Significance
The suprachiasmatic nucleus (SCN) is the principal circadian clock of the mammalian brain. To function effectively, SCN neurons must operate as a synchronized circuit. How cell-autonomous and circuit-level circadian mechanisms interact to achieve this is unclear. Here, we used intersectional genetics to create temporally chimeric mice where both 24-h and 20-h clock neurons were present in the SCN, in different cell populations. The 24-h dopamine receptor-positive cells set the speed of the SCN, imposing their cell-autonomous 24-h period on all cells in the circuit. Exposure to a 20-h lighting cycle, however, inverted this dominance, reprograming the circuit to 20 h. These results show how robust circuit-level signaling underlies complex, nonlinear computations of circadian period that also exhibit a remarkable level of plasticity.
Keywords: period, entrainment, neuropeptide, circuit, oscillator
Abstract
The suprachiasmatic nucleus (SCN) is the master circadian clock controlling daily behavior in mammals. It consists of a heterogeneous network of neurons, in which cell-autonomous molecular feedback loops determine the period and amplitude of circadian oscillations of individual cells. In contrast, circuit-level properties of coherence, synchrony, and ensemble period are determined by intercellular signals and are embodied in a circadian wave of gene expression that progresses daily across the SCN. How cell-autonomous and circuit-level mechanisms interact in timekeeping is poorly understood. To explore this interaction, we used intersectional genetics to create temporally chimeric mice with SCN containing dopamine 1a receptor (Drd1a) cells with an intrinsic period of 24 h alongside non-Drd1a cells with 20-h clocks. Recording of circadian behavior in vivo alongside cellular molecular pacemaking in SCN slices in vitro demonstrated that such chimeric circuits form robust and resilient circadian clocks. It also showed that the computation of ensemble period is nonlinear. Moreover, the chimeric circuit sustained a wave of gene expression comparable to that of nonchimeric SCN, demonstrating that this circuit-level property is independent of differences in cell-intrinsic periods. The relative dominance of 24-h Drd1a and 20-h non-Drd1a neurons in setting ensemble period could be switched by exposure to resonant or nonresonant 24-h or 20-h lighting cycles. The chimeric circuit therefore reveals unanticipated principles of circuit-level operation underlying the emergent plasticity, resilience, and robustness of the SCN clock. The spontaneous and light-driven flexibility of period observed in chimeric mice provides a new perspective on the concept of SCN pacemaker cells.
Daily rhythms of behavior and physiology adapt organisms to the solar cycle. In mammals, these rhythms are coordinated by a central circadian pacemaker: the suprachiasmatic nucleus (SCN) of the hypothalamus (1). The SCN consists of 10,000 neurons that work together to ensure that rhythms are tuned appropriately to anticipate day and night. Circadian dysfunction is increasingly linked to a variety of psychological and metabolic disorders (2). The principal subdivisions of the SCN are the shell, characterized by neurons expressing arginine vasopressin (AVP), and the retinorecipient core, containing vasoactive intestinal peptide (VIP) and gastrin-releasing peptide (GRP) neurons. The core mediates entrainment to light after birth (3), whereas neurons expressing the dopamine 1a receptor (Drd1a) mediate transplacental dopaminergic entrainment of the SCN in utero (4). Drd1a cells are, therefore, a functionally distinct subpopulation that spans elements of both core and shell.
Timekeeping in individual SCN neurons involves a molecular clockwork based on a transcriptional–translational feedback loop (TTFL) whereby PERIOD and CRYPTOCHROME proteins inhibit their own transactivation by CLOCK/BMAL1 heterodimers (5). Almost all of the cells in the body also have this TTFL, but in the absence of SCN input, the amplitude and synchrony of peripheral circadian oscillations are lost. A defining feature of the SCN, therefore, is its intrinsic ability to sustain stable, high-amplitude circadian rhythms (6). Importantly, this is dependent on neuropeptide-mediated interneuronal communication (7–9). This property is embodied in an emergent spatiotemporal wave of gene expression that progresses daily across the SCN, observed in real-time recordings of Per- and Cry-driven bioluminescence in SCN slices (8, 10). The principles that underpin the relationship between cell-autonomous and circuit-level properties are unknown.
To explore these principles, we sought to modulate circadian function in a subset of neurons in the SCN and examine the consequences circuit-wide. We selected a GENSAT (Gene Expression in the Nervous System Atlas) transgenic mouse line that expressed Drd1a promoter-driven Cre recombinase (Cre) with very high expression in the SCN and atypically low expression in other brain areas (11). We used Cre-mediated deletion of the casein kinase 1 epsilon (CK1ε) Tau mutation that accelerates the TTFL (12) to create temporally chimeric mice in which the SCN contained cells with contrasting cell-autonomous periods: 24 h (Tau-deleted) or 20 h (Tau-competent). Importantly, our approach kept intrinsic circadian timekeeping intact in all cells. This contrasts with studies that have selectively removed circadian function in a particular cell population through deletion or overexpression of a core clock gene (13–15). A major limitation of loss/overexpression of circadian transcription factors is that it not only confounds cell-autonomous timing but also compromises expression of neuropeptides that mediate intercellular signaling (14). This renders it difficult to dissociate contributions of cell-autonomous and intercellular mechanisms to circuit-level computations. We therefore considered our enzymatic modulation of cell-autonomous period to provide a valuable complementary perspective on SCN pacemaking. Having created chimeric mice, we could ask whether such a circuit is competent to generate circadian behavioral and molecular rhythms. If so, what is the emergent period? Do Drd1a cells dominate, or are multiple periodicities maintained? Does the altered distribution of cell-autonomous periods in the chimeric circuit affect other circuit-level properties, for example synchrony and generation of the spatiotemporal wave? Finally, what are the limits to the function of such a chimeric circuit? How plastic is it, and how might it be modulated?
Results
Circadian Behavior in Temporally Chimeric Mice.
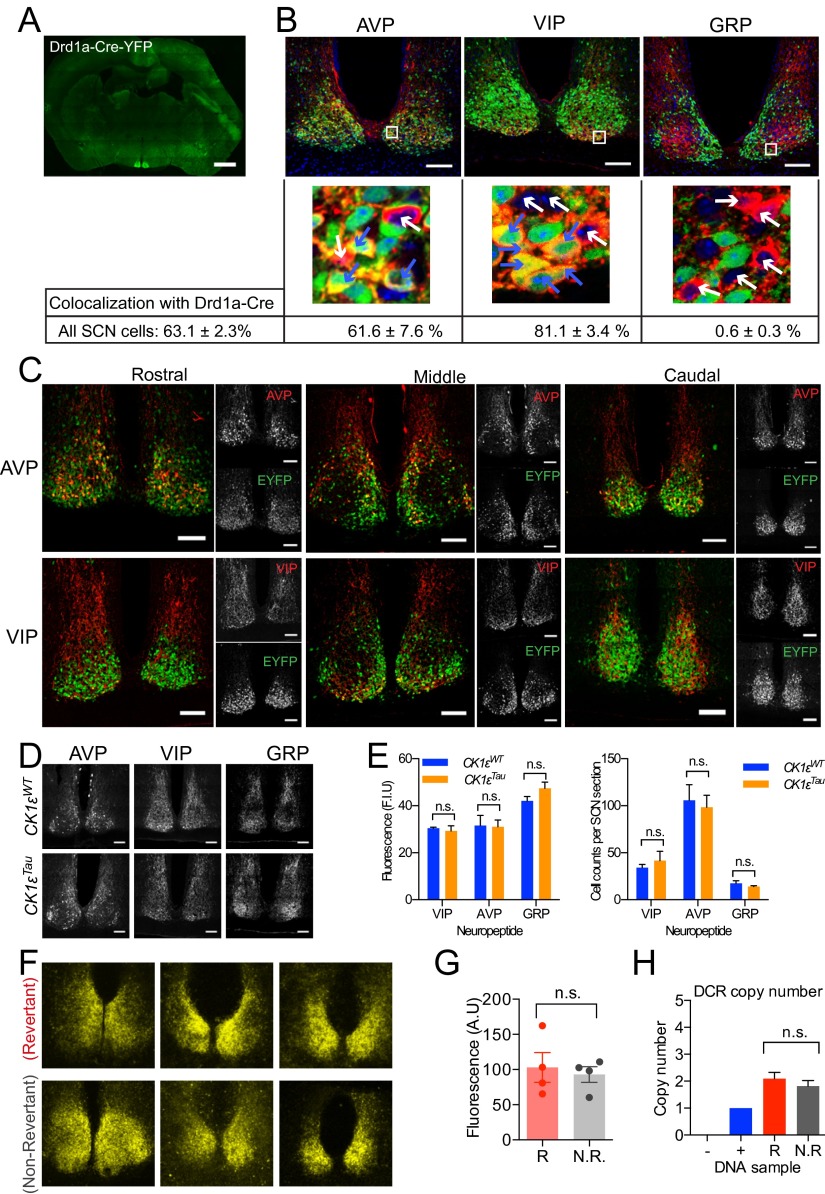
The activity of Cre across the brain of Drd1a-Cre mice was revealed by Cre-mediated constitutive expression of enhanced yellow fluorescent protein (EYFP) (ROSA26-EYFP) (16). Consistent with the GENSAT expression data, this generated strong fluorescence in the SCN (Fig. S1A), with varying levels in the olfactory bulb, amygdala, nucleus accumbens, and scattered striato-nigral neurons. Otherwise, fluorescence across the brain was at background levels. Neuropeptide immunofluorescence revealed that EYFP colocalized with a majority of AVP neurons and VIP neurons (Fig. S1 B and C). EYFP was almost entirely absent from GRP neurons. AVP, VIP, and GRP do not represent the entire SCN, and overall, two-thirds of SCN cells identified by DAPI stain expressed Cre (Fig. S1B). Thus, Drd1a-driven Cre expression is restricted to a neurochemically defined subset of SCN neurons incorporating elements of shell and core.
Fig. S1.
