Significance
Ascomycetes are a prolific source of natural products that are of great significance for human health, yet production of ribosomally synthesized and posttranslationally modified peptides (RiPPs), a ubiquitous class of natural products, have rarely been reported in this fungal phylum. Here we show that phomopsins, a family of antimitotic mycotoxins, have a ribosomal origin and demonstrate the widespread presence of a fungal RiPP pathway for cyclic peptides that we term dikaritins. The framework described herein provides a foundation for mining for additional dikaritin members and investigating the biological activities and biosynthetic chemistry of this family of fungal natural products.
Keywords: RiPP, mycotoxin, posttranslational modification, methyltransferase, biosynthesis
Abstract
Production of ribosomally synthesized and posttranslationally modified peptides (RiPPs) has rarely been reported in fungi, even though organisms of this kingdom have a long history as a prolific source of natural products. Here we report an investigation of the phomopsins, antimitotic mycotoxins. We show that phomopsin is a fungal RiPP and demonstrate the widespread presence of a pathway for the biosynthesis of a family of fungal cyclic RiPPs, which we term dikaritins. We characterize PhomM as an S-adenosylmethionine–dependent α-N-methyltransferase that converts phomopsin A to an N,N-dimethylated congener (phomopsin E), and show that the methyltransferases involved in dikaritin biosynthesis have evolved differently and likely have broad substrate specificities. Genome mining studies identified eight previously unknown dikaritins in different strains, highlighting the untapped capacity of RiPP biosynthesis in fungi and setting the stage for investigating the biological activities and unknown biosynthetic transformations of this family of fungal natural products.
Ascomycetes constitute the largest phylum of the fungal kingdom and are a prolific source of natural products that are of great significance to humankind (1–4). Beneficial ascomycetous natural products include many clinically important drugs and various food and industrial chemicals. Many other compounds produced by ascomycetes are toxic and are known as mycotoxins, which pose substantial threats to human food supplies and health (5). Major classes of natural products from Ascomycetes include alkaloids (6, 7), terpenoids (8), polyketides (PKs) (9–12), nonribosomal peptides (NRPs) (12–14), and PK-NRP hybrids (15, 16). However, ribosomally synthesized and posttranslationally modified peptides (RiPPs), a rapidly growing class of natural products (17), have rarely been found in Ascomycetes. It has been known since 2007 that amatoxins and phallotoxins are RiPPs produced by strains of another fungal phylum, Basidiomycetes (which, together with Ascomycetes, compose the subkingdom Dikarya) (17–19). It also was recently reported that the ustiloxins produced by Aspergillus flavus and Ustilaginoidea virens belong to the RiPP class (20, 21).
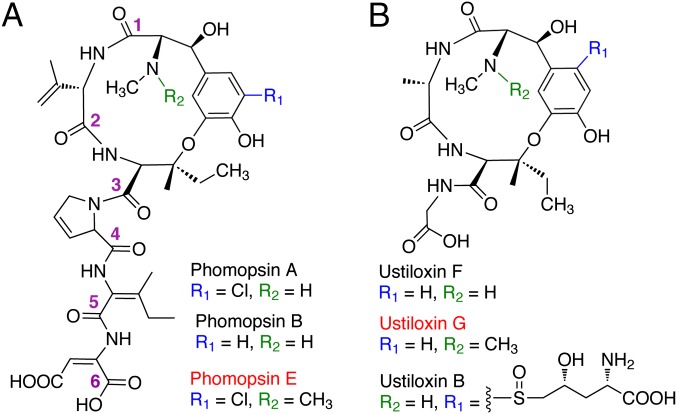
Phomopsins are a group of hexapeptide mycotoxins produced by the pathogenic Ascomycetes Phomopsis leptostromiformis (class Sordariomycetes, order Diaporthales) that infects lupins (Lupinus). Lupin-based animal food contaminated with phomopsins causes lupinosis, a liver disease of sheep and cattle that is particularly problematic in Australia and South Africa but affects other countries as well (22). By targeting the vinca domain of tubulin (23), phomopsins exhibit potent antimitotic activity and thus also represent a potential lead for antitumor drug development. Phomopsins have a 13-member macrocyclic ring formed by an ether bridge linking an Ile residue to the phenyl ring of a Tyr (Fig. 1A). A similar Tyr-containing macrocycle is also present in ustiloxins (Fig. 1B). Unlike ustiloxins, however, phomopsins contain no proteinogenic amino acids and feature a set of dehydroamino acids found only in this group of compounds, including 2,3-dehydroisoleucine, 2,3-dehydroaspartic acid, 3,4-dehydroproline, and 3,4-dehydrovaline (Fig. 1A).
Fig. 1.
Chemical structures of phomopsins (A) and ustiloxins (B). The compounds labeled in red are previously unknown N,N-dimethylated dikaritins identified in this study.
Early isotopic labeling studies showed that Ile, Pro, and Phe were all incorporated into the phomopsin scaffold (24). Because a Phe hydroxylase that can convert Phe to Tyr has not been found in Ascomycetes, the labeling study seems incongruent with a ribosomal route to phomopsins, in which Tyr, not Phe, would need to be incorporated into the phomopsin structure via a tRNA-dependent pathway. Here we report our investigation of the biosynthetic pathway of phomopsins, which explains the paradoxical observations in the early labeling studies and unequivocally demonstrates that phomopsins belong to the family of RiPP natural products. Moreover, our study shows that RiPPs are widespread in fungi, paving the way for a study of their biological activities and biosynthesis.
Results and Discussion
Characterization of the Phomopsin Biosynthetic Gene Cluster.
