the motor (35, 36). Because blotting during the sample vitrification process for cryo-tomographic imaging necessitated imaging in low-viscosity growth medium, we anticipated that a low motor load would result in low stator-complex occupancy and that our subtomogram averaging procedure would not resolve stator complexes. Consistent with this notion, no candidate stator-complex densities were observed in the periplasm in the Salmonella motor structure (Fig. 1 D and G). Although no stator complexes were observed, the outer lobe of the C-ring, which is believed to be the C-terminal domain of FliG upon which stator complexes exert force, was positioned at a 20-nm radius from the flagellar rotational axis.
In contrast, in both V. fischeri and C. jejuni we observed periplasmic densities connecting through the inner membrane to the outer lobe of the C-ring where the stator complexes are expected to reside (Fig. 2 A, B, I, and J) with 13-fold and 17-fold symmetry, respectively, as noted above. This odd-numbered symmetry produced asymmetric cross-sections in both V. fischeri and C. jejuni motors (Figs. 1 and 2). For V. fischeri, these proposed stator complexes were consistent with the size of a recent single-particle reconstruction of the Vibrio stator complex (16). The densities spanned the C-ring to ∼10 nm above the inner membrane, as previously predicted (34), were arranged in a 17-nm radius ring with a with 13-fold rotational symmetry, and contacted the 16-nm radius T-ring (Fig. 2 A–D, red arrows; continuous density is best visualized in Fig. 1H), as previously demonstrated (28). The lever contact point of this putative stator complex with the C-ring, i.e., the FliGC:MotA interface and the radius at which torque would be applied, was at a radius of 21.5 nm from the flagellar axis of rotation (Fig. 2 A–D, blue arrows). Corresponding putative stator-complex densities in C. jejuni formed a wider 30-nm radius ring with 17-fold symmetry extending from the outer lobe of the C-ring to merge with the proximal disk ∼10 nm above the inner membrane (Fig. 2 I–L, red arrows). The putative lever contact point at the FliGC:MotA interface was at a radius of 26.5 nm (Fig. 2 I–L, blue arrows). The 26-nm radius C. jejuni C-ring was correspondingly wider than the 22-nm radius Salmonella or 23-nm radius V. fischeri C-rings, spaced by an additional component on its interior surface (Fig. 2 I and J, yellow arrows). This observation suggests that the C. jejuni C-ring incorporates additional subunits of FliG, FliM, and FliN [and possibly the FliN paralog FliY (37)] to construct a wider C-ring relative to the Salmonella motor.
To test our hypothesis that these densities were the stator complexes, we deleted motB homologs in C. jejuni and V. fischeri [in V. fischeri, the gene motB1, encoding a PomB-type protein from the sodium-driven stator variant (38)]. The proposed MotB densities were lost in both species, supporting our hypothesis (Fig. 2 E–N). In the V. fischeri WT motor, MotB was less dense than the T-ring densities, possibly reflecting partial occupancy of the stator complexes in the Vibrio motor (39). In the C. jejuni motor, deletion of motB resulted in the loss of part of the proximal disk, indicating that MotB is one component of a multiprotein proximal disk assembly (Fig. 2 M–P). Although we cannot rule out the possibility that this loss of density results from the deletion of motB indirectly impacting the assembly of the proximal disk, the fact that this is the expected location of MotB and an unambiguous similar result in V. fischeri overwhelmingly support the assignment of this density as MotB. Loss of MotB did not appear to have as large an impact on the structure of the motor, because the remainder of the proximal disk continues to occupy this location. We also noted that the outer lobes and lever contact points of the Vibrio and C. jejuni C-rings became less well-resolved in ΔmotB mutants, likely indicating C-ring flexibility when not engaged by stator complexes (Fig. 2 F and N). We conclude that V. fischeri and C. jejuni incorporate wider rings of 13 and 17 stator complexes, respectively, into their motors (Fig. 2 C and K). The observation that stator complexes in both organisms associate with their respective disk complexes supports the role of disk complexes as scaffolds for stator rings that are wider than in the model enteric motors of Salmonella and E. coli.
Basal Disks Are Homologous.
Because the fully assembled Campylobacter and Vibrio disk complexes play common roles in binding stator complexes for incorporation and orientation into the flagellar motor, we asked whether these structures evolved convergently or divergently by probing whether some disk-complex components are homologous. Both V. fischeri and C. jejuni disk complexes featured outer membrane-associated basal disks. Consistent with our previous work on Vibrio cholerae (18), the V. fischeri basal disk was ∼28 nm in radius and attached to the H-ring as previously described in work on partial motors from V. alginolyticus (Figs. 2A and 3A) (26). In addition, the Vibrio basal disk is formed of concentric circles and appeared to contact the outer membrane at the base of the flagellar sheath, an unusual extension of the outer membrane present in Vibrio spp. and some other bacteria (40). In comparison, the C. jejuni basal disk attached to the motor via the P-ring and varied in radius among cells, with a mean of 42 nm. We did not resolve discrete concentric circles, suggesting that the C. jejuni disk is an unbounded spiral assembly as noted in close relative W. succinogenes (Fig. 2I) (29).
Fig. 3.
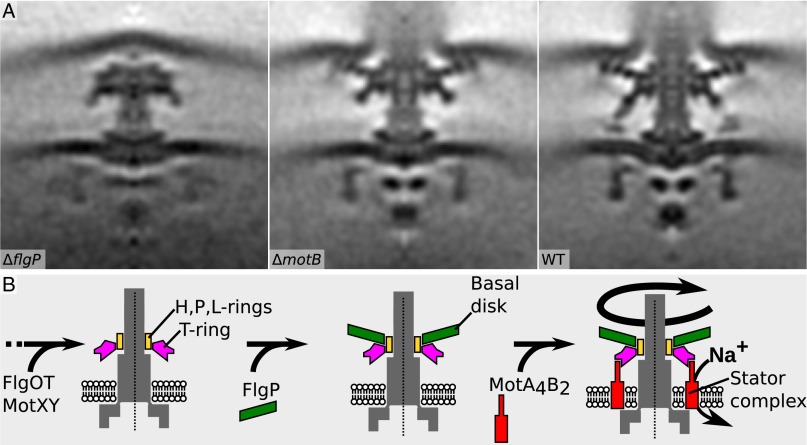
Assembly of the stator complexes in V. fischeri requires FlgP. (A) V. fischeri flagellar motors from ΔflgP (Left), ΔmotB (Center), and WT (Right) strains. (B) Model of assembly of the V. fischeri disk complex. Early-stage disk-complex components MotXY and FlgOT assemble before FlgP incorporation, followed by the assembly of the stator-complex ring. Images show 100 × 100 × 0.81 nm slices through subtomogram averages.
A candidate component of the basal disk in both organisms was FlgP, an abundant outer membrane-associated protein required for motility (but not for flagellation) that we and others previously identified in C. jejuni (41) and Vibrios (38, 42) but absent from Salmonella. To test whether FlgP forms the basal disks in both organisms, we determined structures of ΔflgP flagellar motors. In V. fischeri ΔflgP, motors were very seldom seen and did not assemble the extracellular hook/filament, so deriving an averaged structure was technically challenging. Nevertheless, we were able to collect sufficient data to determine a subtomogram average structure that unambiguously revealed the absence of the basal disk and stator complexes (Fig. 3A). Similarly the C. jejuni ΔflgP motor revealed the loss of the basal disk (Fig. 4) and became flexible between the MS- and P-rings, requiring separate averaging of cytoplasmic and periplasmic parts, as in Salmonella. The medial and proximal disks also were lost in C. jejuni, likely because the basal disk is required for the assembly of these lower disks (Fig. 4). Deletion of flgQ, a gene adjacent to flgP encoding a protein required for FlgP stability and localization to the outer membrane, resulted in a structure indistinguishable from the ΔflgP motor (Fig. 4) (41). The nonfunctional C. jejuni ΔflgP and ΔflgQ motors closely resembled the functional Salmonella motor with the exception of the wider C-ring. These findings suggest that the disk complex is essential for maintaining stator-complex interactions with FliG relocated in a wider C-ring. We conclude that basal disks in V. fischeri and C. jejuni are homologous because FlgP is abundant in the outer membrane of both C. jejuni and V. cholera (Fig. 5A) (41, 42) and the basal disk was lost exclusively in V. fischeri ΔflgP. Nevertheless, the proteins share only 25% sequence identity, which, together with the large-scale differences in the two FlgP basal disks and respective other disk-complex structures, suggests an ancient divergence.