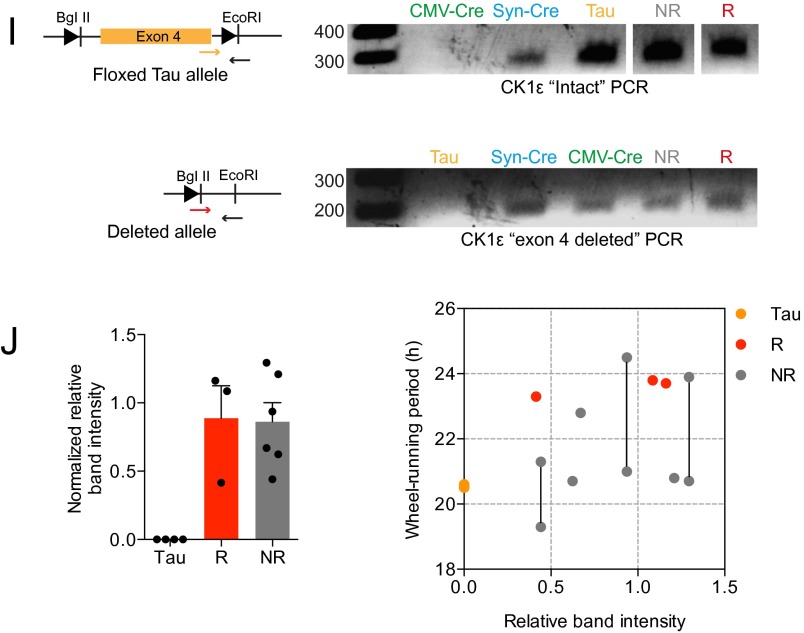
Distribution of Drd1a-mediated Cre recombinase activity reported by ROSA26-EYFP expression (DCR). (A) Representative coronal section at the level of the SCN, showing ROSA26-EYFP fluorescence across the brain. (Scale bar, 1 mm.) (B) Representative immunofluorescent labeling of neuropeptidergic neurons of SCN. Neuropeptides are colored red, EYFP green, and DAPI blue. SCN sections containing high expression of the neuropeptide were chosen. Images of the entire SCN (Top) and zoomed-in snapshots (Bottom, taken from white boxes in the Top panels) are presented. In the zoomed-in images, representative cells that colocalize with Cre activity (blue arrows) or do not have Cre activity (white arrows) are shown. (Scale bar, 100 μm.) Percentage colocalization between Cre and neuropeptides (n = 6 brains, n > 8,000 cells counted) and Cre with all SCN neurons (n = 4 brains, n = 5,700 DAPI-stained cells), was assessed through cell counting (table below images). (C) Representative images showing rostral to caudal distributions of EYFP fluorescence (green) relative to AVP and VIP neuropeptide immunofluorescence (red). (Scale bar, 100 μm.) (D) Representative neuropeptide immunolabeling in the SCN, from DCR-positive animals, either carrying CK1εWT or mutant CK1εTau, the latter being a target for Cre recombinase. (E) Fluorescence intensity and cell counts for neuropeptides were not significantly different in chimeric animals compared with WT (n = 3, two-way ANOVA with Sidak multiple-comparisons test, n.s. P > 0.05). (F) Representative confocal micrographs of EYFP fluorescence in Revertant and Non-Revertant SCN slices. (G) Quantification of EYFP fluorescence intensity measures in Revertant (R) and Non-Revertant (N.R.) SCN slices. (H) Cre-recombinase copy number was detected by digital droplet PCR and compared with WT (–) and one copy VIP-Cre (+) DNA samples. Drd1a-Cre copy number was not significantly different (Student’s t test, P = 0.41) between Revertant (R) and Non-Revertant (N.R.) animals (mean + SEM; Revertant, n = 10; Non-Revertant, n = 7). (I) PCR of genomic DNA to demonstrate successful deletion of CK1ε exon 4 by PCR using DNA from individual SCN slices. Shown are schematic diagrams (Left) and representative electrophoresis gels (Right) showing detection of intact or deleted CK1ε alleles. In the schematic diagrams, LoxP sites are represented by black triangles and positions of primers are marked with small arrows. For the PCR assay, CMV-Cre AAV transduced and floxed Tau SCN samples were generated to give a positive control for CK1ε deletion. Synapsin-Cre (Syn-Cre) AAV-treated floxed Tau SCN samples were generated to give neuronal-specific CK1ε deletion. Representative Tau, Non-Revertant (NR), and Revertant (R) samples are shown. (J) CK1ε deletion was assessed by measuring the relative band intensities from exon 4-deleted PCR, normalized to ActB. Values are calculated relative to CMV-Cre band. Band intensity measures are plotted in a bar chart (Left) or as a scatter plot (Right), where intensities are paired with the corresponding wheel-running period from the same animal. Animals (n = 3) exhibiting multiple periods of wheel-running behavior have both measured periods plotted with lines connecting them.
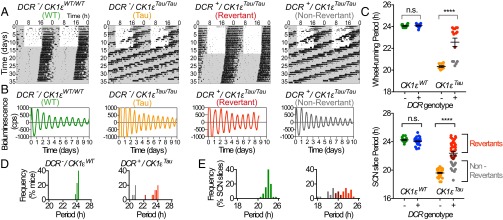
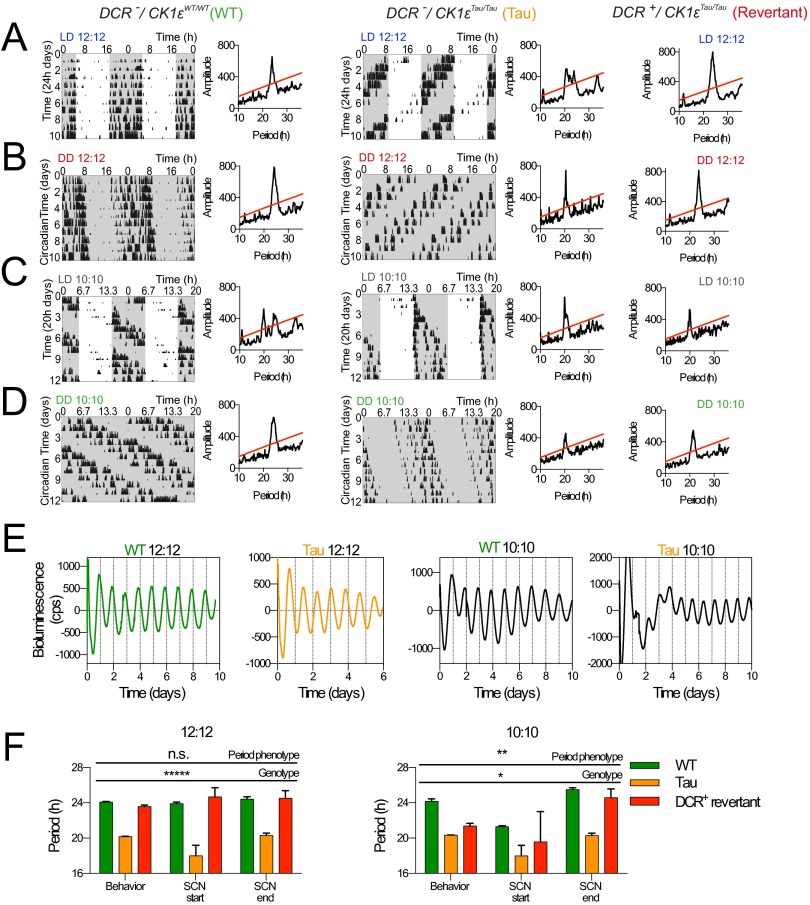
Drd1a-Cre, ROSA26-EYFP mice (DCR) were then crossed with mice carrying floxed CK1εTau alleles (12), along with the PER2::LUCIFERASE bioluminescent reporter (17). Thus, in DCR-CK1εTau mice, the SCN (and potentially other brain regions) should be a chimera of 24-h Drd1a cells alongside 20-h non-Drd1a cells. Deletion of CK1ε was confirmed by PCR (Fig. S1I). Importantly, neuropeptide expression was not significantly different in these animals compared with wild type (WT) (Fig. S1 D and E), indicating that neuropeptide-mediated signaling is likely to be intact in the chimeric SCN. Wheel-running behavior rhythms were monitored to assess the effect of temporal chimerism in vivo (Fig. 1A). DCR+-CK1εWT mice had well-organized activity patterns, comparable to DCR− animals (Fig. 1C and Table S1). Thus, expression of Cre itself did not affect behavior. As expected, the CK1εTau allele shortened the period by ca. 2 h per copy in DCR− (Tau) mice. DCR+ mice carrying CK1εTau alleles also exhibited organized free-running activity rhythms. Chimerism did not, therefore, compromise circadian control of behavior. Chimerism did, however, dramatically lengthen the period of wheel-running behavior compared with DCR− animals (Fig. 1C and Table S1). This effect was not fully penetrant, and two principal phenotypes were apparent: The majority (9/15; 60%) had a period very close to 24 h—i.e., WT-like—consistent with a fully dominant effect of Tau deletion. We describe these animals as “Revertants.” A subset of mice (5/15; 33%), which we refer to as “Non-Revertant,” displayed a shorter period, consistent with their Tau genotype. This dichotomy is clear in the bimodal frequency plot (Fig. 1D). Interestingly, 1 out of fifteen displayed unstable behavioral rhythms (Fig. S2A).
Fig. 1.
Circadian period in temporally chimeric mice. (A) Representative double-plotted actograms from control DCR−/CK1εWT (green), mutant DCR−/CK1εTau (orange), and temporally chimeric DCR+/CK1εTau animals. Chimeric animals displayed a range of phenotypes, including Revertant (red) that phenocopied WT and Non-Revertant (gray) that phenocopied mutant Tau animals. White and gray backgrounds indicate lights on and off, respectively. (B) Representative PER2::LUC bioluminescence traces (de-trended) show circadian oscillations in SCN organotypic slices, where Revertant and Non-Revertant slices phenocopied WT and Tau, respectively. (C) Group data (mean ± SEM), across a CK1εTau mutant background, with (+) and without (–) Drd1a-Cre (DCR) for wheel-running (n > 6 per group) and SCN slices (n > 10 per group). Cre activity significantly lengthened the circadian period of behavior in the presence of CK1εTau (two-way ANOVA with Tukey’s comparison, ****P < 0.0001). The period phenotype in the SCN matched that of behavior. Period was significantly lengthened in the chimeric SCN (Tukey’s post hoc test following two-way ANOVA, n.s. P > 0.05, ****P < 0.0001). (D) Frequency distribution plots of the behavioral period in DCR+ animals illustrate the broad multimodal spread in the CK1εTau population compared with CK1εWT. (E) Frequency distribution for SCN slice circadian period.
Table S1.
Summary of circadian period determination (group data)
| Genotype | n | Mean, h | SEM, h | Median, h | Lower limit, h | Upper limit, h | Range, h | Two-way ANOVA | Kolgomorov–Smirnov |
| Wheel-running behavior | DCR, P****; CK1ε, P****; interaction, P**** | WT/WT, n.s.; Tau/Tau, P** | |||||||
| DCR−/CK1εWT/WT | 12 | 24.1 | 0.043 | 24.1 | 23.9 | 24.3 | 0.4 | ||
| DCR−/CK1εTau/Tau | 9 | 20.3 | 0.083 | 20.2 | 20.0 | 20.7 | 0.7 | ||
| DCR+/CK1εWT/WT | 13 | 24.1 | 0.045 | 24.1 | 23.7 | 24.3 | 0.6 | ||
| DCR+/CK1εTau/Tau | 15 | 22.6 | 0.38 | 23.4 | 20.4 | 23.9 | 3.5 | ||
| SCN slice—mouse age, 10 d | DCR, P****; CK1ε, P****; Interaction, P**** | WT/WT, n.s.; Tau/Tau, P**** | |||||||
| DCR−/CK1εWT/WT | 17 | 24.4 | 0.079 | 24.2 | 23.7 | 24.9 | 1.2 | ||
| DCR−/CK1εTau/Tau | 41 | 19.6 | 0.08 | 19.8 | 18.4 | 20.5 | 2.1 | ||
| DCR+/CK1εWT/WT | 43 | 24.1 | 0.065 | 24.1 | 23.0 | 25.3 | 2.3 | ||
| DCR+/CK1εTau/Tau | 40 | 22.4 | 0.26 | 22.7 | 18.6 | 24.9 | 6.3 | ||
| SCN slice—mouse age, ∼50 d | |||||||||
| DCR−/CK1εWT/WT | 8 | 24.1 | 0.21 | 24.0 | 25.2 | 23.4 | 1.8 | ||
| DCR−/CK1εTau/Tau | 3 | 19.5 | 0.24 | 19.9 | 19.9 | 19.1 | 0.8 | ||
| DCR+/CK1εWT/WT | 4 | 24.1 | 0.35 | 24.1 | 25.0 | 23.4 | 1.6 | ||
| DCR+/CK1εTau/Tau | 9 | 22.1 | 0.83 | 21.2 | 25.6 | 18.4 | 7.2 |
Fig. S2.
