Significance
Many organisms are specialists living within a narrow range of conditions. Pathogens are often adapted to efficiently exploit only a few hosts species, or sometimes, only some genotypes within a species. The genomes of such parasites are predicted to maintain genes critical for host utilization and to lose genes no longer necessary outside their constrained lifestyle. We demonstrate that the genomic content of a fungal pathogen specialized to attack and consume fungus cultivated by ants meets these predictions. Despite a reduced genome size and gene content in comparison with less specialized relatives, the genome of this agricultural pathogen retains genes necessary for production of toxins, a step critical to host attack, and for breaking down nutrients abundant in its host.
Keywords: mycoparasitism, repeat-induced point mutation, Atta cephalotes, attine, genome reduction
Abstract
Many microorganisms with specialized lifestyles have reduced genomes. This is best understood in beneficial bacterial symbioses, where partner fidelity facilitates loss of genes necessary for living independently. Specialized microbial pathogens may also exhibit gene loss relative to generalists. Here, we demonstrate that Escovopsis weberi, a fungal parasite of the crops of fungus-growing ants, has a reduced genome in terms of both size and gene content relative to closely related but less specialized fungi. Although primary metabolism genes have been retained, the E. weberi genome is depleted in carbohydrate active enzymes, which is consistent with reliance on a host with these functions. E. weberi has also lost genes considered necessary for sexual reproduction. Contrasting these losses, the genome encodes unique secondary metabolite biosynthesis clusters, some of which include genes that exhibit up-regulated expression during host attack. Thus, the specialized nature of the interaction between Escovopsis and ant agriculture is reflected in the parasite’s genome.
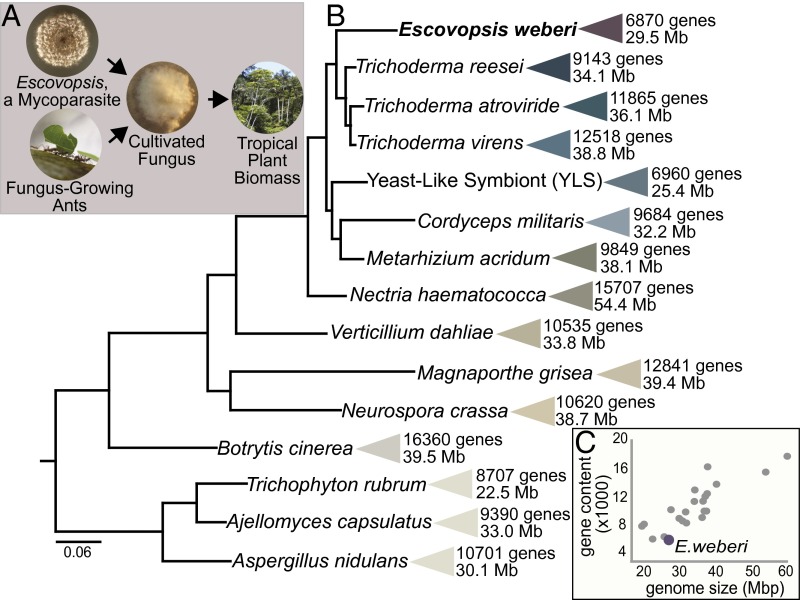
The highly evolved agricultural lifestyle of leaf-cutting ants has attracted particular attention because these ants cultivate a symbiotic fungus that serves as their major food source. These ants cut leaves, preprocess them into small pieces, and feed them to the cultivated fungus (1). The capacity of the cultivated fungus to break down plant material gives ant agriculturalists access to the vast nutrient stores locked within neotropical plants (Fig. 1A) (2–5). The symbiosis between fungus-growing ants and their cultivated fungi has persisted for at least 50 million years (6).
Fig. 1.
Escovopsis weberi, a specialized mycoparasite of the fungus-growing ant symbiosis, has a small genome compared with other Pezizomycotina fungi. (A) Both fungus-growing ants and the mycoparasite E. weberi use the ants’ cultivated fungi as their primary food source. The ability of the cultivated fungi to efficiently break down plant material gives both consumers access to the biomass of neotropical plants. (B) Size and protein-coding gene content of genomes of diverse fungi in the Pezizomycotina. Bayesian phylogeny estimated using partial amino acid alignments of three genes (Rpb1, Rpb2, ef1-α). All posterior probabilities are greater than 0.95. Phylogeny is rooted with Sacchormyces cervesiae (not shown). (C) Relationship between genome size and gene content. A list of genomes included in this panel is in SI Appendix, Table S1.
Like human agriculture, ant agriculture is hampered by disease. The ants’ fungal crops are attacked and consumed by fungal parasites of the genus Escovopsis (Ascomycota, Pezizomycotina: anamorphic Hypocreales) (Fig. 1A) (7), which have evolved in association with the ants and their cultivated fungi (8). Escovopsis infection can have detrimental impacts on garden health and, consequently, on the survival of ant colonies (9, 10). Such mycoparasitism, the phenomenon whereby one fungus is parasitic on another fungus, is rare. It is most well-known for species from the genus Trichoderma, some of which are used as biocontrol agents for fungal diseases and others of which attack human-cultivated fungi (11–13). In contrast to Trichoderma species, however, Escovopsis species grow poorly in their hosts’ absence (SI Appendix, Figs. S1 and S2).
Escovopsis species have never been isolated outside of fungus-growing ant colonies, and different strains of Escovopsis are capable of attacking the fungi grown by different fungus-growing ant species (8, 14, 15). The long-term, specialized evolutionary history of the association between Escovopsis and their hosts provides a unique venue to explore the consequences of host specialization on pathogen genome evolution. Here, we assemble and annotate the genome of a strain of Escovopsis weberi. Consistent with expectations under an evolutionary transition toward using a narrow host range, and similar to many other specialized, host-associated microbes (16, 17), E. weberi exhibits gene loss. Contrasting other fungal pathogens, the large genomes of which are expanded with genetic elements that influence host adaptation (18), the genome size of Escovopsis is small compared with those of its closest sequenced relatives.