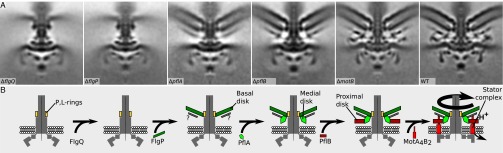
Fig. 4.
Protein location within the C. jejuni disk complex and hierarchical assembly pathway revealed by subtomogram averaging of WT and mutant motors. (A) Subtomogram averages arranged to illustrate the hierarchical assembly dependencies of the C. jejuni disk complex. Note that the outer membrane is poorly resolved in ΔflgQ and ΔflgP mutants because without the basal disk as an anchor the membrane is in a different position in each component subtomogram. Images are 100 × 100 × 0.81 nm slices through subtomogram averages. (B) Model of the hierarchical assembly of the C. jejuni disk complex.
Fig. 5.
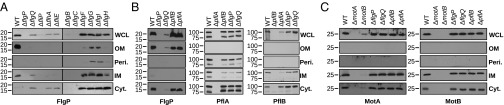
Localization interdependencies of five components of the C. jejuni disk complex. Immunoblot analysis of localization of FlgP, PflA, PflB, MotA, and MotB in various WT C. jejuni and mutant strains. (A) FlgP subcellular localization in flagellar T3SS, rod, and ring mutants. (B) Subcellular localization of FlgP, PflA, and PflB in specific flagellar disk mutants. (C) Localization of MotA and MotB stator proteins in flagellar disk-complex mutants. All proteins were detected with specific antiserum. Cyt, cytoplasm; IM, inner membrane; OM, outer membrane; Peri, periplasm; WCL, whole-cell lysate. For all immunoblots, equivalent amounts of cell cultures were lysed and subcellular compartments fractionated.
The V. fischeri Disk Complex Has Evolved to Become Essential.
Previous work identified other components of the Vibrio disk complex in addition to FlgP (26). In Vibrio species, additional structures composed of FlgO and FlgT (the H-ring) and MotX and MotY (the T-ring) are present in the flagellar motor (Fig. 1 E and H), and the T-ring is required for binding stator complexes for assembly into the motor (Figs. 1 E and H, 2 A and B, and 3A). A comparison of the tomograms of flagellar motors of different Vibrio disk and stator mutants suggests the order of the biogenesis of disk-complex assembly (Fig. 3B). We propose that in Vibrio spp. a core flagellar motor structurally similar to the Salmonella motor assembles first, followed by the incorporation of the H- and T-rings on the L- and P-rings, respectively (Fig. 3B). Formation of the H-ring likely provides a platform for assembly of FlgP to form the basal disk, which attaches to the periplasmic face of the outer membrane (Fig. 3B). Because the T-ring contacts the stator complex, but no stators are evident in the motor of the ΔflgP mutant, we suspect that the formation of the basal disk may alter H- and T-ring architecture that ultimately influences the ability of the T-ring to interact with stator complexes for incorporation into the motor.
The C. jejuni Disk Complex Is Required for Incorporation of the Stator Complexes.
To understand better the mechanism of stator scaffolding by the previously uncharacterized C. jejuni disk complex, we sought to identify its remaining components. We used transposon (Tn) mutagenesis to identify proteins not present in Salmonella that are required for flagellar motility in C. jejuni but not for biogenesis of an extracellular flagellar filament (indicating that flagellar biogenesis is disrupted) (Table 1). In this approach, we conducted a three-step screening process with a C. jejuni Tn mutant library carrying the astA transcriptional reporter to a nonessential flagellar gene (Materials and Methods). We first identified Tn mutants that expressed the transcriptional reporter as an indicator of normal flagellar gene expression. Next, we identified mutants that were nonmotile in motility agar. Finally, we collected the nonmotile mutants that aggregated in standing broth cultures, a property of flagellated C. jejuni strains, to identify nonmotile mutants that retained flagellation. This screen identified 117 genes, with the greatest number of Tn insertions in two genes, pflA and pflB (Table 1). PflA and PflB previously had been identified as interacting proteins essential for C. jejuni motility, but the reason they are required for motility was unknown (43). PflA contains a predicted secretion signal for periplasmic localization, whereas PflB is a predicted bitopic inner membrane protein with a small N-terminal cytoplasmic domain and a 700-amino acid periplasmic domain. Intriguingly, PflA and PflB contain multiple TPR motifs, which are associated with scaffolding roles in multiprotein complexes (44).
Table 1.
C. jejuni Tn mutants that produce flagella that do not support motility
| General classification of factors | Gene | Number of independent Tn mutants isolated in this gene | Verified or proposed function |
| Flagellar motor protein | motA | 9 | Motor stator protein |
| motB | 2 | Motor stator protein | |
| fliM | 6 | Motor switch protein | |
| fliY | 1 | Motor switch protein | |
| Chemotaxis system | cheA | 1 | Chemosensory histidine kinase |
| cheY | 2 | Chemosensory response regulator | |
| Flagellar structural components | flaA | 1 | Major flagellin |
| flaB | 1 | Minor flagellin | |
| flaG | 1 | Flagellin-like protein | |
| flgH | 2 | L-ring protein | |
| fliW | 1 | Putative flagellar chaperone | |
| flgN | 1 | Flagellar protein chaperone | |
| pseE | 3 | Pseudoaminic acid biosynthetic protein; flagellin glycosylation | |
| Flagellar gene or biosynthesis regulators | fliA | 6 | σ28 |
| flgM | 1 | Anti-σ factor for σ28 | |
| flhF | 1 | Flagellar biosynthesis GTPase | |
| Motility determinants (unknown functions) | pflA | 23 | Paralyzed flagellar protein |
| pflB | 19 | TPR-containing inner membrane protein | |
| Cjj81176_0276 | 3 | Hypothetical protein | |
| Other factors | clpA | 1 | ATP-binding component of ClgAP protease |
| comEA | 1 | Competence protein | |
| fedA | 1 | Putative hemerythrin | |
| fedC | 1 | DnaJ-domain containing protein | |
| polA | 1 | DNA polymerase (This Tn is at 3′ of polA and immediately upstream of motAB, which encode the stator proteins required for motility) | |
| pglE | 1 | Capsule biosynthesis protein | |
| Cjj81176_0246 | 1 | Hypothetical protein | |
| Cjj81176_0294 | 1 | Hypothetical protein | |
| Cjj81176_0481 | 4 | Hypothetical protein | |
| miaB | 1 | Adenosine tRNA methyltransferase | |
| selA | 1 | Selenocysteine synthase | |
| Unknown | 19 |
We next used immunoblot analysis to compare the motor structures of C. jejuni mutants lacking a specific disk protein, tracking the subcellular location of disk proteins to provide insights into the codependencies of disk and core flagellar motor proteins. Consistent with our visualization of C. jejuni FlgP forming a basal disk attached to the P-ring and outer membrane, FlgP stability and outer membrane association was dependent on most core motor components. In the type III secretion system (e.g., fliP and flhA) and proximal rod (fliE, flgB, and flgC) mutants, FlgP was generally absent or was present only at low levels in lysates (Fig. 5A). In the cytoplasm of some of these mutants, we detected a low level of FlgP in the cytoplasm; however, FlgP was not detected in the cytoplasm of WT C. jejuni, presumably the protein was targeted efficiently to the outer membrane (Fig. 5A). In P-ring (flgI), L-ring (flgH), or distal rod (flgF and flgG) mutants that do not assemble L- or P-rings, FlgP was stable and accumulated in the cytoplasm and inner membrane but did not fully localize to the outer membrane (Fig. 5A). FlgP localization was unaffected by the deletion of pflA or pflB (Fig. 5B), although these mutants were nonmotile.