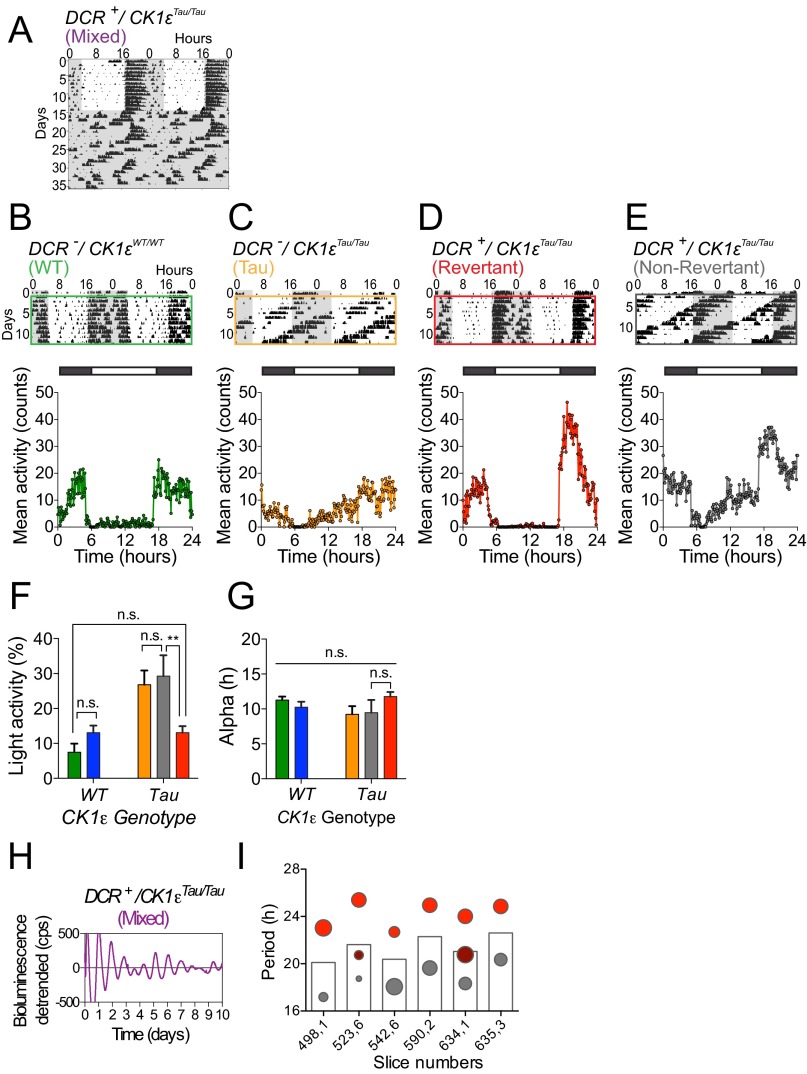
Analyses of behavioral and SCN rhythms. (A) A representative double-plotted actogram from a DCR+/CK1εTau animal exhibiting a multiple period phenotype (mixed). (B–E) Representative, wheel-running actograms and their respective mean activity profiles of animals kept in LD 12:12 conditions. White and black bars above the activity profile show times of lights on and off, respectively. Colored boxes on actograms indicate the time frame analyzed to produce the mean activity profile. (F) Group data (mean ± SEM, n > 6 per group) for light phase activity. There were significant differences in light phase activity across the different genotypic groups (ordinary one-way ANOVA, ****P < 0.0001). Revertant animals displayed significantly less activity than Non-Revertant and Tau animals during the light phase (Student’s t test, **P < 0.01). (G) Group data (mean ± SEM, n > 6 per group) for alpha (duration of activity phase) show that activity durations were not significantly different across all genotypes (ordinary one-way ANOVA, P = 0.25) nor between DCR+/CK1εTau phenotypes (Student’s t test, P = 0.16). (H) A representative PER2::LUC bioluminescence trace (de-trended) illustrating the rare event (n = 7) of multiple period circadian rhythms (“mixed”). (I) Group data for SCN slices showing multiple, “unstable” periods. Mean period is shown as a bar and the determined multiple periods as circles, where the size of the circle represents the relative robustness of that particular period determination; that is, a larger circle represents a low RAE and thus more robust.
The consequences of chimerism were reflected in entrained daily activity profiles of wheel running (Fig. S2 B–E). Revertant animals entrained to the light–dark (LD) cycle, with a clear onset of consolidated nocturnal activity (Fig. S2D), whereas entrainment to 24-h cycles was variable in Non-Revertant animals (Fig. S2E), similar to Tau mice. Thus, Revertants showed minimal activity in the light phase, comparable to WT, whereas Tau and some Non-Revertant mice failed to entrain, exhibiting significantly higher levels of photophase activity (Fig. S2F). Importantly, chimerism did not affect the overall duration of wheel-running (alpha), which was comparable across all genotypes in LD (Fig. S2G). This suggests that neuropeptide-mediated coupling of SCN subcircuits regulating evening and morning activity bouts was intact, in contrast to the disruptive effects of TTFL-based chimerism (14). In summary, targeted manipulation of the cell-autonomous period in Drd1a neurons did not compromise coherent timekeeping but did respecify the period of behavior in the majority of mice and, by inference, the period of the SCN circuit.
Molecular Pacemaking in SCN of Temporally Chimeric Mice.
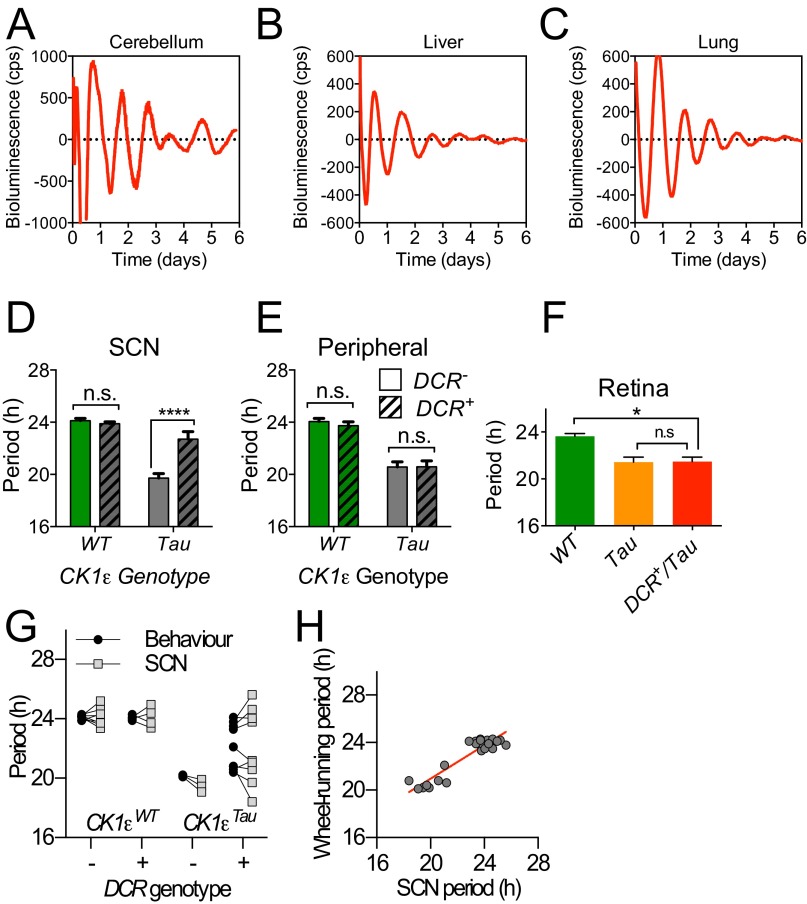
PER2::LUC bioluminescence was monitored in organotypic SCN slices. The majority of chimeric SCNs exhibited very clear circadian cycles of bioluminescence (Fig. 1B). As with behavior, temporal chimerism did not destabilize circadian function of the SCN. It did, however, significantly lengthen the period compared with Tau SCN slices (Fig. 1C). Importantly, this effect was tissue-specific because a range of tissues that do not express Drd1a-Cre (cerebellum, liver, and lung) faithfully reported their Tau genotype, irrespective of Cre genotype (Fig. S3 and Table S2). Furthermore, the period of retinal explants remained ca. 20 h (Fig. S3F). Nevertheless, a dichotomy in chimeric SCN phenotype was again seen, with both Revertant and Non-Revertant SCNs at ratios comparable to those seen for behavior. A potential cause of the spectrum of the chimeric phenotype could be differing levels of Cre activity between animals, but we did not find clear evidence that this was the case. There were no detectable differences in the intensity or distribution of Cre-dependent EYFP expression (Fig. S1 F and G) or the copy number of Cre transgene between Revertant and Non-Revertant SCN (Fig. S1H). Moreover, assessment of CK1ε deletion in SCN genomic DNA did not reveal any obvious differences in the efficiency of deletion (Fig. S1 I and J). Paired measurements of period in wheel-running and SCN PER2::LUC oscillations from the same animal were highly correlated (Fig. S3G and Table S1): Mice identified as Revertants or Non-Revertants in vivo had SCNs of the corresponding phenotype (Fig. S3H). Thus, the ensemble SCN period was predictive, and likely the source, of chimeric behavior in vivo.
Fig. S3.
Circadian period of SCN is predictive of behavior. (A–C) Representative PER2::LUC bioluminescence rhythms for cerebellum, liver, and lung slices. (D) Group data for SCN slices taken from the same animals used for collecting peripheral slices. DCR+/CK1εTau SCN slices had significantly longer periods than their DCR− counterparts (two-way ANOVA; DCR genotype, P < 0.0001; CK1ε genotype, P < 0.0001, with Tukey’s multiple comparisons test, ****P < 0.0001). (E) Group data for peripheral slices. DCR+/CK1εTau peripheral slices did not have significantly longer periods than their DCR− counterparts (two-way ANOVA; DCR genotype, P = 0.76; CK1ε genotype, P < 0.0001, with Tukey’s multiple comparisons test, n.s. P > 0.05). For all group data, mean ± SEM, n > 3 for each group. (F) The circadian period of ex vivo retina slices from DCR+/CK1εTau was significantly shorter than from WT retina, but not significantly different from Cre-negative Tau mutant retina (n ≥ 3, one-way ANOVA with Tukey’s multiple comparisons test, *P < 0.05). (G) Group data for wheel-running and SCN bioluminescence rhythms from individual control and chimeric mice were not significantly different (n = 24, repeated-measures two-way ANOVA, P = 0.41). (H) The circadian period of behavior in vivo was predictive of the circadian period of SCN bioluminescence in vitro, across all mice. Red line is the linear regression fit of the data (y = 0.70x + 6.88; R2 = 0.85; P < 0.0001).
Table S2.
Summary of period determination of peripheral tissues, by tissue type
| Cerebellum | Liver | Lung | |||||||
| Genotype | N | Period, h | SEM, h | n | Period, h | SEM, h | n | Period, h | SEM, h |
| Peripheral tissues—mouse age, 10 d | |||||||||
| DCR−/CK1εWT/WT | 1 | 23.8 | N/A | 1 | 24.1 | N/A | 1 | 24.3 | N/A |
| DCR−/CK1εTau/Tau | 2 | 20.8 | 0.65 | 3 | 20.2 | 0.78 | 8 | 20.6 | 0.45 |
| DCR+/CK1εWT/WT | 6 | 24.1 | 0.71 | 11 | 23.4 | 0.39 | 12 | 23.7 | 0.28 |
| DCR+/CK1εTau/Tau | 0 | 0 | 0 | 3 | 20.9 | 0.72 | 4 | 20.8 | 0.82 |
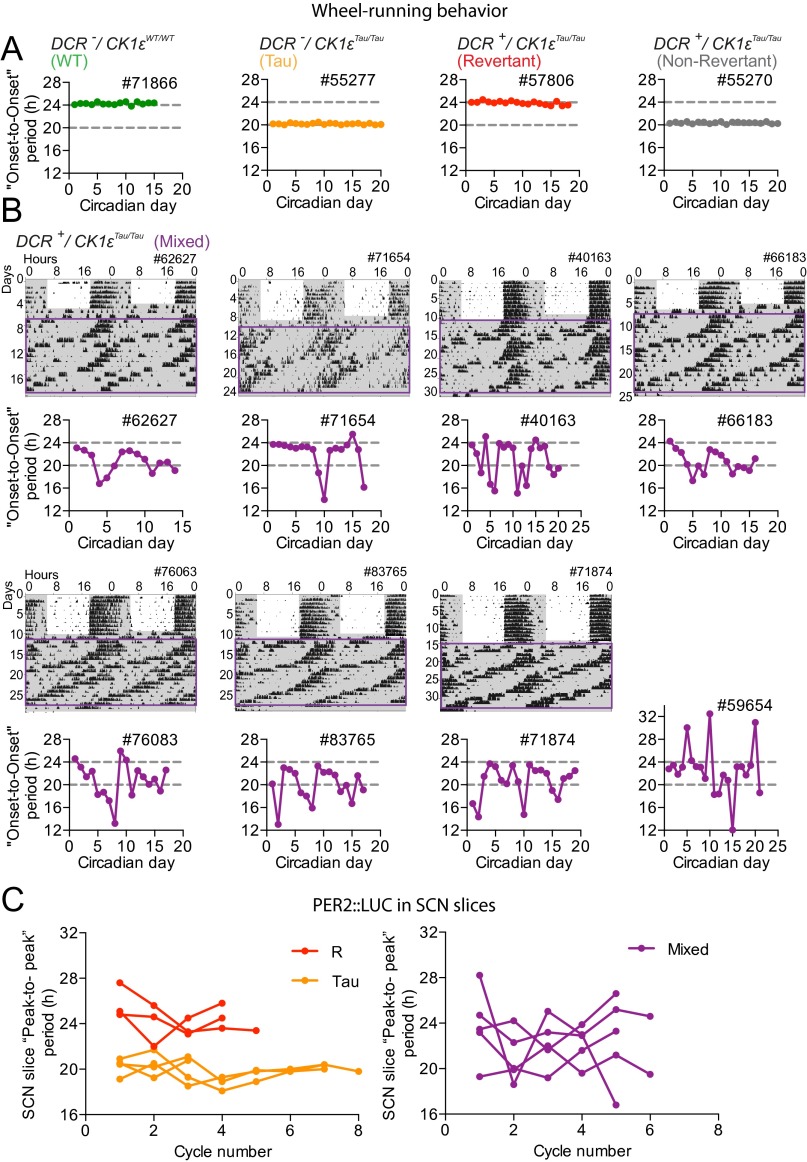
Seven of the 46 SCN slices had rhythms with multiple periods (Fig. S2 H and I) reminiscent of the single mouse from the wheel-running cohort that exhibited unstable behavioral rhythms (Fig. S2A). To investigate this further, we screened for animals that displayed multiple behavioral periods (Fig. S4). Eight “Mixed-period” animals showed behavior that switched back and forth between short (<21 h) and long (∼24 h) periods (Fig. S4B), in contrast to Revertant and Non-Revertant mice, which displayed a very stable 24-h and 20-h period, respectively. Furthermore, this was reflected in PER2::LUC SCN rhythms. Peak-to-peak period varied greatly in SCNs from Mixed-period mice, whereas the period remained stable in Tau and Revertant SCN slices (Fig. S4C). This suggests that spontaneous plasticity of period-setting can occur in individual SCN networks and the direction of spontaneous change from long (Revertant) to short (Non-Revertant) cannot be explained by differences in Cre-mediated excision, which only causes short to long transitions.