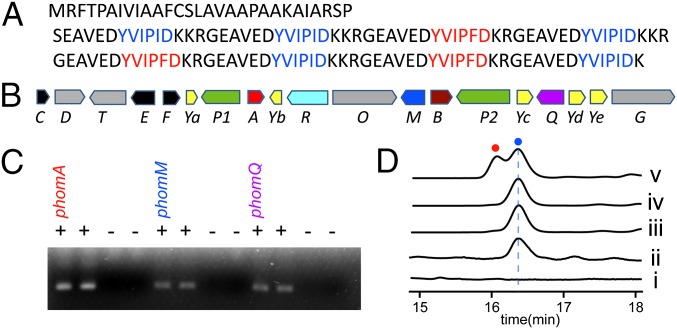
To study phomopsin biosynthesis, we sequenced the genome of P. leptostromiformis ATCC 26115, a potential phomopsin-producer strain. The sequenced genome was found to encode several putative nonribosomal synthases, but none of these appeared to be a suitable candidate for phomopsin biosynthesis. Instead, a protein-to-nucleotide Blast (tBlastn) search against the sequenced genome using the YVIPID sequence (the putative precursor peptide sequence of phomopsin) as the query resulted in identification of a precursor peptide gene, phomA, whose product contains five repeats of short peptides with the sequence YVIPID (Fig. 2A). Examination of the DNA sequence adjacent to phomA led to identification of a gene cluster (here designated the phom gene cluster; GenBank accession nos. KU645826–KU645844), whose predicted protein products include a copper-containing tyrosinase (PhomQ), an S41 family peptidase (PhomP1), an S-adenosylmethionine (SAM)-dependent methyltransferase (PhomM), and several functionally unknown proteins (Fig. 2B and SI Appendix, Table S1). (The nomenclature used for the phomopsin biosynthetic enzymes in this study is aimed at maintaining consistency with that used for ustiloxin biosynthesis.) Based on the sequence of the putative PhomA precursor peptide and the type of activities catalyzed by these enzymes, this gene cluster is likely responsible for phomopsin biosynthesis. (A hypothetical biosynthetic pathway toward phomopsin A is shown in SI Appendix, Fig. S1.) We then performed a transcriptional analysis of P. leptostromiformis ATCC 26115 by RT-PCR, which showed that phomA, phomM, and phomQ were all transcribed by the culture in the late log phase (Fig. 2C), indicating that the phom gene cluster was actively expressed.
Fig. 2.
Biosynthesis of phomopsins. (A) The sequence of PhomA. The two different core peptide sequences are shown in blue and red. (B) Schematic representation of the phomopsin biosynthetic gene cluster. The gene nomenclature is aimed to maintain consistency with that for ustiloxin biosynthesis, and the boundary of the cluster is not clearly defined. The genes are shown in different colors according to the network shown in Fig. 4B. (C) Transcription of phomA, phomM, and phomQ genes by the cells in a late log-phase culture. RT-PCR was performed for two parallel samples; “+” or “−” indicates whether the reverse transcriptase was added or omitted in the assay, respectively. (D) HPLC analysis of the fermentation cultures of (i) phomQ-knockout mutant and (ii) P. leptostromiformis ATCC 26115 wild-type strain; HPLC analysis of PhomM control assays using (iii) the supernatant of the boiled enzyme and (iv) lacking SAM in the reaction; and (v) HPLC analysis of the PhomM reaction with all of the required components. Phomopsin A and phomopsin E are shown by a blue circle and a red circle, respectively.
The tyrosinase PhomQ is likely responsible for formation of the cyclic scaffold of phomopsins (SI Appendix, Fig. S1), and was hypothesized to be essential for phomopsin biosynthesis. Thus, we generated a phomQ-knockout strain by protoplast-mediated transformation and single homologous insertion. Comparison of the HPLC profiles of cultures of the wild-type strain and mutant led to identification of a compound in the former whose production is apparently phomQ-dependent (Fig. 2D, traces i and ii). This compound exhibited a protonated molecular ion at m/z = 789.2859 in high-resolution (HR) LC-MS analysis, and thus closely corresponds to phomopsin A ([M + H]+ calc. 789.2862; 0.4 ppm error). Large-scale fermentation (4 L) and purification yielded 1.5 mg of material, and detailed structural characterization using HR-MS/MS, 1H NMR, 13C NMR, and 2D NMR analyses unambiguously confirmed this product as phomopsin A (SI Appendix, Figs. S2–S7).
In Vitro Methylation of Phomopsin A by PhomM.
Posttranslational methylation of N-terminal α-amino groups (α-N-methylation) has been documented for several RiPP natural products, including the thiazole/oxazole-modified peptide plantazolicin (25, 26) and the linaridin family of compounds (27–29). In all of these cases, the α-amino groups are dimethylated. In contrast, currently known phomopsins (phomopsins A and B) are only monomethylated at the N-terminal α-amino group (Fig. 1).