To determine whether PflA and PflB formed part of the C. jejuni disk complex, we determined the motor structures for each mutant using electron cryo-tomography and subtomogram averaging (Fig. 4A). Each motor exhibited flexibility, necessitating the merging of separate periplasmic and cytoplasmic structures. As reported above, the C. jejuni ΔmotB motor assembled a partial proximal disk because of the loss of MotB (Figs. 2 M and N and 4A). In the C. jejuni ΔpflB mutant, the entire proximal disk was lost, including MotB (Fig. 4A). Further supporting evidence that PflB could form the proximal disk that appears to contact the inner membrane toward its center was our fractionation of PflB with inner membrane proteins in C. jejuni (Fig. 5B). These results suggest that PflB forms a majority of the proximal disk and functions to incorporate stator complexes into the peripheral region of the proximal disk in the C. jejuni motor, thus explaining the requirement of PflB for motility.
We next compared the structures of the C. jejuni ΔpflB and ΔpflA motors. The only difference between these motors was the absence of the inner lobe of the medial disk associated with the periplasm in ΔpflA. Consistent with this finding, we were able to localize PflA to the periplasm of C. jejuni (Fig. 5B). A previous study detected interactions between PflA and PflB in C. jejuni (43). We also were able to identify a codependence in stability between PflA and PflB. Although PflB was not required for WT levels of PflA stability or for localization to the periplasm, PflA was required for full stability and inner membrane localization of PflB (Fig. 5B). These data suggest that PflA polymerization into a part (or all) of the medial disk is required to form a platform for PflB to form the proximal disk and subsequently incorporate stator complexes. This requirement is reflected by the loss of both disks in the ΔpflA motor. Of note, we observed a disordered vestige of the medial disk beneath the basal disk in both ΔpflA and ΔpflB motors (Fig. 4A). Currently, the identity of this density is unknown. This density could be FlgQ that assists FlgP assembly into the basal disk, an as yet unidentified protein that forms part of the medial disk with PflA, or peptidoglycan that becomes ordered upon formation of the FlgP basal disk. If this component is peptidoglycan, it could be bound by MotB upon the incorporation of stator complexes into the proximal disk and the flagellar motor. Relevant areas of the V. fischeri and C. jejuni MotB proteins conform to the peptidoglycan-binding consensus sequence.
Intriguingly, the stability of the MotA and MotB proteins of the stator complex did not appear to be affected by the mutation of any disk component (Fig. 5C), and these proteins localized to the inner membrane in each mutant. These findings further support the notion that the function of the disk complex is to incorporate stator complexes into the flagellar motor and that the disk complex is not simply required for stability or for the localization of stator complexes into the inner membrane.
Discussion
In this work we sought to determine whether the generation of high torque by the flagellar motors of specific bacterial species could be functionally attributed to the unique disk complexes associated with their respective motors. For this approach, we analyzed the flagellar motors of two bacterial species, V. fischeri and C. jejuni, with a combination of genetic and biochemical analysis of bacterial mutants and electron cryo-tomographic imaging and compared these motors with the enteric motor assembled in Salmonella. Our studies indicate that the disk complexes of these two organisms, although displaying structural diversity and composed of both homologous and unrelated proteins, share a common role in forming a platform for the integration of different numbers of stator complexes into the flagellar motor. Furthermore, these disk complexes not only integrate more stator complexes into a motor but also place them at wider radii from the center of the motor’s axis of rotation than seen in the enteric motor. As discussed in detail below, we propose that this creation of a wider stator ring with an increased number of stator complexes inevitably results in increased torque in these motors, directly affecting the swimming ability of the respective bacteria by conferring higher swimming velocities and the ability to migrate through environments with increased viscosity.
A Proposed Biogenesis Order for Assembly of the C. jejuni Disk Complex.
Although parts of the purified Vibrio motor have been studied previously, no components of the C. jejuni disk complex had been identified. Identifying the components of the C. jejuni motor and examining the structures of mutants of these components enable us to propose an assembly pathway for the biogenesis of a functional C. jejuni flagellar motor (Fig. 4B). This biogenesis pathway is likely divided into two stages. The first stage is the formation of a nonfunctional C. jejuni core flagellar motor that resembles the (functional) Salmonella motor, as evidenced by the nonfunctional ΔflgP or ΔflgQ C. jejuni motors that fail to incorporate any disk-complex components (Fig. 4A). The second stage involves the sequential assembly of each disk in the disk complex, ultimately enabling correct incorporation of the stator complexes to produce a functional motor. Consistent with our findings that FlgP requires core flagellar components for outer membrane association, we suspect that FlgP polymerizes into the basal disk (with assistance from FlgQ) only after the core flagellar motor forms to provide an attachment site for the basal disk (possibly at the P- and/or L-ring). Besides FlgQ, no other disk-complex proteins are required for FlgP to localize to the outer membrane or form the basal disk (Figs. 4 and 5 A and B). Formation of the basal disk provides a platform and site of interaction with PflA to form the inner node of the medial disk (Fig. 4). As the medial disk subsequently forms, PflA serves as a platform for PflB together with the stator complexes to assemble into the proximal disk (Fig. 4). The disk complex therefore forms a separate and previously unappreciated additional biogenesis step in ε-proteobacteria that occurs after the conserved assembly of the flagellar type III secretion system, rotor, switch, rod, hook, and filament.
FlgP-Based Disk Complexes Are Widespread.
Because C. jejuni and V. fischeri FlgP proteins share low sequence identity, FlgP may represent an ancient recruitment to the ancestral motor, followed by later independent evolutionary recruitment of the remaining Vibrio or ε-proteobacterial disk proteins to create motors with disk complexes that incorporate additional stator units into the motor. The speculation that FlgP recruitment was the first step in the evolution of disk complexes is supported further by the morphological divergence and mutually exclusive sets of additional accessory proteins between the two structural variants. Widespread presence of FlgP and other C. jejuni disk-complex proteins (i.e., FlgQ, PflA, and PflB) across the ε-proteobacteria, together with previous observations of flagellar motors incorporating disk complexes (18, 29), confirm that other ε-proteobacteria such as Helicobacter and Wolinella genera also assemble C. jejuni-type homologous disk complexes. FlgP also is widespread among many γ-proteobacterial genera including Vibrio, Shewanella, and Rhodobacter, but in these genera components of the Vibrio disk complex MotX, MotY, FlgO, and FlgT are encoded, as is consistent with assembly of Vibrio-type disk-complex homologs (45, 46).
Intriguingly, however, the widespread FlgP basal disks do not interact directly with the stator complexes in either the Vibrio or ε-proteobacterial disk-complex variants. In V. fischeri, FlgP connects the T-ring to the outer membrane, and components of the H-ring interact with the stator complexes. In the absence of FlgP and the basal disk, the T- and H-rings are produced, but we suspect that their architecture may be altered so that these rings cannot interact with a stator complex. In C. jejuni, the FlgP basal disk is required to assemble the remainder of the disk complex, with the proximal disk responsible for incorporation of stator complexes. Nevertheless, the common role of FlgP in ultimately creating scaffolds for stator-complex incorporation in both C. jejuni and V. fischeri suggests that their common ancestor had the same function of providing scaffolding for the stator complex, a function that was retained during divergence of the two forms of disk complexes by the incorporation of the additional proteins. Alternatively the two disk-complex lineages may have independently evolved roles to scaffold additional stator complexes. We now are pursuing a follow-up study to understand the phylogeny and probe the origins of these disk complexes.
Interactions of Stator Complexes with Flagellar Motors Vary in Motile Bacterial Species.
The interactions of stator complexes with the disk complexes of the Vibrio and Campylobacter motors are different from those of stator-complex interactions with the enteric motor. In the Salmonella enteric motor, the periplasmic region of MotB in a stator complex contacts the P-ring to anchor to the flagellar structure and contacts the peptidoglycan to tether the motor to the cell wall (13). The Vibrio stator complex also interacts with peptidoglycan, but additional interactions with the T-ring allow the stators to be incorporated at a wider position in the Vibrio flagellar motor than in Salmonella. Currently, it is unknown if the C. jejuni stator complexes also interact with peptidoglycan, although the core OmpA peptidoglycan-binding domain motif is intact (47), and MotB from closely related H. pylori has been cocrystallized with a bound glycan strand (48). Our work suggests that MotB also must interact with PflB, because these two proteins colocalize in subtomogram averages, and deletion of pflB results in the failure of MotB to incorporate into the motor, further supporting our conclusion that the disk complex functions to scaffold stator complexes.