Fig. S4.
Intrinsic flexibility in temporally chimeric animals. (A) Onset-to-Onset circadian period determination of wheel-running behavior, in DD, from representative individual animals categorized as WT, Tau, Revertant, and Non-Revertant. Animals in these groups showed a stable period from day to day. (B) Wheel-running actograms and accompanying Onset-to-Onset circadian period determination for a representative selection of Mixed-period animals, where blocks of both 20-h and 24-h period free-running behavior are observed in a single animal. These animals displayed unstable period, which tended to “bounce” between long and short phenotypes. Note that the actogram for animal 59654 already appears in Fig. S2A and thus is not shown here. (C) Peak-to-peak period determination of PER2::LUC bioluminescence of individual SCN slices taken from animals previously screened for their behavioral circadian phenotype. Tau and Revertant SCN slices (Left) showed stable periods, whereas animals selected as having a mixed phenotype in behavior (Right) also displayed a mixed phenotype in the SCN.
Cell-Autonomous Circadian Pacemaking in Temporally Chimeric SCN.
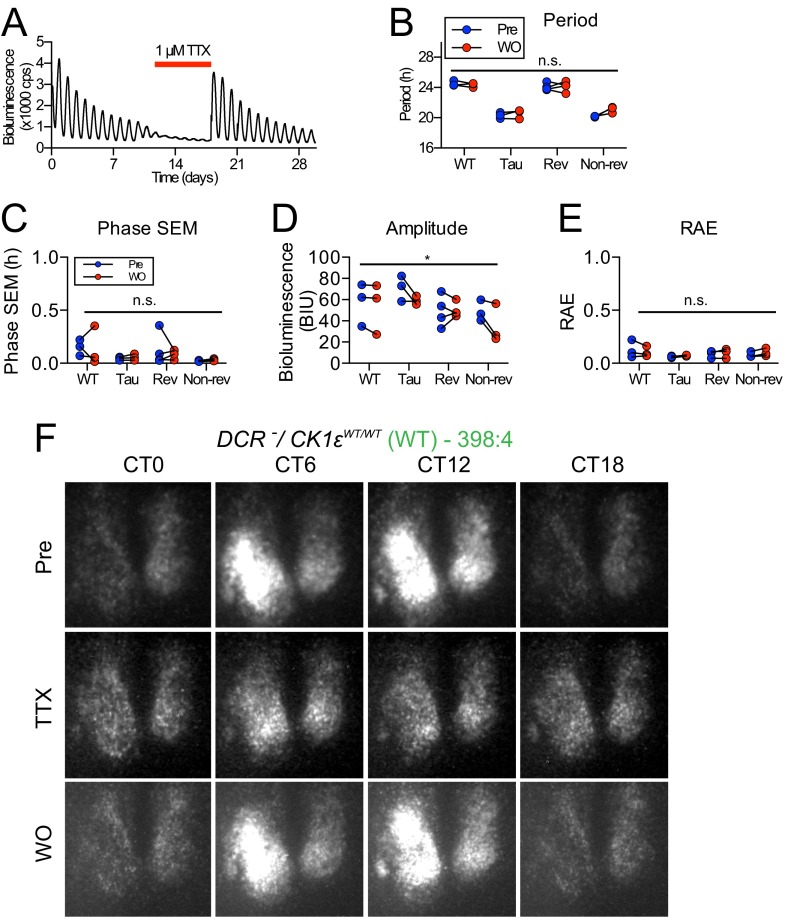
The variable effect of chimerism between animals raised the possibility that ensemble period is a stochastic property of the network. Tetrodotoxin (TTX), a disruptor of neuronal communication (8), was applied to SCN slices to observe whether on removal of TTX the circuit reassembled with the same phenotype (Fig. S5A). By monitoring PER2::LUC, it was clear that this was the case (Fig. S5 B–E). Thus, the Non-Revertant/Revertant nature of a chimeric SCN was a self-organizing, determinative property of each circuit.
Fig. S5.
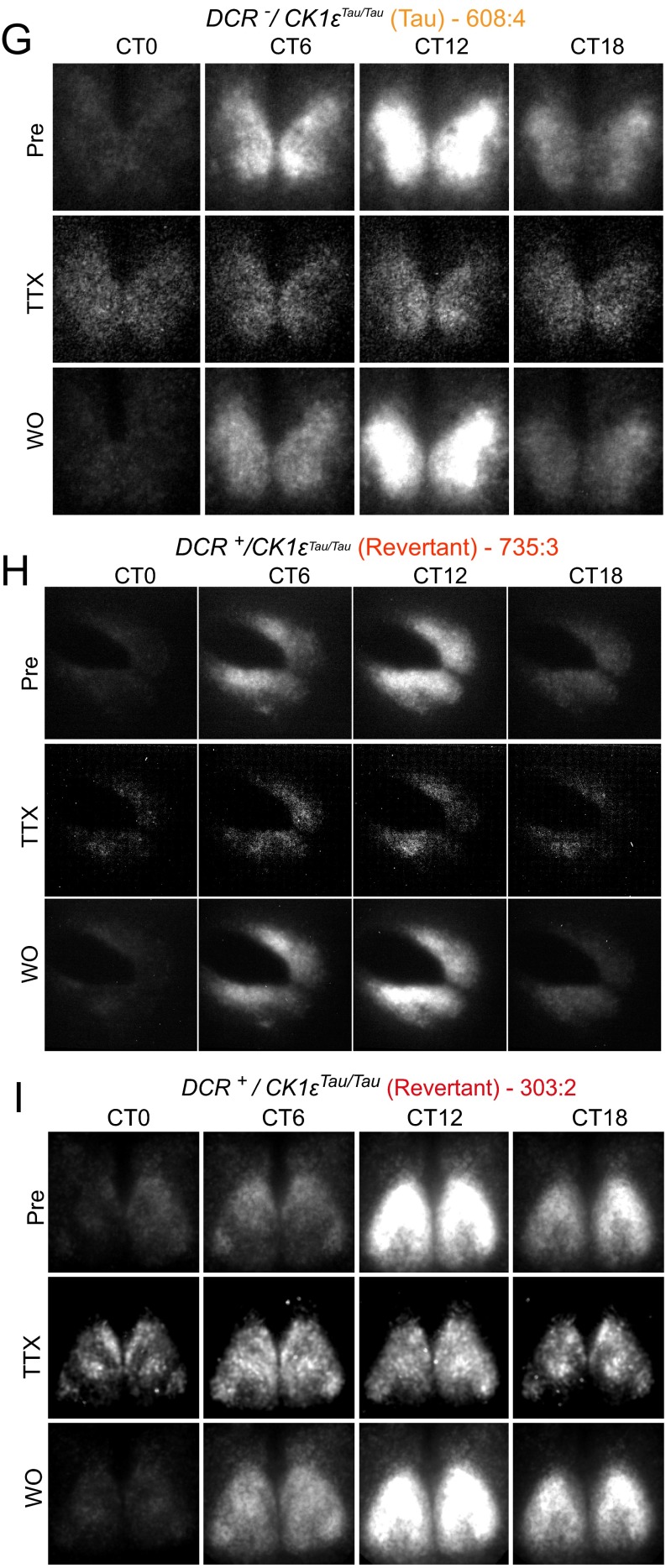
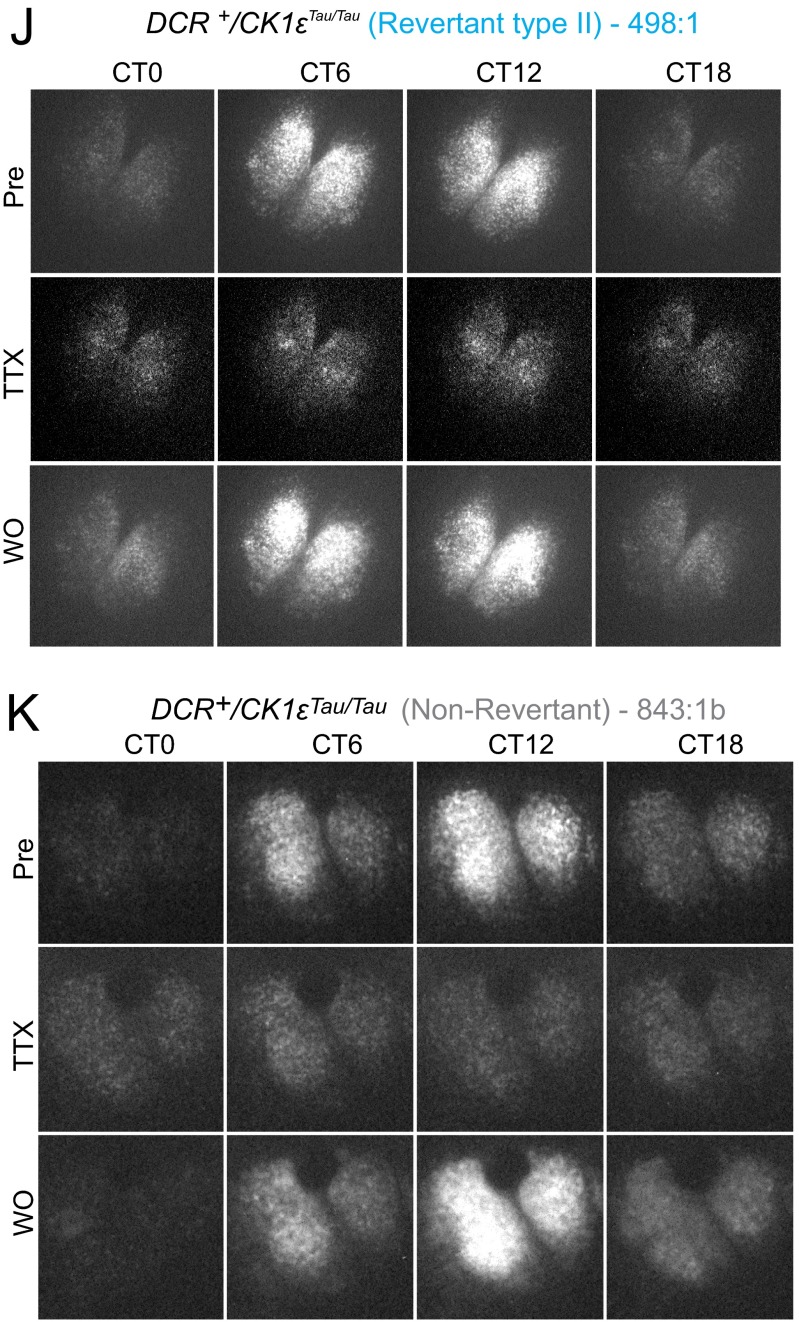
Effect of TTX treatment on a temporally chimeric SCN circuit. (A) Representative bioluminescence recording from Revertant SCN slice before, during, and after treatment with TTX (1 μM). (B–E) Group data for circadian parameters of period, phase SEM, amplitude, and RAE, comparing before and after washout (WO) of TTX treatment. There was no significant effect of TTX treatment on subsequent period, phase SEM, or RAE (two-way ANOVA, n.s. P > 0.5), with a small amount of variability in amplitude (two-way ANOVA, *P < 0.05). (F–K) Representative frames from CCD camera recording of SCN PER2::LUC bioluminescence before (Pre), during, and after WO of TTX (1 μM) treatment for WT, Tau, Revertant, and Non-Revertant SCN slices.