Basic Features of the Small Escovopsis Genome
We sequenced the genome of E. weberi strain CC031208-10 isolated from a fungal garden of the leaf-cutting ant Atta cephalotes, a widely distributed fungus-growing ant species, the genome of which has been recently sequenced (19). This strain is closely related to Escovopsis strains isolated from other leaf-cutting ant colonies (SI Appendix, Fig. S3). Sequencing performed with the 454 FLX Titanium pyrosequencing platform generated ∼4.4 million reads, which assembled into 29 scaffolds with a N50 of 2.58 Mbp and an overall genome assembly length of 27.20 Mbp. The G+C content of the Escovopsis genome is 55.74%, similar to other fungi in the Hypocreales (SI Appendix, Table S1). We identified 204 tRNA genes in association with 44 codons and all 20 amino acids (Dataset S1). Approximately 4% of the assembly consists of repetitive elements, including simple sequence repeats such as microsatellites (Dataset S1) and transposable elements (SI Appendix, Fig. S4). The genome can be viewed through the Genome Browser at gb2.fungalgenomes.org/.
Using k-mer frequency analysis as an assembly-independent estimate of genome size (Dataset S2) (20), we estimate the true genome size to be 29.45 ± 2 Mb in length, among the smallest known genomes of all Pezizomycotina (Fig. 1), the largest and most diverse group of ascomycete fungi. Indicative of a complete genome assembly, we identified 239 of 248 super-conserved Core Eukaryotic Genes (CEGs) (21, 22). Escovopsis has 6,870 predicted protein-coding genes (Dataset S3), substantially fewer than other Pezizomycotina (Fig. 1 and SI Appendix, Table S1). The average gene length (1,623 bp) and mean content of exons per gene (2.74) are similar to estimates from closely related Pezizomycotina (SI Appendix, Table S2). Fifty-five percent of the encoded proteins were assigned to Gene Ontology terms, and 76% contain a protein family (PFAM) domain (Dataset S4). Although the number of predicted genes is greatly reduced compared with most other Pezizomycotina, PFAM analysis as well as manual functional annotation of all genes against the National Center for Biotechnology Information (NCBI) nonredundant database (Dataset S3) indicate that the largest gene families in Escovopsis are also common in closely related fungi (SI Appendix, Table S3).
Potential Loss of Sex
An inability of E. weberi to undergo sexual reproduction is suggested by the striking absence of a functional Mating-Type (MAT) locus, as no complete MAT1-2 and MAT1-1 loci were identified (see SI Appendix, Fig. S5, for details). E. weberi also has no homologs to the small peptide pheromones necessary for sexual reproduction in Trichoderma reesei (23). These findings are consistent with the fact that there is no described teleomorph for E. weberi and suggest that this fungus—unlike most others that are predominantly found in their anamorphic form (24)—is asexual. Identification of STE2 and STE3 genes in the genome, homologs of T. reesei receptor proteins necessary for sexual reproduction (23), does suggest that E. weberi—or an ancestor—has a history of sexual reproduction.
Loss of sex in E. weberi would be surprising because Escovopsis presumably must adapt to an array of defenses mounted by its fungal host and associated symbionts, and sexual recombination can provide an advantage in terms of facilitating the generation of variants that are able to counter changing defenses (25, 26). In response to Escovopsis infection, the cultivar can use antibiotics that inhibit Escovopsis growth (14), the ant agriculturalists mount a number of behavioral defenses to remove the pathogen (27), and the ants support bacteria that produce Escovopsis-inhibiting antibiotics (28). All of these defenses could potentially change (either plastically or evolutionarily) in response to Escovopsis infection. There are several important considerations in the case of complex symbiotic systems such as that of the fungus-growing ant symbiosis, however. First, the cultivar, likely under the strongest selection to evolve defenses to counteract Escovopsis attack, reproduces mostly asexually, and somatic incompatibilities limit genetic exchange between strains (29, 30); the cultivar may be constrained to not evolve rapidly so as to maintain a mutualism with the ants [i.e., Red King hypothesis for slow evolution of mutualistic partners (31)]. Second, Escovopsis too can benefit from symbionts [e.g., black yeast that inhibit growth of antibiotic-producing bacteria (32)], which in turn themselves could evolve in response to changing defenses. These combined features may lessen selection to maintain sexual recombination.
Lack of Repeat-Induced Point Mutation
One important consequence of the loss of sex for the genome would be the hindrance of continued Repeat-Induced Point mutation (RIP), which requires sexual recombination (33). RIP, originally described in Neurospora crassa (33) and later shown to be active in Trichoderma (34), a genus of fungi closely related to Escovopsis (Fig. 1B and SI Appendix, Fig. S6), is a common (SI Appendix, Table S4), irreversible fungal-defense mechanism that preferentially alters C:G to T:A nucleotides and acts mainly on transposable elements but also on protein-coding genes (35), potentially leading to gene inactivation and genome reduction. The E. weberi genome lacks four genes involved in RIP: qip, qde1, qde3, and sad1 (SI Appendix, Table S5). Similar RIP deactivation in Blumeria graminis and other specialized plant pathogens is postulated to have led to extensive retrotransposon proliferation and genome-size expansion (16), contrasting the genomic architecture of E. weberi, which along with its reduced genome size, has a paucity of transposable elements (SI Appendix, Fig. S4) and gene paralogs (only four; SI Appendix, Table S6). One possibility is that RIP deactivation in E. weberi is a fairly recent phenomenon: the genome does contain footprints of past RIP operation (SI Appendix, Table S7), suggesting that RIP may have limited genome expansion in the past.