We did not observe stator-complex densities in the Salmonella motor despite evidence that Salmonella minicells swim (31); this finding is consistent with previous studies that failed to visualize Salmonella stator complexes (18, 31). Because Salmonella stator complexes are dynamic (49), we suspect that under our low-load conditions few are engaged and thus will be difficult to resolve without increasing stator-complex occupancy, using classification approaches, or using larger datasets. Comparing the Salmonella motor with V. fischeri and C. jejuni further suggests that Salmonella incorporates no more than 11 stator complexes, because a stator ring immediately around the rod would be too narrow to incorporate more. This limitation has been noted previously in a model of stator-complex incorporation into the V. alginolyticus motor when the additional stator radius provided by the scaffolding role of the T-ring is not taken into account (16).
A Structural Rationale That Quantitatively Accounts for Higher Torque Generation.
Considering this work and interpreting previous work (18, 19), we see a compelling correlation between the torque of a flagellar motor and its architecture. In addition to the results reported here, the spirochete flagellar motor with a torque of ∼4,000 pN⋅nm has 16 proposed stator-complex densities (17, 19) in a 30-nm radius ring (although it is curious that the spirochetes have convergently evolved an alternative stator scaffold, the P-collar, which shares no homologous components with the Vibrio or ε-proteobacterial disk complexes).
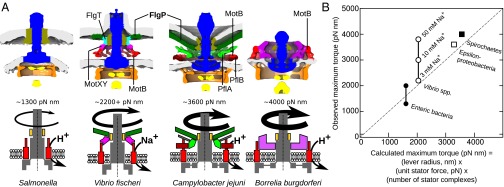
Can these data quantitatively predict observed motor torques? Given that the torque of a single stator complex in enteric bacteria is 146 pN⋅nm (14), our data, consistent with others, show that the lever contact point at which MotA contacts the outer lobe of FliG to apply force is ∼20 nm from the motor axis of rotation (31, 33), leading to the estimation of the force exerted by a single stator complex as ∼7.3 pN. There is good evidence for the assumption that stator complexes exert a constant force in all bacteria, because proton motive forces across bacteria are consistently reported to be between −100 and −200 mV (50); however it should be noted that the sodium motive force in sodium-driven motors may be higher than the proton motive force. Although sodium motive force increases with increasing external sodium ion concentration, it plateaus at an external sodium concentration of ∼250 mM, and the a maximum sodium motive force does not exceed −200 mV (25, 51). Thus, given the radius of stator complexes around the axis of rotation and the number of stator complexes, we can predict the torque of a motor by making the approximation that the torque from multiple stator complexes is roughly additive (14). Our structural data are sufficient to make this prediction for four groups of organisms (Fig. 6A): enteric bacteria such as Salmonella (in which 11 stator complexes exert force at a radius of 20 nm); Vibrio spp. (in which 13 stator complexes exert force at a lever contact point of 21.5 nm); ε-proteobacteria (in which 17 stator complexes exert force at a lever contact point of 26.5 nm); and spirochetes (in which 16 stator complexes exert force at a lever contact point of 30.5 nm). Multiplying the number of stator complexes by the radius of the contact point lever and by the estimated 7.3-pN force exerted per stator complex, we observe excellent correlation between our predicted and the observed torque magnitudes in all four groups (Fig. 6B): a predicted torque of 1,606 pN⋅nm for enteric bacteria vs. an observed torque of 1,260 pN⋅nm (14), with some estimates of 2,000 pN⋅nm (20); a predicted torque of 2,040 pN⋅nm for Vibrio vs. an observed torque of 2,200 pN⋅nm at low sodium concentrations, increasing with higher concentrations; a predicted torque of 3,288 pN⋅nm for ε-proteobacteria vs. an observed torque of 3,600 pN⋅nm; and a predicted torque of 3,562 pN⋅nm for spirochetes vs. an observed torque of 4,000 pN⋅nm. Although this model is clearly a simplification of the process, and biophysical studies reveal that additional stator complexes provide incrementally smaller torque contributions (14), we believe the salient features of our model's predictions are compelling. Another possibility is that ε-proteobacteria assemble more stator complexes at wider radii to accommodate a lower proton motive force. However, the proton motive force of H. pylori has been reported to be relatively high, −220 mV (52). Alternatively, additional stator complexes may act as more sensitive mechanosensors.
Fig. 6.
Wider stators featuring more stator complexes quantitatively account for torque diversity. (A) Disk complexes scaffold stators to increase the number of stators and the radius of the stator-complex ring. The stator number and increasing wider C-ring in each flagellar motor correlate directly with the torque produced by each motor. (B) Comparison of predicted and observed torques of flagellar motors from various species. Filled circles represent enteric bacteria; open circles represent Vibrio spp.; open squares represent ε-proteobacteria; and filled squares represent spirochetes. Where there was no direct torque measurement, C. jejuni and B. burgdorferi torques are inferred from closely related species with similar motors (H. pylori and Leptospira, respectively). Relative torque strengths are validated by swimming speeds and the ability of different bacteria to swim through viscous fluids (1, 4, 7). The dotted line represents a perfect prediction.
In all organisms the C-ring radius tends to scale with both stator-ring radius (as would be expected to maintain MotA:FliGC interaction) and motor torque. Curiously, however, although this correlation is clear, it is not tightly constrained. For example, ε-proteobacteria and spirochetes have wide stator- and C-rings, but the C-ring is wider in spirochetes than in ε-proteobacteria; conversely the stator ring is wider in ε-proteobacterial than in spirochetes. The observation that these variables scale together, but without strict correlation, provides a mechanistic insight: No strict stoichiometric or symmetric correspondence is necessary for motor function. This lack of strict correlation also suggests a clear reducibly complex pathway for the evolution of higher-torque motors, because the increases in C-ring and stator-ring radii can be staggered progressively and asynchronously over evolutionary time through the addition of spacers while a functional motor is maintained.
Together these results indicate that different bacteria have modified their motors to produce torques suited to their environments. C. crescentus has evolved a low-torque efficient motor suited to its low-nutrient, low-viscosity freshwater habitat, whereas the ε-proteobacteria and spirochetes have evolved high-torque motors that require high energy expenditure to bore through viscous mucus and tissue. Such modification of the mechanical output of a molecular machine is not without precedent, because similar observations have been made in the unrelated F-ATPase rotor ring (53).
Our observations propose a mechanistic model for how different bacterial flagellar motors produce different torques. By assembling wider stator rings using a proteinaceous scaffold, individual stator complexes can exert greater leverage on the axial driveshaft of the motor, producing greater torque. Additionally, more stator complexes can be accommodated around the motor, further increasing torque. In addition to describing the evolution of higher torque, this study illustrates a mechanism for adapting mechanical output that might be capitalized on in future synthetic repurposing of molecular machinery.
Materials and Methods
Construction of C. jejuni Mutants.
All strains are listed in Table S1, and plasmids used are listed in Table S2. C. jejuni mutants were constructed by electroporation as previously described (54). For removal from the C. jejuni 81-176 SmR (DRH212) chromosome, genes were first amplified by PCR using primers containing 5′ BamHI restriction sites. Each amplified fragment contained the gene of interest with ∼750 flanking regions. Cloning of fragments into pUC19 resulted in plasmids pLKB652 (pUC19::pflB), pDRH2526 (pUC19::pflA), and pDRH2312 (pUC19::motAB). An SmaI-digested cat-rpsL cassette was ligated into the SwaI site of pflB in pLKB652 to create pLKB658, into the EcoRV site of pflA in pDRH2526 to create pDRH2532, and into the SpeI site of motA in pDRH2312 to create pDRH3330. To insert the cat-rpsL cassette in motB, site-directed mutagenesis was used to create an EcoRV site within motB to form pDRH2324 followed by the insertion of cat-rspL to generate pDRH3331.
Table S1.