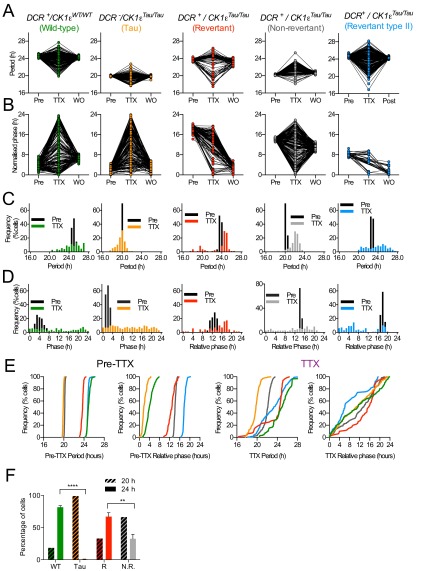
To characterize the relationship between circuit-level and cell-autonomous circadian pacemaking, PER2::LUC rhythms were monitored by CCD in individual cells (Fig. S5 F–J) of SCNs treated with TTX. Before treatment, all Revertant and Non-Revertant SCNs, alongside corresponding WT and Tau controls, displayed tight, unimodal frequency distributions of period and phase, with steep cumulative frequency slopes (Fig. S6 A–E and Table S3). TTX increased the spread of cellular periods and relative phases for all genotypes, as disconnected cells expressed cell-autonomous properties. This reverted to the pretreatment configuration on TTX washout. Measures of cellular period and relative phase during TTX treatment confirmed temporal chimerism at the cell-autonomous level. For DCR− control SCNs (both WT and Tau), the dispersals of cellular period and phase were symmetrical about the pre-TTX distribution, with no skewness (Fig. S6C). In four of six Revertant SCNs, however, cellular circadian periods under TTX became bimodal, showing two distinct plateaus in cumulative frequency plots (Fig. 2A and Table S3). One cell cluster remained at ca. 24 h, whereas a second emerged with a mean period of 20 h. In the remaining two Revertant SCNs, TTX produced a phase realignment with a bimodal distribution (Fig. S6B and Table S3). In the case of 20-h Non-Revertant SCNs, TTX again dispersed cellular period, and the distribution was asymmetrical with a larger proportion of cells expressing periods greater than 20 h: The cumulative frequency curves were right-shifted compared with Tau slices (Fig. 2A and Fig. S6E). Under TTX, therefore, both the Revertant and Non-Revertant SCNs contained a mix of short (putative Tau-competent) and long (Tau-deleted) period cells, but the Non-Revertant had relatively fewer cells with an autonomous period significantly longer than 20 h, which may reflect marginally incomplete Tau deletion and is consistent with the shorter ensemble period (Fig. S6F).
Fig. S6.
Effect of TTX on individual cells of the SCN in a temporally chimeric mouse. (A) Individual cell data from representative SCN slices of the five different groups for the circadian period before, during, and after TTX (1 μM) treatment. (B) Same as in A but for relative circadian phase. (C) Frequency distributions of the circadian period before and during TTX treatment. (D) Same as in B but for the relative circadian phase, where phase has a narrow distribution before treatment and a wide distribution during treatment for all four conditions. (E) Mean cumulative frequency distribution of circadian period and relative phase for the five different phenotypic groups, before (Pre) TTX treatment (Left two panels) and during TTX treatment (Right two panels). Overall, for the TTX data, the slopes are less steep than the pre-TTX condition. In particular, the Revertant type I period curve shows two plateaus, indicative of a bimodal distribution, whereas the Revertant type II phase curve shows a plateau before reaching the peak 100% frequency, also indicative of a bimodal distribution. (F) Estimation of the proportion of intrinsic 20-h and 24-h cells in DCR SCN slices. Individual SCN neurons were grouped into 20-h cells (striped bars; <22-h period) or 24-h cells (solid bars; >22-h period) for each slice (during TTX treatment) and percentages for each group calculated. Both WT and Revertant SCN slices had significantly more 24-h cells than Tau and Non-Revertant slices, respectively (one-way ANOVA with multiple comparisons, **P < 0.01, ****P < 0.0001).
Table S3.
Summary of area under the curve analyses for frequency distributions of cellular circadian period and phase
| Peak 1 | Peak 2 | Peak 1 | Peak 2 | |||||||||
| Slice | Genotype | Phenotype | Peaks | X = | % area | X = | % area | Peaks | X = | % area | X = | % area |
| Pre-TTX period, h | TTX period, h | |||||||||||
| 398:4 | DCR − /CK1ε WT/WT | WTe | 1 | 24.0 | 100 | N/A | N/A | 1 | 24.5 | 100 | N/A | N/A |
| 398:6 | DCR − /CK1ε WT/WT | WT | 1 | 25.0 | 100 | N/A | N/A | 1 | 25.5 | 100 | N/A | N/A |
| 412:4 | DCR − /CK1ε WT/WT | WT | 1 | 24.0 | 100 | N/A | N/A | 1 | 24.0 | 100 | N/A | N/A |
| 608:4 | DCR − /CK1ε Tau/Tau | Tau | 1 | 20.0 | 100 | N/A | N/A | 1 | 20.0 | 100 | N/A | N/A |
| 639:3 | DCR − /CK1ε Tau/Tau | Tau | 1 | 20.5 | 100 | N/A | N/A | 1 | 20.5 | 100 | N/A | N/A |
| 639:5 | DCR − /CK1ε Tau/Tau | Tau | 1 | 20.5 | 100 | N/A | N/A | 1 | 20.5 | 100 | N/A | N/A |
| 303:2 | DCR + /CK1ε Tau/Tau | Revertant type I | 1 | 23.5 | 100 | N/A | N/A | 2 | 19.0 | 18.1 | 24.5 | 81.9 |
| 608:5 | DCR + /CK1ε Tau/Tau | Revertant type I | 1 | 23.5 | 100 | N/A | N/A | 2 | 19.5 | 19.6 | 24.5 | 80.4 |
| 510:6 | DCR + /CK1ε Tau/Tau | Revertant type I | 1 | 24.0 | 100 | N/A | N/A | 2 | 21.5 | 38.7 | 25.5 | 61.3 |
| 735:3 | DCR + /CK1ε Tau/Tau | Revertant type I | 1 | 24.0 | 100 | N/A | N/A | 2 | 20.0 | 46.4 | 25.5 | 53.6 |
| 498:1 | DCR + /CK1ε Tau/Tau | Revertant type II | 2 | 25.0 | 52.9 | 25.4 | 47.1 | 1 | 24.5 | 100 | N/A | N/A |
| 303:3 | DCR + /CK1ε Tau/Tau | Revertant type II | 1 | 22.5 | 100 | N/A | N/A | 1 | 25.0 | 100 | N/A | N/A |
| 843:1b | DCR + /CK1ε Tau/Tau | Non-Revertant | 1 | 20.0 | 100 | N/A | N/A | 1 | 22.0 | 100 | N/A | N/A |
| 874:4b | DCR + /CK1ε Tau/Tau | Non-Revertant | 1 | 20.0 | 100 | N/A | N/A | 2 | 20.5 | 79.2 | 24.0 | 20.8 |
| 874:5a | DCR + /CK1ε Tau/Tau | Non-Revertant | 1 | 20.0 | 100 | N/A | N/A | 1 | 20.5 | 100 | N/A | N/A |
| Pre-TTX phase, h | TTX phase, h | |||||||||||
| 398:4 | DCR − /CK1ε WT/WT | WT | 1 | 2.0 | 100 | N/A | N/A | 1 | 4.0 | 100 | N/A | N/A |
| 398:6 | DCR − /CK1ε WT/WT | WT | 1 | 3.0 | 100 | N/A | N/A | 1 | 22.0 | 100 | N/A | N/A |
| 412:4 | DCR − /CK1ε WT/WT | WT | 1 | 18.0 | 100 | N/A | N/A | 1 | 2.0 | 100 | N/A | N/A |
| 608:4 | DCR – /CK1ε Tau/Tau | Tau | 1 | 2.0 | 100 | N/A | N/A | 1 | 3.0 | 100 | N/A | N/A |
| 639:3 | DCR − /CK1ε Tau/Tau | Tau | 1 | 7.0 | 100 | N/A | N/A | 1 | 12.0 | 100 | N/A | N/A |
| 639:5 | DCR − /CK1ε Tau/Tau | Tau | 1 | 10.0 | 100 | N/A | N/A | 1 | 4.0 | 100 | N/A | N/A |
| 303:2 | DCR + /CK1ε Tau/Tau | Revertant type I | 1 | 12.0 | 100 | N/A | N/A | 1 | 8.0 | 100 | N/A | N/A |
| 608:5 | DCR + /CK1ε Tau/Tau | Revertant type I | 1 | 13.0 | 100 | N/A | N/A | 1 | 18.0 | 100 | N/A | N/A |
| 510:6 | DCR + /CK1ε Tau/Tau | Revertant type I | 1 | 2.0 | 100 | N/A | N/A | 1 | 12 | 100 | N/A | N/A |
| 735:3 | DCR + /CK1ε Tau/Tau | Revertant type I | 1 | 3.0 | 100 | N/A | N/A | 1 | 20.0 | 100 | N/A | N/A |
| 498:1 | DCR + /CK1ε Tau/Tau | Revertant type II | 1 | 8.0 | 100 | N/A | N/A | 3 | 0.0 | 74.2 | 8.0 | 25.8 |
| 303:3 | DCR + /CK1ε Tau/Tau | Revertant type II | 1 | 18.0 | 100 | N/A | N/A | 2 | 6.0 | 76.1 | 18.0 | 23.9 |
| 843:1b | DCR + /CK1ε Tau/Tau | Non-Revertant | 1 | 14.0 | 100 | N/A | N/A | 1 | 17.0 | 100 | N/A | N/A |
| 874:4b | DCR + /CK1ε Tau/Tau | Non-Revertant | 1 | 5.0 | 100 | N/A | N/A | 1 | 10.0 | 100 | N/A | N/A |
| 874:5a | DCR + /CK1ε Tau/Tau | Non-Revertant | 1 | 11.0 | 100 | N/A | N/A | 1 | 2.0 | 100 | N/A | N/A |
Fig. 2.