Genomic Similarities to Closely Related Fungi
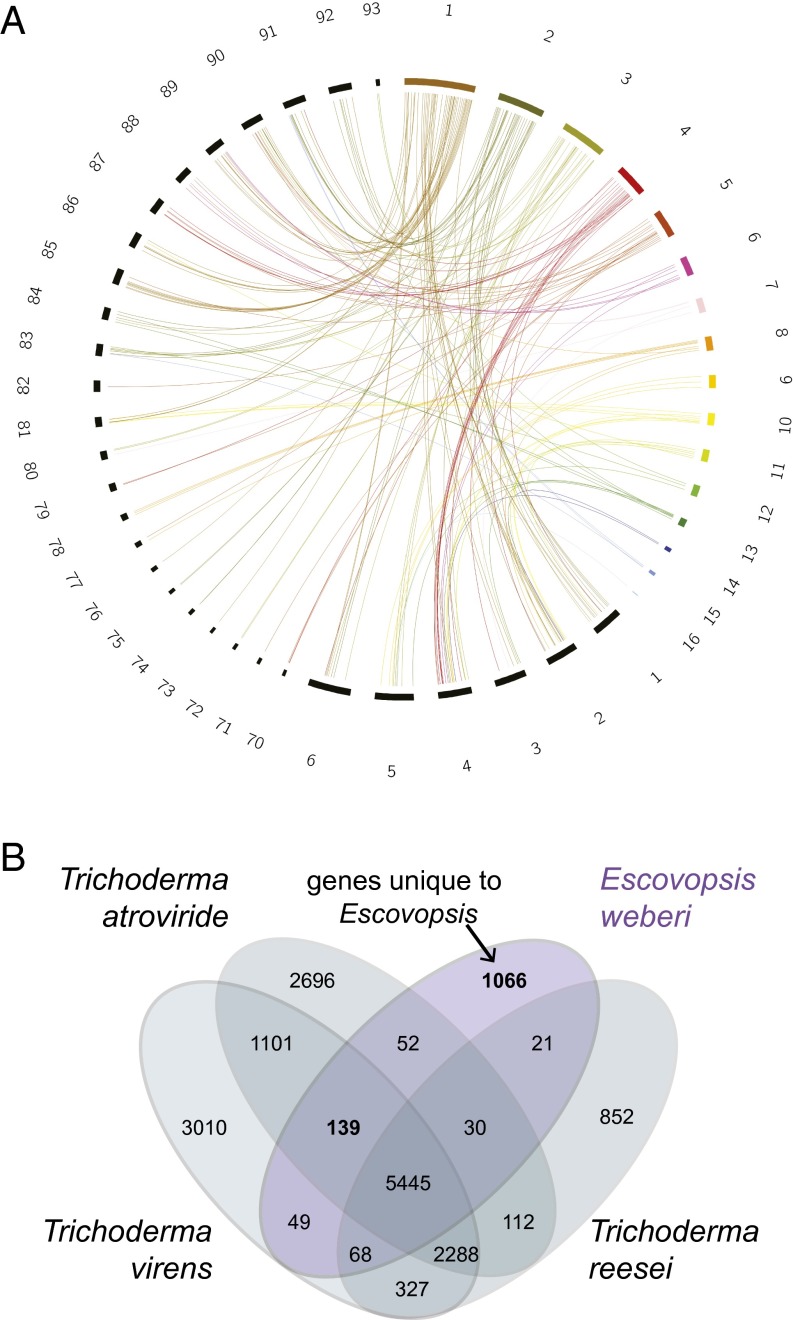
Phylogenetic placement of Escovopsis within the Hypocreales (SI Appendix, Fig. S6) confirms that the most closely related fungi to Escovopsis with available genome sequences are within the genus Trichoderma, which diversified from a mycoparasitic ancestor (36). Escovopsis diverged from Trichoderma ∼50 million years ago (SI Appendix, Fig. S7), coincident with the evolution of ant fungiculture (6, 37). Pairwise sequence comparison of the genome sequences of E. weberi with both Trichoderma atroviride and Trichoderma virens (Fig. 2A) revealed a high degree of micro mesosynteny, indicating that genome segments have a similar gene content but shuffled order and orientation, likely due to intrachromosomal rearrangements (38). Compared with T. virens, only 6% of E. weberi’s genes are located outside of shared syntenic blocks; 42% of these nonsyntenic genes are species-specific to E. weberi and encode proteins of unknown function. Similarities to Trichoderma will facilitate further functional analyses of this ant agricultural pathogen.
Fig. 2.
Similarities between Escovopsis and Trichoderma. (A) Mesosynteny between E. weberi and T. virens. Scaffolds of E. weberi are multicolored. T. virens scaffolds are black. Only scaffolds containing syntenic regions are shown. (B) Gene content overlap between E. weberi and three Trichoderma species. Like Escovopsis spp., Trichoderma spp. are mycoparasites (fungi that attack and consume other fungi), although they are less specialized and are also able to obtain nutrients from dead organic matter. Orthologs were assigned using all-against-all BLASTP for amino acids and inparanoid/multiparanoid (sequence overlap coverage ≥50%).
Orthology analysis, based on bidirectional best BLAST hits, between the E. weberi gene set and those of three Trichoderma spp. (T. atroviride, T. reesei, T. virens), which, like Escovopsis spp., are mycoparasites, revealed that 80% of E. weberi’s genes have homologs in all three Trichoderma genomes, and an additional 5% are found in at least one of the Trichoderma genomes (Fig. 2B and Dataset S3). E. weberi shares more orthologs with T. virens and T. atroviride (Fig. 2B), which may be driven by their substantially higher gene content than T. reesei (Fig. 1B). Most of the 1,066 genes unique to E. weberi relative to Trichoderma spp. are of unknown function, and only 128 of these genes exhibit homology to proteins in other Pezizomycotina (Dataset S5), including Metarhizium, Fusarium, and Colletotrichum species (SI Appendix, Table S8). The latter is intriguing as Colletotrichum is not closely related to the genus Escovopsis but is a genus containing obligate pathogens (39).
Genomic Similarities to Other Specialized Fungi with Small Genomes
Although some specialized, host-associated fungi exhibit genome expansion, in part due to proliferation of retrotransposons [e.g., B. graminis (16)], other specialized, host-associated fungi have small genomes. For example, the Yeast Like Symbiont (YLS), an obligate, specialized fungal endosymbiont of the aphid Cerataphis brasiliensis (40), is predicted to have had strict association with its host insects for millions of years, replacing the role of Buchnera aphidicola, an obligate bacterial symbiont found in other aphid species (41). Trichophyton rubrum, another example, is a human skin-specific fungal pathogen and causative agent of athlete’s foot (42). Like E. weberi, YLS and T. rubrum have two of the smallest estimated genome sizes among the Pezizomycotina (∼25 and 22 Mb, respectively). Fifty-one percent of E. weberi’s 6,870 protein-coding genes have orthologs in both YLS and T. rubrum (SI Appendix, Fig. S8), indicative of a core gene set for these host-associated, although ecologically distinct, taxa. This overlapping core set consists mostly of housekeeping genes involved in central metabolism and in DNA, RNA, protein, and organelle biosynthesis. Genes unique to E. weberi, relative to those shared between the three genomes, are enriched in transcription factors (Zn2Cys6 and C2H2 type) and glycosyl hydrolases, which assist in the hydrolysis of glycosidic bonds in complex sugars (SI Appendix, Table S9). Of the 1,834 genes unique to E. weberi relative to YLS and T. rubrum, 1,064 are found in the mycoparasite T. virens of which 459 encode uncharacterized putative proteins. Of note, the glycosyl hydrolases present in the core set (i.e., those shared among YLS, Trichophyton, and Escovopsis) and those shared only between T. virens and E. weberi exhibit a clear bias: whereas the core set contains all of the GH13 amylolytic and GH16 β-glucanolytic hydrolases, the 1,064 genes shared with T. virens are strongly enriched in GH3, GH5, and GH12 endo- and exo–β-glucanases and particularly in GH18 chitinases (SI Appendix, Table S9), which may play a role in Escovopsis breaking down the chitin within the cell walls of host fungi.