Strains used in the study
| Strain | Genotype | Source/reference |
| DH5α | E. coli supE44 ΔlacU169 (ϕ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| TH16943 | Salmonella enterica Para ftsZ | Kelly Hughes, University of Utah, Salt Lake City, UT |
| 81-176 | Wild-type C. jejuni clinical isolate | (66) |
| DRH212 | 81-176 rpsLSm | (54) |
| DRH979 | 81-176 rpsLSm ΔastA ΔflhA | (63) |
| DRH1016 | 81-176 rpsLSm ΔastA ΔfliP | (63) |
| DRH2070 | 81-176 rpsLSm ΔflgP | (41) |
| DRH2071 | 81-176 rpsLSm ΔflgQ | (41) |
| DRH2449 | 81-176 rpsLSm ΔflgH | This study |
| DRH2550 | 81-176 rpsLSm flgF::cat-rpsL | This study |
| DRH2553 | 81-176 rpsLSm flgI::cat-rpsL | This study |
| DAR866 | 81-176 rpsLSm pflA::cat-rpsL | This study |
| DAR981 | 81-176 rpsLSm ΔpflB | This study |
| DAR1066 | 81-176 rpsLSm ΔmotB | This study |
| DAR1124 | 81-176 rpsLSm ΔpflA | This study |
| DAR1131 | 81-176 rpsLSm ΔmotA | This study |
| ABT337 | 81-176 rpsLSm pflB::cat-rpsL | This study |
| MB1225 | 81-176 rpsLSm motA::cat-rpsL | This study |
| MB1226 | 81-176 rpsLSm motB::cat-rpsL | This study |
| SNJ915 | 81-176 rpsLSm ΔastA ΔfliE | (67) |
| SNJ922 | 81-176 rpsLSm ΔflgF | This study |
| SNJ925 | 81-176 rpsLSm ΔastA ΔflgG | (68) |
| SNJ1046 | 81-176 rpsLSm ΔastA ΔflgB | (69) |
| SNJ1048 | 81-176 rpsLSm ΔastA ΔflgC | (69) |
| ALU103 | 81-176 rpsLSm flgH::cat-rpsL | This study |
| MJM1100 | V. fischeri ES114, sequenced wild-type light organ isolate | (70) |
| MB06265 | MJM1100 flgP::Tnerm | (38) |
| MB06357 | MJM1100 motB1::Tnerm | (38) |
Table S2.
Plasmids used in this study
| Plasmid | Genotype/description | Source/reference |
| pET21a | AmpR | Novagen |
| pT7-7 | AmpR | (71) |
| pUC19 | AmpR | New England Biolabs |
| pMalP2X | AmpR | New England Biolabs |
| pMalC2C | AmpR | New England Biolabs |
| pDRH265 | pUC19::cat-rpsL | (54) |
| pDRH1955 | pUC19::flgP containing cat-rpsL in the NheI site of flgP | (41) |
| pDRH1957 | pUC19::ΔflgP | (41) |
| pDRH1958 | pUC19::ΔflgQ | (41) |
| pDRH2029 | pUC19::flgQ containing cat-rpsL in the MscI site of flgQ | (41) |
| pDRH2312 | pUC19 containing motAB with 750 bases of upstream and downstream sequence cloned into the BamHI site | This study |
| pDRH2324 | pUC19::motB with the creation of an EcoRV site within the coding sequence of motB | This study |
| pDRH2422 | pUC19::ΔflgH | (69) |
| pDRH2504 | pUC19::ΔflgF | (69) |
| pDRH2526 | pUC19 containing pflA with 750 bases of upstream and downstreamsequence cloned into the BamHI site | This study |
| pDRH2532 | pUC19::pflA with a cat-rpsL cassette cloned into the EcoRV site of pflA | This study |
| pDRH2534 | pUC19::flgF containing cat-rpsL in the EcoRV site of flgF | (69) |
| pDRH2536 | pUC19::flgI containing cat-rpsL in the EcoRV site of flgI | (69) |
| pDRH2745 | pUC19 with fusion of DNA upstream and downstream of pflA to delete the entire coding sequence | This study |
| pDRH3330 | pUC19::motA with a cat-rpsL cassette cloned into the SpeI site of motA | This study |
| pDRH3331 | pDRH2324 with a cat-rpsL cassette cloned into the EcoRV site within motB | This study |
| pDAR906 | pT7-7 with motB coding sequence containing 3′ in-frame fusion of codons for 6×His tag cloned into the NdeI and PstI sites | This study |
| pDAR1012 | pUC19 with an in-frame deletion of motA fusing the start codon to codon 252 | This study |
| pDAR1013 | pUC19 with an in-frame deletion of motB fusing codon 58 to codon 212 | This study |
| pDAR1180 | pET21a with motA coding sequence cloned into the BamHI and XhoI sites | This study |
| pDAR2107 | pMalP2X with pflA coding sequence and 3′ sequence encoding for 6×His tag cloned into the BamHI and PstI sites | This study |
| pDAR2226 | pMAL-c2X with pflB coding sequence from codons 103–304 cloned into the BamHI and PstI sites | This study |
| pLKB652 | pUC19 containing pflB with 750 bases of upstream and downstream sequence cloned into the BamHI site | This study |
| pLKB658 | pUC19::pflB with a cat-rpsL cassette cloned into the SwaI site of pflB | This study |
| pABT324 | pUC19 with an in-frame deletion of pflB fusing codons 5–808 | This study |
| pALU101 | pUC19::flgH containing cat-rspL in the PmeI site of flgH | (69) |
Plasmids were electroporated into C. jejuni 81-176 SmR (DRH212) to inactivate each respective gene on the chromosome by insertion of the cat-rpsL cassette. In addition, the previously constructed plasmids pDRH2534, pDRH2536, and pALU101 were used to interrupt flgF, flgI, and flgH, respectively, with a cat-rpsL cassette on the C. jejuni 81-176 SmR (DRH212) chromosome. Transformants were recovered on Mueller Hinton (MH) (Difco) agar containing chloramphenicol and mutants were verified by colony PCR to recover the following isogenic mutants: ABT337 (pflB::cat-rpsL), ALU103 (flgH::cat-rpsL), DAR866 (pflA::cat-rpsL), DRH2550 (flgF::cat-rpsL), DRH2553 (flgI::cat-rpsL), MB1225 (motA::cat-rpsL), and MB1226 (motB::cat-rpsL).
In-frame deletions of genes were created with specific primers using PCR-mediated mutagenesis or gene splicing by overlap extension mutagenesis (55, 56). Resulting plasmids were verified by sequencing to form pABT324 (pUC19::ΔpflB), pDRH2745 (pUC19::ΔpflA), pDAR1012 (pUC19::ΔmotA), and pDAR1013 (pUC19:: ΔmotB). Gene deletions were achieved by electroporating corresponding plasmids into strains containing genes interrupted by the insertion of the cat-rpsL cassette. Transformants were recovered on MH agar containing 0.5, 1, or 2 mg/mL streptomycin and then were screened for chloramphenicol sensitivity. Deletions were verified by PCR, which resulted in the creation of isogenic 81-176 SmR mutants containing deletions: ΔpflB (DAR981), ΔpflA (DAR1124), ΔmotA (DAR1131), ΔmotB (DAR1066), ΔflgH (DRH2449), and ΔflgF (SNJ922).
Bacterial Growth.
Salmonella strain TH16943 (ParaftsZ) was grown aerobically overnight in LB broth from freezer stocks at 37 °C. After overnight culture, 50 μL of 5% (wt/vol) l-arabinose were added to 5 mL fresh LB broth for each culture, and cultures were incubated for an additional 5 h. To enrich for minicells, 3 mL of this culture were spun at 6,000 × g for 5 min, and the supernatant was recovered. The supernatant was spin-concentrated by centrifugation at 18,000 × g for 5 min, and the resultant pellet was resuspended in ∼50 μL LB broth and was immediately plunge-frozen. V. fischeri was grown aerobically overnight at 28 °C in salt-supplemented LB medium (38) (LBS) with 35 mM MgSO4. Cultures were reinoculated into fresh LBS, harvested in early log phase, spin-concentrated to an OD600 of ∼20, and immediately plunge-frozen. C. jejuni were grown for 48–60 h on MH agar under microaerobic conditions using CampyPak sachets (Oxoid) at 37 °C. Cultures were generously restreaked and returned to incubate for a further 16 h. Bacteria were harvested by resuspension into ∼1 mL MH broth to an OD600 of ∼10.0 and were immediately plunge-frozen.