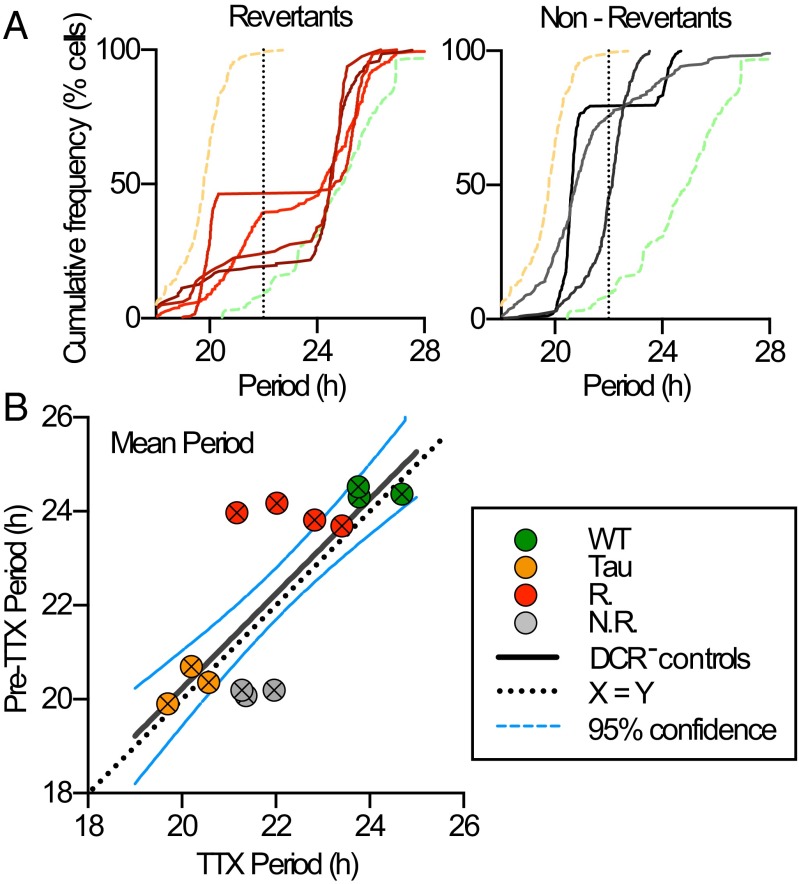
Nonlinear computation of SCN period in temporally chimeric mice. (A) Cumulative frequency plots showing the distribution of circadian periods of neurons during treatment of SCN slices with TTX (1 μM). Revertant SCN slices (red traces; Left panel) show bimodal distribution, whereas Non-Revertant slices (gray traces; Right panel) display an even distribution from 20 h up to 24 h. For comparison, both graphs show average plots for Tau (n = 3, yellow dashed line) and WT slices (n = 3, green dashed line). The black dotted line indicates 22 h. Note the bimodal distribution of Revertant slice periods, on either side of the 22 h line. (B) Relationship between observed SCN ensemble period (Pre-TTX) and period predicted by distribution of cell-autonomous periods under TTX (mean of single-cell data, n > 100 cells per SCN and n ≥ 3 SCN per group). Regression plot using data from control WT and Tau SCNs (solid line, y = 1.008x + 0.73) is not significantly different from line of equality (dotted). Note that the data from Revertant and Non-Revertant temporally chimeric mice fall off the control line. In addition, three of four Revertant and three of three Non-Revertant slices fall outside the 95% confidence intervals (blue dashed line), indicating nonlinear computation of the ensemble circadian period. The periods predicted from TTX data are longer than (Non-Revertant) or shorter than (Revertant SCN) those observed, although the predicted periods do not differ between the two phenotypic categories (Student’s t test, P = 0.23).
To test whether the emergent period of the chimeric SCN was simply the average of all of the individual cellular periods, the SCN ensemble period before treatment with TTX was compared with the predicted average intrinsic period calculated from the individual cellular periods during TTX. For control WT and Tau SCN, these measures were in direct register (Fig. 2B), but this was not the case for the chimeric SCN. The periods of three of four Revertant SCNs and three of three Non-Revertant SCNs fell outside the 95% confidence interval set about the linear regression of the control SCN slices (Fig. 2B). Thus, Revertant SCNs oscillated with a period longer than predicted by their constituent cells, suggesting that the 24-h cells carried more “weight” within the Revertant circuit. Conversely, the Non-Revertant SCNs had a period shorter than predicted, and thus 20-h cells dominated. Furthermore, the mean periods under TTX were comparable between Revertants and Non-Revertants, consistent with them having comparable genetic status (mean ± SEM—Revertants, 22.4 ± 0.40 h; Non-Revertants, 21.5 ± 0.22 h; n = 4 and 3; P = 0.23, t test). Thus, nonlinear computations based on cell-autonomous periods determined the ensemble period of the SCN. Pace-setting can therefore be unequally weighted in favor of particular cells in the circuit: In the majority of SCNs, it was the Drd1a cells, but in Non-Revertant SCNs, the non-Drd1a cells held sway. In both cases, the heterochronic circuit was nevertheless synchronized and functional.
Circuit-Level Circadian Pacemaking in Temporally Chimeric SCN.
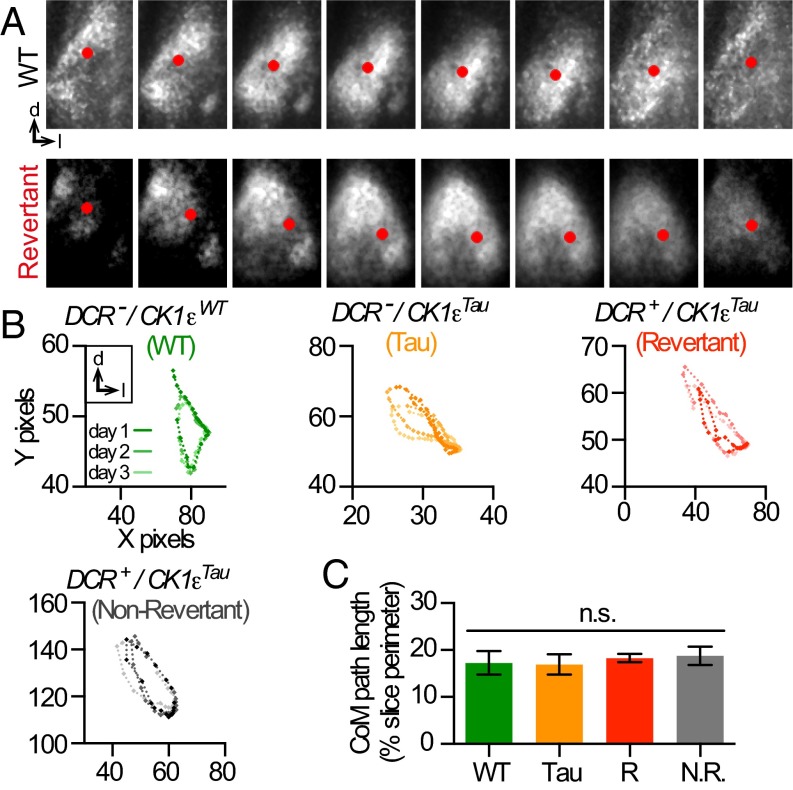
Having confirmed cellular chimerism in the DCR-CK1εTau SCN, we examined the circuit-level behavior by focusing on the wave of PER2::LUC bioluminescence, which follows a stereotypical spatiotemporal orbit across the SCN (Fig. 3A). In CCD time-lapse recordings, this can be described by center-of-mass (CoM) analysis (Fig. 3B) (18). In WT SCN, the CoM followed the trajectory described previously (18–20), sweeping from dorso-medial to ventral SCN and back again. The 20-h Tau SCN exhibited comparable geometry in the wave CoM (Fig. 3B). An attractive hypothesis with experimental support is that the leading edge of the wave arises in the dorsomedial SCN because cells in this region have a shorter cell-autonomous period (8, 21). In the chimeric SCN, short-period (20 h) DCR− cells are distributed predominantly in the central SCN, not the dorsomedial SCN, and so if this hypothesis were correct, the geometry of the wave should be different. In both Revertant and Non-Revertant SCNs, however, the geometry and path lengths of the trajectories of the wave CoM were comparable to control slices (Fig. 3 B and C). Chimerism did not, therefore, affect this higher level organization of circadian timekeeping: circuit-based mechanisms were able to impose a common, stereotypical spatiotemporal order even though the mix of cell-autonomous and ensemble periods was different between SCNs. Thus, the wave is not a product of the relative mix of cell-autonomous periods.
Fig. 3.
Spatiotemporal waves of circadian PER2::LUC bioluminescence are conserved in temporally chimeric SCN. (A) Frames from representative CCD recordings of WT and Revertant SCNs to illustrate wave-like dorso-medial to ventral-lateral movement of CoM of bioluminescence (red dot). (B) Poincaré plots of circadian CoM wave trajectory over 3 d (dark-, medium-, and light-colored lines for each consecutive day) from representative control WT and Tau, and Revertant and Non-Revertant temporally chimeric SCNs. (C) Group data (n ≥ 3, mean ± SEM) illustrate path length of CoM trajectory. The extent of the spatiotemporal wave in the DCR+ chimeric SCN, for both Revertant (R) and Non-Revertant (NR) SCN, is not significantly different from control WT and Tau SCN (one-way ANOVA, P = 0.89).
Temporal Chimerism in the SCN Confers Plasticity to Behavioral Circadian Rhythms in Mice.
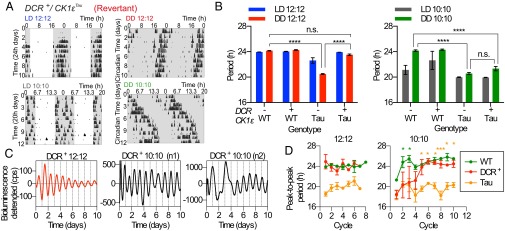
In Mixed-period mice, the behavioral period changed spontaneously. To test whether the computation of the ensemble period could be systematically reprogramed, WT, Tau, and Revertant mice were held on 24-h 12 L:12 D or 20-h 10 L:10 D lighting schedules before transfer to continuous dim red light (continuous darkness, DD). As noted earlier, both WT and Revertant mice entrained very effectively to the 24-h cycle (Fig. 4A and Fig. S7A), whereas Tau mice did not, and on transfer to DD, all groups exhibited their intrinsic periods (Fig. 4A and Fig. S7B). Conversely, Tau but not WT mice were able to entrain to the 20-h cycle (Fig. S6 C and D). Unexpectedly, Revertant mice were also able to express stable 20-h behavioral rhythms on the 20-h lighting cycle (Fig. 4A). Furthermore, on transfer to DD, they had a period close to 20 h (Fig. 4 D and E), whereas WT mice instead ran at ca. 24 h. The exposure to a 20-h lighting cycle had therefore brought about a long-term reprogramming of the chimeric SCN, enabling it to sustain a 20-h period. This unanticipated flexibility of period-setting is inconsistent with any change in Cre-mediated excision and must reflect reorganized circuit-level computations.
Fig. 4.
Photic reprogramming of the circadian period in Revertant temporally chimeric mice. (A) Representative, double-plotted actograms for a Revertant mouse subjected to different lighting schedules. The mouse was exposed to a LD 12:12 schedule (Top Left) and then transferred to DD (Top Right), both plotted on a 24-h time base. The mouse was then exposed to LD 10:10 (Bottom Left) and then transferred to DD (Bottom Right), both plotted on a 20-h time base. (B) Period of wheel-running rhythm (group mean + SEM) under LD 12:12, succeeding DD (Left) and LD 10:10, succeeding DD (Right). When kept in LD 12:12, chimeric mice subsequently had a mean period not different from WT (P = 0.40), but after LD 10:10, they were then not significantly different from Tau mice (P = 0.24). Periods for WT and Tau mice were significantly different from each other after both lighting schedules (two-way ANOVA, ****P < 0.0001 by Tukey’s comparison). (C) PER2::LUC bioluminescence traces (de-trended) from representative Revertant SCNs from animals kept in LD 12:12 (red) or LD 10:10 (black), immediately before slice preparation and recording. (D) Group data (n = 4 per group, mean ± SEM) of peak-to-peak period of each cycle for SCNs from animals kept in LD 12:12 or LD 10:10. Note the trend for period increase in the Revertant SCNs from LD 10:10 mice but no change in Revertant SCNs from LD 12:12 mice. Revertant animals had significantly shorter periods than WT at the start of the recordings (green; *P < 0.05) but then significantly longer periods than Tau animals after the fifth cycle (two-way ANOVA with Tukey’s comparison; orange; *P < 0.05, ***P < 0.001).
Fig. S7.