Specialization and Gene Loss
In some respects, fungus-growing ants and Escovopsis occupy a similar niche, obtaining nutrients from the cultivated fungus, which has the capacity to break down diverse, abundant plant material into nutrients that the ants and parasite can use (Fig. 1A) (2, 3). Thus, there should be many degradation capacities of the cultivated fungi that Escovopsis spp. do not require. E. weberi is able to grow on several carbon sources in absence of its fungal host (SI Appendix, Fig. S2 and Dataset S6), and a specific search for presence of genes encoding enzymes of primary metabolism (i.e., carbohydrate, amino acid, lipid, and nucleic acid anabolism and catabolism) revealed that the E. weberi genome contains all genes required for growth on media containing an organic carbon source and salts except for genes required for the synthesis of dehydroascorbic acid, an oxidized form of ascorbic acid. When E. weberi growth was compared with that of T. atroviride using phenotype microarray plates, however, it exhibited much slower growth on most carbon sources (SI Appendix, Fig. S2 and Dataset S6). In these assays, E. weberi grew most rapidly on the α-glucans trehalose and maltose, which is consistent with the findings that E. weberi has retained genes encoding α-glucan–degrading enzymes and that the associated genes are up-regulated when E. weberi is growing toward and overgrowing the fungal cultivar (SI Appendix, Fig. S9 and Dataset S7). It is possible that E. weberi may have specialized in the utilization of these simple and unbranched α-glucans as these are the most abundant carbohydrates in its host fungus (43).
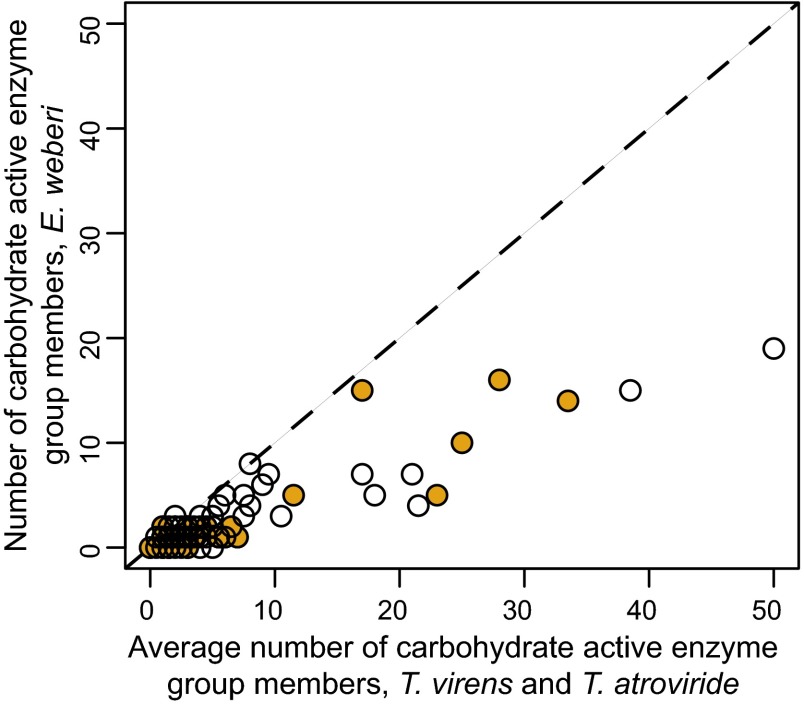
In contrast to T. reesei and T. virens, E. weberi is depleted in genes encoding amino acid transporters and major facilitator superfamily transporters, which transport small solutes. It also contains many fewer cytochrome P450 proteins, flavin-dependent monooxygenases, ankyrins, and PTH11 receptors, which have been implicated in host recognition by fungal pathogens (44) (SI Appendix, Table S3 and Dataset S8). Most interestingly, relative to Trichoderma spp., E. weberi exhibits strong reduction in several gene families encoding polysaccharide depolymerizing enzymes (a.k.a., carbohydrate active enzymes, CAZmyes) (Fig. 3 and SI Appendix, Table S10). E. weberi lacks all cellobiohydrolases [Glycoside Hydrolase family 6 (GH6) and GH7], all xylanases (GH10, GH11, GH30), and also auxiliary proteins like polysaccharide monooxygenases (GH61) and the expansin-like protein swollenin. Consistent with the fact that Escovopsis breaks down the ants’ cultivated fungus but not leaves collected by the ants to feed to their fungus (7), cellulose-binding domains, which are a hallmark of fungi that use plant material for nutrients (34), are also strongly reduced and present only in two endo–β-1,4-glucanases (GH5, GH7; orthologous to T. reesei endo–β-1,4-glucanases EGL and EGL1) and in two chitinases (GH18). The genome of E. weberi also contains only one GH family member that encodes enzymes for hydrolysis of α-galactosides and of α-arabinofuranosides (GH27, GH51); these glycoside hydrolases are expanded in Trichoderma (34, 36). This reduction is reminiscent to that found in some plant pathogens that also lack some GH enzymes (45). On the other hand, E. weberi has a similar number of chitinases (GH18, GH20) and of β–1,3/β-1,4-glucanases (GH16) as T. reesei, indicating that the potential for attacking the host fungus’ cell wall has been maintained. Interestingly, proteomic, transcriptomic, and draft genome sequencing have identified some of these missing enzymes to be present and highly expressed in the ant-cultivated fungus Leucoagaricus gongylophorus (Fig. 3 and SI Appendix, Table S10) (2, 3). Taken together, these losses are consistent with previous findings that the specialized mycoparasite Escovopsis breaks down fungal but not plant material (7) and suggest that E. weberi has lost the ability to feed on lignocellulosic plant material, an ability retained by other microbial members of fungus-growing ant gardens (46).
Fig. 3.
The E. weberi genome encodes a reduced number of carbohydrate active enzymes. Carbohydrate active enzymes are divided into families. Each point represents the relation between the number of members of a given CAZmye family for E. weberi plotted against the average number of family members for the less specialized mycoparasites T. virens and T. atroviride. Members of some of these families, indicated in orange, are known to be highly expressed in E. weberi’s host fungus (2, 3). Additional details are in SI Appendix, Table S10.