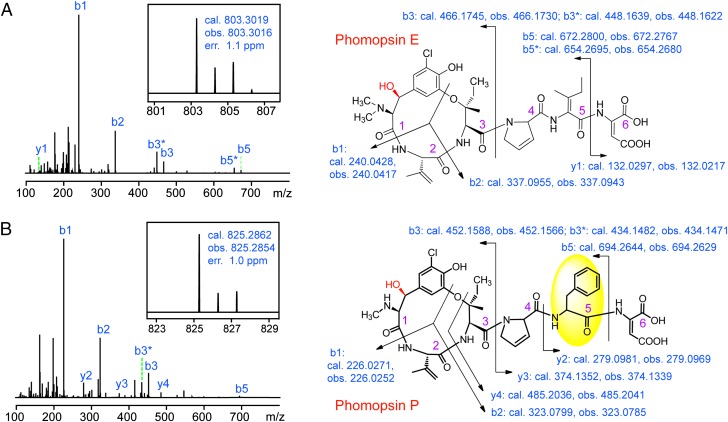
To interrogate whether dimethylated phomopsin congeners could be produced in vitro, we overexpressed PhomM in Escherichia coli with an N-terminal hexa-histidine tag and purified it by Ni2+-affinity chromatography (SI Appendix, Methods). We then tested the methyltransferase activity of PhomM by incubating the enzyme with SAM, phomopsin A, and S-adenosylhomocysteine (SAH) hydrolase; the latter was included to minimize potential product inhibition (30). HPLC analysis of the reaction mixture clearly showed production of a different compound with a retention time very close to that of phomopsin A (Fig. 2D, trace v), and this compound was absent in the control reactions in which either SAM was omitted or the supernatant of the boiled enzyme was used (Fig. 2D, traces iii and iv). HR-LC-MS analysis showed that this compound has a protonated molecular ion at m/z = 803.3010, which is consistent with the expected formula of the dimethylated phomopsin (phomopsin E) ([M + H]+ calc. m/z 803.3019; 1.1 ppm error) (Fig. 3A). HR-MS/MS analysis and a comparison of its mass spectrum (Fig. 3A) with that of phomopsin A (SI Appendix, Fig. S2) clearly demonstrated that the newly produced compound is a phomopsin A analog featuring a dimethylated amino group at the N terminus. This result shows that PhomM is an α-N-methyltransferase involved in phomopsin biosynthesis. We note that the LC-MS analysis showed that phomopsin E was also produced in the fermentation culture of P. leptostromiformis; however, the yield was significantly (∼50-fold) lower than that of phomopsin A and phomopsin E could not be detected by absorption analysis (Fig. 2D, trace ii).
Fig. 3.
HR-MS/MS analysis of phomopsins identified in this study, showing the MS and MS/MS spectra, and the collision-induced dissociation (CID) ion fragments of (A) phomopsin E produced in the PhomM in vitro assay and (B) phomopsin P produced in the culture of P. leptostromiformis. Here b* represents the [b – 18]+ ion, likely derived from dehydration of the hydroxyl group, highlighted in red. The Phe motif in phomospin P is highlighted by a yellow circle.
Unusual Phe-Containing Phomopsins.
The product of phomA contains five repeats of short peptides with the sequence YVIPID (Fig. 2A) and thus is reminiscent of the precursor peptides of cyanobactins (31) and several plant-derived RiPPs (32, 33), which also contain multiple putative core peptides and a leader peptide before the start of the repeats (Fig. 2A). The posttranslational modifications in these peptides are very different from those found in the phomopsins, however.
An intriguing observation is that PhomA contains three repeats of another short peptide with the sequence YVIPFD in addition to the five repeats of YVIPID (Fig. 2A). This observation raised the possibility that the phom gene cluster also encodes peptides in which a Phe (or a Phe-derived residue) replaces the 2,3-dehydroisoleucine in phomopsin A and B (Fig. 1). To test our proposal, we analyzed the fermentation culture of P. leptostromiformis by HR-LC-MS, which revealed two low-abundance products with protonated molecular ions at m/z = 825.2862 and 791.3252 (Fig. 3B and SI Appendix, Fig. S8). These two products (here termed phomopsin P and phomopsin Q, respectively) closely correspond (1.0 and 0.8 ppm error, respectively) to the chlorinated and nonchlorinated peptides containing a Phe residue at position 5 (Fig. 1A), and this postulate was supported by detailed comparative HR-MS/MS analysis (Fig. 3B and SI Appendix, Fig. S8).
In contrast to the corresponding dehydroisoleucine residue in phomopsin A and B, the MS/MS data suggest that the Cα-Cβ bond of the Phe residue in phomopsin P and Q is not desaturated, indicating that the Ile-to-Phe permutation in PhomA likely precludes the action of the dehydrogenase at this site. Furthermore, we observed an additional low-abundance product corresponding to the N,N-dimethylated analog of phomopsin P, and the structure of this peptide (here termed phomopsin R) was also supported by MS/MS analysis (SI Appendix, Fig. S9).
The perfect correlation between the repeat sequences in the PhomA peptide and the newly characterized compounds further established phomopsins as a family of ascomycetous RiPPs. These results also provide an explanation for the seemingly paradoxical observation that radiolabeled Phe appeared to be incorporated into phomopsin A when this compound does not contain any Phe residue (24). It is highly likely that the radiolabeled Phe was not incorporated into any dehydroisoleucine-containing phomopsins (i.e., phomopsins A, B, and E), but instead was incorporated into Phe-containing phomopsins (i.e., phomopsins P, Q, and R), which were identified in this study based on the genetic information of the producer strain. Notably, the isotopic labeling studies showed that the labeling efficiency of Phe was significantly lower than that of Ile (24), which is consistent with our results showing that the Phe-containing phomopsins were produced only in very low yields (<20 μg/L, estimated according to their intensities in LC-MS analysis), which is likely why these Phe-containing congeners escaped previous identification.
Search for Additional Gene Clusters in Fungi.