Photic reprogramming of the circadian period of wheel-running behavior of Revertant temporally chimeric mice is reversible. (A) Representative, double-plotted actograms and associated χ2 periodograms from control WT (Left), Tau (Center), and Revertant chimeric mice (Right; actograms shown in Fig. 4) held under a LD 12:12 lighting schedule. (B) As above, but mice now transferred to DD. Actograms in A and B are plotted on a 24-h time base. (C) As above, but mice now held on LD 10:10 lighting schedule. (D) As above, but mice now transferred to DD after the LD 10:10 schedule. Actograms in C and D are plotted on a 20-h time base. (E) De-trended bioluminescence traces measured by PMTs from representative WT and DCR−/CK1εTau control SCN slices taken from mice exposed to 12:12 or 10:10 LD cycles. (F) Group data (mean + SEM, n ≥ 3 per group) for the circadian period calculated for animals entrained to either LD12:12 or LD10:10 conditions, where period was assessed across different phenotypic measures. Period was calculated in vivo (wheel-running behavior) followed by ex vivo PER2::LUC recordings in SCN slices, where period was determined at the start and end of the bioluminescence recording. In animals exposed to LD 12:12 lighting schedules, only genotype effects on period were observed, not phenotype effects (two-way ANOVA with repeated measures, ****P < 0.0001, n.s. P > 0.05); thus, the circadian period was stable across all three period measures. In animals exposed to LD 10:10 lighting schedules, there were significant phenotype-dependent changes to period (two-way ANOVA with repeated measures, *P < 0.05, **P < 0.01).
To determine the origin of this 20-h period, SCNs were dissected from mice that had previously entrained to either a 20-h or 24-h LD cycle (Fig. 4C). Slices taken from mice kept on a 24-h cycle all maintained their respective intrinsic periods in culture (Fig. 4D). This was also the case for SCN slices taken from mutant Tau and WT animals kept on a 20-h cycle, where WT SCNs reverted to a ca. 24-h period after the first cycle (Fig. 4D and Fig. S7E). This, however, was not the case for the SCN of chimeric mice. For the first 3 d in culture, the SCN oscillated with a period close to 20 h, and then the period lengthened progressively to ca. 24 h by the sixth cycle (Fig. 4D and Fig. S7F). It should also be noted that the rhythms were very unstable during this adjustment phase, where there were large fluctuations of period, before settling down to 24 h (Fig. 4 C and D). In conclusion, the 20-h LD cycle reprogrammed the computation of behavioral and SCN periods of temporally chimeric animals. This recomputation was stable in DD, but once the SCN was isolated in culture, it progressively reverted to its earlier 24-h period. Thus, even though under steady-state conditions the circuit-level specification of circadian period is stable and self-organizing, appropriate retinal activation of the SCN can reversibly alter the computation of the ensemble period and reconfigure the circuit and its control over circadian behavior.
Discussion
To explore how cell-autonomous properties relate to circuit-level circadian functions of the SCN, we created DCR-CK1εTau temporally chimeric mice, which contained a neurochemically and functionally distinct subpopulation of Drd1a cells with a 24-h period, alongside non-Drd1a cells with a 20-h period. This revealed a series of circuit-level operating principles of the SCN. First, synchrony and a common period can be imposed on a temporally heterogeneous population of cells: marked divergence of cell-autonomous periods did not disable the circuit. Second, determination of the ensemble circadian period of the SCN is a nonlinear computation, rather than simple averaging. Third, spatiotemporal waves of circadian gene expression are generated by circuit-level mechanisms indifferent to contrasting cell-autonomous periods. Finally, circadian period-setting is flexible, such that under particular circumstances Drd1a or non-Drd1a cells can set the ensemble period and that this relative dominance can be reversibly reprogrammed by the LD cycle.
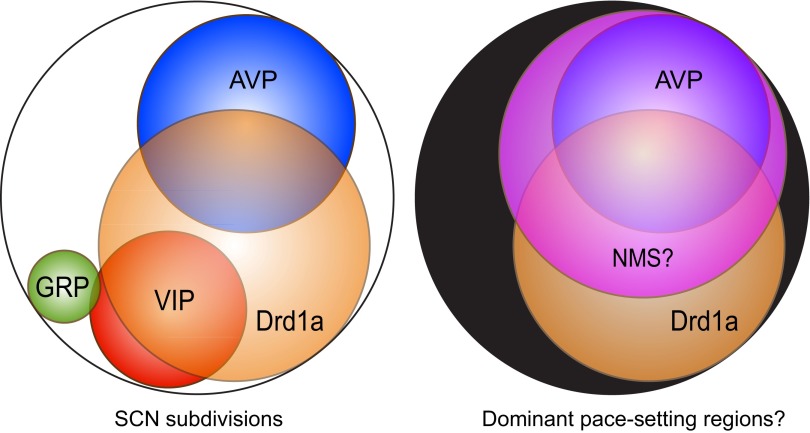
The Revertant phenotype demonstrated that targeting a neuronal subpopulation in an otherwise competent SCN circuit could dramatically alter the circadian period of the entire animal. Hence, 24-h Drd1a cells dominated the 20-h non-Drd1a cells. Similar pace-setting effects have been reported for AVP cells in AVP-Bmal−/− mice (14) and for neuromedin-S (NMS) cells in NMS-Clockdelta19 mice (15), although the effects on period were less pronounced (<1 h) than with DCR-CK1εTau mice. The effects seen in AVP-Bmal−/− mice likely arose from localized loss of neuropeptide expression and thus disruption and loosening of circuit-level coupling. The SCN of DCR-CK1εTau mice had normal neuropeptide expression, and so period effects arose in the presence of effective coupling. Interestingly, expression of Clockdelta19 solely in VIP neurons did not affect in vivo period (15). Given that essentially all AVP neurons are NMS+ (15), the simplest interpretation is that AVP cells alongside non-VIP Drd1a cells and perhaps unidentified, non-VIP, NMS+ cells form the top level of a pace-setting hierarchy (Fig. S8).
Fig. S8.
Venn diagram schematic representation of cell distributions of the SCN. (Left) The overlapping but stereotypical distributions of cell populations based on functionally relevant neuropeptides (AVP, blue; GRP, green; VIP, red) and Drd1a (orange). (Right) Our hypothesis that the Drd1a (orange) population, including AVP neurons, is normally dominant in pacemaking. If we also consider other recent circadian chimera studies (15, 16), this pacemaker population would not include the VIP neurons but would also include NMS-positive cells (purple; indicating hypothesized overlap with Drd1a-positive cells).
Pace-setting is clearly not an absolute cellular property, however. Rather, it reflects the circuit context: Non-Drd1a cells dominated in Non-Revertant animals. Moreover, Mixed-period mice showed regular transitions between long and short behavioral periods. The origin of the period spectrum is unclear. We found no evidence of a genetic cause: transgene copy number, EYFP-reported Cre activity in the SCN, and deletion efficiency did not differ between mice of contrasting phenotypes. We cannot exclude, however, the possibility of subtle variability in the efficacy of Tau excision, and indeed single-cell analysis highlighted a trend for fewer cells of longer period in Non-Revertant SCNs. Several lines of evidence argue, however, for a circuit-level, nongenetic origin to the spectrum. First, Mixed-period mice showed spontaneous changes of behavioral period from short to long and long to short. The latter is incompatible with a genetic model: excision of the Tau allele is irreversible. Second, Revertant mice exhibited unanticipated plasticity of period in response to nonresonant lighting cycles. Again, conversion from a long to short period is incompatible with a genetic, excision-based model. Finally, in both Revertant and Non-Revertant SCNs, single-cell analysis under TTX showed that the computation of ensemble period of an intact circuit, irrespective of its absolute value, was nonlinear. Thus, pace-setting may not be based on irreversible cellular identities but instead based on complex and flexible computations (22). The power of these computations is revealed by the surprising robustness within the SCN, which incorporated a wide range of cell-intrinsic periods into a functional and synchronous circuit. Theoretical data from a generic, coupled-oscillator mathematical model (23) and experimental data from the Clockdelta19 chimera mouse (24) support a linear computation model of ensemble period. These studies, however, involved a random, evenly distributed mixture of WT and mutant cells that do not consider cellular heterogeneity. Here we specifically targeted the Drd1a cells, and consistent with some formal models (25), our results support hierarchical, nonlinear period determination within the SCN, with the Drd1a intrinsic period dominating pace-setting in the majority of cases. Thus, circuit-level mechanisms, likely neuropeptidergic (9), can override the genetically specified period of subordinate SCN cells when they are embedded in a fully functional circuit.
Contrary to previous speculation that the spatiotemporal wave arises through short-period cell phase leading (8, 21), our results show that this is not the case. The chimeric SCN exhibited comparable wave geometry, despite having a very different spatial mapping of intrinsic circadian periods. Thus, the wave is generated by circuit-level mechanisms indifferent to contrasting cell-autonomous periods. Peptidergic control is likely a key component of its generation: For example, chemogenetic activation of VIP cells can reprogram the wave (18).
Finally, the retinally mediated reprogramming of the behavioral period has echoes of the “aftereffects” described in classical circadian biology, where non–24-h lighting regimes can modulate period. In WT mice (26, 27), these effects are small (<1 h), whereas the behavioral periods of chimeric mice were reprogrammed by 2.2 ± 0.3 h. This recoding was an intrinsic property of the SCN because immediately on recording, SCNs from Revertant animals exposed to 20-h LD had a short period. This was not sustained, however, and the period lengthened progressively over 3–4 d, “relaxing” back to 24 h. Similar relaxation is seen in the SCNs of animals exposed to “long day” photoperiodic cycles (20 L:4 D), where phase-splitting in PER2 rhythms of core and shell regions gradually resynchronized in vitro (19). Our data not only show that photic input to the SCN can alter the relative contributions of Drd1a and non-Drd1a cells to the computation of ensemble period, but also they present the possibility that extra-SCN signals are required to sustain this new program in vivo. SCN grafting studies (9) suggest that even though SCN slices do not capture all SCN neurons, sufficient network communication is present to support reprogramming in vitro. A potential origin of extra-SCN signals is the retina: retinal innervation is necessary to sustain a circadian rhythm of MAP kinase activity restricted to the SCN core of hamsters (28). Although the retina does not normally affect circadian pacemaking, it has a clock. We hypothesize that non-Drd1a cells in the retinorecipient core of the SCN, such as GRP neurons, could acquire dominance under 20-h LD conditions through resonance with the photic cues from the 20-h retina, whereas 24-h VIP cells receive heterochronic stimulation on a 20-h schedule and so lose coherence. In contrast, once the slice is made, this direct, resonant stimulation is lost, and progressively, the 24-h cells in the circuit come to dictate period.
In conclusion, we have demonstrated an unanticipated flexibility in the determination of ensemble period, providing a new perspective on the concept of pacemaker cells in the SCN. Moreover, we have revealed a series of operating principles that govern the interactions between cell-autonomous and circuit-level functions of the SCN. Elucidation of the mechanisms that implement these principles offers an opportunity to understand how cell interactions support neuronal computations to generate flexible emergent programs of behavior. This has strong parallels with studies in Drosophila, where genetic manipulation of kinase or TTFL components in subgroups of fly neurons can impose particular cell-autonomous periods over the circuit and behavior (29). The fly circuit does not, therefore, operate in a simple linear manner. The logic of circadian computations may therefore be a conserved feature of flies and mammals.
Materials and Methods
Animal work was conducted in accordance with the UK Animals (Scientific Procedures) Act 1986, with Medical Research Council Laboratory of Molecular Biology Local Ethical Review Committee. Wheel-running patterns were analyzed using ClockLab (ActiMetrix Inc.). SCN and peripheral tissue slices were prepared as previously described (30). Bioluminescence emission was recorded using photon multipliers (Hamamatsu), CCD cameras (Hamamatsu), or LV200 bioluminescence imaging systems (Olympus). Graphs were plotted, data analyses performed, and statistical tests calculated using Prism 6 (GraphPad). A more detailed description of the materials and methods are provided in SI Materials and Methods.
SI Materials and Methods
Animals.
Drd1a-Cre mice [B6.Cg-Tg(Drd1a-cre)266Gsat/Mmcd, MMRRC:03425] were purchased from the GENSAT (Gene Expression in the Nervous System Atlas) project (Rockefeller University) (12), through the Mutant Mouse Regional Resource Centers. ROSA-YFP mice were provided by A. McKenzie, Medical Research Council Laboratory of Molecular Biology, Cambridge, United Kingdom (17).
Wheel-Running.
Mice were housed individually and their activity patterns were assessed using running wheels and passive infrared movement detectors. Mice were entrained to a LD cycle for at least 10 d before being put onto a schedule of DD for 14 d. Food and water were provided ad libitum. Data were analyzed using ClockLab (ActiMetrix Inc.), running within Matlab (Mathworks). Circadian period (periodogram analysis) and mean LD activity profiles were calculated for each animal, where activity was averaged over 8–10 d of activity and organized into 0.1-h bins. “Onset-to-Onset” circadian period was calculated by defining onset of wheel-running activity to be 4 h of inactivity followed by 4 h of activity. For each calendar day, one or two onset points were detected automatically by the software, manually checked, and then the time between onsets was recorded.