Further Genomic Signatures of Exploitation of a Fungal Symbiosis
E. weberi has been shown to kill the fungi cultivated by the ants from a distance (7), a process that likely involves the secretion of toxins. Using SignalP (47), a secretion-specific signal peptide was predicted for 4.8% of E. weberi’s proteins (Dataset S9), about half the percentage found for Trichoderma (9.0, 8.6, and 8.7% in T. reesei, T. atroviride, and T. virens, respectively) (48). The E. weberi secretome is dominated by genes with no known function, particularly in comparison with Trichoderma (55.8% for E. weberi versus 25–30% for Trichoderma spp.).
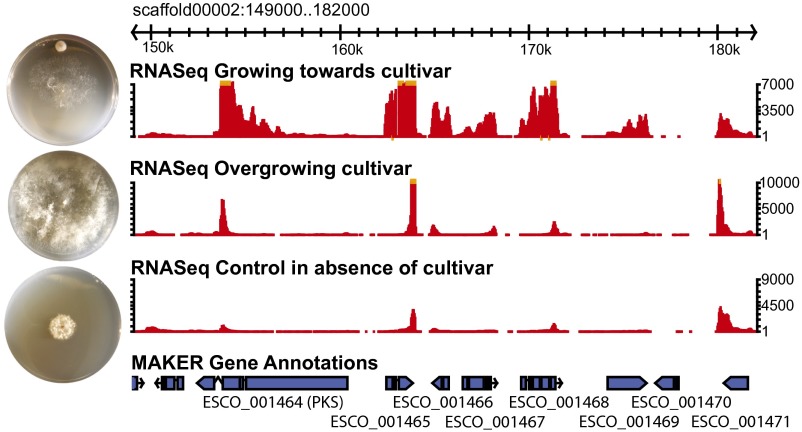
Toward identification of low-molecular-weight toxins, we used antismash 2.0 (49) to identify 17 putative secondary metabolite biosynthesis clusters in the genome (SI Appendix, Table S11), three of which are unique to E. weberi. All three unique clusters are predicted to code for terpene synthases, metabolites known to be involved in the production of mycotoxins (50). Other clusters are predicted to code for polyketide synthases (PKS). Expression of some genes within these PKS clusters was significantly up-regulated when E. weberi was growing toward its host (Fig. 4 and Dataset S7). One such gene (ESCO_001469) encodes a protein with an amino-terminal extracellular cysteine-rich EGF-like (a.k.a. CFEM, or Common in several Fungal Extracellular Membrane proteins) domain (51). Proteins bearing this domain in the rice pathogen Magnaporthe grisea are involved in virulence (51), and CFEM proteins in the human pathogen Candida albicans influence cell-surface characteristics and biofilm formation (52). InterProScan analysis revealed seven CFEM-domain proteins in E. weberi (SI Appendix, Table S12), the largest domain family in those genes that are unique to E. weberi relative to Trichoderma spp. based on orthology analysis (SI Appendix, Table S13).
Fig. 4.
Up-regulation of gene expression within a secondary metabolite cluster during interaction with cultivated host fungi. Gbrowse genome browser view of 1 of 16 secondary metabolite clusters in the E. weberi genome. Below the scaffold are three tracks illustrating RNAseq-based gene expression when E. weberi is growing toward its host (Top), when it has overgrown its host (Middle), and in the absence of its host (Bottom); MAKER2 gene model predictions are illustrated below. Photographs next to each RNAseq track illustrate the growth of E. weberi under each condition. Each Petri dish was inoculated with the cultivated fungus near the top (when present) and E. weberi near the center 1 wk later; photographs were taken 3–4 d after E. weberi inoculation. Note that E. weberi grows much more rapidly in the presence than in the absence of its host. See SI Appendix, Fig. S1 for additional images.
There is also evidence of retention of nonribosomal peptide synthases (NRPSs), enzymes known to synthesize a multitude of secondary metabolites (53). The E. weberi genome encodes two peptaibol synthases (ESCO_001464 and ESCO_003769), NRPSs that have been found only in Trichoderma and a few close relatives (54). These enzymes have been shown to inhibit cell-wall resynthesis by Trichoderma hosts when they are being attacked by Trichoderma’s cell-wall hydrolases (55). Finally, the E. weberi genome encodes three β-lactamases (ESCO_006545, ESCO_005342, ESCO_005794) and one tetracycline resistance gene (ESCO_002770), which inactivate antibiotics. The observation that these genes have been maintained in E. weberi despite general genome reduction suggests a similar mycoparasitic mechanism for Escovopsis when attacking the ants’ cultivated fungi and maintenance of mechanisms to combat antagonists within the complex microbial community of fungus-growing ant gardens.
Conclusions
Specialization over evolutionary timescales can facilitate gene loss and genome reduction. Fungus-growing ants and Escovopsis use the same fungus as a primary food source, and this obligate dependency is reflected in genetic modifications relative to the closest relatives of each. The genome of the ant A. cephalotes, for example, is depleted of genes related to nutrient acquisition, including serine proteases, genes involved in arginine biosynthesis, and a hexamerin involved in amino acid sequestration during development in other insects (19). Here we show that E. weberi has a small genome and reduced gene content relative to its closest sequenced relatives with broader host ranges. The E. weberi genome is depleted in genes associated with plant degradation yet has retained genes associated with attacking fungal hosts. Thus, dependence on the cultivated fungus shapes the genomes of the ants and Escovopsis, unrelated but ecologically linked organisms.
Although the reduced functional capacity of E. weberi is consistent with loss of genes no longer necessary given its highly specialized, mycoparasitic lifestyle, its relatively small genome, with few mobile elements and duplications, is harder to attribute to specific evolutionary processes, particularly given the inactivation of RIP, the loss of which should allow for genome expansion. Although specialized bacteria, and in particular obligate symbionts, consistently exhibit genome reduction, which is facilitated by several evolutionary processes (17), specialized fungi vary greatly in genome size. Some obligately parasitic fungi have large genomes with many transposable elements (16, 18). This is hypothesized to be in part because eukaryotes with small effective population sizes can tolerate accumulation of slightly deleterious transposable elements, multiple introns, and gene duplications (56) and in part because mobile elements can facilitate rapid adaptation in some organisms (18, 57). However, some obligately parasitic fungi, such as the microsporidium Encephalitozoon cuniculi, have reduced genomes with few mobile elements, which is likely due to sustained drift-influenced genome reduction (58). Interestingly, Pezizomycotina fungi with genomes less than 75 Mbp, such as E. weberi, exhibit a pattern of decreased genome size with increased drift (59). This may be coupled with selectively beneficial loss of genes and other genomic content no longer essential for a host-associated, specialized lifestyle.