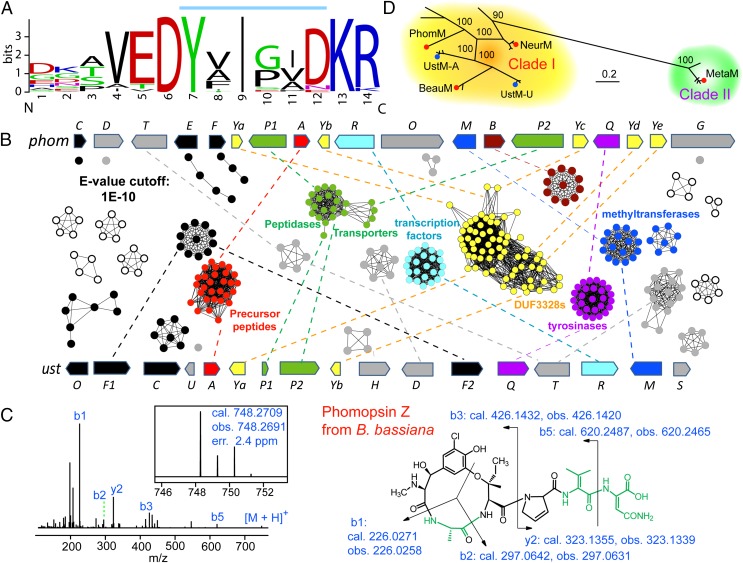
The identification and functional validation of the phom gene cluster prompted us to further assess whether the phomopsin-like RiPPs are widespread in nature. To this end, we searched for PhomA, PhomQ, and PhomM homologous proteins in the National Center for Biotechnology Information database, resulting in the identification of 27 gene clusters similar to that for phomopsin biosynthesis. (Additional information on these gene clusters, including their accession numbers, is summarized in SI Appendix, Table S2.) The precursor peptides of these gene clusters consist of an N-terminal leader peptide that shares no sequence similarity with any conserved protein domains, along with several repeats of short peptides containing mostly semiconserved core sequences (SI Appendix, Table S2) that always have a Tyr at the first position and an Ile at the third position. These core sequences are flanked by stretches of 7∼11 amino acids that are generally variable but have highly conserved ED and KR/KK motifs next to the N and C termini of the putative core peptides (Fig. 4A), which likely are necessary for recognition by posttranslationally modifying enzymes. The KR motif is a known recognition site for KEX proteases in fungi (34), and these enzymes could be involved in the required proteolysis to excise the phomopsins from the precursor peptide. Notably, the precursor peptides of the cyanobactins, amatoxins, and cyclotides, three structurally unrelated RiPP families, also contain highly conserved sequences flanking the core peptides, and these conserved sequences are recognized by the proteases that excise the core peptides from their larger precursor peptides (35–40).
Fig. 4.
Biosynthesis of dikaritins. (A) Representative logo of the repeated sequence of dikaritin precursor peptides. The logo was constructed using the repeated sequence from all of the precursor peptides identified in this study. The putative core peptide is shown by a blue bar above the residues. Note that the C termini of the core peptides could be truncated, as observed for ustiloxins and metarisins. (B) A global network of dikaritin biosynthetic proteins, constructed by an all-to-all BlastP comparison of each sequence against each other sequence. Each node in the network represents a protein sequence encoded by a dikaritin gene cluster, and each edge represents the pairwise connection between two sequences with a BlastP E value <1E-10. The nodes are shown in different colors according to their functions; oxidoreductases and other functionally unknown proteins are shown in black and gray, respectively. The proteins not encoded by the phom and ust clusters but encoded in some other putative dikaritin gene clusters are shown as unfilled nodes. For these proteins, only conserved proteins are shown, and any singletons present only in individual clusters were omitted for clarity. Note that the borders of the gene clusters are not well defined, and we selected 15 genes upstream and downstream of the precursor peptide genes in the analysis. Because genes outside of the clusters are not conserved and in most cases appear as singletons, their protein products were removed from the network. (C) Structural characterization of phomopsin Z produced by B. bassiana, showing the MS and MS/MS spectra, and the CID ion fragments. The amino acids that differ from phomopsin A are shown in green. The position of the double bond in the dehydrovaline is not defined by the data and could also be between the β and γ carbons. (D) MCMC tree of the methyltransferases involved in dikaritin biosynthesis. UstMs involved in ustiloxin biosynthesis are shown by blue circles (UStM-A and UstM-U are enzymes from A. flavus and U. virens, respectively). Enzymes expressed and analyzed in this study are shown as red circles.
Identification of these phom-like gene clusters suggests the presence of a family of fungal RiPP natural products. We propose the name “dikaritins” for these peptides, because they appear to associate with strains of the subkingdom Dikarya. They include ustiloxins and phomopsins, and likely additional compounds as well, given the diversity of encoded proteins and core peptide sequences. All of the gene clusters encode a precursor peptide, a tyrosinase, and a methyltransferase, whereas most of the other proteins are functionally unknown. Because many putative dikaritin biosynthetic enzymes are small, lack biochemically/genetically characterized precedents, and may contain introns, it proved difficult to unambiguously predict the ORFs of these gene clusters. A case in point is Beauveria bassiana, which produces a compound similar to phomopsin A but has a different gene cluster than that of P. leptostromiformis (see below).