Preparation of Electron Microscopy Samples, Data Collection, and Tomogram Reconstruction.
Quantifoil R2/2 grids (200-mesh) (Quantifoil Micro Tools GmbH) were glow-discharged for 60 s at 10 mA. A solution of 10-nm colloidal gold was added to cells immediately before plunge freezing. A 3-μL droplet of the sample solution was applied to the glow-discharged electron microscopy grid; then the grid was blotted and plunge-frozen into a liquid ethane-propane mixture using a Vitrobot plunge-freezing robot (FEI Company) using a wait time of 60–180 s, a blot time of 5–10 s, and blot offsets between −2 and −5 mm. Grids were stored under liquid nitrogen until data collection.
Tilt series were collected on a 200-kV FEI Tecnai TF20 FEG transmission electron microscope (FEI Company) equipped with a Falcon II direct electron detector camera (FEI Company) using Gatan 914 or 626 cryo-holders. Tilt series were recorded from −61° to +61° with an increment of 3° collected defocus between −4 μm and −10 μm using Leginon automated data-collection software (57) at a nominal magnification of 50,000× and were binned four times to final pixel size of 0.81nm. Cumulative doses of ∼120 e−/Å2 were used. Overnight data collection was facilitated by the addition of a 3-L cold-trap Dewar flask and automated refilling of the Dewar cryo-holder triggered by a custom-written Leginon node interfaced with a computer-controlled liquid nitrogen pump (Norhof LN2 Systems).
Tomograms were reconstructed automatically using RAPTOR (58) and the IMOD package (59). Low-defocus images were low-pass filtered to remove data beyond 3.5 nm−1. High-defocus datasets were contrast transfer function (CTF)-corrected using TomoCTF (60).
Subtomogram Extraction, Alignment, and Averaging.
Positions of flagellar motors in tomograms were initially aligned manually along their rotational axes. Datasets were halved, and the particle estimation for electron tomography (PEET) package (61) was used for iterative subtomogram extraction, fine alignment, and averaging of the two halves independently. Resolution was estimated by gold-standard Fourier shell correlation (FSC) by correlating the two halves of the dataset using FSC (Fig. S1). Salmonella motors and C. jejuni ΔpflA, ΔpflB, ΔflgP, and ΔflgQ exhibited flexibility between the top and bottom parts of the motor. In these cases individual averages were constructed for the top and bottom segments and ultimately were merged into a composite structure in analogy to recent single-particle studies (62).
Tn Mutagenesis and Screening of C. jejuni 81-176.
To identify potential disk mutants of C. jejuni 81-176, we conducted a three-step Tn mutagenesis screening procedure to identify flagellated but nonmotile mutants. C. jejuni 81-176 SmR ΔastA flaB::astA chromosomal DNA was used in in vitro transposition reactions with the darkhelmet Tn as described previously (54, 63, 64). Tn mutants were recovered on MH agar containing chloramphenicol, kanamycin, and 35 μg/mL 5-bromo-4-chloro-3-indolyl sulfate. Approximately 8,000 colonies displaying a blue phenotype indicating an intact transcriptional pathway for the expression of flagellar genes were stabbed in 0.4% MH motility agar and incubated for 24 h in microaerobic conditions. Mutants with impaired motility were recovered and grown for an additional 48 h. Mutants then were restreaked, grown for another 16 h, and resuspended to an OD600 of 0.8 in MH broth in 15-mL conical tubes. After at least 1-h incubation at room temperature, mutants that aggregated (indicating production of flagella), as determined by visual inspection of tubes, were saved to identify the location of the Tn insertion by direct DNA sequencing with a primer annealing to the end of the Tn.
Antisera Generation.
Primers were designed to amplify the coding sequence of motA from codon 2 through the stop codon with in-frame BamHI and XhoI restriction sites added to the 5′ ends of the primers. PCR-amplified motA was ligated into pET21a digested with BamHI and XhoI, which allowed expression of MotA with a C-terminal 6×His -tag in pDAR1180.
Primers were designed to amplify the coding sequence of motB from codon 2 through the penultimate codon with in-frame NdeI and PstI restriction sites added to the 5′ end of the primers. In addition, one primer contained codons for a C-terminal 6×His -tag to be added to the penultimate codon on motB. PCR-amplified motB was ligated into pT7-7 digested with NdeI and PstI, which allowed the expression of MotB-6×His and created pDAR906.
Primers were designed to amplify the coding sequence of pflB from codons for V103 to L304 with in-frame BamHI and PstI sites added to the 5′ end of the primers. Additionally, one primer contained codons for a C-terminal 6×His tag to be added after codon 304. After digestion, the PCR-amplified pflB fragment was ligated into BamHI- and PstI-digested pMAL-c2X, allowing expression of a fragment of PflBV103-L304 fused to maltose-binding protein (MBP) with a C-terminal 6×His tag to create pDAR2226.
Primers were designed to amplify the coding sequence of pflA from codon 2 to the stop codon with in-frame BamHI and PstI sites added to the 5′ end of the primers. In addition one primer also contained codons for a C-terminal 6×His tag to be added to the last codon. After digestion, PCR-amplified pflA was ligated into BamHI- and PstI-digested pMal-p2X, which allowed expression of PflA fused to MBP with a C-terminal 6×His tag to create pDAR2111.
For the induction of MotA-6×His, MotB-6×His, PflBV103-L304-MBP-6×His, and PflA-MBP-6×His, the respective plasmids were transformed into BL21 (DE3). Bacteria were grown in 500 mL LB to an OD600 of 0.4. For induction, isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 1 mM, and the cultures were incubated for an additional 3 h.
Bacteria expressing recombinant proteins were harvested by centrifugation and lysed with an EmulsiFlex C5 cell disrupter (Avestin). Lysis buffer for purification of MotB-6×His contained 0.5 M sucrose and 0.2% Zwittergent (Calbiochem). Lysis buffer for MotA-6×His purification contained 0.5 M sucrose, 0.2% Zwittergent, and 8 M urea. PflA-MBP-6×His and PflBV103-L304-MBP-6×His proteins were purified in denaturing conditions with 8 M urea. All proteins were purified by Ni-NTA chromatography. Purified proteins were used to immunize mice or guinea pigs for the generation of polyclonal antisera.
Fractionation of C. jejuni Strains and Analysis of Proteins.
Recovery of whole-cell lysates and fraction of C. jejuni strains into subcellular compartments for analysis of localization of proteins from the inner membrane, outer membrane, periplasm, and cytoplasm were performed as previously described (65). Proteins from each fraction representing equivalent cell numbers (∼200 μL of a C. jejuni culture at an OD600 0.8) were loaded onto 10% SDS/PAGE gels and then were transferred to membranes for immunoblot analysis.
For immunoblotting analyses, the following primary antisera were used at the stated dilutions: FlgP M4 or M5 (1:2,000) (41), MotA M219 (1:500), MotB M195 (1:500), PflA M231 (1:500), and PflB GP143 (1:500). Secondary antisera were used at a dilution of 1:15,000.
Acknowledgments
We thank Anchi Cheng for advice on programming an additional Leginon node; Kelly Hughes for the generous gift of the Salmonella minicell strain TH16943; Tillmann Pape and Amanda Wilson for technical assistance during electron microscopy data collection; and Bonnie Chaban, Velocity Hughes, Ariane Briegel, Alain Filloux, and Richard Berry for critical reading of the manuscript. This work was supported by Biotechnology and Biological Sciences Research Council Grant BB/L023091/1 (to M.B.), Howard Hughes Medical Institute funding (G.J.J.), and National Institutes of Health Grants 5R01AI065539 and 5R21AI103643 (to D.R.H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The electron cryo-tomography subtomogram average density maps reported in this paper have been deposited in the Electron Microscopy Data Bank (EMD) (accession nos. Salmonella WT: EMD-3154; Vibrio fischeri WT: EMD-3155; Campylobacter jejuni WT: EMD-3150; V. fischeri motB: EMD-3156; C. jejuni motB: EMD-3157; C. jejuni flgQ: EMD-3158; C. jejuni flgP: EMD-3159; C. jejuni pflA: EMD-3160; C. jejuni pflB: EMD-3161; and V. fischeri flgP: EMD-3162).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518952113/-/DCSupplemental.