Specialization of Escovopsis spp. goes beyond just specializing on fungus-growing ant fungi in general. Different Escovopsis spp. have different host ranges. For example, strains isolated from colonies of Atta spp. ants, like the strain genomically described here, are typically able to infect fungi cultivated by Atta and other leaf-cutting ant species but have narrow abilities to attack fungi grown by non–leaf-cutting ant species (60). In fact, even within a symbiosis involving a single ant species and its associated fungi, there can be variation in host range, suggesting genotype-by-genotype specificity (14, 15, 60). Therefore, the annotation of this first Escovopsis genome provides a starting point to investigate the genomic changes underlying a dynamically evolving host–pathogen system.
Materials and Methods
Detailed descriptions of materials and methods are provided in SI Appendix. In brief, we sequenced the genome of a single strain of E. weberi isolated from an A. cephalotes colony from Gamboa, Panama, using the 454 FLX Titanium pyrosequencing platform with both fragment and paired-end approaches (2.5 whole-genome shotgun fragment run, one 8-kpb insert paired-end library run). We assembled the genome using the De Novo GS Assembler v 2.6 from the Newbler software package developed by Roche. The raw dataset is deposited at DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank under PRJNA253870, and the whole-genome assembly is deposited under accession LGSR00000000. The version described here is version LGSR01000000. RNA-seq reads are deposited in the Sequence Read Archive under accession SRP049545.
We assessed genome assembly completeness using three independent methods: (i) we calculated basic statistics, including total length and fragmentation of the assembled sequences; (ii) we identified CEGs in our genome assembly using CEGMA 2.4 (22); and (iii) we took a K-mer–based genome size estimation approach. For the latter, we generated a frequency distribution of unique 31-mers in the raw sequencing reads with Jellyfish 1.1.11 (61) and included K-mers with more than 12 copies in the genome, those located to the right of the inflection point (Dataset S2), for computation of genome size.
We used MAKER 2.28 for gene discovery with exon support provided by alignment of RNA-seq transcripts from E. weberi grown in the presence and absence of its host and by available Trichoderma ESTs and proteomes, other fungal proteomes, and NCBI’s NR database. Protein-coding genes were predicted with the ab initio gene predictors Augustus 2.7 (62), SNAP 0.15.4 (63), and GeneMark 2.5 (64) using exon hits from the protein and RNAseq transcript evidence. We functionally annotated all predicted proteins using InterProScan 5–44.0 (65). The genome annotation can be visualized at gb2.fungalgenomes.org/ with GBrowse (66).
We assessed evidence for RIP by computing RIP indices [TA/AT > 0.89 and CA+TG/AC+GT < 1.03 are considered evidence for RIP (67)] for the five most prevalent repeat families within the E. weberi genome and the unmapped reads using RIPCAL 1.0 (68). We also searched for orthologs of genes known to be involved in the RIP process in N. crassa (69).
Supplementary Material
Acknowledgments
We thank the sequencing and production team at the 454 Sequencing Center for their help and expertise during this project, especially L. Li; the Smithsonian Tropical Research Institute in Panama for logistical support during sample collection, especially M. Paz, O. Arosemena, Y. Clemons, L. Seid, and R. Urriola; the Panamanian Autoridad Nacional del Ambiente y el Mar for collection and export permits; and F. Stewart for access to the Georgia Tech PACE server. Genome annotation and comparisons were performed using the University of California at Riverside Institute for Integrative Biology bioinformatics high performance cluster. This research was supported by a 454 Life Sciences’ 10GB sequencing grant to N.M.G., C.R.C., G.S., G. Weinstock, J. Taylor, and S. Slater. Vienna laboratory research (C.P.K., K.C., L.A., I.S.D.) was supported by the Austrian Science Foundation (Grant FWF-P 25613 to I.S.D.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The raw dataset has been deposited at DNA Data Bank of Japan/European Molecular Biology Laboratory/GenBank under accession PRJNA253870, and the whole-genome assembly has been deposited under accession LGSR00000000. The version described here is version LGSR01000000. RNA-seq reads have been deposited in the Sequence Read Archive under accession SRP049545.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1518501113/-/DCSupplemental.
References
- 1.Weber NA. Fungus-growing ants. Science. 1966;153(3736):587–604. doi: 10.1126/science.153.3736.587. [DOI] [PubMed] [Google Scholar]
- 2.Grell MN, et al. The fungal symbiont of Acromyrmex leaf-cutting ants expresses the full spectrum of genes to degrade cellulose and other plant cell wall polysaccharides. BMC Genomics. 2013;14:928. doi: 10.1186/1471-2164-14-928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aylward FO, et al. Leucoagaricus gongylophorus produces diverse enzymes for the degradation of recalcitrant plant polymers in leaf-cutter ant fungus gardens. Appl Environ Microbiol. 2013;79(12):3770–3778. doi: 10.1128/AEM.03833-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schiøtt M, De Fine Licht HH, Lange L, Boomsma JJ. Towards a molecular understanding of symbiont function: Identification of a fungal gene for the degradation of xylan in the fungus gardens of leaf-cutting ants. BMC Microbiol. 2008;8(1):40. doi: 10.1186/1471-2180-8-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moller IE, De Fine Licht HH, Harholt J, Willats WGT, Boomsma JJ. The dynamics of plant cell-wall polysaccharide decomposition in leaf-cutting ant fungus gardens. PLoS One. 2011;6(3):e17506–e17509. doi: 10.1371/journal.pone.0017506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schultz TR, Brady SG. Major evolutionary transitions in ant agriculture. Proc Natl Acad Sci USA. 2008;105(14):5435–5440. doi: 10.1073/pnas.0711024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Reynolds HT, Currie CR. Pathogenicity of Escovopsis weberi: The parasite of the attine ant-microbe symbiosis directly consumes the ant-cultivated fungus. Mycologia. 2004;96(5):955–959. [PubMed] [Google Scholar]