To identify the fully and partially conserved enzymes encoded in the gene clusters, we constructed a global sequence similarity network in which all of the putative proteins from the identified gene clusters were compared with one another (Fig. 4B; details of network construction are provided in the figure legend and in SI Appendix). The biosynthetic enzymes of both phomopsins and ustiloxins were included in the network for comparative analysis. The network analysis showed that these gene clusters feature a set of highly conserved proteins, including a PhomA-like precursor peptide, a PhomQ-like tyrosinase, a PhomR-like zinc finger transcription-regulating protein, and S41 family peptidases. A conspicuous observation was the presence of multiple DUF3328 proteins (a group of proteins belonging to the PFAM11807 family) in all of the gene clusters (Fig. 4B), whose role in dikaritin biosynthesis is not clear at present. The gene clusters all encode a SAM-dependent methyltransferase that likely is responsible for α-N-methylation of the Tyr-derived residue (Fig. 1A). Unlike other protein groups in the network that are relatively homogeneous, the methyltransferases form two distinct clusters (Fig. 4B). We also noted a functionally unknown gene, phomB, in the phom cluster, with its homologous genes present in a subgroup of gene clusters but absent in others (Fig. 4B). Overall, our analysis highlights the widespread occurrence of dikaritins and sets the stage for investigation of the unprecedented biosynthetic chemistry of this family of fungal RiPPs.
To demonstrate that dikaritin biosynthetic pathways are not silent, we fermented three other strains from different genera, B. bassiana, Metarhizium anisopliae, and Aspergillus oryzae, and screened their cultures by HR-LC-MS. We observed the production of a phomopsin A analog from B. bassiana (phomopsin Z; Fig. 4C), two dikaritins from M. anisopliae (metarisin A and B; SI Appendix, Figs. S10 and S11), and a previously unidentified ustiloxin (ustiloxin G; Fig. 1B) from A. oryzae. All three compounds were produced at very low levels, as judged from a comparison of UV and MS analyses of the HPLC traces. The assigned structures of these three peptides were supported by comparative HR-MS/MS analysis and the sequences of the precursor peptides encoded in the respective gene clusters (Fig. 4C and SI Appendix, Table S2 and Figs. S10–S12). These experiments serve as a proof of concept of the ability to identify additional dikaritins by genome mining.
Based on the results presented herein and extrapolation of future genome sequencing projects, it is apparent that many more dikaritins await discovery. It is noteworthy that phomopsin Z from B. bassiana is structurally similar to phomopsin A, containing a full set of dehydroamino acids and chlorination of the phenyl ring (Fig. 4C). However, the predicted gene cluster of phomopsin Z is very different than that from P. leptostromiformis (SI Appendix, Fig. S13), illustrating the significant variation among dikaritin gene clusters.
Methyltransferases Involved in Dikaritin Biosynthesis.
An unexpected observation in the sequence similarity network analysis was the splitting of the methyltransferases into two separate clusters at a low BlastP E-value cutoff (1E-10). To study the phylogeny of the methyltransferases involved in dikaritin biosynthesis, we constructed a Bayesian Markov chain Monte Carlo (MCMC) tree. The MCMC tree was consistent with the observations in the sequence similarity network and showed that the methyltransferases fall into two major clades that are clearly separated (Fig. 4D and SI Appendix, Fig. S14). Clade I contains most of the currently available dikaritin methyltransferases, including the UstMs (involved in ustiloxin biosynthesis) and PhomM, whereas clade II consists of enzymes from Metarhizium and Ophiocordyceps.
It has been reported that the enzyme structure required for SAM binding and methyl transfer is highly flexible (41, 42), and it is likely that the methyltransferases involved in dikaritin biosynthesis have evolved differently but perform similar activities. To test this hypothesis, we expressed two additional clade I methyltransferases from B. bassiana (BeauM) and Neurospora crassa (NeurM) and a clade II methyltransferase from M. anisopliae (MetaM) (Fig. 4D). All three enzymes were expressed in E. coli with an N-terminal hexa-histidine tag and purified by Ni2+-affinity chromatography. Assays were performed by incubation of each enzyme with phomopsin A, SAM, and SAH hydrolase. HPLC analysis of each reaction mixture showed that, like PhomM, all three methyltransferases converted phomopsin A to phomopsin E (Fig. 5), suggesting that the dikaritin methyltransferases catalyze similar activities on their cognate substrates. Given that enzymes from different gene clusters presumably act on related, yet different substrates (SI Appendix, Table S2), these results also suggest that the methyltransferases involved in dikaritin biosynthesis likely have broad substrate specificities.
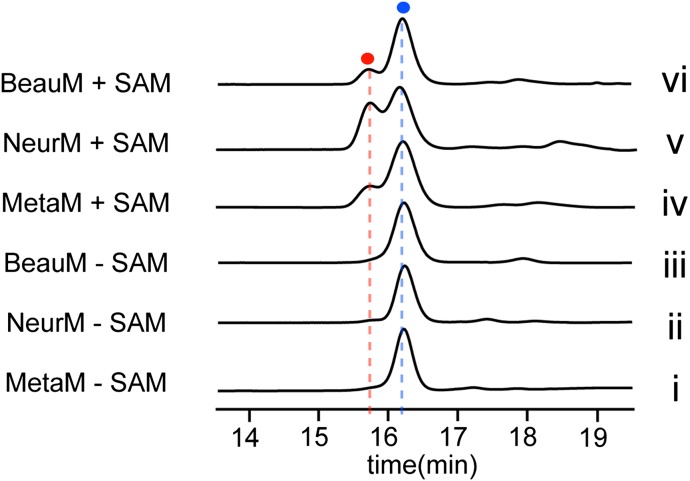
Fig. 5.
In vitro methylation of phomopsin A to produce phomopsin E, showing the HPLC profiles of the control assays of MetaM (i), NeurM (ii), and BeauM (iii), in which SAM was omitted in the reaction, and the assays of MetaM (iv), NeurM (v), and BeauM (vi) with all of the required components. Phomopsin A and phomopsin E are shown by a blue circle and a red circle, respectively. Mass spectra of the peaks are shown in SI Appendix, Fig. S15.