References
- 1.Gao Y, et al. Altered motility of Caulobacter crescentus in viscous and viscoelastic media. BMC Microbiol. 2014;14(1):322–332. doi: 10.1186/s12866-014-0322-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li G, Tang JX. Low flagellar motor torque and high swimming efficiency of Caulobacter crescentus swarmer cells. Biophys J. 2006;91(7):2726–2734. doi: 10.1529/biophysj.106.080697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li N, Kojima S, Homma M. Sodium-driven motor of the polar flagellum in marine bacteria Vibrio. Genes to Cells. 2011;16:985–999. doi: 10.1111/j.1365-2443.2011.01545.x. [DOI] [PubMed] [Google Scholar]
- 4.Ferrero RL, Lee A. Motility of Campylobacter jejuni in a viscous environment: Comparison with conventional rod-shaped bacteria. J Gen Microbiol. 1988;134(1):53–59. doi: 10.1099/00221287-134-1-53. [DOI] [PubMed] [Google Scholar]
- 5.Hazell SL, Lee A, Brady L, Hennessy W. Campylobacter pyloridis and gastritis: Association with intercellular spaces and adaptation to an environment of mucus as important factors in colonization of the gastric epithelium. J Infect Dis. 1986;153(4):658–663. doi: 10.1093/infdis/153.4.658. [DOI] [PubMed] [Google Scholar]
- 6.Beeby M. Motility in the epsilon-proteobacteria. Curr Opin Microbiol. 2015;28:115–121. doi: 10.1016/j.mib.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Kaiser GE, Doetsch RN. Letter: Enhanced translational motion of Leptospira in viscous environments. Nature. 1975;255(5510):656–657. doi: 10.1038/255656a0. [DOI] [PubMed] [Google Scholar]
- 8.Berg HC, Turner L. Movement of microorganisms in viscous environments. Nature. 1979;278(5702):349–351. doi: 10.1038/278349a0. [DOI] [PubMed] [Google Scholar]
- 9.Erhardt M, Namba K, Hughes KT. Bacterial nanomachines: The flagellum and type III injectisome. Cold Spring Harb Perspect Biol. 2010;2(11):a000299. doi: 10.1101/cshperspect.a000299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Asai Y, Kawagishi I, Sockett RE, Homma M. Hybrid motor with H(+)- and Na(+)-driven components can rotate Vibrio polar flagella by using sodium ions. J Bacteriol. 1999;181(20):6332–6338. doi: 10.1128/jb.181.20.6332-6338.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blair DF, Kim DY, Berg HC. Mutant MotB proteins in Escherichia coli. J Bacteriol. 1991;173(13):4049–4055. doi: 10.1128/jb.173.13.4049-4055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kojima S, et al. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol Microbiol. 2009;73(4):710–718. doi: 10.1111/j.1365-2958.2009.06802.x. [DOI] [PubMed] [Google Scholar]
- 13.Hizukuri Y, Kojima S, Homma M. Disulphide cross-linking between the stator and the bearing components in the bacterial flagellar motor. J Biochem. 2010;148(3):309–318. doi: 10.1093/jb/mvq067. [DOI] [PubMed] [Google Scholar]
- 14.Reid SW, et al. The maximum number of torque-generating units in the flagellar motor of Escherichia coli is at least 11. Proc Natl Acad Sci USA. 2006;103(21):8066–8071. doi: 10.1073/pnas.0509932103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Khan S, Dapice M, Reese TS. Effects of mot gene expression on the structure of the flagellar motor. J Mol Biol. 1988;202(3):575–584. doi: 10.1016/0022-2836(88)90287-2. [DOI] [PubMed] [Google Scholar]
- 16.Yonekura K, Maki-Yonekura S, Homma M. Structure of the flagellar motor protein complex PomAB: Implications for the torque-generating conformation. J Bacteriol. 2011;193(15):3863–3870. doi: 10.1128/JB.05021-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy GE, Leadbetter JR, Jensen GJ. In situ structure of the complete Treponema primitia flagellar motor. Nature. 2006;442(7106):1062–1064. doi: 10.1038/nature05015. [DOI] [PubMed] [Google Scholar]
- 18.Chen S, et al. Structural diversity of bacterial flagellar motors. EMBO J. 2011;30(14):2972–2981. doi: 10.1038/emboj.2011.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu J, et al. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: Evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol. 2009;191(16):5026–5036. doi: 10.1128/JB.00340-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Che Y-S, et al. Load-sensitive coupling of proton translocation and torque generation in the bacterial flagellar motor. Mol Microbiol. 2014;91(1):175–184. doi: 10.1111/mmi.12453. [DOI] [PubMed] [Google Scholar]
- 21.Sowa Y, Berry RM. Bacterial flagellar motor. Q Rev Biophys. 2008;41(2):103–132. doi: 10.1017/S0033583508004691. [DOI] [PubMed] [Google Scholar]
- 22.Celli JP, et al. Helicobacter pylori moves through mucus by reducing mucin viscoelasticity. Proc Natl Acad Sci USA. 2009;106(34):14321–14326. doi: 10.1073/pnas.0903438106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura S, Leshansky A, Magariyama Y, Namba K, Kudo S. Direct measurement of helical cell motion of the spirochete leptospira. Biophys J. 2014;106(1):47–54. doi: 10.1016/j.bpj.2013.11.1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sowa Y, Hotta H, Homma M, Ishijima A. Torque-speed relationship of the Na+-driven flagellar motor of Vibrio alginolyticus. J Mol Biol. 2003;327(5):1043–1051. doi: 10.1016/s0022-2836(03)00176-1. [DOI] [PubMed] [Google Scholar]
- 25.Lo C-J, Sowa Y, Pilizota T, Berry RM. Mechanism and kinetics of a sodium-driven bacterial flagellar motor. Proc Natl Acad Sci USA. 2013;110(28):E2544–E2551. doi: 10.1073/pnas.1301664110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terashima H, et al. Insight into the assembly mechanism in the supramolecular rings of the sodium-driven Vibrio flagellar motor from the structure of FlgT. Proc Natl Acad Sci USA. 2013;110(15):6133–6138. doi: 10.1073/pnas.1222655110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Terashima H, Koike M, Kojima S, Homma M. The flagellar basal body-associated protein FlgT is essential for a novel ring structure in the sodium-driven Vibrio motor. J Bacteriol. 2010;192(21):5609–5615. doi: 10.1128/JB.00720-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Okabe M, Yakushi T, Homma M. Interactions of MotX with MotY and with the PomA/PomB sodium ion channel complex of the Vibrio alginolyticus polar flagellum. J Biol Chem. 2005;280(27):25659–25664. doi: 10.1074/jbc.M500263200. [DOI] [PubMed] [Google Scholar]
- 29.Engelhardt H, Schuster SC, Baeuerlein E. An archimedian spiral: The basal disk of the Wolinella flagellar motor. Science. 1993;262(5136):1046–1048. doi: 10.1126/science.8235620. [DOI] [PubMed] [Google Scholar]
- 30.Coulton JW, Murray RGE. Membrane-associated components of the bacterial flagellar apparatus. Biochim Biophys Acta. 1977;465(2):290–310. doi: 10.1016/0005-2736(77)90080-3. [DOI] [PubMed] [Google Scholar]
- 31.Kawamoto A, et al. Common and distinct structural features of Salmonella injectisome and flagellar basal body. Sci Rep. 2013;3:3369. doi: 10.1038/srep03369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nakamura S, Kami-ike N, Yokota JP, Minamino T, Namba K. Evidence for symmetry in the elementary process of bidirectional torque generation by the bacterial flagellar motor. Proc Natl Acad Sci USA. 2010;107(41):17616–17620. doi: 10.1073/pnas.1007448107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thomas DR, Francis NR, Xu C, DeRosier DJ. The three-dimensional structure of the flagellar rotor from a clockwise-locked mutant of Salmonella enterica serovar Typhimurium. J Bacteriol. 2006;188(20):7039–7048. doi: 10.1128/JB.00552-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu S, et al. Conformational change in the periplamic region of the flagellar stator coupled with the assembly around the rotor. Proc Natl Acad Sci USA. 2014;111(37):13523–13528. doi: 10.1073/pnas.1324201111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tipping MJ, Delalez NJ, Lim R, Berry RM, Armitage JP. Load-dependent assembly of the bacterial flagellar motor. MBio. 2013;4(4):e00551-13. doi: 10.1128/mBio.00551-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lele PP, Hosu BG, Berg HC. Dynamics of mechanosensing in the bacterial flagellar motor. Proc Natl Acad Sci USA. 2013;110(29):11839–11844. doi: 10.1073/pnas.1305885110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lertsethtakarn P, Ottemann KM, Hendrixson DR. Motility and chemotaxis in Campylobacter and Helicobacter. Annu Rev Microbiol. 2011;65(1):389–410. doi: 10.1146/annurev-micro-090110-102908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brennan CA, Mandel MJ, Gyllborg MC, Thomasgard KA, Ruby EG. Genetic determinants of swimming motility in the squid light-organ symbiont Vibrio fischeri. MicrobiologyOpen. 2013;2(4):576–594. doi: 10.1002/mbo3.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukuoka H, Wada T, Kojima S, Ishijima A, Homma M. Sodium-dependent dynamic assembly of membrane complexes in sodium-driven flagellar motors. Mol Microbiol. 2009;71(4):825–835. doi: 10.1111/j.1365-2958.2008.06569.x. [DOI] [PubMed] [Google Scholar]