Immunofluorescence.
For Fig. S1B and cell counts, adult male mice were stereotaxically injected with colchicine (2 μg/μL; Tocris Biosciences) into the third ventricle (5.2 mm deep to dural surface, on Bregma) under halothane anesthesia. Animals received a 1-μL (0.9% NaCl) injection delivered over 1 min. Mice were then killed 55 h postinjection by transcardial perfusion of 0.01 M PBS followed by 4% (vol/vol) PFA fixative before immunohistochemical processing of the brains for the detection of neuropeptides in the SCN. For Fig. S1 C and D, brains were postfixed in 4% (vol/vol) PFA. Confocal microscopy (Zeiss 780 with 63× oil immersion objective) was used to image immunofluorescent staining of rabbit anti-AVP (1:1,000; Bachem), guinea pig anti-VIP (1:1,000, Bachem; 1:1,000, Immunostar), and rabbit anti-GRP (1:1,000; Immunostar), alongside intrinsic EYFP signal, in 40 μm PFA-fixed SCN brain sections, mounted with DAPI (Vectorlabs).
Organotypic Slices.
Slices were kept for 7 d in culture before recording bioluminescence rhythms, except for peripheral tissues and adult SCN slices, which were recorded from the day of preparation. Whole-slice emissions were measured by photon multiplier tubes (Hamamatsu), whereas imaging of bioluminescence of individual cells within the slice was recorded with Orca CCD cameras (Hamamatsu) mounted on an inverted microscope. For TTX treatment, SCN slices were recorded for 7 d, followed by addition of 1 μM TTX (Abcam) for 7 d and final washout for a further 7 d.
CK1ε Deletion Detection by PCR.
Genomic DNA was extracted from SCN slices using the DNeasy Blood and Tissue Extraction Kit (Qiagen). Three PCR reactions were carried out. CK1ε-specific primers were designed to detect either “intact” or “exon 4 deletion” in the CK1ε gene (Fig. S1I), and primers against ActinB were made to act as a reference. The intact-specific CK1ε PCR used a forward primer that binds within exon 4 and a reverse primer that binds just downstream of exon 4. When the intact CK1ε gene is present, a ∼300-bp amplicon is produced. When exon 4 is deleted by Cre-recombinase, the binding site for the intact-specific forward primer is lost, so no product is formed. The deletion-specific PCR uses a forward primer that is upstream of exon 4 and the same reverse primer as used in the intact-specific reaction. When the intact CK1ε gene is present, the deletion-specific forward primer binds at a distant site compared with the reverse primer, so efficient amplification does not occur. When exon 4 is deleted by Cre-recombinase, the forward and reverse primers are brought close together to produce a ∼200-bp amplicon. Primers were used at 0.5 μM, with 4 μL of DNA in a 25-μL Taq polymerase reaction (Bioline). Control SCN slices were prepared to run alongside the DCR samples:
-
•
floxed Tau SCN slices (n = 4) transduced with CMV-Cre-GFP AAV to generate bands arising by deletion in all cell types of exon 4 of CK1ε,
-
•
floxed-Tau SCN slices (n = 4) to generate bands representative of intact CK1ε in all cells, and
-
•
floxed-Tau SCN slices (n = 4) transduced with Synapsin-Cre-mCherry AAV to generate bands arising by deletion of exon 4 of CK1ε in neurons.
CMV-Cre-GFP AAV was sourced from Penn Vector Core (University of Pennsylvania), and hSyn-Cre-mCherry was obtained from UNC Vector Core (University of North Carolina). The following PCR conditions were used: 2 min at 94 °C followed by 38 cycles of 94 °C for 20 s, 54 °C for 10 s, and 68 °C for 30 s. The relative signal of exon 4 deletion amplicons was calculated from intensity of the bands relative to the ActB reference and normalized to the CMV-Cre sample. Band intensity measures were carried out using Image Lab software (Bio-Rad).
The following primer sequences were used: CK1ε intact forward, GTC CAC AGG CAG ATA TGA ACA C; CK1ε KO forward, TGG TGT ATT TGG CCT ACT GG; CK1ε reverse, ACG ATG GCT GGA GTC AAA AG; Actin B forward, GTC CAC CTT CCA GCA GAT GT; and ActinB reverse, GAA AGG GTG TAA AAC GCA.
Data Analyses and Statistical Tests.
Parameters for bioluminescence rhythms [period, relative phase, amplitude, and relative amplitude error (RAE)] were calculated by nonlinear fast-Fourier transform analysis, as part of the circadian analysis package running on BioDare (A. Millar, University of Edinburgh, Edinburgh, United Kingdom; https://www.biodare.ed.ac.uk/). The first 24 h of the recordings were omitted from the analysis to avoid potential experimental artifacts. Phase was calculated relative to the start of each dataset. Assessment of period evolution during SCN imaging recordings, from animals subjected to different light schedules, was made by measuring “peak-to-peak” time intervals of PER2 bioluminescence. Peak-to-peak measurements were also used to assess the period of peripheral tissues, due to them not being prolonged enough to be processed by BioDare. CoM analysis to describe the trajectory of the wave of bioluminescence was conducted in ImageJ as described (18). The perimeter of the CoM excursion was expressed as a percentage of the perimeter of the temporally integrated bioluminescence signal. All graphs and statistics were carried out using Prism (Graphpad), unless specified. All group-based comparative tests between three or more groups were made by ordinary one-way ANOVA, and those also with a treatment variable were analyzed by two-way ANOVA, with Tukey’s post hoc multiple comparisons test. Correlations were made using two-tailed Pearson’s correlation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1511351113/-/DCSupplemental.
Change History
August 16, 2021: The SI Appendix has been updated to coincide with a formal Correction.
References
- 1.Mohawk JA, Takahashi JS. Cell autonomy and synchrony of suprachiasmatic nucleus circadian oscillators. Trends Neurosci. 2011;34(7):349–358. doi: 10.1016/j.tins.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asher G, Schibler U. Crosstalk between components of circadian and metabolic cycles in mammals. Cell Metab. 2011;13(2):125–137. doi: 10.1016/j.cmet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 3.Chen SK, Badea TC, Hattar S. Photoentrainment and pupillary light reflex are mediated by distinct populations of ipRGCs. Nature. 2011;476(7358):92–95. doi: 10.1038/nature10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Viswanathan N, Weaver DR, Reppert SM, Davis FC. Entrainment of the fetal hamster circadian pacemaker by prenatal injections of the dopamine agonist SKF 38393. J Neurosci. 1994;14(9):5393–5398. doi: 10.1523/JNEUROSCI.14-09-05393.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418(6901):935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- 6.Hastings MH, Brancaccio M, Maywood ES. Circadian pacemaking in cells and circuits of the suprachiasmatic nucleus. J Neuroendocrinol. 2014;26(1):2–10. doi: 10.1111/jne.12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu AC, et al. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129(3):605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi S, et al. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302(5649):1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 9.Maywood ES, Chesham JE, O’Brien JA, Hastings MH. A diversity of paracrine signals sustains molecular circadian cycling in suprachiasmatic nucleus circuits. Proc Natl Acad Sci USA. 2011;108(34):14306–14311. doi: 10.1073/pnas.1101767108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maywood ES, et al. Analysis of core circadian feedback loop in suprachiasmatic nucleus of mCry1-luc transgenic reporter mouse. Proc Natl Acad Sci USA. 2013;110(23):9547–9552. doi: 10.1073/pnas.1220894110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heintz N. Gene expression nervous system atlas (GENSAT) Nat Neurosci. 2004;7(5):483. doi: 10.1038/nn0504-483. [DOI] [PubMed] [Google Scholar]
- 12.Meng QJ, et al. Setting clock speed in mammals: The CK1 epsilon tau mutation in mice accelerates circadian pacemakers by selectively destabilizing PERIOD proteins. Neuron. 2008;58(1):78–88. doi: 10.1016/j.neuron.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Husse J, Zhou X, Shostak A, Oster H, Eichele G. Synaptotagmin10-Cre, a driver to disrupt clock genes in the SCN. J Biol Rhythms. 2011;26(5):379–389. doi: 10.1177/0748730411415363. [DOI] [PubMed] [Google Scholar]
- 14.Mieda M, et al. Cellular clocks in AVP neurons of the SCN are critical for interneuronal coupling regulating circadian behavior rhythm. Neuron. 2015;85(5):1103–1116. doi: 10.1016/j.neuron.2015.02.005. [DOI] [PubMed] [Google Scholar]
- 15.Lee IT, et al. Neuromedin s-producing neurons act as essential pacemakers in the suprachiasmatic nucleus to couple clock neurons and dictate circadian rhythms. Neuron. 2015;85(5):1086–1102. doi: 10.1016/j.neuron.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Srinivas S, et al. Cre reporter strains produced by targeted insertion of EYFP and ECFP into the ROSA26 locus. BMC Dev Biol. 2001;1:4. doi: 10.1186/1471-213X-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoo SH, et al. PERIOD2:LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci USA. 2004;101(15):5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brancaccio M, Maywood ES, Chesham JE, Loudon AS, Hastings MH. A Gq-Ca2+ axis controls circuit-level encoding of circadian time in the suprachiasmatic nucleus. Neuron. 2013;78(4):714–728. doi: 10.1016/j.neuron.2013.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans JA, Leise TL, Castanon-Cervantes O, Davidson AJ. Dynamic interactions mediated by nonredundant signaling mechanisms couple circadian clock neurons. Neuron. 2013;80(4):973–983. doi: 10.1016/j.neuron.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaguchi Y, et al. Mice genetically deficient in vasopressin V1a and V1b receptors are resistant to jet lag. Science. 2013;342(6154):85–90. doi: 10.1126/science.1238599. [DOI] [PubMed] [Google Scholar]
- 21.Doi M, et al. Circadian regulation of intracellular G-protein signalling mediates intercellular synchrony and rhythmicity in the suprachiasmatic nucleus. Nat Commun. 2011;2:327. doi: 10.1038/ncomms1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: Retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916(1-2):172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- 23.Abraham U, et al. Coupling governs entrainment range of circadian clocks. Mol Syst Biol. 2010;6:438. doi: 10.1038/msb.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Low-Zeddies SS, Takahashi JS. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell. 2001;105(1):25–42. doi: 10.1016/s0092-8674(01)00294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bernard S, Gonze D, Cajavec B, Herzel H, Kramer A. Synchronization-induced rhythmicity of circadian oscillators in the suprachiasmatic nucleus. PLOS Comput Biol. 2007;3(4):e68. doi: 10.1371/journal.pcbi.0030068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aton SJ, Block GD, Tei H, Yamazaki S, Herzog ED. Plasticity of circadian behavior and the suprachiasmatic nucleus following exposure to non-24-hour light cycles. J Biol Rhythms. 2004;19(3):198–207. doi: 10.1177/0748730404264156. [DOI] [PubMed] [Google Scholar]
- 27.Azzi A, et al. Circadian behavior is light-reprogrammed by plastic DNA methylation. Nat Neurosci. 2014;17(3):377–382. doi: 10.1038/nn.3651. [DOI] [PubMed] [Google Scholar]
- 28.Lee HS, Nelms JL, Nguyen M, Silver R, Lehman MN. The eye is necessary for a circadian rhythm in the suprachiasmatic nucleus. Nat Neurosci. 2003;6(2):111–112. doi: 10.1038/nn1006. [DOI] [PubMed] [Google Scholar]
- 29.Yao Z, Shafer OT. The Drosophila circadian clock is a variably coupled network of multiple peptidergic units. Science. 2014;343(6178):1516–1520. doi: 10.1126/science.1251285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hastings MH, Reddy AB, McMahon DG, Maywood ES. Analysis of circadian mechanisms in the suprachiasmatic nucleus by transgenesis and biolistic transfection. Methods Enzymol. 2005;393:579–592. doi: 10.1016/S0076-6879(05)93030-9. [DOI] [PubMed] [Google Scholar]