Conclusion
This investigation establishes that phomopsins are members of the RiPP family. Disruption of the tyrosinase gene phomQ abolished production of phomopsins, and characterization of the precursor gene phomA led to the identification of three previously unknown Phe-containing phomopsin congeners. Characterization of PhomM as a SAM-dependent methyltransferase allowed further identification of several N,N-dimethylated dikaritins (phomopsins E and R, metarisin B, and ustiloxin G). This study also demonstrates that gene clusters generating similar fungal cyclic peptides are widespread, hinting at a larger untapped capacity of RiPP biosynthesis in fungi and the potential for discovering previously unknown dikaritins by genome mining. These fungal RiPPs likely are potent antimitotic agents and may have potential pharmaceutical applications. This study also sets the stage for a detailed investigation of dikaritin biosynthetic pathways, which, based on the structures of phomopsins, almost certainly involve unprecedented and intriguing biosynthetic transformations.
Materials and Methods
Detailed information on instrumental settings, culture conditions, gene cloning, mutant construction, protein expression and purification, and bioinformatical analysis is provided in SI Appendix.
P. leptostromiformis and A. oryzae were purchased from the American Type Culture Collection. N. crassa, B. bassiana, and M. anisopliae were obtained from the China General Microbiological Culture Collection Center (SI Appendix). P. leptostromiformis genome sequencing was performed by the Biotechnology Center at the University of Illinois at Urbana–Champaign. High-resolution mass spectra were acquired using either a SYNAPT ESI Quadrupole Time-of-Flight Mass Spectrometry System (Waters) equipped with an Acquity Ultra Performance Liquid Chromatography System (Waters), or a Q-Exactive Focus Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher) equipped with a Dionex UltiMate 3000 HPLC System (Thermo Fisher). RNA extraction and cDNA synthesis were performed using RNA extraction reagent and the HiScript II First-Strand cDNA Synthesis Kit (Vazyme Biotech).
Production of Dikaritins.
DMSO stocks of different strains were prepared by mixing 600 µL of liquid culture with an equal volume of sterilized 20% DMSO. The resulting solution was mixed, flash-frozen, and stored at −80 °C. A tube with 3 mL of PD medium (24 g of potato dextrose dissolved in 1 L of Milli-Q filtered deionized water) was inoculated with 200 µL of DMSO stocks and then incubated at 28 °C for 3–5 d. The resulting culture was used to inoculate 200 mL of production medium as described previously (43), which is a modification of the well-known Czapek–Dox formulation containing the following (in grams per liter of solution): 10.0 sucrose, 10.0 yeast extract, 2.0 NaNO3, 1.0 KCl, 0.8 sodium glycerophosphate, 0.4 MgSO4, and 0.01 FeSO4. The pH was adjusted to 6.0 before sterilization. After incubation at 25 °C for 25–30 d (for P. leptostromiformis and M. anisopliae) or 14–16 d (for A. oryzae), the cultures were processed in a high-pressure homogenizer (FB-110X; Shanghai Litu Mechanical Equipment Engineering or JN-02HC; Guangzhou Juneng Biology & Technology). The supernatant obtained after centrifugation (23,800 × g for 30 min) was passed through an Amberlite XAD-4 column. The column was then washed with distilled water, followed by elution with methanol. The eluate was taken to dryness on a rotary evaporator, and the residue was redissolved in methanol before HPLC/LCMS analysis. Phomopsin A (1.5 mg) was purified from 4 L (20 × 200 mL) of production culture.
In Vitro Methyltransferase Activity Assay.
The in vitro methyltransferase activity assay contained 50 μM phomopsin A, 500 μM SAM, 20–40 μM methyltransferase, 5 μM SAH hydrolase, and 5 μM NAD+ in 50 mM Tris⋅HCl buffer (pH 8.0). Reaction volumes were typically 50 μL and were maintained at 30 °C for 3–4 h before quenching by addition of an equal volume of methanol. Negative controls were performed by using the supernatant of the boiled enzyme, or by omitting SAM in the reaction. After the protein precipitates were removed by centrifugation, the supernatant was subjected to HPLC analysis. The HPLC analysis was performed using a Thermo Fisher BDS-HYPERSIL-C18 column (particle size, 5 μm; dimensions, 4.6 × 250 mm; SN: 12151272TDI) at a flow rate of 1 mL/min and UV detection at 280 nm. The column was equilibrated with 80% solvent A (H2O, 0.1% TFA) and 20% solvent B (CH3CN), and the analysis was developed with an isocratic elution program (0–25 min, 80% A/20%). Phomopsins were normally eluted around 16 min. The same HPLC program was also used for analyzing the fermentation culture of P. leptostromiformis wild-type strain and phomQ-knockout mutant.
Bioinformatics.
Putative dikaritin biosynthetic gene clusters were identified by BlastP searches (SI Appendix). The global sequence similarity network of dikaritin biosynthetic enzymes was constructed by an all-to-all BlastP comparison of each putative enzyme against another. An in-house Matlab script was used to keep the most stringent comparisons and remove all of the duplicates, and the result was imported into the Cytoscape software package (44). Each node in the network represents a protein sequence, and each edge represents the pairwise connection between two proteins with a BlastP e-value better than a cutoff of 1E-10. The nodes were arranged using the yFiles organic layout provided with Cytoscape version 2.8.3. Bayesian MCMC inference analyses were performed using the program MrBayes version 3.2 (45), as detailed in SI Appendix.
Note Added in Proof.
While this work was under review, the Umemura group reported a complementary study that shows that dikaritins are an even larger family of RiPPs (46).