- 8.Currie CR, et al. Ancient tripartite coevolution in the attine ant-microbe symbiosis. Science. 2003;299(5605):386–388. doi: 10.1126/science.1078155. [DOI] [PubMed] [Google Scholar]
- 9.Currie CR. Prevalence and impact of a virulent parasite on a tripartite mutualism. Oecologia. 2001;128(1):99–106. doi: 10.1007/s004420100630. [DOI] [PubMed] [Google Scholar]
- 10.Currie CR, Mueller UG, Malloch D. The agricultural pathology of ant fungus gardens. Proc Natl Acad Sci USA. 1999;96(14):7998–8002. doi: 10.1073/pnas.96.14.7998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harman GE, Howell CR, Viterbo A, Chet I, Lorito M. Trichoderma species: Opportunistic, avirulent plant symbionts. Nat Rev Microbiol. 2004;2(1):43–56. doi: 10.1038/nrmicro797. [DOI] [PubMed] [Google Scholar]
- 12.Bailey BA, et al. Antibiosis, mycoparasitism, and colonization success for endophytic Trichoderma isolates with biological control potential in Theobroma cacao. Biol Control. 2008;46(1):24–35. [Google Scholar]
- 13.Druzhinina IS, et al. Trichoderma: The genomics of opportunistic success. Nat Rev Microbiol. 2011;9(10):749–759. doi: 10.1038/nrmicro2637. [DOI] [PubMed] [Google Scholar]
- 14.Gerardo NM, Jacobs SR, Currie CR, Mueller UG. Ancient host-pathogen associations maintained by specificity of chemotaxis and antibiosis. PLoS Biol. 2006;4(8):e235. doi: 10.1371/journal.pbio.0040235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gerardo NM, Mueller UG, Price SL, Currie CR. Exploiting a mutualism: Parasite specialization on cultivars within the fungus-growing ant symbiosis. Proc Biol Sci. 2004;271(1550):1791–1798. doi: 10.1098/rspb.2004.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Spanu PD, et al. Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science. 2010;330(6010):1543–1546. doi: 10.1126/science.1194573. [DOI] [PubMed] [Google Scholar]
- 17.McCutcheon JP, Moran NA. Extreme genome reduction in symbiotic bacteria. Nat Rev Microbiol. 2012;10(1):13–26. doi: 10.1038/nrmicro2670. [DOI] [PubMed] [Google Scholar]
- 18.Raffaele S, Kamoun S. Genome evolution in filamentous plant pathogens: Why bigger can be better. Nat Rev Microbiol. 2012;10(6):417–430. doi: 10.1038/nrmicro2790. [DOI] [PubMed] [Google Scholar]
- 19.Suen G, et al. The genome sequence of the leaf-cutter ant Atta cephalotes reveals insights into its obligate symbiotic lifestyle. PLoS Genet. 2011;7(2):e1002007. doi: 10.1371/journal.pgen.1002007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu B, et al. 2013. Estimation of genomic characteristics by analyzing k- mer frequency in de novo genome projects. arXivorg:1–47. Available at arxiv.org/abs/1308.2012.
- 21.Chain PSG, et al. Genomic Standards Consortium Human Microbiome Project Jumpstart Consortium Genomics. Genome project standards in a new era of sequencing. Science. 2009;326(5950):236–237. doi: 10.1126/science.1180614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parra G, Bradnam K, Korf I. CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–1067. doi: 10.1093/bioinformatics/btm071. [DOI] [PubMed] [Google Scholar]
- 23.Seibel C, Tisch D, Kubicek CP, Schmoll M. The role of pheromone receptors for communication and mating in Hypocrea jecorina (Trichoderma reesei) Fungal Genet Biol. 2012;49(10):814–824. doi: 10.1016/j.fgb.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hibbett DS, Taylor JW. Fungal systematics: Is a new age of enlightenment at hand? Nat Rev Microbiol. 2013;11(2):129–133. doi: 10.1038/nrmicro2963. [DOI] [PubMed] [Google Scholar]
- 25.Morran LT, Parmenter MD, Phillips PC. Mutation load and rapid adaptation favour outcrossing over self-fertilization. Nature. 2009;462(7271):350–352. doi: 10.1038/nature08496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith JM. 1978. The Evolution of Sex (Cambridge Univ Press, Cambridge, UK)
- 27.Currie CR, Stuart AE. Weeding and grooming of pathogens in agriculture by ants. Proc Biol Sci. 2001;268(1471):1033–1039. doi: 10.1098/rspb.2001.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Currie CR, Poulsen M, Mendenhall J, Boomsma JJ, Billen J. Coevolved crypts and exocrine glands support mutualistic bacteria in fungus-growing ants. Science. 2006;311(5757):81–83. doi: 10.1126/science.1119744. [DOI] [PubMed] [Google Scholar]
- 29.Mikheyev AS, Mueller UG, Abbot P. Cryptic sex and many-to-one coevolution in the fungus-growing ant symbiosis. Proc Natl Acad Sci USA. 2006;103(28):10702–10706. doi: 10.1073/pnas.0601441103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kooij PW, Poulsen M, Schiøtt M, Boomsma JJ. Somatic incompatibility and genetic structure of fungal crops in sympatric Atta colombica and Acromyrmex echinatior leaf-cutting ants. Fungal Ecol. 2015;18:10–17. doi: 10.1016/j.funeco.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bergstrom CT, Lachmann M. The Red King effect: When the slowest runner wins the coevolutionary race. Proc Natl Acad Sci USA. 2003;100(2):593–598. doi: 10.1073/pnas.0134966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Little AEF, Currie CR. Black yeast symbionts compromise the efficiency of antibiotic defenses in fungus-growing ants. Ecology. 2008;89(5):1216–1222. doi: 10.1890/07-0815.1. [DOI] [PubMed] [Google Scholar]
- 33.Selker EU, Cambareri EB, Jensen BC, Haack KR. Rearrangement of duplicated DNA in specialized cells of Neurospora. Cell. 1987;51(5):741–752. doi: 10.1016/0092-8674(87)90097-3. [DOI] [PubMed] [Google Scholar]
- 34.Martinez D, et al. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat Biotechnol. 2008;26(5):553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- 35.Galagan JE, et al. The genome sequence of the filamentous fungus Neurospora crassa. Nature. 2003;422(6934):859–868. doi: 10.1038/nature01554. [DOI] [PubMed] [Google Scholar]
- 36.Kubicek CP, et al. Comparative genome sequence analysis underscores mycoparasitism as the ancestral life style of Trichoderma. Genome Biol. 2011;12(4):R40. doi: 10.1186/gb-2011-12-4-r40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mikheyev AS, Mueller UG, Abbot P. Comparative dating of attine ant and lepiotaceous cultivar phylogenies reveals coevolutionary synchrony and discord. Am Nat. 2010;175(6):E126–E133. doi: 10.1086/652472. [DOI] [PubMed] [Google Scholar]