- 40.Brennan CA, et al. A model symbiosis reveals a role for sheathed-flagellum rotation in the release of immunogenic lipopolysaccharide. eLife. 2014;3:e01579. doi: 10.7554/eLife.01579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sommerlad SM, Hendrixson DR. Analysis of the roles of FlgP and FlgQ in flagellar motility of Campylobacter jejuni. J Bacteriol. 2007;189(1):179–186. doi: 10.1128/JB.01199-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morris DC, Peng F, Barker JR, Klose KE. Lipidation of an FlrC-dependent protein is required for enhanced intestinal colonization by Vibrio cholerae. J Bacteriol. 2008;190(1):231–239. doi: 10.1128/JB.00924-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao B, Lara-Tejero M, Lefebre M, Goodman AL, Galán JE. Novel components of the flagellar system in epsilonproteobacteria. MBio. 2014;5(3):e01349–e14. doi: 10.1128/mBio.01349-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blatch GL, Lässle M. The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. BioEssays. 1999;21(11):932–939. doi: 10.1002/(SICI)1521-1878(199911)21:11<932::AID-BIES5>3.0.CO;2-N. [DOI] [PubMed] [Google Scholar]
- 45.Wu L, Wang J, Tang P, Chen H, Gao H. Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis. PLoS One. 2011;6(6):e21479. doi: 10.1371/journal.pone.0021479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fabela S, et al. A distant homologue of the FlgT protein interacts with MotB and FliL and is essential for flagellar rotation in Rhodobacter sphaeroides. J Bacteriol. 2013;195(23):5285–5296. doi: 10.1128/JB.00760-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.De Mot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12(2):333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 48.Roujeinikova A. Crystal structure of the cell wall anchor domain of MotB, a stator component of the bacterial flagellar motor: Implications for peptidoglycan recognition. Proc Natl Acad Sci USA. 2008;105(30):10348–10353. doi: 10.1073/pnas.0803039105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tipping MJ, Steel BC, Delalez NJ, Berry RM, Armitage JP. Quantification of flagellar motor stator dynamics through in vivo proton-motive force control. Mol Microbiol. 2013;87(2):338–347. doi: 10.1111/mmi.12098. [DOI] [PubMed] [Google Scholar]
- 50.Kashket ER. 2003 The proton motive force in bacteria: A critical assessment of methods. Available at www.annualreviews.org/doi/abs/10.1146/annurev.mi.39.100185.001251. Accessed July 16, 2015.
- 51.Willey JM, Waterbury JB, Greenberg EP. Sodium-coupled motility in a swimming cyanobacterium. J Bacteriol. 1987;169(8):3429–3434. doi: 10.1128/jb.169.8.3429-3434.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meyer-Rosberg K, Scott DR, Rex D, Melchers K, Sachs G. The effect of environmental pH on the proton motive force of Helicobacter pylori. Gastroenterology. 1996;111(4):886–900. doi: 10.1016/s0016-5085(96)70056-2. [DOI] [PubMed] [Google Scholar]
- 53.Pogoryelov D, et al. Engineering rotor ring stoichiometries in the ATP synthase. Proc Natl Acad Sci USA. 2012;109(25):E1599–E1608. doi: 10.1073/pnas.1120027109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hendrixson DR, Akerley BJ, DiRita VJ. Transposon mutagenesis of Campylobacter jejuni identifies a bipartite energy taxis system required for motility. Mol Microbiol. 2001;40(1):214–224. doi: 10.1046/j.1365-2958.2001.02376.x. [DOI] [PubMed] [Google Scholar]
- 55.Makarova O, Kamberov E, Margolis B. Generation of deletion and point mutations with one primer in a single cloning step. Biotechniques. 2000;29(5):970–972. doi: 10.2144/00295bm08. [DOI] [PubMed] [Google Scholar]
- 56.Innis MA, Gelfand DH, Sninsky JJ, White TJ, editors. PCR Protocols: A Guide to Methods and Applications. SPI edition Academic; San Diego: 1990. [Google Scholar]
- 57.Suloway C, et al. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol. 2009;167(1):11–18. doi: 10.1016/j.jsb.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Amat F, et al. Markov random field based automatic image alignment for electron tomography. J Struct Biol. 2008;161(3):260–275. doi: 10.1016/j.jsb.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 59.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 60.Fernandez JJ, Li S, Crowther RA. CTF determination and correction in electron cryotomography. Ultramicroscopy. 2006;106:587–596. doi: 10.1016/j.ultramic.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 61.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313(5789):944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 62.Low HH, et al. Structure of a type IV secretion system. Nature. 2014;508(7497):550–553. doi: 10.1038/nature13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hendrixson DR, DiRita VJ. Transcription of σ54-dependent but not σ28-dependent flagellar genes in Campylobacter jejuni is associated with formation of the flagellar secretory apparatus. Mol Microbiol. 2003;50(2):687–702. doi: 10.1046/j.1365-2958.2003.03731.x. [DOI] [PubMed] [Google Scholar]
- 64.Hendrixson DR. Regulation of Flagellar Gene Expression and Assembly. Campylobacter. 3rd Ed. ASM; Washington, DC: 2008. pp. 545–558. [Google Scholar]
- 65.Bingham-Ramos LK, Hendrixson DR. Characterization of two putative cytochrome c peroxidases of Campylobacter jejuni involved in promoting commensal colonization of poultry. Infect Immun. 2008;76(3):1105–1114. doi: 10.1128/IAI.01430-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Korlath JA, Osterholm MT, Judy LA, Forfang JC, Robinson RA. A point-source outbreak of campylobacteriosis associated with consumption of raw milk. J Infect Dis. 1985;152(3):592–596. doi: 10.1093/infdis/152.3.592. [DOI] [PubMed] [Google Scholar]
- 67.Balaban M, Hendrixson DR. Polar flagellar biosynthesis and a regulator of flagellar number influence spatial parameters of cell division in Campylobacter jejuni. PLoS Pathog. 2011;7(12):e1002420. doi: 10.1371/journal.ppat.1002420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Balaban M, Joslin SN, Hendrixson DR. FlhF and its GTPase activity are required for distinct processes in flagellar gene regulation and biosynthesis in Campylobacter jejuni. J Bacteriol. 2009;191(21):6602–6611. doi: 10.1128/JB.00884-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boll JM, Hendrixson DR. A regulatory checkpoint during flagellar biogenesis in Campylobacter jejuni initiates signal transduction to activate transcription of flagellar genes. MBio. 2013;4(5):e00432–e13. doi: 10.1128/mBio.00432-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boettcher KJ, Ruby EG. Depressed light emission by symbiotic Vibrio fischeri of the sepiolid squid Euprymna scolopes. J Bacteriol. 1990;172(7):3701–3706. doi: 10.1128/jb.172.7.3701-3706.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tabor S, Richardson CC. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci USA. 1985;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]