Supplementary Material
Acknowledgments
We thank Professor Yi Yu (Wuhan University) for help in creating the phomQ-knockout mutant, and Dr. Mark Walker (University of Illinois at Urbana–Champaign) for helpful discussions. Q.Z. thanks the Thousand Talents Program for support. This work was supported by grants from the National Institutes of Health (GM R01 058822, to W.A.v.d.D.) and from the Chinese National Natural Science Foundation (31500028, to Q.Z.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. KU645826–KU645844).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1522907113/-/DCSupplemental.
References
- 1.Keller NP, Turner G, Bennett JW. Fungal secondary metabolism: From biochemistry to genomics. Nat Rev Microbiol. 2005;3(12):937–947. doi: 10.1038/nrmicro1286. [DOI] [PubMed] [Google Scholar]
- 2.Zhong JJ, Xiao JH. Secondary metabolites from higher fungi: Discovery, bioactivity, and bioproduction. Adv Biochem Eng Biotechnol. 2009;113:79–150. doi: 10.1007/10_2008_26. [DOI] [PubMed] [Google Scholar]
- 3.Schueffler A, Anke T. Fungal natural products in research and development. Nat Prod Rep. 2014;31(10):1425–1448. doi: 10.1039/c4np00060a. [DOI] [PubMed] [Google Scholar]
- 4.Evidente A, et al. Fungal metabolites with anticancer activity. Nat Prod Rep. 2014;31(5):617–627. doi: 10.1039/c3np70078j. [DOI] [PubMed] [Google Scholar]
- 5.Marroquín-Cardona AG, Johnson NM, Phillips TD, Hayes AW. Mycotoxins in a changing global environment: A review. Food Chem Toxicol. 2014;69:220–230. doi: 10.1016/j.fct.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 6.Xu W, Gavia DJ, Tang Y. Biosynthesis of fungal indole alkaloids. Nat Prod Rep. 2014;31(10):1474–1487. doi: 10.1039/c4np00073k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jakubczyk D, Cheng JZ, O’Connor SE. Biosynthesis of the ergot alkaloids. Nat Prod Rep. 2014;31(10):1328–1338. doi: 10.1039/c4np00062e. [DOI] [PubMed] [Google Scholar]
- 8.Quin MB, Flynn CM, Schmidt-Dannert C. Traversing the fungal terpenome. Nat Prod Rep. 2014;31(10):1449–1473. doi: 10.1039/c4np00075g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vederas JC. Explorations of fungal biosynthesis of reduced polyketides: A personal viewpoint. Nat Prod Rep. 2014;31(10):1253–1259. doi: 10.1039/c4np00091a. [DOI] [PubMed] [Google Scholar]
- 10.Crawford JM, Townsend CA. New insights into the formation of fungal aromatic polyketides. Nat Rev Microbiol. 2010;8(12):879–889. doi: 10.1038/nrmicro2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto M, Nonaka T, Fujii I. Fungal type III polyketide synthases. Nat Prod Rep. 2014;31(10):1306–1317. doi: 10.1039/c4np00096j. [DOI] [PubMed] [Google Scholar]
- 12.Wang H, Fewer DP, Holm L, Rouhiainen L, Sivonen K. Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc Natl Acad Sci USA. 2014;111(25):9259–9264. doi: 10.1073/pnas.1401734111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Finking R, Marahiel MA. Biosynthesis of nonribosomal peptides. Annu Rev Microbiol. 2004;58:453–488. doi: 10.1146/annurev.micro.58.030603.123615. [DOI] [PubMed] [Google Scholar]
- 14.Bills G, et al. New insights into the echinocandins and other fungal non-ribosomal peptides and peptaibiotics. Nat Prod Rep. 2014;31(10):1348–1375. doi: 10.1039/c4np00046c. [DOI] [PubMed] [Google Scholar]
- 15.Sheridan KJ, Dolan SK, Doyle S. Endogenous cross-talk of fungal metabolites. Front Microbiol. 2014;5:732. doi: 10.3389/fmicb.2014.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boettger D, Hertweck C. Molecular diversity sculpted by fungal PKS-NRPS hybrids. ChemBioChem. 2013;14(1):28–42. doi: 10.1002/cbic.201200624. [DOI] [PubMed] [Google Scholar]
- 17.Arnison PG, et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat Prod Rep. 2013;30(1):108–160. doi: 10.1039/c2np20085f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Walton JD, Hallen-Adams HE, Luo H. Ribosomal biosynthesis of the cyclic peptide toxins of Amanita mushrooms. Biopolymers. 2010;94(5):659–664. doi: 10.1002/bip.21416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hallen HE, Luo H, Scott-Craig JS, Walton JD. Gene family encoding the major toxins of lethal Amanita mushrooms. Proc Natl Acad Sci USA. 2007;104(48):19097–19101. doi: 10.1073/pnas.0707340104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Umemura M, et al. Characterization of the biosynthetic gene cluster for the ribosomally synthesized cyclic peptide ustiloxin B in Aspergillus flavus. Fungal Genet Biol. 2014;68:23–30. doi: 10.1016/j.fgb.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Tsukui T, et al. Ustiloxins, fungal cyclic peptides, are ribosomally synthesized in Ustilaginoidea virens. Bioinformatics. 2015;31(7):981–985. doi: 10.1093/bioinformatics/btu753. [DOI] [PubMed] [Google Scholar]
- 22.Battilani P, et al. Phomopsins: An overview of phytopathological and chemical aspects, toxicity, analysis and occurrence. World Mycotoxin J. 2011;4:345–359. [Google Scholar]