- 38.Hane JK, et al. A novel mode of chromosomal evolution peculiar to filamentous Ascomycete fungi. Genome Biol. 2011;12(5):R45. doi: 10.1186/gb-2011-12-5-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.O’Connell RJ, et al. Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat Genet. 2012;44(9):1060–1065. doi: 10.1038/ng.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vogel KJ, Moran NA. Functional and evolutionary analysis of the genome of an obligate fungal symbiont. Genome Biol Evol. 2013;5(5):891–904. doi: 10.1093/gbe/evt054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baumann P, et al. Genetics, physiology, and evolutionary relationships of the genus Buchnera: Intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 42.Martinez DA, et al. Comparative genome analysis of Trichophyton rubrum and related dermatophytes reveals candidate genes involved in infection. MBio. 2012;3(5):e00259. doi: 10.1128/mBio.00259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Martin MM, Carman RM, Macconnell JG. Nutrients derived from the fungus cultured by the fungus-growing ant Atta colombica tonsipes. Ann Entomol Soc Am. 1969;62(1):11–13. doi: 10.1093/aesa/62.6.1386. [DOI] [PubMed] [Google Scholar]
- 44.DeZwaan TM, Carroll AM, Valent B, Sweigard JA. Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell. 1999;11(10):2013–2030. doi: 10.1105/tpc.11.10.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhao Z, Liu H, Wang C, Xu J-R. Comparative analysis of fungal genomes reveals different plant cell wall degrading capacity in fungi. BMC Genomics. 2013;14(1):274. doi: 10.1186/1471-2164-14-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Aylward FO, Currie CR, Suen G. The evolutionary innovation of nutritional symbioses in leaf-cutter ants. Insects. 2012;3:41–61. doi: 10.3390/insects3010041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 48.Druzhinina IS, Shelest E, Kubicek CP. Novel traits of Trichoderma predicted through the analysis of its secretome. FEMS Microbiol Lett. 2012;337(1):1–9. doi: 10.1111/j.1574-6968.2012.02665.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blin K, et al. antiSMASH 2.0: A versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res. 2013;41(Web Server issue):W204–W212. doi: 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rynkiewicz MJ, Cane DE, Christianson DW. Structure of trichodiene synthase from Fusarium sporotrichioides provides mechanistic inferences on the terpene cyclization cascade. Proc Natl Acad Sci USA. 2001;98(24):13543–13548. doi: 10.1073/pnas.231313098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kulkarni RD, Kelkar HS, Dean RA. An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem Sci. 2003;28(3):118–121. doi: 10.1016/S0968-0004(03)00025-2. [DOI] [PubMed] [Google Scholar]
- 52.Pérez A, et al. Some biological features of Candida albicans mutants for genes coding fungal proteins containing the CFEM domain. FEMS Yeast Res. 2011;11(3):273–284. doi: 10.1111/j.1567-1364.2010.00714.x. [DOI] [PubMed] [Google Scholar]
- 53.Strieker M, Tanović A, Marahiel MA. Nonribosomal peptide synthetases: Structures and dynamics. Curr Opin Struct Biol. 2010;20(2):234–240. doi: 10.1016/j.sbi.2010.01.009. [DOI] [PubMed] [Google Scholar]
- 54.Mukherjee PK, Horwitz BA, Kenerley CM. Secondary metabolism in Trichoderma: A genomic perspective. Microbiology. 2012;158(Pt 1):35–45. doi: 10.1099/mic.0.053629-0. [DOI] [PubMed] [Google Scholar]
- 55.Lorito M, Farkas V, Rebuffat S, Bodo B, Kubicek CP. Cell wall synthesis is a major target of mycoparasitic antagonism by Trichoderma harzianum. J Bacteriol. 1996;178(21):6382–6385. doi: 10.1128/jb.178.21.6382-6385.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lynch M, Conery JS. The origins of genome complexity. Science. 2003;302(5649):1401–1404. doi: 10.1126/science.1089370. [DOI] [PubMed] [Google Scholar]
- 57.Stukenbrock EH, Croll D. The evolving fungal genome. Fungal Biol Rev. 2014;28(1):1–12. [Google Scholar]
- 58.Katinka MD, et al. Genome sequence and gene compaction of the eukaryote parasite Encephalitozoon cuniculi. Nature. 2001;414(6862):450–453. doi: 10.1038/35106579. [DOI] [PubMed] [Google Scholar]
- 59.Kelkar YD, Ochman H. Causes and consequences of genome expansion in fungi. Genome Biol Evol. 2012;4(1):13–23. doi: 10.1093/gbe/evr124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Birnbaum SSL, Gerardo NM. Patterns of specificity of the pathogen Escovopsis across the fungus-growing ant symbiosis. Am Nat. 2016 doi: 10.1086/686911. in press. [DOI] [PubMed] [Google Scholar]
- 61.Marçais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27(6):764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stanke M, Waack S. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(Suppl 2):ii215–ii225. doi: 10.1093/bioinformatics/btg1080. [DOI] [PubMed] [Google Scholar]
- 63.Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5:59. doi: 10.1186/1471-2105-5-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lomsadze A, Ter-Hovhannisyan V, Chernoff YO, Borodovsky M. Gene identification in novel eukaryotic genomes by self-training algorithm. Nucleic Acids Res. 2005;33(20):6494–6506. doi: 10.1093/nar/gki937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zdobnov EM, Apweiler R. InterProScan: An integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 66.Stein LD, et al. The generic genome browser: A building block for a model organism system database. Genome Res. 2002;12(10):1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Margolin BS, et al. A methylated Neurospora 5S rRNA pseudogene contains a transposable element inactivated by repeat-induced point mutation. Genetics. 1998;149(4):1787–1797. doi: 10.1093/genetics/149.4.1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hane JK, Oliver RP. RIPCAL: A tool for alignment-based analysis of repeat-induced point mutations in fungal genomic sequences. BMC Bioinformatics. 2008;9:478. doi: 10.1186/1471-2105-9-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Borkovich KA, et al. Lessons from the genome sequence of Neurospora crassa: Tracing the path from genomic blueprint to multicellular organism. Microbiol Mol Biol Rev. 2004;68(1):1–108. doi: 10.1128/MMBR.68.1.1-108.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.