- 23.Cormier A, Marchand M, Ravelli RBG, Knossow M, Gigant B. Structural insight into the inhibition of tubulin by vinca domain peptide ligands. EMBO Rep. 2008;9(11):1101–1106. doi: 10.1038/embor.2008.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Payne AL. Biosynthesis of radiolabeled phomopsin by Phomopsis leptostromiformis. Appl Environ Microbiol. 1983;45(2):389–392. doi: 10.1128/aem.45.2.389-392.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Piwowarska NA, Banala S, Overkleeft HS, Süssmuth RD. Arg-Thz is a minimal substrate for the N(α),N(α)-arginyl methyltransferase involved in the biosynthesis of plantazolicin. Chem Commun (Camb) 2013;49(91):10703–10705. doi: 10.1039/c3cc45898a. [DOI] [PubMed] [Google Scholar]
- 26.Lee J, et al. Structural and functional insight into an unexpectedly selective N-methyltransferase involved in plantazolicin biosynthesis. Proc Natl Acad Sci USA. 2013;110(32):12954–12959. doi: 10.1073/pnas.1306101110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Q, van der Donk WA. Catalytic promiscuity of a bacterial α-N-methyltransferase. FEBS Lett. 2012;586(19):3391–3397. doi: 10.1016/j.febslet.2012.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Claesen J, Bibb M. Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides. Proc Natl Acad Sci USA. 2010;107(37):16297–16302. doi: 10.1073/pnas.1008608107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rateb ME, et al. Legonaridin, a new member of linaridin RiPP from a Ghanaian Streptomyces isolate. Org Biomol Chem. 2015;13(37):9585–9592. doi: 10.1039/c5ob01269d. [DOI] [PubMed] [Google Scholar]
- 30.Hendricks CL, Ross JR, Pichersky E, Noel JP, Zhou ZS. An enzyme-coupled colorimetric assay for S-adenosylmethionine–dependent methyltransferases. Anal Biochem. 2004;326(1):100–105. doi: 10.1016/j.ab.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 31.Donia MS, et al. Natural combinatorial peptide libraries in cyanobacterial symbionts of marine ascidians. Nat Chem Biol. 2006;2(12):729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 32.Jennings C, West J, Waine C, Craik D, Anderson M. Biosynthesis and insecticidal properties of plant cyclotides: The cyclic knotted proteins from Oldenlandia affinis. Proc Natl Acad Sci USA. 2001;98(19):10614–10619. doi: 10.1073/pnas.191366898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Condie JA, et al. The biosynthesis of Caryophyllaceae-like cyclic peptides in Saponaria vaccaria L. from DNA-encoded precursors. Plant J. 2011;67(4):682–690. doi: 10.1111/j.1365-313X.2011.04626.x. [DOI] [PubMed] [Google Scholar]
- 34.Douglas CM, Sturley SL, Bostian KA. Role of protein processing, intracellular trafficking, and endocytosis in production of and immunity to yeast killer toxin. Eur J Epidemiol. 1988;4(4):400–408. doi: 10.1007/BF00146389. [DOI] [PubMed] [Google Scholar]
- 35.Donia MS, Ravel J, Schmidt EW. A global assembly line for cyanobactins. Nat Chem Biol. 2008;4(6):341–343. doi: 10.1038/nchembio.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koehnke J, et al. The mechanism of patellamide macrocyclization revealed by the characterization of the PatG macrocyclase domain. Nat Struct Mol Biol. 2012;19(8):767–772. doi: 10.1038/nsmb.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sardar D, Pierce E, McIntosh JA, Schmidt EW. Recognition sequences and substrate evolution in cyanobactin biosynthesis. ACS Synth Biol. 2015;4(2):167–176. doi: 10.1021/sb500019b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nguyen GKT, et al. Butelase 1 is an Asx-specific ligase enabling peptide macrocyclization and synthesis. Nat Chem Biol. 2014;10(9):732–738. doi: 10.1038/nchembio.1586. [DOI] [PubMed] [Google Scholar]
- 39.Luo H, et al. Peptide macrocyclization catalyzed by a prolyl oligopeptidase involved in α-amanitin biosynthesis. Chem Biol. 2014;21(12):1610–1617. doi: 10.1016/j.chembiol.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saska I, et al. An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J Biol Chem. 2007;282(40):29721–29728. doi: 10.1074/jbc.M705185200. [DOI] [PubMed] [Google Scholar]
- 41.Schubert HL, Blumenthal RM, Cheng X. Many paths to methyltransfer: A chronicle of convergence. Trends Biochem Sci. 2003;28(6):329–335. doi: 10.1016/S0968-0004(03)00090-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Struck AW, Thompson ML, Wong LS, Micklefield J. S-adenosyl-methionine–dependent methyltransferases: Highly versatile enzymes in biocatalysis, biosynthesis, and other biotechnological applications. ChemBioChem. 2012;13(18):2642–2655. doi: 10.1002/cbic.201200556. [DOI] [PubMed] [Google Scholar]
- 43.Lanigan GW, Payne AL, Smith LW, Wood PMR, Petterson DS. Phomopsin A production by Phomopsis leptostromiformis in liquid media. Appl Environ Microbiol. 1979;37(2):289–292. doi: 10.1128/aem.37.2.289-292.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cline MS, et al. Integration of biological networks and gene expression data using Cytoscape. Nat Protoc. 2007;2(10):2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ronquist F, et al. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61(3):539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nagano N, et al. Class of cyclic ribosomal peptide synthetic genes in filamentous fungi. Fungal Genet Biol. 2016;86:58–70. doi: 10.1016/j.fgb.2015.12.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.